Abstract
Hepatic Stellate Cells (HSCs) are the major source of the excessive extracellular matrix (ECM) production that replaces liver parenchyma with fibrous tissue during liver fibrosis. The signal transducer and activator of transcription 3 (STAT3) promotes HCSs survival, proliferation, and activation contributing to fibrogenesis. We have previously used a fragment-based drug design approach and have discovered a novel STAT3 inhibitor, HJC0123. Here, we explored the biological effects of HJC0123 on the fibrogenic properties of HSCs. HJC0123 treatment resulted in the inhibition of HSCs proliferation at submicromolar concentrations. HJC0123 reduced the phosphorylation, nuclear translocation, and transcriptional activity of STAT3. It decreased the expression of STAT3-regulated proteins, induced cell cycle arrest, promoted apoptosis and downregulated SOCS3. HJC0123 treatment inhibited HSCs activation and downregulated ECM protein fibronectin and type I collagen expression. In addition, HJC0123 increased IL-6 production and decreased TGF-β induced Smad2/3 phosphorylation. These results demonstrate that HJC0123 represents a novel STAT3 inhibitor that suppresses the fibrogenic properties of HSCs, suggesting its therapeutic potential in liver fibrosis.
Keywords: hepatic fibrosis, STAT3, fragment-based drug design, stellate cell, collagen
1. Introduction
Liver fibrogenesis is a wound-healing response to a variety of injuries, including toxic, metabolic and viral insults. In acute or self-limited injury, fibrotic changes are transient and the normal hepatic architecture is typically restored. However, in chronic or recurrent injury, prolonged inflammation and accumulation of extracellular matrix (ECM) proteins result in a progressive replacement of liver parenchyma by fibrotic tissue. Fibrosis can then progress to liver cirrhosis and eventually lead to organ failure and death.1–4
Liver fibrosis is the result of excessive production and decreased turnover of ECM due to the accumulation of myofibroblasts in response to repetitive liver damage. Myofibroblasts can originate from different cells; however, hepatic stellate cells (HSCs) are identified as their major source.1,5,6 Following injury, HSCs undergo activation, an orchestrated transformation process from a quiescent state to a myofibroblast-like phenotype with simultaneous changes in cell behavior.2,7 The signal transducer and activator of transcription 3 (STAT3) is a transcription factor that regulates cell growth and survival. STAT3 is associated with liver injury, inflammation and regeneration.8,9 STAT3 activation in HSCs promotes their survival, proliferation, and activation, contributing to liver fibrogenesis.10–12
Emerging data suggest that fragment-based drug design (FBDD) is a successful approach for the generation of molecules against therapeutic targets.13 This method allows for synthesis of new molecules with enhanced drug-likeness by chemically merging privileged fragments from small molecule libraries.14–17 Using FBDD, our team previously designed, synthesized and characterized a new STAT3 inhibitor, compound HJC0123 (Figure 1A). This small molecule induces growth arrest and apoptosis in MDA-MB-231 breast cancer cells. Its antitumor properties have been successfully tested in a murine model, demonstrating that HJC0123 is a potent, efficacious, and orally bioavailable drug candidate.18
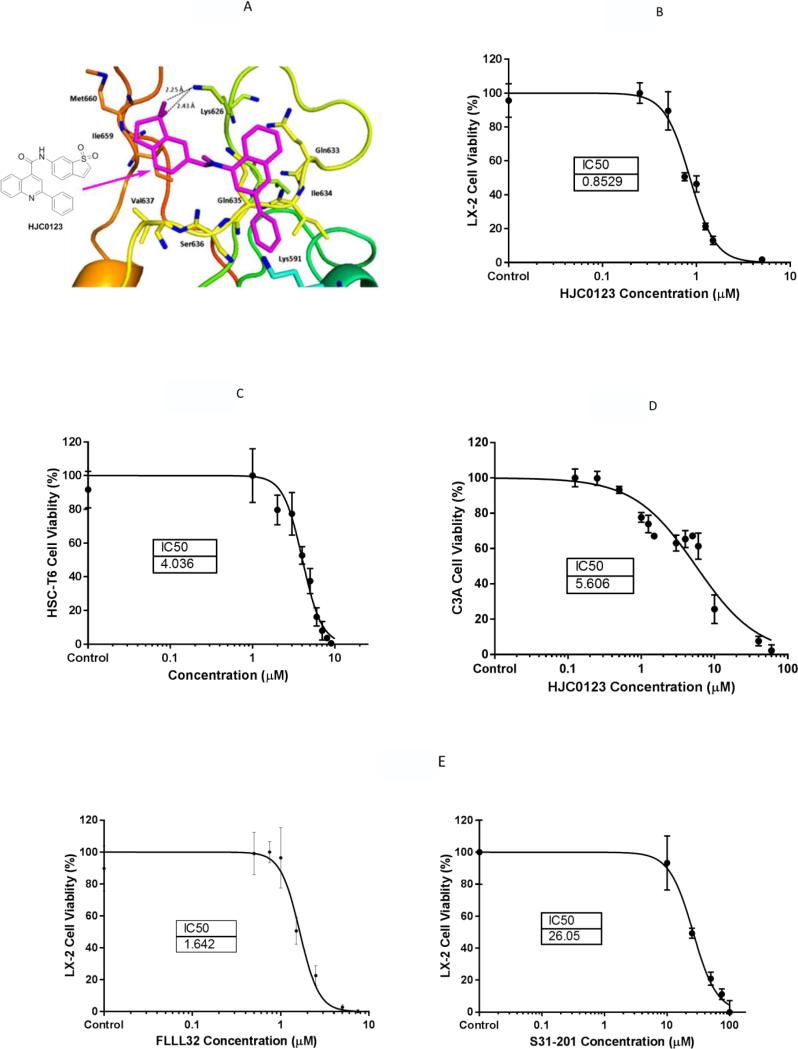
Figure 1. HJC0123 inhibits human and rat hepatic stellate cells proliferation.
(A) Interactions of HJC0123 with the residues of STAT3. HJC0123 is shown in small sticks and in pink color with the chemical structure depicted. Hydrogen bonds are indicated by dashed lines. LX-2 cells (B), HSC-T6 cells (C) and C3A (D) cells were treated with incremental concentrations of HJC0123 for 48 hours, and cell viability was determined using Alamar Blue assay. Similarly, LX-2 cells were treated with increasing concentrations of commercially available STAT3 inhibitors (E) FLLL32 (left) and S31-201 (right). Dose response curves were calculated using GraphPad Prism 6.0. The results are representative of at least three independent experiments.
We hypothesized that the novel small molecule HJC0123 will inhibit STAT3 activation in human HSCs resulting in decreased fibrogenesis, unveiling a potential role for its use as a therapeutic agent for the treatment of liver fibrosis.
2. MATERIALS AND METHODS
2.1. Cell Lines and Culture and Reagents
The immortalized human HSC line LX-2 and rat HSC line HSC-T6 were used (American Type Cell Culture, Manassas, VA). Both cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) (Life Technologies Corporation, Grand Island, NY) with a high glucose concentration (4.5 g/L) supplemented with 5% fetal bovine serum (FBS), 100U/ml penicillin G and 100 μg/ml streptomycin (Gibco BRL, Gaithersburg, MD). Human hepatic hepatocytes C3A were also used and cultured in Eagle's Minimum Essential Medium (American Type Cell Culture, Manassas, VA) supplemented with 10% FBS. All cells were incubated at 37°C in a humidified atmosphere containing 5% CO2. The following antibodies were used: Alpha-smooth muscle actin (α-SMA) (A5228) and SAA2 (SAB1401350) (Sigma-Aldrich, St. Louis, MO); Fibronectin (sc-6952), STAT3 (sc-482), SOCS3 (sc-9023), Lamin-B (sc-6216), and alpha-Tubulin (sc-8035)(Santa Cruz Biotechnology, Santa Cruz, CA); phospho-STAT3 (Y705) (ab34716)(Abcam, Cambridge, MA); Collagen Type I (600-401-103) (Rockland Immunochemicals, Gilbertsville, PA); GAPDH (5174), Smad2/3 (8685) and phospho-Smad2/3 (8828) (Cell Signaling Technology, Danvers, MA; 5174); and Topoisomerase Iiβ (611492) (BD Biosciences, Franklin Lakes, NJ). The STAT3 inhibitors S31-201 (CAS501919-59-1) (Santa Cruz Biotechnology, Santa Cruz, CA) and FLLL32 (530153) (Millipore, Darmstadt, Germany) were used.
2.2. Alamar Blue Cell Viability Assay
LX-2 cells (3×103 cells/well) and HSC-T6 cells (4 × 103 cells/well) in 100 μL of medium were seeded into 96-well plates. After reaching 50-60% confluence, media was replaced and treated as indicated. Alamar Blue stock solution (Life Technologies Corporation, Grand Island, NY) was diluted 1:1 with culture medium and a volume of 25 μL/well was transferred into the assay plate for final concentration of 10% Alamar Blue. Cells were incubated for 4 additional hours. Fluorescence intensity was measured using a SpectraMax M2 microplate reader (Molecular Devices, LLC, Sunnyvale, CA) with excitation and emission wavelengths set at 540 and 590 nm, as previously described.19 Assay was performed in triplicate and repeated at least three times. Viability was assessed by Alamar Blue assay (Life Technologies, Grand Island, NY) following manufacturer's instructions.
2.3. Western Immunoblotting
Whole cell extracts were prepared as previously described.19 Protein (10-30 μg) was fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Life Technologies Corporation, Grand Island, NY) under denaturing conditions and then electro-transferred to a polyvinylidene fluoride (PVDF) membrane. After incubation with Blocking buffer (LI-COR, Inc., Lincoln, NE), membrane was probed with the indicated primary antibody (Ab). Membranes were washed with phosphate buffered saline with 0.1% Tween 20 (PBST), and incubated for 1 hour with IRDye (infrared fluorescent dye) 680-conjugated anti-mouse or IRDye 800-conjugated anti-rabbit Ab (LI-COR, Inc., Lincoln, NE). Finally, the membranes were washed with PBST, and signals were scanned and visualized by Odyssey Infrared Imaging System (LICOR, Inc., Lincoln, NE).
2.4. Luciferase Reporter Gene Activity Assay
We used the luciferase reporter assay (the Dual-Luciferase Reporter Assay System, Promega, UK, #E1910) to investigate the transcriptional activity of STAT3. Transient transfection was performed in 96-well plates at a cell density of 60-70% confluence per well. The pSTAT3 luciferase reporter plasmid was co-transfected with a constitutively expressing Renilla construct, which acts as internal control for transfection efficiency (Qiagen, Hilden, Germany; CCS-9028L) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The transfected cells were treated with HJC0123 for 24 hours, and the cell lysates were prepared for assessment of luciferase activity. Firefly and Renilla luciferase activities were measured using a luminometer (Centro XS3 LB960, Berthold, Germany) according to the manufacturer's instructions. Relative firefly luciferase activity normalized by Renilla luciferase activity was compared with dimethyl sulfoxide (DMSO), used as control.
2.5. Cell Cycle Analysis by Flow Cytometry
Nuclear DNA content was measured using propidium iodide staining and fluorescence-activated cell sorter analysis, as previously described.20 Briefly, 2 × 106 adherent cells were trypsinized, washed with PBS, resuspended in a low-salt stain solution (3% polyethylene glycol 8000, 50 μg of PI per mL, 0.1% Triton X-100, 4 mM sodium citrate, 180 units of RNase A per mL), and incubated at 37°C for 20 minutes. An equal volume of high-salt stain solution (3% polyethylene glycol 8000, 50 μg of propidium iodide per ml, 0.1% Triton X-100, and 400 mM sodium chloride) was then added to the cell suspension. Propidium iodide-stained nuclei were stored at 4°C at least 3 hours before fluorescence-activated cell sorter analysis using BD FACSCanto II flow cytometer (BD Biosciences, Franklin Lakes, NJ) at the University of Texas Medical Branch Flow Cytometry and Cell Sorting Core Facility. ModFit LT for Win32 software was used for data analysis (Verity Software House, Topsham, ME).
2.6. Immunofluorescence Staining
Cells were grown in chamber slides with supplemented medium. After 24 hours of incubation, the cells were treated with 1 μM HJC0123 for 48 hours. Cells were fixed with 4% formaldehyde diluted in phosphate buffered saline (PBS) for 15 minutes at room temperature and washed with PBS. After incubating for 60 minutes with blocking buffer, cells were incubated for 2 hours at room temperature with the corresponding primary antibody. Next, samples were rinsed with PBS for 5 minutes each, followed by a 2-hour incubation with corresponding secondary antibody at room temperature in the dark. Sections were counterstained with hematoxylin for 15 seconds before examined under microscope. Untreated cells were used as control. Only cytoplasmic or cytoplasmic and nuclear reactivity was considered as a positive staining. To quantify immunoreactivity, a single in-focus plane was acquired. Immunofluorescence was determined by Nikon Eclipse Ti confocal microscope at 20x magnification (Nikon Instruments, Inc, Melville, NY). Using Image J (v1.50b, NIH), mean corrected cellular fluorescence was calculated and equalized, with results presented as fold increase as previously described.21
2.7. Detection of Yo-Pro-1 and PI uptake
Cell death detection was performed as previously described.22 Briefly, 2 × 105 cells were seeded in 24-well plates. Cells were treated as indicated for 24 hours. After being washed with PBS, cell were incubated for 1 hour with Yo-Pro-1 and Propidium iodide (PI) mixed solutions according to the manufacturer's instructions. Yo-Pro-1 and PI uptake were determined using the Nikon Eclipse Ti confocal microscope at 20x magnification (Nikon Instruments, Inc, Melville, NY) and total corrected cellular fluorescence was calculated and equalized, with results presented as fold increase using Image J (v1.50b, NIH).
2.8. TUNEL Assay
Cells were fixed with 4% paraformaldehyde for 20 minutes after the treatment in triplicate and were stained using the DeadEnd™ Fluorometric TUNEL System (Promega G3250). The DeadEnd™ Fluorometric TUNEL System is a standard TUNEL assay, able to detect apoptosis in many cell types. The Fluorometric TUNEL system measures the fragmented DNA of apoptotic cells by incorporating flurorecein-12 dUTP at 3’-OH DNA ends using Terminal Deoxynucleotiyl Transferase, recombinant, enzyme (rTdT). rTdT forms a polymeric tail using the principle of the TUNEL (TdT-mediated dUTP Nick-End labeling) assay. The flurorecein-12 dUTP incorporation results in localized green fluorescence within the nucleus of apoptotic cells only. Total corrected cellular fluorescence was calculated and equalized, with results presented as fold increase using Image J (v1.50b, NIH).
2.9. Measurement of IL-6 secretion
The concentrations of secreted IL-6 in the cell culture supernatant were determined using a specific human IL-6 enzyme-link immunosorbent assay (ELISA) kit according to the manufacturer's instructions.
2.10. Data and statistical analysis
The data and statistical analysis comply with the recommendations on experimental design and analysis in pharmacology.23 Statistical analysis was performed using GraphPad Prism 6.0 (GraphPad Software Inc. La Jolla, CA). Technical replicates were used to ensure the reliability of single values. The results are representative of at least three independent experiments. Data presented as mean ± standard error of the mean (SEM), with significance defined as p < 0.05.
3. RESULTS
3.1. HJC0123 decreases HSCs Proliferation
In order to assess the effects of HJC0123 on hepatic stellate cell proliferation, we measured cell viability using the Alamar Blue assay. Cells were treated with varying concentrations of HJC0123 or its diluent (DMSO) as control. Compared to the non-treated control groups, HJC0123 treatment for 48 hours significantly decreased cell viability of human LX-2 and murine HSC-T6 cell lines in a dose-dependent manner. After construction of a dose-response curve, IC50s were calculated. In the LX-2 cell line HJC0123 IC50 was 0.85 μM (95% CI 0.80-0.90; Figures 1B). In the HSC-T6 cell line HJC0123 IC50 was 4.04 μM (95% CI 3.78-4.31; Figures 1C). Similar effects on LX-2 cells proliferation were observed with the use of STAT3 inhibitors, FLLL32 and S31-201, and respective IC50's are shown in Figure 1D. In addition, we examined the cytotoxicity of HJC0123 in parenchymal cells by assessing cell viability after treatment with HJC0123 in the human C3A cell line, a well differentiated clonal derivative of Hep G2 cells and used extensively for in vitro hepatic toxicity assays.24,25 The calculated IC50 was 5.61 μM (95% CI 4.84-6.49; Figure 1E), more than 6 times higher than the IC50 in human HSCs, indicating that HJC0123 lacks significant cytotoxic effects on hepatocytes at the proposed dose of 1.0 μM.
3.2. HCJ0123 inhibits the STAT3 signaling pathway
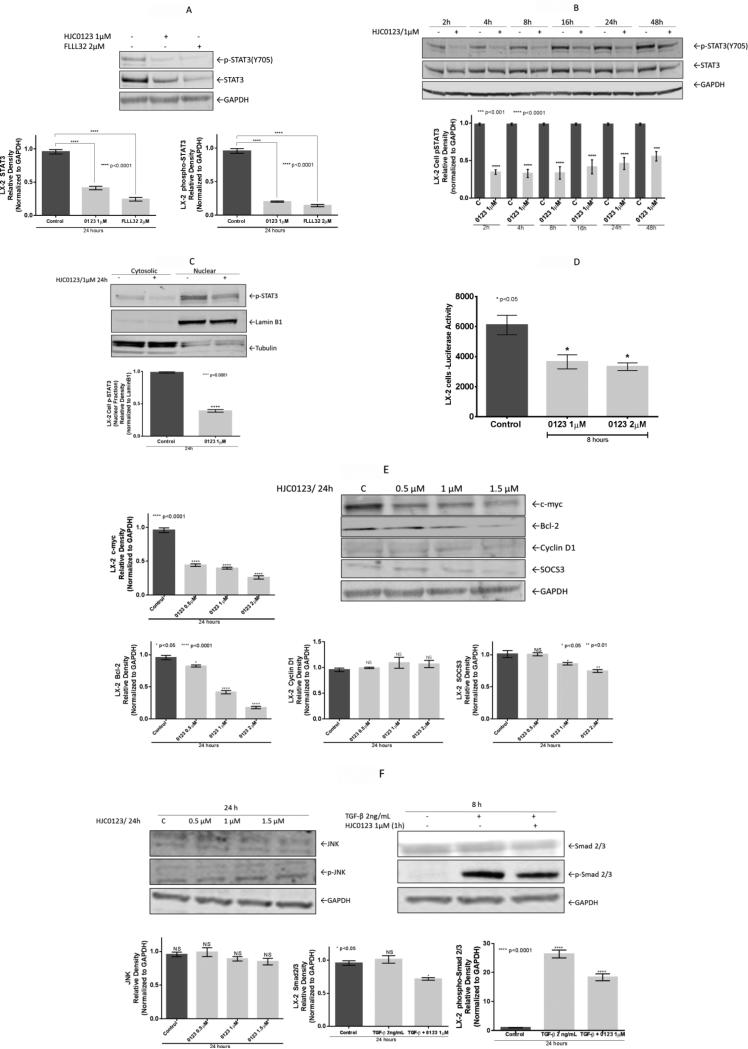
The effects of HJC0123 on the STAT3 signaling pathway were examined. Protein expression was measured, LX-2 cells levels of STAT3 and pSTAT3(Y705) decreased significantly after 24 hours of HJC0123 treatment. A similar response was demonstrated with FLLL32 and S31-201 treatment (data not shown), commercially available STAT3 inhibitors (Figure 2A). Next, we assessed the time-dependent effects of HJC0123 on STAT3 phosphorylation; STAT3 phosphorylation was inhibited as early as 2 hours and remained downregulated up to 48 hours after treatment (Figure 2B). Levels of pSTAT3(S727) were not affected by HJC0123 treatment (data not shown). After fractionation, nuclear and cytosolic protein expression was measured; HJC0123 downregulated nuclear pSTAT3(Y705) indicating that pSTAT3(Y705) nuclear translocation was inhibited by HJC0123 (Figure 2C). STAT3 transcriptional activity was measured via the luciferase reporter assay, HJC0123 decreased transcriptional activity by 40% after treatment for 8 hours (Figure 2D). Next, the expression of STAT3-regulated proteins was analyzed; HJC0123 significantly decreased the expression of c-myc and Bcl-2. However, expression levels of cyclin D1 did not change significantly. Further, we examined the effects of HJC0123 treatment on the expression of the suppressor of cytokine signaling-3 (SOCS3), an endogenous modulator of the activation of STAT3. HJC0123 treatment downregulated SOCS3 expression in a dose-dependent fashion after 24 hours treatment (Figure 2E).
Figure 2. Treatment with HJC0123 impairs the STAT3 signaling pathway in LX-2 cells.
(A) FLLL32 and HJC0123 treatment inhibited STAT3 expression and STAT3 phosphorylation HJC0123; protein expression was measured by Western immunoblotting. (B) HJC0123 treatment (1 μM) significantly decreased the expression of pSTAT3(Y705) as early as 2 hours and remained downregulated up to 48 hours; expression was measured by Western immunoblotting. Levels of non-phosphorylated STAT3 were not significantly altered after HJC0123 treatment. (C) HJC0123 (1 μM for 24 hours) significantly reduced nuclear translocation of pSTAT3(Y705), assessed by nuclear fractionation and Western blotting. (D) HJC0123 treatment for 8 hours significantly decreased the transcriptional activity of STAT3 in a dose-dependent manner, measured by Luciferase reporter gene activity assay. (E) LX-2 cells were treated with HJC0123 for 24 hours, Western blotting analysis demonstrated a significant reduction in the expression levels of the STAT3-regulated proteins c-myc, Bcl-2 and SOCS3 in a dose-dependent fashion; HJC0123 did not alter the expression of Cyclin D1. (F) LX- cells were treated with HJC0123, proteins expression measured via Western blotting, expression of JNK and p-JNK did not change significantly after 24 hours treatment (left). TGF-β stimulation (2 ng/mL) did not affect the expression of Smad2/3 but induced Smad2/3 phosphorylation. HJC0123 1 hour pre-treatment with HJC0123 (1 μM) followed by TGF-β stimulation for 8 hours significantly downregulated TGF- β-induced Smad2/3 phosphorylation (right). Experiments were repeated three times and representative results are shown. Error bars indicate ± SE.
Molecular crosstalk between STAT3 and other signaling pathways is well recognized. We examined the levels of the phosphorylated and non-phosphorylated forms of JNK (c-Jun N-terminal kinase), an upstream activator of the STAT3 pathway; HJC0123 did not alter significantly the expression of JNK or p-JNK (Figure 2F). Next, we analyzed the effects of HJC0123 treatment on the TGF-β-induced Smad2/3 activation, a key process in HSC fibrogenesis.2,26 TGF-β stimulation (2 ng/mL) did not change the expression levels of Smad2/3. However, TGF-β stimulation significantly increased Smad2/3 phosphorylation (~26 fold increase). Further, HJC0123 pre-treatment (1 μM, 1 hour) significantly decreased TGF-β stimulated Smad2/3 phosphorylation in LX-2 cells. (Figure 2F).
3.3. HJC0123 promotes cell cycle arrest and apoptosis
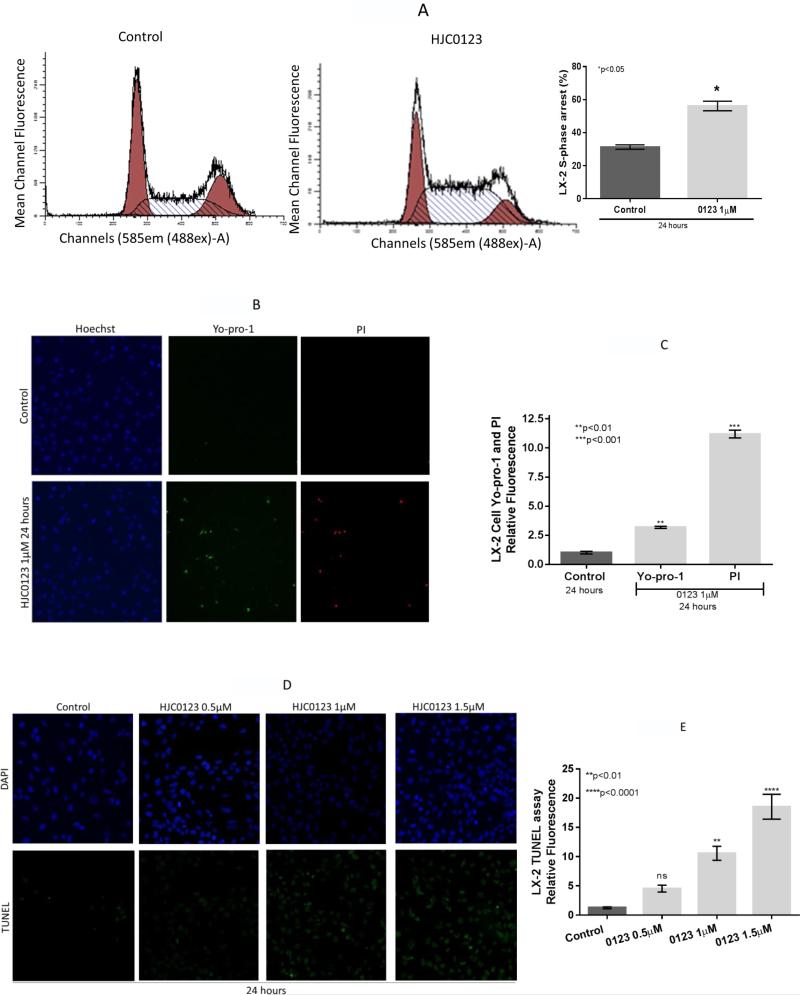
The pro-apoptotic effects of HJC0123 treatment in LX-2 cells were assessed. First, cell cycle was examined using flow cytometry. HJC0123 significantly increased LX-2 S-phase cell cycle arrest when compared to the control (56.19 ± 4.10% versus 31.30 ± 1.86%, p=0.016; Figure 3A). Second, immunofluorescence staining was used to detect cell death, after 1 μM HJC0123 treatment for 24 hours, LX-2 cells showed a three-fold increase of Yo-pro-1 staining, a marker of early apoptosis and an eleven-fold increase of PI staining, a marker of late apoptosis (Figures 3B, 3C). Third, cell death was also assessed with TUNEL assay; HJC0123 treatment induced LX-2 cell death in a dose-dependent manner (Figures 3D, 3E). In a similar fashion, the anti-apoptotic protein Bcl-2 was downregulated after HJC0123 treatment (Figure 2E). Caspase 3 and cleaved caspase 3 levels were not affected by HJC0123 treatments. Taken together, these findings seem to indicate that HJC0123 promotes caspase-independent apoptosis of HSCs.
Figure 3. HJC0123 induces LX-2 cell cycle arrest and promotes apoptosis.
(A) LX-2 cells were treated with HJC0123 (1 μM) for 24 hours. Cells were stained with propidium iodide as described in Methods and analyzed by flow cytometry; HJC0123 treatment significantly increased S-phase arrest. (B, C) Apoptosis was assessed by immunofluorescence staining using Yo-Pro-1 and PI as markers of early and late apoptosis, respectively; Hoechst 3342 was used as nuclear stain. HJC0123 treatment (1 μM for 24 hours) significantly induced early and late apoptosis. Immunofluorescence results were quantified as described in Methods. (D,E) Fluorometric TUNEL assay showed increased cell death in a dose-dependent fashion. The results are representative of at least three independent experiments. Error bars indicate ± SE.
3.4. HJC0123 inhibits HSC activation and downregulates ECM proteins expression
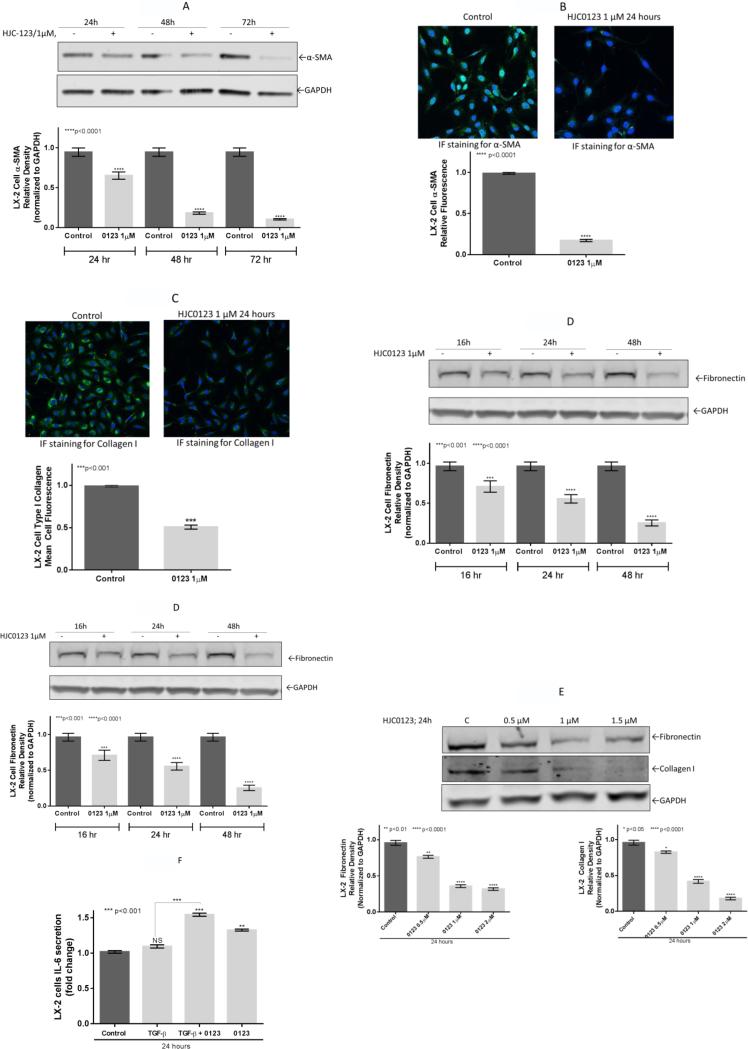
To explore the antifibrogenic properties of HJC0123, we measured the expression of α-SMA, an activation marker of HSCs, and the production of the ECM proteins, type I collagen and fibronectin. LX-2 cells expression of α-SMA, was significantly reduced after 24, 48 and 72 hours of HJC0123 treatment compared to the control (36, 83 and 89% respectively; Figure 4A). Next, these findings were confirmed using immunofluorescence staining, which consistently showed reduction of α-SMA expression after HJC0123 treatment (83% decrease compared to control; Figure 4B). Further, type I collagen was significantly downregulated by 50% after HJC0123 treatment (Figure 4C). Similarly, Western immunoblotting revealed that HJC0123 treatment decreased the endogenous expression of fibronectin in a time-dependent manner (33, 43, and 74%, respectively; Figure 4D). Both collagen and fibronectin expression were downregulated in a dose-dependent fashion (Figure 4E).
Figure 4. HJC0123 inhibits hepatic stellate cells activation and decreases endogenous ECM proteins expression.
The expression levels of α-SMA, a marker of HSCs activation, were measured by (A) Western immunoblotting and (B) immunofluorescence analysis. HJC0123 treatment (1 μM for 24 hours) significantly inhibited the expression of α-SMA in a dose-dependent manner. (C) LX-2 cells treated with HJC0123 (1 μM for 24 hours) showed a significant decrease of expression of Collagen type I, assessed by Immunofluorescence staining. (D) HJC0123 treatment (1 μM) significantly reduced the expression of fibronectin in a time-dependent manner and (E) significantly decreased the endogenous expression of both ECM proteins, fibronectin and collagen type I, in a dose-dependent fashion (24 hours treatment); proteins levels were measured by Western immunoblotting. (F) Secretion of IL-6 was not significantly affected by TGF-β stimulation (2 ng/mL). However, HJC0123 (1μM) used alone or as pre-treatment followed by TGF-β stimulation significantly increased IL-6 production. Experiments were repeated three times and representative results are shown. Error bars indicate ± SE.
IL-6 is a known positively regulator of STAT3 phosphorylation mediated by the activation of receptor-associated kinases.27 However, the results of STAT3 inhibition on the production and autocrine effects of IL-6 in HSCs remain poorly explored. We examined the effects of HJC0123 treatment on IL-6 production in LX-2 cells via ELISA. Secretion of IL-6 was not significantly affected by TGF-β stimulation (2 ng/mL). However, HJC0123 (1μM) used alone or as pre-treatment followed by TGF-β stimulation significantly increased IL-6 production. (Figure 4F).
4. DISCUSSION
We have demonstrated that HJC0123 inhibits the STAT3 signaling pathway in hepatic stellate cells. HJC0123 treatment impairs STAT3 phosphorylation, nuclear translocation, and STAT3-regulated gene expression. Subsequently, HJC0123 inhibits HSCs proliferation, decreases activation, and downregulates the endogenous production of ECM proteins. These results indicate that HJC0123 represents a novel molecule that impairs STAT3 activation and can be considered as a lead candidate for further pharmacological development in the treatment of liver fibrosis.
STAT3 overexpression has been detected in the majority of the rodent models of liver injury and in human liver diseases. However, the role of the STAT3 pathway in liver fibrosis is controversial due to its model-dependent and cell-specific functions.28,29 Studies in murine models of carbon tetrachloride (CCl4) induced fibrosis and sclerosing cholangitis have shown that STAT3 activation prevents liver fibrosis by enhancing hepatocyte survival and liver regeneration.30–33 Conversely, activation of STAT3 in the dimethylnitrosamine (DMN) induced fibrosis model increases transforming growth factor β-1 (TGF-β1) production in hepatocytes, promoting fibrosis.29 In contrast, STAT3 activation in HSCs results in cell activation and increased proliferation leading to fibrogenesis.9,34–37 The STAT3-dependent mechanisms involved in fibrogenesis include an increase in TGF-β1 expression, downregulation of SREBP-1c, and decrease in MMP-1 expression. These mechanisms, and likely others not yet elucidated, lead to HSC activation and increased production and accumulation of ECM proteins, the hallmark of liver fibrosis.29,37,38The observed antifibrogenic response in our study correspond with the pivotal role of STAT3 in liver fibrogenesis, which upon activation promotes the fibrogenic behavior of HSCs. STAT3 is activated by diverse signaling proteins, including cytokines (e.g., Il-6, IL-17, leptin) and growth factors (via receptor tyrosine kinases). In addition, crosstalk between signaling pathways (e.g., JNK [c-Jun N-terminal kinase], NFκB [nuclear factor kappa-light-chain-enhancer of activated B cells]) also can result in STAT3 activation; such convergence makes STAT3 activation an optimal therapeutic target.
Fragment-based drug discovery is an attractive approach to screening for the generation of chemical leads for drug targets.13 HJC0123 was designed using six privileged fragments from non-peptidic STAT3 inhibitors in order to identify orally bioavailable inhibitors.16,18 In the present study, we have demonstrated that HJC0123 inhibits proliferation of HSCs at low (micromolar to submicromolar) concentrations, and decreases the phosphorylation of STAT3 in HSCs. In addition, by assessing cell viability in C3A cells after treatment with increasing doses of HJC0123, a method used by others to assess hepatotoxicity in vitro24,25, our data show that HJC0123 lacks significant cytotoxicity in hepatocytes at the dose range used to affect HSCs. Furthermore, we have shown that HJC0123 induces cell cycle arrest and promotes apoptosis. These findings are consistent with the well-known functions of STAT3, which include cell cycle progression, cell survival, and mitogenesis.28,39,40 Our observations are supported by evidence of increased expression of markers of early and late apoptosis (Yo-pro-I and PI, respectively) as demonstrated by immunofluorescence staining, decreased expression of the anti-apoptotic protein Bcl-2, downregulation of c-myc, a known cell cycle and apoptosis regulator. Caspases activation is not observed during HJC0123-induced apoptosis in LX-2 cells. Previous studies examining apoptosis after STAT3 inhibition in malignant glioma cells found neither PARP cleavage nor caspase activation while caspase inhibitor treatment did not affect cell death index, suggesting mechanisms of caspase-independent apoptosis.41 Chen et al recently described caspase-independent apoptosis in HSCs while studying the antifibrogenic properties of Saikosaponin d.42 Although we did not fully explore the involved apoptotic mechanisms, taking together with previous literature, our data suggest that HJC0123 treatment and subsequent STAT3 inhibition might lead to caspase-independent apoptosis.
Recent studies have improved our understanding of STAT3 activity. In addition to the originally described activation by phosphorylation STAT3(Y705), it is recognized that dimers of nonphosphorylated STAT3 can exist and be active, and phosphorylation of either STAT3(Y705), STAT3(S727) or both can produce transcriptional activity.43–45 HJC0123 treatment inhibits phosphorylation and also decreases STAT3 nuclear translocation, further diminishing its transcriptional activity as confirmed by the downregulation of STAT3-regulated genes. SOCS3, induced by STAT3, inhibits STAT3 activation and modulates the expression of inflammatory genes triggered by the IL-6-type cytokines, acting as a negative feedback mechanism regulator.46,47 Expectedly, the inhibition of STAT3 phosphorylation results in decreased expression of SOCS3. Similarly to STAT3, the biological functions of SOCS3 are cell-dependent. We speculate that its downregulation is not a direct effect of HJC0123 treatment but it is most likely secondary to the exogenous inhibition of STAT3. Recently, Chen et al described that SOCS3 contributes to LX-2 senescence by interacting with p53 and enhancing p21 expression leading to cell cycle arrest in LX-2 cells exposed to soluble egg antigens from Schistosoma japonicum.48,49 Notably, the authors found that SOCS3 increased expression was accompanied by augmented STAT3 phosphorylation48 suggesting that diverse mechanisms affecting STAT3 (and simultaneously SOCS3) activity, either by enhancement or inhibition, may lead to an antifibrogenic response.
Other groups have also explored STAT3 activation as a strategy to ameliorate liver fibrogenesis. The anti-inflammatory cytokine IL-22 has been identified to affect HSCs activation and senescence. IL-22 induces cellular senescence by upregulating the expression of p53 and p21 via STAT3.50 However, these effects require the exogenous administration of IL-22. During liver injury or hepatic fibrosis, HSCs are exposed to a pro-inflammatory microenvironment due to autocrine and paracrine effects of parenchymal and non-parenchymal cells.51–53 Proinflammatory cytokines activate STAT3 eliciting a fibrogenic cell behavior. Thus, we consider that the inhibition of the STAT3 pathway is a valid approach to modulate fibrogenesis. Therefore, it must be considered that the dichotomy of STAT3 role in HSCs depends on the inflammatory context (endotoxin/cytokines exposure) and other stimuli that trigger or perpetuate fibrogenesis suggesting that these factors must be accounted for when utilizing interventions targeting STAT3.
Receptor tyrosine kinase (RTK) inhibitors use has been explored in liver fibrogenesis.54 Kim et al demonstrated that imatinib has antifibrogenic properties and reported an unexpected link between imatinib treatment and increased secretion of IL-6.55 HJC0123, with similar suppressive effects of HSC activation, also enhances IL-6 secretion. IL-6 is involved in the activation of DNA repair enzymes in hepatocytes and is needed for hepatic regeneration;56,57 its increased production in the setting of decreased HSC activation and suppressed fibrogenesis could represent a restorative response to increase hepatocyte mass and function. However, these observations need to be supported by evidence from in vivo studies, as concern might exist regarding the oncogenic potential of IL-6.58
The activation of the JNK pathway results in increased fibrogenesis and explained the profibrotic profile of several cytokines (e.g., IL-17A) and acute phase proteins (e.g., Serum Amyloid A).59,60 Crosstalk between the JNK and STAT3 pathway has been studied in different cells. Gkouveris et al reported that JNK decreases STAT3(Y705) phosphorylation leading to inhibition of STAT3 activity in cell lines of oral squamous cell carcinoma.61 Conversely, Kim et al found that JNK-dependent STAT3 activation is associated with doxorubicin resistance in different breast cancer cell lines. 62 We did not find significant changes in the activity of the JNKs signaling pathway.
The TGF-β-induced activation of Smad2/3 phosphorylation triggers a strong fibrogenic response in HSCs.63–65 Crosstalk between the STAT and Smad pathway has been reported.66,67 Both pathways can function cooperatively to modulate gene expression and also modulate each other. HJC0123 partially inhibited the TGF-β induced phosphorylation of Smad2/3. As previously exposed by others, the role of JNK in STAT3 modulation and STAT interaction with Smads vary significantly according to the cell type and pathological process.68 The understanding of the underlying crosstalk between other molecules is helpful to design therapeutic interventions.
We have demonstrated that HJC0123 downregulates α-SMA, a surrogate marker of HSC activation. Furthermore, we have shown that HJC0123 treatment inhibits fibronectin and type I collagen production. These antifibrogenic properties of HJC0123 can potentially be explained by the inhibition of TGF-β1 dependent collagen I production as a consequence of STAT3 inhibition.
5. CONCLUSION
HJC0123 inhibits STAT3 activity in HSCs resulting in decreased fibrogenesis. The present study demonstrated the use of FBDD for the development of a novel compound, with reproducible biological activity and enhanced potency. HJC0123 can be administered orally with favorable bioavailability and excellent tolerance to high doses in a murine model.18 However, further work including in vivo studies are warranted to elucidate the detailed mechanisms that link STAT3 to the fibrogenic cellular events, allowing for the development of effective therapeutic strategies.
ACKNOWLEDGEMENTS
This work was supported by grants P50 CA097007, P30 DA028821, R21 MH098344 (J.Z.) and T32-GM8256 (F.J.B.) from the National Institutes of Health, Cancer Prevention Research Institute of Texas award, R. A. Welch Foundation Chemistry and Biology Collaborative Grant (J.Z.) from the Gulf Coast Consortia, and John Sealy Memorial Endowment Fund and the Center for Addiction Research (J.Z.) from the University of Texas Medical Branch. The authors thank Dr. Arline Kandathiparampil for her technical help, Dr. Celeste C. Finnerty for her critical and Steve Schuenke and Eileen Figueroa for their editorial assistance.
Footnotes
7. DISCLOSURE
The authors have no commercial or financial relationships that could represent a potential conflict of interest.
REFERENCES
- 1.Hernandez-Gea V, Friedman SL. Pathogenesis of Liver Fibrosis. Annu Rev Pathol Mech Dis. 2011;6(1):425–456. doi: 10.1146/annurev-pathol-011110-130246. doi:10.1146/annurev-pathol-011110-130246. [DOI] [PubMed] [Google Scholar]
- 2.Elpek GÖ. Cellular and molecular mechanisms in the pathogenesis of liver fibrosis: An update. World J Gastroenterol. 2014;20(23):7260. doi: 10.3748/wjg.v20.i23.7260. doi:10.3748/wjg.v20.i23.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puche JE, Saiman Y, Friedman SL. Hepatic stellate cells and liver fibrosis. Compr Physiol. 2013;3(4):1473–1492. doi: 10.1002/cphy.c120035. doi:10.1002/cphy.c120035. [DOI] [PubMed] [Google Scholar]
- 4.Ellis EL, Mann DA. Clinical evidence for the regression of liver fibrosis. J Hepatol. 2012;56(5):1171–1180. doi: 10.1016/j.jhep.2011.09.024. doi:10.1016/j.jhep.2011.09.024. [DOI] [PubMed] [Google Scholar]
- 5.Kisseleva T, Brenner D a. Role of hepatic stellate cells in fibrogenesis and the reversal of fibrosis. J Gastroenterol Hepatol. 2007;22(Suppl 1)(May):S73–8. doi: 10.1111/j.1440-1746.2006.04658.x. doi:10.1111/j.1440-1746.2006.04658.x. [DOI] [PubMed] [Google Scholar]
- 6.Bataller R, Brenner D a. Hepatic Stellate Cells as a Target for the Treatment of Liver Fibrosis. Semin Liver Dis. 2001;21(3):437–452. doi: 10.1055/s-2001-17558. doi:10.1055/s-2001-17558. [DOI] [PubMed] [Google Scholar]
- 7.Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134(2):1655–1669. doi: 10.1053/j.gastro.2008.03.003. doi:10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong X, Horiguchi N, Mori M, Gao B. Cytokines and STATs in Liver Fibrosis. Front Physiol. 2012;3(April):69. doi: 10.3389/fphys.2012.00069. doi:10.3389/fphys.2012.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, Lafdil F, Kong X, Gao B. Signal transducer and activator of transcription 3 in liver diseases: A novel therapeutic target. Int J Biol Sci. 2011;7(5):536–550. doi: 10.7150/ijbs.7.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang JX, Mikami K, Venugopal S, Li YTN. Apoptotic body engulfment by hepatic stellate cells promotes their survival by the JAK/STAT and Akt/NF-kappaB-dependent pathways. J Hepatol. 2009;51(1):139–148. doi: 10.1016/j.jhep.2009.03.024. doi:10.1016/j.jhep.2009.03.024.Apoptotic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parola M, Marra F. Adipokines and redox signaling: impact on fatty liver disease. Antioxid Redox Signal. 2011;15(2):461–483. doi: 10.1089/ars.2010.3848. doi:10.1089/ars.2010.3848. [DOI] [PubMed] [Google Scholar]
- 12.Su T-H, Shiau C-W, Jao P, et al. Sorafenib and its derivative SC-1 exhibit antifibrotic effects through signal transducer and activator of transcription 3 inhibition. Proc Natl Acad Sci. 2015;112(23):7243–7248. doi: 10.1073/pnas.1507499112. doi:10.1073/pnas.1507499112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H, Zhou X, Wang A, Zheng Y, Gao Y, Zhou J. Evolutions in fragment-based drug design: the deconstruciton-reconstruction approach. Drug Discov Today. 2015;20(1):105–113. doi: 10.1016/j.drudis.2014.09.015. doi:10.1016/j.surg.2006.10.010.Use. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erlanson DA. Fragment-based lead discovery: a chemical update. Curr Opin Biotechnol. 2006;17(6):643–652. doi: 10.1016/j.copbio.2006.10.007. doi:10.1016/j.copbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Leach AR, Hann MM. Molecular complexity and fragment-based drug discovery: Ten years on. Curr Opin Chem Biol. 2011;15(4):489–496. doi: 10.1016/j.cbpa.2011.05.008. doi:10.1016/j.cbpa.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 16.Murray CW, Verdonk ML, Rees DC. Experiences in fragment-based drug discovery. Trends Pharmacol Sci. 2012;33(5):224–232. doi: 10.1016/j.tips.2012.02.006. doi:10.1016/j.tips.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Chen H, Yang Z, Ding C, et al. Discovery of Potent Anticancer Agent HJC0416, an Orally Bioavailable Small Molecule Inhibitor of Signal Transducer and Activator of Transcription 3 (STAT3). Eur J Med Chem. 2014;82:195–203. doi: 10.1016/j.ejmech.2014.05.049. doi:10.1016/j.surg.2006.10.010.Use. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H, Yang Z, Ding C, et al. Fragment-based drug design and identification of HJC0123, a novel orally bioavailable STAT3 inhibitor for cancer therapy. Eur J Med Chem. 2013;62:498–507. doi: 10.1016/j.ejmech.2013.01.023. doi:10.1016/j.ejmech.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohanon FJ, Wang X, Graham BM, et al. Enhanced anti-fibrogenic effects of novel oridonin derivative CYD0692 in hepatic stellate cells. Mol Cell Biochem. 2015;410(1-2):293–300. doi: 10.1007/s11010-015-2562-4. doi:10.1007/s11010-015-2562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bohanon FJ, Wang X, Ding C, et al. Oridonin inhibits hepatic stellate cell proliferation and fibrogenesis. J Surg Res. 2014;190(1):55–63. doi: 10.1016/j.jss.2014.03.036. doi:10.1016/j.jss.2014.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A. Partial inhibition of Cdk1 in G2 phase overrides the SAC and decouples mitotic events. Cell Cycle. 2014;13(9):1400–1412. doi: 10.4161/cc.28401. doi:10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bohanon FJ, Wang X, Graham BM, et al. Enhanced effects of novel oridonin analog CYD0682 for hepatic fibrosis. J Surg Res. 2015:1–9. doi: 10.1016/j.jss.2015.07.042. doi:10.1016/j.jss.2015.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Curtis MJ, Bond RA, Spina D, et al. Experimental design and analysis and their reporting: new guidance for publication in BJP. Br J Pharmacol. 2015;172:3461–3471. doi: 10.1111/bph.12856. doi:10.1111/bph.12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bandele OJ, Santillo MF, Ferguson M, Wiesenfeld PL. In vitro toxicity screening of chemical mixtures using HepG2/C3A cells. Food Chem Toxicol. 2012;50(5):1653–1659. doi: 10.1016/j.fct.2012.02.016. doi: http://dx.doi.org/10.1016/j.fct.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 25.Flynn TJ, Ferguson MS. Multiendpoint mechanistic profiling of hepatotoxicants in HepG2/C3A human hepatoma cells and novel statistical approaches for development of a prediction model for acute hepatotoxicity. Toxicol In Vitro. 2008;22(6):1618–1631. doi: 10.1016/j.tiv.2008.04.016. doi:10.1016/j.tiv.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 26.Dooley S, Delvoux B, Streckert M, et al. Transforming growth factor-beta and platelet-derived growth factor signal via c-Jun N-terminal kinase-dependent Smad2/3 phosphorylation in rat hepatic stellate cells after acute liver injury. FEBS Lett. 2001;502(1-2):4–10. doi: 10.1016/s0014-5793(01)02656-4. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, van Boxel-Dezaire AHH, Cheon H, Yang J, Stark GR. STAT3 activation in response to IL-6 is prolonged by the binding of IL-6 receptor to EGF receptor. Proc Natl Acad Sci U S A. 2013;110(42):16975–16980. doi: 10.1073/pnas.1315862110. doi:10.1073/pnas.1315862110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu M-Y, Hu J-J, Shen J, et al. Stat3 signaling activation crosslinking of TGF-β1 in hepatic stellate cell exacerbates liver injury and fibrosis. Biochim Biophys Acta. 2014;1842(11):1–9. doi: 10.1016/j.bbadis.2014.07.025. doi:10.1016/j.bbadis.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 29.Ogata H, Chinen T, Yoshida T, et al. Loss of SOCS3 in the liver promotes fibrosis by enhancing STAT3-mediated TGF-beta1 production. Oncogene. 2006;25(17):2520–2530. doi: 10.1038/sj.onc.1209281. doi:10.1038/sj.onc.1209281. [DOI] [PubMed] [Google Scholar]
- 30.Gao B, Wang H, Lafdil F, Feng D. STAT proteins - Key regulators of anti-viral responses, inflammation, and tumorigenesis in the liver. J Hepatol. 2012;57(2):430–441. doi: 10.1016/j.jhep.2012.01.029. doi:10.1016/j.jhep.2012.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mair M, Zollner G, Schneller D, et al. Signal Transducer and Activator of Transcription 3 Protects From Liver Injury and Fibrosis in a Mouse Model of Sclerosing Cholangitis. Gastroenterology. 2010;138(7):2499–2508. doi: 10.1053/j.gastro.2010.02.049. doi:10.1053/j.gastro.2010.02.049. [DOI] [PubMed] [Google Scholar]
- 32.Plum W, Tschaharganeh DF, Kroy DC, et al. Lack of glycoprotein 130/signal transducer and activator of transcription 3-mediated signaling in hepatocytes enhances chronic liver injury and fibrosis progression in a model of sclerosing cholangitis. Am J Pathol. 2010;176(5):2236–2246. doi: 10.2353/ajpath.2010.090469. doi:10.2353/ajpath.2010.090469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horiguchi N, Lafdil F, Miller AM, et al. Dissociation of Liver Inflammation and Hepatocellular Damage Induced by Carbon Tetrachloride in Myeloid Cell-Specific STAT3 Knockout Mice. Hepatology. 2010;51(5):1724–1734. doi: 10.1002/hep.23532. doi:10.1073/pnas.1222384110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saxena NK, Ikeda K, Rockey DC, Friedman SL, Anania FA. Leptin in Hepatic Fibrosis: Evidence for Increased Collagen Production in Stellate Cells and Lean Littermates of ob/ob Mice. 2002;35(4):762–771. doi: 10.1053/jhep.2002.32029. doi:10.1053/jhep.2002.32029.Leptin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saxena NK, Saliba G, Floyd J, Anania FA. Leptin Induces Increased α2(I) Collagen Expression in Cultured Rat Hepatic Stellate Cells. J Cell Biochem. 2003;89(2):311–320. doi: 10.1002/jcb.10494. doi:10.1016/j.surg.2006.10.010.Use. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meng F, Wang K, Aoyama T, et al. IL-17 signaling in inflammatory cells, Kupffer cells and Hepatic Stellate cells exacerbates liver fibrosis Fanli. Gastroenterology. 2012;143(3):1–18. doi: 10.1053/j.gastro.2012.05.049. doi:10.1053/j.gastro.2012.05.049.IL-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang W, Niu M, Yan K, et al. Stat3 pathway correlates with the roles of leptin in mouse liver fibrosis and sterol regulatory element binding protein-1c expression of rat hepatic stellate cells. Int J Biochem Cell Biol. 2013;45(3):736–744. doi: 10.1016/j.biocel.2012.12.019. doi:10.1016/j.biocel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 38.Schon H-T, Bartneck M, Borkham-Kamphorst E, et al. Pharmacological Intervention in Hepatic Stellate Cell Activation and Hepatic Fibrosis. Front Pharmacol. 2016 Feb;7 doi: 10.3389/fphar.2016.00033. doi:10.3389/fphar.2016.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Leclercq I, Brymora JM, et al. Kupffer Cells Mediate Leptin-Induced Liver Fibrosis. 2009;137(2):713–723. doi: 10.1053/j.gastro.2009.04.011. doi:10.1053/j.gastro.2009.04.011.KUPFFER. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirano T, Ishihara K, Hibi M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene. 2000;19(21):2548–2556. doi: 10.1038/sj.onc.1203551. doi:10.1038/sj.onc.1203551. [DOI] [PubMed] [Google Scholar]
- 41.Premkumar DR, Jane EP, Pollack IF. Cucurbitacin-I inhibits Aurora kinase A, Aurora kinase B and survivin, induces defects in cell cycle progression and promotes ABT-737-induced cell death in a caspase-independent manner in malignant human glioma cells. Cancer Biol Ther. 2015;16(2):233–243. doi: 10.4161/15384047.2014.987548. doi:10.4161/15384047.2014.987548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen M-F, Huang SJ, Huang C-C, et al. Saikosaponin d induces cell death through caspase-3-dependent, caspase-3-independent and mitochondrial pathways in mammalian hepatic stellate cells. BMC Cancer. 2016;16:532. doi: 10.1186/s12885-016-2599-0. doi:10.1186/s12885-016-2599-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li C, Iness A, Yoon J, et al. Noncanonical STAT3 activation regulates excess TGF-β1 and collagen I expression in muscle of stricturing Crohn's disease. J Immunol. 2015;194(7):3422–3431. doi: 10.4049/jimmunol.1401779. doi:10.4049/jimmunol.1401779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Y, Qiu J, Dong S, et al. Stat3 isoforms, α and β, demonstrate distinct intracellular dynamics with prolonged nuclear retention of Stat3β mapping to its unique C-terminal end. J Biol Chem. 2007;282(48):34958–34967. doi: 10.1074/jbc.M704548200. doi:10.1074/jbc.M704548200. [DOI] [PubMed] [Google Scholar]
- 45.Wake MS, Watson CJ. STAT3 the oncogene - Still eluding therapy? FEBS J. 2015;282(14):2600–2611. doi: 10.1111/febs.13285. doi:10.1111/febs.13285. [DOI] [PubMed] [Google Scholar]
- 46.Rottenberg ME, Carow B. SOCS3 and STAT3, major controllers of the outcome of infection with Mycobacterium tuberculosis. Semin Immunol. 2014;26(6):518–532. doi: 10.1016/j.smim.2014.10.004. doi:10.1016/j.smim.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 47.Mahony R, Ahmed S, Diskin C, Stevenson NJ. SOCS3 revisited: a broad regulator of disease, now ready for therapeutic use? Cell Mol Life Sci. 2016;73(17):3323–3336. doi: 10.1007/s00018-016-2234-x. doi:10.1007/s00018-016-2234-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J, Pan J, Wang J, et al. Soluble egg antigens of Schistosoma japonicum induce senescence in activated hepatic stellate cells by activation of the STAT3/p53/p21 pathway. Sci Rep. 2016;6:30957. doi: 10.1038/srep30957. doi:10.1038/srep30957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sitko JC, Yeh B, Kim M, et al. SOCS3 regulates p21 expression and cell cycle arrest in response to DNA damage. Cell Signal. 2008;20(12):2221–2230. doi: 10.1016/j.cellsig.2008.08.011. doi:10.1016/j.cellsig.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kong X, Feng D, Wang H, et al. Interleukin-22 induces hepatic stellate cell senescence and restricts liver fibrosis in mice. Hepatology. 2012;56(3):1150–1159. doi: 10.1002/hep.25744. doi:10.1002/hep.25744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Casini A, Ceni E, Salzano R, et al. Neutrophil-derived superoxide anion induces lipid peroxidation and stimulates collagen synthesis in human hepatic stellate cells: role of nitric oxide. Hepatology. 1997;25(2):361–367. doi: 10.1053/jhep.1997.v25.pm0009021948. doi:10.1053/jhep.1997.v25.pm0009021948. [DOI] [PubMed] [Google Scholar]
- 52.Canbay A, Friedman S, Gores GJ. Apoptosis: The Nexus of Liver Injury and Fibrosis. Hepatology. 2004;39(2):273–278. doi: 10.1002/hep.20051. doi:10.1002/hep.20051. [DOI] [PubMed] [Google Scholar]
- 53.Maher JJ. Interactions between hepatic stellate cells and the immune system. Semin Liver Dis. 2001;21(3):417–426. doi: 10.1055/s-2001-17555. [DOI] [PubMed] [Google Scholar]
- 54.Qu K, Huang Z, Lin T, et al. New Insight into the Anti-liver Fibrosis Effect of Multitargeted Tyrosine Kinase Inhibitors: From Molecular Target to Clinical Trials. Front Pharmacol. 2015;6:300. doi: 10.3389/fphar.2015.00300. doi:10.3389/fphar.2015.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim Y, Fiel MI, Albanis E, et al. Anti-fibrotic activity and enhanced interleukin-6 production by hepatic stellate cells in response to imatinib mesylate. Liver Int. 2012;32(6):1008–1017. doi: 10.1111/j.1478-3231.2012.02806.x. doi:10.1111/j.1478-3231.2012.02806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cressman DE, Greenbaum LE, DeAngelis RA, et al. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science. 1996;274(5291):1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 57.Tachibana S, Zhang X, Ito K, et al. Interleukin-6 is required for cell cycle arrest and activation of DNA repair enzymes after partial hepatectomy in mice. Cell Biosci. 2014;4(1):6. doi: 10.1186/2045-3701-4-6. doi:10.1186/2045-3701-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ji T, Li G, Chen J, et al. Distinct role of interleukin-6 and tumor necrosis factor receptor-1 in oval cell-mediated liver regeneration and inflammation-associated hepatocarcinogenesis. Oncotarget. August. 2016 doi: 10.18632/oncotarget.11365. doi:10.18632/oncotarget.11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siegmund SV, Schlosser M, Schildberg FA, et al. Avila MA, editor. Serum Amyloid A Induces Inflammation, Proliferation and Cell Death in Activated Hepatic Stellate Cells. PLoS One. 2016;11(3):e0150893. doi: 10.1371/journal.pone.0150893. doi:10.1371/journal.pone.0150893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fabre T, Kared H, Friedman SL, Shoukry NH. IL-17A enhances the expression of profibrotic genes through upregulation of the TGF-beta receptor on hepatic stellate cells in a JNK-dependent manner. J Immunol. 2014;193(8):3925–3933. doi: 10.4049/jimmunol.1400861. doi:10.4049/jimmunol.1400861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gkouveris I, Nikitakis N, Karanikou M, Rassidakis G, Sklavounou A. JNK1/2 expression and modulation of STAT3 signaling in oral cancer. Oncol Lett. 2016;12(1):699–706. doi: 10.3892/ol.2016.4614. doi:10.3892/ol.2016.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim J-H, Lee SC, Ro J, Kang HS, Kim HS, Yoon S. Jnk signaling pathway-mediated regulation of Stat3 activation is linked to the development of doxorubicin resistance in cancer cell lines. Biochem Pharmacol. 2010;79(3):373–380. doi: 10.1016/j.bcp.2009.09.008. doi:10.1016/j.bcp.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 63.Yoshida K, Matsuzaki K, Mori S, et al. Transforming Growth Factor-β and Platelet-Derived Growth Factor Signal via c-Jun N-Terminal Kinase-Dependent Smad2/3 Phosphorylation in Rat Hepatic Stellate Cells after Acute Liver Injury. Am J Pathol. 2005;166(4):1029–1039. doi: 10.1016/s0002-9440(10)62324-3. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1602385/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y, Meyer C, Muller A, et al. IL-13 induces connective tissue growth factor in rat hepatic stellate cells via TGF-beta-independent Smad signaling. J Immunol. 2011;187(5):2814–2823. doi: 10.4049/jimmunol.1003260. doi:10.4049/jimmunol.1003260. [DOI] [PubMed] [Google Scholar]
- 65.Fang L, Huang C, Meng X, et al. TGF-beta1-elevated TRPM7 channel regulates collagen expression in hepatic stellate cells via TGF-beta1/Smad pathway. Toxicol Appl Pharmacol. 2014;280(2):335–344. doi: 10.1016/j.taap.2014.08.006. doi:10.1016/j.taap.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 66.Long J, Matsuura I, He D, Wang G, Shuai K, Liu F. Repression of Smad transcriptional activity by PIASy, an inhibitor of activated STAT. Proc Natl Acad Sci U S A. 2003;100(17):9791–9796. doi: 10.1073/pnas.1733973100. doi:10.1073/pnas.1733973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nakashima K, Yanagisawa M, Arakawa H, et al. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284(5413):479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- 68.Luo K. Regulation of the Smad Pathway by Signaling Cross-Talk. 2008 https://cshmonographs.org/index.php/monographs/article/view/3847.