Abstract
Decline of transcription factor FoxN1, which predominantly regulates thymic epithelial cell (TEC) differentiation and homeostasis lifelong, is demonstrated to be casually related to age-related thymic involution. Whereas, a global role of microRNAs (miRNAs) has also been demonstrated to control and maintain TEC-constituting thymic microenvironment and to be changed in expression profile in the aged thymus. Therefore, it is urgently necessary to build knowledge regarding whether and which miRNAs regulate FoxN1 gene in the aged thymus. We primarily compared changes in miRNA expression profile between young and aged murine TECs with Mus musculus miRBase-V20 arrays (containing 1892 unique probes), and clearly identified and validated that at least one miRNA, miR-125a-5p, was increased in aged thymus. Applying miR-125a-5p mimics was able to inhibit FoxN1 3′UTR luciferase activity in a 293T cell line and to suppress FoxN1 expression in murine TEC Z210 cells. Since a single miRNA can play a fine-tuning role to regulate expression of multiple genes and a single gene can be regulated by multiple miRNAs, our result adds a single miRNA, miR-125a-5p, into the panel of FoxN1-regulating miRNAs associated with thymic aging.
Keywords: Thymic aging, Foxn1 gene, microRNA-125a-5p, negative fine tune
Age-related thymic involution is due to a disruption in homeostasis of thymic epithelial cells (TECs), which is regulated by transcription factor FoxN1 gene [1-3]. Whereas, like most other genes, FoxN1 gene should be regulated by multiple microRNAs (miRNAs). This is a currently accepted principle that miRNAs act as powerful modulators to regulate a wide array of biological processes in development, aging, cancer generation, and immunological responses in eukaryotic organisms [4-6]. Recently, ample evidence shows that a global role of miRNAs indeed directly regulates TEC’s function. This work was demonstrated by blocking miRNA developing pathways in TECs (with a FoxN1 promoter-driven Cre-induced gene knockout approach) either from primary miRNAs to intermediate miRNAs (knocking out enzyme DGCR8) [7] or from intermediate miRNAs to mature miRNAs (knocking out enzyme Dicer) [8]. Additionally, in the naturally-aged thymic situation, miRNA expression profile was altered [9, 10], and single specific miRNAs, such as miR-18b and miR-518b, were reported to lead to down-regulation of FOXN1 in differentiation from a stem cell line to epithelial lineage [11]. These emerging findings provide a regulatory axis from miRNAs’ fine-tuning FoxN1 to age-related disruption of TEC homeostasis in thymic involution. However, based on the principle that a single miRNA can regulate multiple genes (usually several hundreds), while a single gene can be regulated by multiple miRNAs [4, 12], single miRNAs in the pool of FoxN1-regulating miRNAs associated with thymic aging still remains largely unknown.
FoxN1 is an epithelial cell-autonomous gene, predominantly regulating development of TECs and skin keratinocytes [13]. Inborn FoxN1 mutation results in thymic and hair follicle epithelial development failure [14-16] associated with primary immune deficiency [17-19] and hairless nude skin [20, 21], respectively. In postnatal life, FoxN1 is decreased with age, which is casually related to age-related thymic involution [1-3]. Mouse FoxN1 gene contains nine exons with about one kilo-base pairs (1kb)-long 3′UTR (untranslated region), which is a potential region targeted by miRNAs for post-transcriptional regulation.
Mature miRNAs are a large group of conserved, single-stranded, around 22 nucleotide (nt)-long, non-coding RNAs that act as post-transcriptional regulators of gene expression by binding to a target coding mRNA through imperfect complementarity at multiple sites of 3′UTR of a gene, resulting in coding mRNA cleavage, translational repression, or chromatin modification [22-24] to interfere a gene expression [25], which is mediated by more than one miRNAs [4, 12]. It is demonstrated that although a single miRNA, miR-205, is highly and preferentially expressed in medullary TECs (mTECs), genetic knockout of it is not sufficient to induce a deficient phenotype [26].
In the current study, we primarily compared changes in miRNA expression profile between young and aged TECs with miRBase-V20 arrays (containing 1892 unique probes). We found there are 341 of them giving detectable signals in TEC from young and old mouse groups. Among the miRNAs with signal intensity over 500 units, we found 8 of them up-regulated and 4 of them down-regulated significantly in TECs from aged, compared to young, murine thymuses. Among those 8 up-regulated miRNAs, we identified that miR-125a-5p negatively fine-tunes FoxN1 expression by validation through a real-time RT-PCR, and confirmed by a FoxN1 3′UTR luciferase assay in a 293T cell line and a miR-125a-5p mimic to FoxN1 suppressive assay in a TEC line: Z210. Since a single miRNA can play a fine-tuning role in regulation of multiple gene expression, while a single gene can be regulated by multiple miRNAs [4, 12], a single miRNA, miR-125a-5p is not the sole miRNA in the panel of FoxN1-regulating miRNAs, but we indeed add a single miRNA into the panel associated with thymic aging.
MATERIAL AND METHODS
Mice and animal care
Thymus samples were freshly isolated from either young (2 months) or aged (19-21 months) mice (C57BL/6 genetic background). All animal experiments were performed according to the protocols approved by the Institutional Animal Care and Use Committee of the University of North Texas Health Science Center, in accordance with guidelines from the National Institutes of Health, USA.
miRNA microarray and Real-time RT-PCR
Total RNAs for miRNA microarray were isolated from enriched TECs (based on our previously published enzymatic method [27]) of young and aged murine thymuses using QIAGEN miRNeasy Mini Kit (QIAGEN Cat#217004) according to the manufacturer’s instructions. The miRNA microarray assay was performed through a service by LC Sciences, LLC (www.lcsciences.com) with miRBase-V20 arrays containing 1892 unique probes. Total RNAs for real-time RT-PCRs from enriched TECs of freshly isolated murine thymuses or from TEC line Z210 were isolated with TRIzol reagent (Invitrogen) and reverse transcribed to cDNA with the SuperScriptIII cDNA kit (Invitrogen). Real-time RT-PCRs for miRNAs were performed with TaqMan® MicroRNA Assay kits of individual miRNA primers and probes (Applied Biosystems). Real-time RT-PCR of FoxN1 primers and probe (TaqMan method) were as described previously [27]. Real-time RT-PCR was run on a Step-One-Plus thermal cycler system (Applied Biosystems). The microRNA results were internally normalized to U6snRNA control levels, while the relative expression levels of FoxN1 mRNAs from the Z210 were internally normalized to GAPDH levels. The average ΔΔCT value from multiple young animals was always arbitrarily set as 1.0 in each real-time PCR reaction.
Bioinformatics prediction of the targets of miRNAs
The targets of the miRNAs were predicted using multiple databases, including the TargetScan (www.Targetscan.org), miRanda (www.microrna.org), microcosm Targets (www.ebi.ac.uk/enright-srv/microcosm), and Segal Lab of Computational Biology (http://genie.weizmann.ac.il/index.html). The predicted hits from each algorithm were sorted as per the scores.
FoxN1 3′UTR plasmid construction and transfection
To generate a FoxN1 3′UTR luciferase reporter construct, we amplified a 1257-base pair (bp) DNA fragment consisting of the last 50bps of FoxN1 coding region and 1203bps of the 3'UTR of FoxN1 mRNA from a mouse bacterial artificial chromosome (BAC) clone RP24-137A13 DNA with PCR primers: Sense 5'-CCTG CCGTGTACCTCAGTC-3'; Antisense 5'-GGGACTG AAGGTCCCAAAAA-3'. This 1.257Kb DNA fragment of FoxN1 containing the predicted target sites of miR-125a-5p was then subcloned into downstream of Gaussia luciferase (GLuc) that is driven by a phosphoglycerate kinase (PGK) promoter. Meanwhile, we also constructed plasmids with mutated target site on this DNA fragment to miR-125a-5p by replacing the seed sequences of miR-125a-5p (7-8 nucleotides at the 3' end of its target site, CTCAGGG to GAGTCAT) with scrambled sequences, which is termed m(364-387).
293T cells were maintained in RPMI 1640 supplemented with 1% penicillin/streptomycin and 10% fetal bovine serum. Lipofectamine 2000 (Invitrogen) was used in transient transfection of FoxN1 3′UTR luciferase reporter plasmids into 293T cells based on the manufacturer protocol. The medium was replaced with normal culture medium 6hr post transfection.
Luciferase reporter assay
Each well of 293T cells cultured on a 12-well plate was transfected with 50ng of wild-type FoxN1 3′UTR luciferase reporter plasmids or m(364-387) mutated reporter plasmids, and miR-125a-5p mimics (with a various of concentrations ). Aliquots of medium from the transfected wells were collected at 24h, 48h, and 72h post transfection to measure luciferase activity. 50μl of the medium (diluted if necessary) was mixed with 100μl of substrate solution containing 0.5μg/ml of coelenterazine (CTZ), 200mM NaCl, 50mM Tris-HCl and 0.01% Triton X-100, at pH 8.7. The light emission was measured at a wavelength of 480 nm and normalized with the SEAP expression.
miRNA mimics
Mmu-miR-125a-5p mimics was synthesized by GenePharma, Inc. (Shanghai, China), with the following sequences: miR-125a-5p, UCCCUGAGACCCUUUAA CCUGUGA; and negative control (miR-CON): UGACAACCUGGUAGAAAGAGACUUC.
Statistics
Data are presented as mean±SEM. Statistical significance was analyzed by unpaired Student’s t-test and two-way ANOVA test. Differences were considered statistically significant at values of p < 0.05.
RESULTS
Different expression profiles of miRNAs in TECs from young and aged murine thymuses
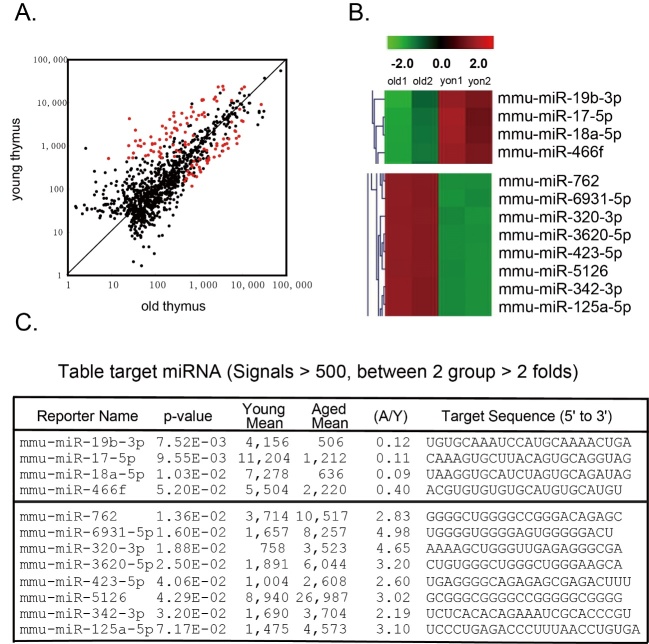
To screen age-related miRNAs in murine thymic epithelial cells (TECs), we assessed the miRNA expression profiles of total RNAs from enriched TECs isolated from the thymuses of 2-month-old and 21-months-old mice, respectively, with miRNA microarray (miRBase-V20 arrays containing 1892 unique probes, performed by LC Sciences). The results show that 341 miRNAs were detectably expressed in TECs with signal intensity over 500 in any of each (young or aged) or both (young and aged) groups (Red dots in Figure 1A). After future analysis by focusing on those with signaling strength ≥ 500, we found there are 4 miRNAs decreased (fold change ≤ -2.0) and 8 miRNAs increased (fold change ≥ 2.0) in aged TECs compared to young TECs. A hierarchical clustering/dendrogram analysis of these 12 miRNAs between 2 old (Old 1 & 2) and young (Young1 & 2) mice is shown in Figure 1B, and their detailed information is shown in Figure 1C, suggesting that TECs from young and old mice indeed have a certain of miRNA expression distinctly, and these twelve differently expressed miRNAs are potentially related to TEC-conducted thymic aging, associated with FoxN1 expression.
Figure 1. Microarray results of miRNAs expression from enriched TECs of young and old mice.
(A) A dot plot shows the positions of signals detectable miRNAs in TECs of both young and aged groups. Among these miRNAs there are 341 miRNAs (Red dots represent these miRNAs) with signal intensity over 500. (B) Heatmap shows 12 miRNAs which have signaling strength ≥ 500, decreased (≤ 2 folds) or increased (≥ 2 folds) in aged TECs compared to young TECs. (C) Detailed information about these 12 miRNAs.
Validation of expression of miRNAs in TECs potentially associated with thymic aging
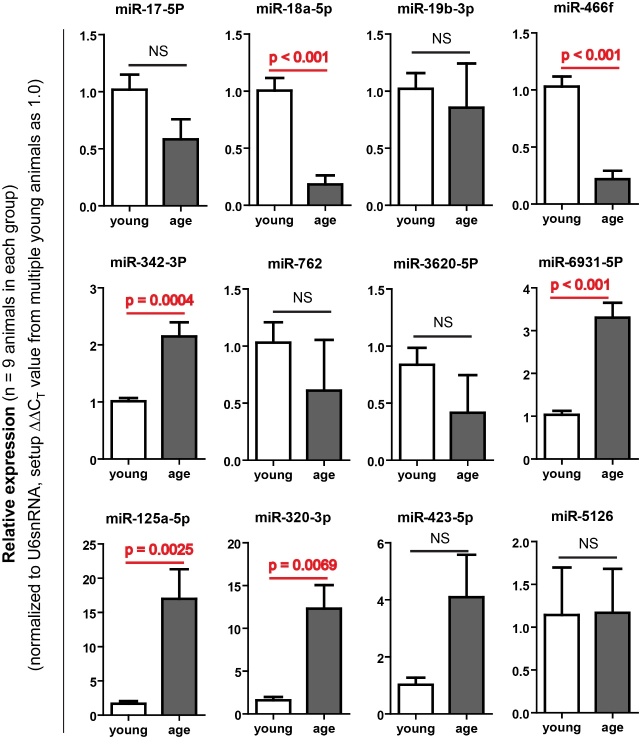
To validate the results obtained from the microarray assay (Figure 1), we chose those screened 12 miRNAs to perform a more in-depth assay on their expression in TECs with more numbers of 2-month-old and 19-month-old mice by a real-time RT-PCR approach. Results are shown in Figure 2. In those 12 miRNAs, there were 6 miRNAs with statistically different expression, among which two (miR-18a-5p, miR-466f) were down-regulated (< -2.0 folds) in aged TECs which were consistent the result with ones in the microarray (Fig. 2 top panels), and four (miR-342-3p, miR-6931-5p, miR-125a-5p, miR320-5p) were up-regulated (> 2.0-fold), confirmed the trend in the microarray data (Fig. 2 middle and bottom panels). The real-time RT-PCR utilized more numbers of animals, which provide reliable biological standard deviation, confirm that microarray clues are informational.
Figure 2. Validating 12 microRNA expression with Real-time RT-PCR.
The microRNA expression levels of miR-18a-5p, miR-466f, miR342-3p, miR-6931-5p, miR-125a-5p and miR-320-5p were consistent with the miRNA microarray results. The microRNA expression levels of miR-18a-5p and miR-466f were down-regulated in the aged thymus, while the microRNA expression levels of miR342-3p, miR-6931-5p, miR-125a-5p and miR-320-5p were up-regulated in the aged thymus. Data are presented as mean ± SEM (n = 9, i.e. Animal numbers in each group are 9 in three independent experiments). *p < 0.05, **p < 0.01 and ***p < 0.001, versus control (Student’s t-test). p-values are showed in each graph.
Predicting conserved target sites of potential miRNAs in the FoxN1 3′UTR
Since we focused on miRNAs with directly-inhibiting FoxN1 role, we particularly aimed those up-regulated 4 miRNAs and predicted their potential target sites on FoxN1 3′UTR using the online target prediction programs [28] of TargetScan Microcosm Targets, and miRanda. We did not find targeting sites of miR-6931-5p and miR-342-3p in FoxN1 3′UTR, while we found that miR-320-3p and miR-125a-5p have 3 and 5 targeting sites in FoxN1 3′UTR, respectively (Fig. 3). Because miR-125a-5p has 5 targeting sites in FoxN1 3′UTR, in which nucleotide positions 364-387 were aligned with each other (as shown in Figure 3A top panel), miR-125a-5p attracts us. This clue suggests that miR-125a-5p has a higher potential to target FoxN1 to play a negative fine-tune for inhibition of FoxN1 expression in aged thymus.
Figure 3. Computative analysis of predicted target sites of miR-320-3p and miR-125a-5p in FoxN1 3′UTR.
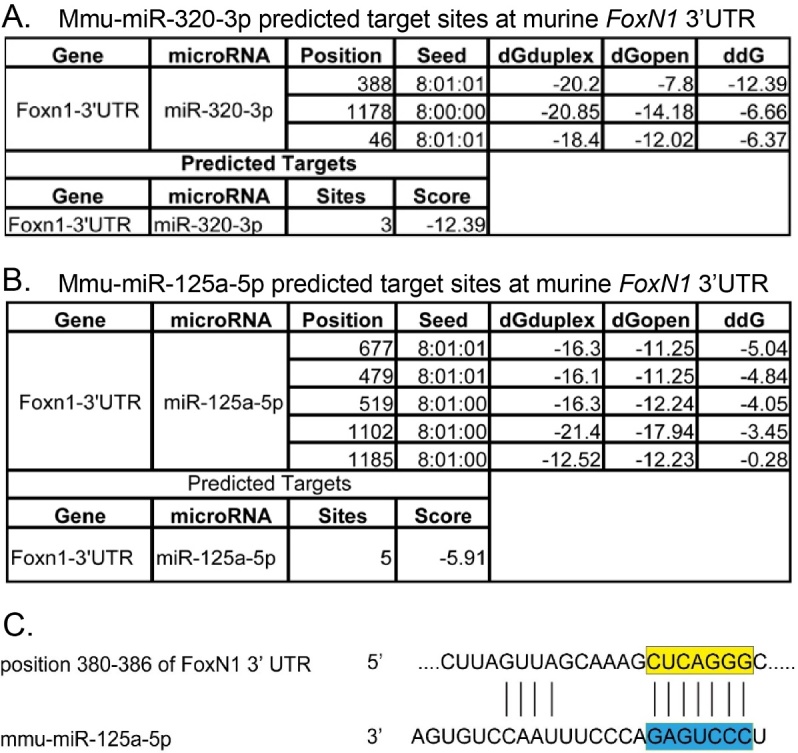
(A). Result of scanning the FoxN1 3′UTR for miR-320-3p target sites based on databases. (B). Result of scanning the FoxN1 3′UTR for miR-125a-5p target sites based on databases. (C). Alignment of one predicted miR-125a-5p target site in the FoxN1 3′UTR.
To further explore the potential role of miR-125a-5p in repressing FoxN1, we constructed a FoxN1 3′UTR luciferase reporter plasmid. A ~1.25Kb FoxN1 3′UTR DNA fragment [containing the last 50bps of FoxN1 translated region, via PCR from a mouse bacterial artificial chromosome (BAC) clone RP24-137A13] was sub-cloned into downstream of Gaussia luciferase (Gluc) that is driven by PGK promoter (kindly provided by Dr. Sui) [29] (Figure 4). Meanwhile, we also made a mutant 3′UTR of FoxN1 [a 7bp mutation on the first target site for miR-125a-5p, termed m(364 - 387)], as a control vector (Fig. 4A bottom panel), along with wide-type (WT) 3′UTR of FoxN1 vector (Fig. 4A top penal). NIH293T cells were transfected with an either WT FoxN1 3′UTR luciferase reporter plasmid followed by adding synthetic mimics of miR-125a-5p with a final concentration of 0 - 200nM (Fig. 4B). A significant dose-dependent repression in the WT FoxN1 3′UTR luciferase reporter activity was observed at 24h, 48h and 72h post co-culture (Fig. 4B). In addition, a mutant FoxN1 3′UTR luciferase reporter plasmid m (364-387) (at 100nM) was also transfected into 293T cells. However, the suppression, seen in WT FoxN1 3′UTR transfection (Fig. 4B), could not be observed in the 24hr post co-culture compared with miR-CON (a negative control miRNA), which is a random sequence of a miRNA molecule that has been extensively tested in human cell lines and tissues not to produce identifiable effects on known miRNA function. (Fig. 4C). Taking together, the result that miR-125a-5p mimic was able to suppress post-transcription activity of WT 3′UTR of FoxN1 at a dose-dependent manner, but could not suppress this activity of mutant 3′UTR of FoxN1, which indicates that miR-125a-5p specifically targets 3′UTR of FoxN1.
Figure 4.
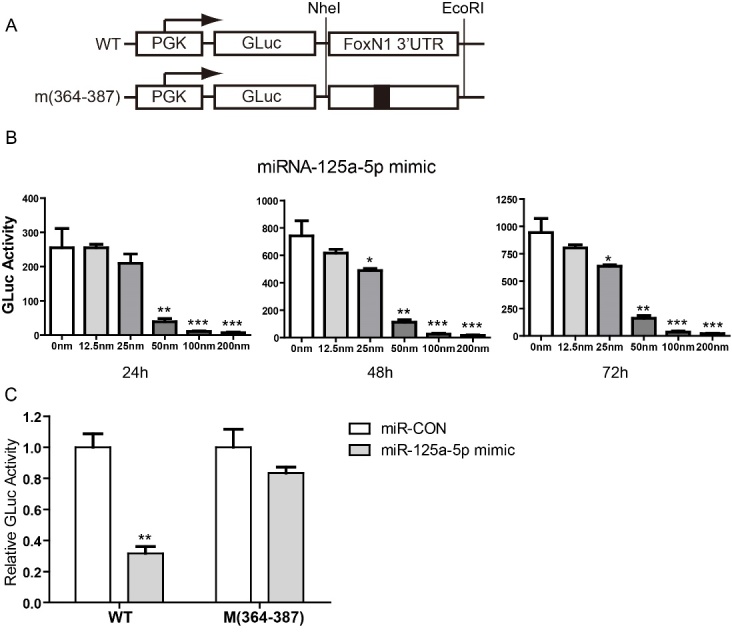
Results of miR125a-5p mimics inhibiting FoxN1 3′UTR luciferase reporter assay. (A) Schematic diagrams of construction of wild type (WT) FoxN1 3′UTR luciferase reporter (top panel), in which the last 50bp of FoxN1 coding region and the first 1203 bps of FoxN1 3′UTR embracing the predicted miR125a-5p target sites is subcloned into downstream of GLuc driven by PGK promoter, and mutant FoxN1 3′UTR luciferase reporter (bottom panel), in which FoxN1 3′UTR at the first miR-125a-5p target site (364-387) was scrambled. (B) Results of inhibition to GLuc activity by miR-125a-5p mimics with a dose-dependent manner and time-course associated. GLuc activity was determined 24h, 48h, 72h post transfection and normalized by SEAP activity (see Materials and Methods for details). (C) One hundred nM of miR-CON (mimic negative control) or miR-125a-5p was co-transfected with 50ng of the wild type (WT) or mutant FoxN1 3′UTR luciferase reporter plasmids, and the SEAP-expressing plasmid. GLuc activity was measured 24h post transfection and normalized as described above. Data are presented as mean ± SEM (n = 9, i.e. nine independent experiments). *p < 0.05, **p < 0.01, and ***p < 0.001, versus control (two-way ANOVA test).
In vitro effects of miR-125a-5p on FoxN1 expression in a TEC line
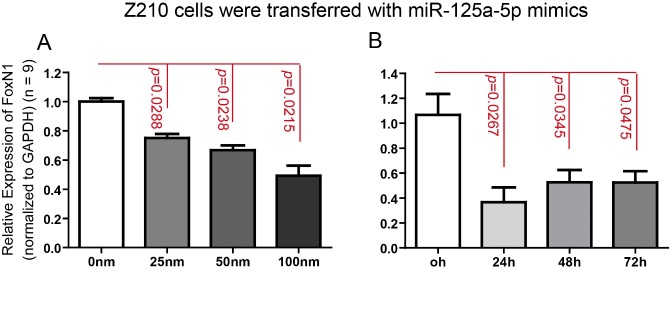
Since FoxN1 is endogenously expressed in epithelial cells, to determine whether miR-125a-5p is able to fine-tune endogenous FoxN1 expression, we transfected the synthetic miR-125a-5p mimics into murine TEC line Z210 (a murine thymic stromal cell line with a medullary phenotype, generated by Dr. Farr) [30] and observed inhibiting effects of miR-125a-5p on the TEC endogenous FoxN1 expression with Real-time RT-PCR. As shown in Figure 5, increasing concentration of miR-125a-5p mimics (25, 50, 100nM) negatively regulates the expression of endogenous FoxN1 in TECs at 48hrs post transfection (Fig. 5A). We further analysis the time-course inhibiting effect at a highest concentration of miR-125a-5p mimics (100 nM) and found that there is not a time-course change, but the inhibiting effect cannot be reversed even by 72hrs (Fig. 5B).
Figure 5.
Inhibition of FoxN1 expression by miR-125a-5p mimics on thymic epithelial cells at dose-dependent and time-course manners. (A) TEC line Z210 cells were treated with increasing miR-125a-5p (0, 25, 50 and 100nM, compensated with miR-CON to 100nM, if necessary) for 48h. FoxN1 expression in Z210 cells was analyzed by Real-Time RT-PCR; (B) Z210 cells were treated with 100nM miR-125a-5p for 24h, 48h and 72h. FoxN1 expression in Z210 cells was analyzed by Real-Time RT-PCR. Data are presented as mean ± SEM (n = 9, i.e. nine independent experiments). *p < 0.05, **p < 0.01, and ***p < 0.001, versus control (two-way ANOVA test).
DISCUSSION
The thymus is a unique organ to be responsible for T cell development and central tolerance establishment. Age-related thymic involution has a severe clinical outcome, which results not only in reduced output of naïve T cells but also in increased output of self-reactive T cells in the elderly [31]. The thymus consists of two major cellular components: thymocytes from hematopoietic origin and stromal cells, mostly thymic epithelial cells (TECs), from non-hematopoietic origin. TECs mainly constitute thymic microenvironment to support thymocyte development. However, TEC development and homeostasis are mainly regulated by an epithelial cell-autonomous transcription factor, FoxN1 gene, through the p63-FoxN1 regulatory axis [32]. Age-related thymic involution [33-35] has been demonstrated to be causally associated with decline of FoxN1 [1-3]. However, decline of FoxN1 with age could be mostly regulated by epigenetic alteration, since mounting evidence indicates that changes in many transcription factors are epigenetically regulated in the context of the aged microenvironment [36-40], where epigenetic modifications are associated with remodeling of chromatin structure to play a central role in controlling molecular expression [37-40] via alteration of transcriptional activity. Epigenetic regulation includes three major mechanisms at three layers: DNA methylation (DNA layer), histone modifications (chromatin layer), and non-coding microRNA expression (post-transcription layer). However, limited information is available on how changes in organism affect FoxN1 expression in the aged thymus. Our study suggests a new target at post-transcription level in the aged thymic context. We identified a potential FoxN1 negative regulator, miR-125a-5p, which is able to fine-tune FoxN1 expression based on (1) increased miR-125a-5p expression in the aged thymuses, (2) an inhibiting effect on FoxN1 3′UTR luciferase reporter by transferring miR-125a-5p mimics, and (3) a dose-dependent decrease of endogenous FoxN1 expression in a TEC line by transferring miR-125a-5p mimics.
Regulation of TEC biological processes by miRNAs has been recognized in many context, particularly in TEC development [7, 8, 11]. FoxN1 gene is critical in the regulation and maintenance of TEC biological function and it also controls postnatal TEC homeostasis. Therefore, miRNAs appear to be involved in the epigenetic regulation of TEC aging through repressing FoxN1 post-transcription in the aged thymus. However, the regulation of thymic aging by miRNAs’ suppressive function to FoxN1 gene is the lack of evidence, while studying on miRNA profiles between young and aged thymus just began [9, 10]. It is well-known that a single gene can be regulated by multiple miRNAs. However, specific miRNAs on regulation of age-related decline of FoxN1 has been less well investigated. Although some miRNA candidates, such as miR-18b and miR-518b, were reported to lead to downregulation of FOXN1 in stem cells [11], herein, we add one more candidate miR-125a-5p leading to downregulation of FoxN1 in the aged thymus. A steady-state up-regulation of miR-125a-5p was also observed in 10- and 19-month-old murine thymuses by other group [9], which the possibility that miR-125a-5p is indeed involved in thymic aging associated with a decline of FoxN1. Because a single miRNA may only play a role for a fine-tune, we do not expect that totally abolishing FoxN1 expression can be achieved by knocking out or inhibiting single miR-125a-5p.
MiR-125 is a group of miRNA family, whose members include miR-125a-5p and miR-125b etc. MiR-125 play important roles in development and cell differentiation [41]. A significant down-regulation in miR-125a-5p and miR-125b was observed in malignant and aggressive breast cancer tissues [42]. An important role for miR-125a-5p in angiogenesis regulation via RTEF-1 was elucidated in aging mice [43]. Evidence also shows that miR-125a-5p may be as a potential tumor-suppressor in gastric cancer [44, 45], and acts as an inhibitor to epidermal growth factor receptor signaling to block migration and invasion of lung cancer cells [46]. These findings of mir-125a-5p as a suppressor of proliferation and migration in cancer cells may provide a clue that an increase of mir-125a-5p in the aged thymus can suppress thymic epithelial cell homeostasis during aging. Additionally, in non-cancer cells, miR-125a-5p also has a role in suppressing classical activation of macrophages while promoting alternative activation by targeting Kruppel-like transcription factor 13 (KLF13) [41], which negatively regulates inflammation associated with T cell activation. An important role for miR-125a-5p in angiogenesis regulation via RTEF-1 was elucidated in aging mice [43]. In a recent study, miR-125a-5p appears to cause inflammatory activation of THP-1 cells [47], and activates NF-κB activity in diffuse large B-cell lymphoma cells [48]. These findings may be reminiscent of increased inflammation in the aged thymus.
In conclusion, our finding suggests that miR-125a-5p plays a role as a negative regulator of FoxN1 in thymic involution during aging. Targeting miR-125a-5p in aged thymus should be one of therapeutic strategies to delay age-related thymic involution and/or improve thymic function in the elderly.
Acknowledgements
We sincerely thank Dr. Guang-Qiao Sui (College of Life Science, Northeast Forestry University, China/Wake Forest University School of Medicine, USA) for kindly providing vector.
This work was partially supported by an NIH/NIAID USA grant (R01AI121147) to D-M. S., and by the Science and Technology Project Foundation of Education Department of Jiangxi Provincial (GJJ150961) to Xu, the Higher Education Foundation of Jiangxi Provincial (KJLD2090) to Wang, the Natural Science Foundation of Jiangxi Province (20132BAB205032) to Wang and National Natural Science Foundation of China (31260279 and 31660256) to Wang.
References
- [1].Sun L, Guo J, Brown R, Amagai T, Zhao Y, Su DM (2010). Declining expression of a single epithelial cell-autonomous gene accelerates age-related thymic involution. Aging Cell, 9: 347-357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zook EC, Krishack PA, Zhang S, Zeleznik-Le NJ, Firulli AB, Witte PL, et al. (2011). Overexpression of Foxn1 attenuates age-associated thymic involution and prevents the expansion of peripheral CD4 memory T cells. Blood, 118: 5723-5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bredenkamp N, Nowell CS, Blackburn CC (2014). Regeneration of the aged thymus by a single transcription factor. Development, 141: 1627-1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bartel DP (2009). MicroRNAs: target recognition and regulatory functions. Cell, 136: 215-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Tomankova T, Petrek M, Gallo J, Kriegova E (2011). MicroRNAs: emerging regulators of immune-mediated diseases. Scand J Immunol [DOI] [PubMed] [Google Scholar]
- [6].Pritchard CC, Cheng HH, Tewari M (2012). MicroRNA profiling: approaches and considerations. Nat Rev Genet, 13: 358-369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Khan IS, Taniguchi RT, Fasano KJ, Anderson MS, Jeker LT (2014). Canonical microRNAs in thymic epithelial cells promote central tolerance. Eur J Immunol, 44: 1313-1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zuklys S, Mayer CE, Zhanybekova S, Stefanski HE, Nusspaumer G, Gill J, et al. (2012). MicroRNAs control the maintenance of thymic epithelia and their competence for T lineage commitment and thymocyte selection. J Immunol, 189: 3894-3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ye Y, Li D, Ouyang D, Deng L, Zhang Y, Ma Y, et al. (2014). MicroRNA expression in the aging mouse thymus. Gene, 547: 218-225 [DOI] [PubMed] [Google Scholar]
- [10].Guo Z, Chi F, Song Y, Wang C, Yu R, Wei T, et al. (2013). Transcriptome analysis of murine thymic epithelial cells reveals ageassociated changes in microRNA expression. Int J Mol Med, 32: 835-842 [DOI] [PubMed] [Google Scholar]
- [11].Kushwaha R, Thodima V, Tomishima MJ, Bosl GJ, Chaganti RS (2014). miR-18b and miR-518b Target FOXN1 during epithelial lineage differentiation in pluripotent cells. Stem cells and development, 23: 1149-1156 [DOI] [PubMed] [Google Scholar]
- [12].Kim VN, Han J, Siomi MC (2009). Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol, 10: 126-139 [DOI] [PubMed] [Google Scholar]
- [13].Su DM, Navarre S, Oh WJ, Condie BG, Manley NR (2003). A domain of Foxn1 required for crosstalk-dependent thymic epithelial cell differentiation. Nat Immunol, 4: 1128-1135 [DOI] [PubMed] [Google Scholar]
- [14].Pantelouris EM (1968). Absence of thymus in a mouse mutant. Nature, 217: 370-371 [DOI] [PubMed] [Google Scholar]
- [15].Pantelouris EM, Hair J (1970). Thymus dysgenesis in nude (nu nu) mice. J Embryol Exp Morphol, 24: 615-623 [PubMed] [Google Scholar]
- [16].Nehls M, Kyewski B, Messerle M, Waldschutz R, Schuddekopf K, Smith AJ, et al. (1996). Two genetically separable steps in the differentiation of thymic epithelium. Science, 272: 886-889 [DOI] [PubMed] [Google Scholar]
- [17].Frank J, Pignata C, Panteleyev AA, Prowse DM, Baden H, Weiner L, et al. (1999). Exposing the human nude phenotype. Nature, 398: 473-474 [DOI] [PubMed] [Google Scholar]
- [18].Cunningham-Rundles C, Ponda PP (2005). Molecular defects in T- and B-cell primary immunodeficiency diseases. Nat Rev Immunol, 5: 880-892 [DOI] [PubMed] [Google Scholar]
- [19].Amorosi S, D'Armiento M, Calcagno G, Russo I, Adriani M, Christiano AM, et al. (2008). FOXN1 homozygous mutation associated with anencephaly and severe neural tube defect in human athymic Nude/SCID fetus. Clin Genet, 73: 380-384 [DOI] [PubMed] [Google Scholar]
- [20].Schlake T (2001). The nude gene and the skin. Exp Dermatol, 10: 293-304 [DOI] [PubMed] [Google Scholar]
- [21].Mecklenburg L, Tychsen B, Paus R (2005). Learning from nudity: lessons from the nude phenotype. Exp Dermatol, 14: 797-810 [DOI] [PubMed] [Google Scholar]
- [22].Bartel DP (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116: 281-297 [DOI] [PubMed] [Google Scholar]
- [23].He L, Hannon GJ (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet, 5: 522-531 [DOI] [PubMed] [Google Scholar]
- [24].Ambros V (2004). The functions of animal microRNAs. Nature, 431: 350-355 [DOI] [PubMed] [Google Scholar]
- [25].Hobert O (2007). miRNAs play a tune. Cell, 131: 22-24 [DOI] [PubMed] [Google Scholar]
- [26].Khan IS, Park CY, Mavropoulos A, Shariat N, Pollack JL, Barczak AJ, et al. (2015). Identification of MiR-205 As a MicroRNA That Is Highly Expressed in Medullary Thymic Epithelial Cells. PLoS One, 10: e0135440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cheng L, Guo J, Sun L, Fu J, Barnes PF, Metzger D, et al. (2010). Postnatal tissue-specific disruption of transcription factor FoxN1 triggers acute thymic atrophy. J Biol Chem, 285: 5836-5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E (2007). The role of site accessibility in microRNA target recognition. Nat Genet, 39: 1278-1284 [DOI] [PubMed] [Google Scholar]
- [29].Cao P, Deng Z, Wan M, Huang W, Cramer SD, Xu J, et al. (2010). MicroRNA-101 negatively regulates Ezh2 and its expression is modulated by androgen receptor and HIF-1alpha/HIF-1beta. Mol Cancer, 9: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Friend SL, Hosier S, Nelson A, Foxworthe D, Williams DE, Farr A (1994). A thymic stromal cell line supports in vitro development of surface IgM+ B cells and produces a novel growth factor affecting B and T lineage cells. Exp Hematol, 22: 321-328 [PubMed] [Google Scholar]
- [31].Coder B, Su DM (2015). Thymic involution beyond T-cell insufficiency. Oncotarget, 6: 21777-21778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Burnley P, Rahman M, Wang H, Zhang Z, Sun X, Zhuge Q, et al. (2013). Role of the p63-FoxN1 regulatory axis in thymic epithelial cell homeostasis during aging. Cell Death Dis, 4: e932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chidgey A, Dudakov J, Seach N, Boyd R (2007). Impact of niche aging on thymic regeneration and immune reconstitution. Semin Immunol, 19: 331-340 [DOI] [PubMed] [Google Scholar]
- [34].Fry TJ, Mackall CL (2002). Current concepts of thymic aging. Springer Semin Immunopathol, 24: 7-22 [DOI] [PubMed] [Google Scholar]
- [35].Taub DD, Longo DL (2005). Insights into thymic aging and regeneration. Immunol Rev, 205: 72-93 [DOI] [PubMed] [Google Scholar]
- [36].Jaenisch R, Bird A (2003). Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet, 33 Suppl: 245-254 [DOI] [PubMed] [Google Scholar]
- [37].Berdasco M, Esteller M (2012). Hot topics in epigenetic mechanisms of aging: 2011. Aging Cell, 11: 181-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sedivy JM, Banumathy G, Adams PD (2008). Aging by epigenetics--a consequence of chromatin damage? Exp Cell Res, 314: 1909-1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Fraga MF (2009). Genetic and epigenetic regulation of aging. Curr Opin Immunol, 21: 446-453 [DOI] [PubMed] [Google Scholar]
- [40].Rando TA, Chang HY (2012). Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell, 148: 46-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Banerjee S, Cui H, Xie N, Tan Z, Yang S, Icyuz M, et al. (2013). miR-125a-5p regulates differential activation of macrophages and inflammation. J Biol Chem, 288: 35428-35436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hsieh TH, Hsu CY, Tsai CF, Long CY, Chai CY, Hou MF, et al. (2015). miR-125a-5p is a prognostic biomarker that targets HDAC4 to suppress breast tumorigenesis. Oncotarget, 6: 494-509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Che P, Liu J, Shan Z, Wu R, Yao C, Cui J, et al. (2014). miR-125a-5p impairs endothelial cell angiogenesis in aging mice via RTEF-1 downregulation. Aging Cell, 13: 926-934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Xu Y, Huang Z, Liu Y (2014). Reduced miR-125a-5p expression is associated with gastric carcinogenesis through the targeting of E2F3. Mol Med Rep, 10: 2601-2608 [DOI] [PubMed] [Google Scholar]
- [45].Song C, Wu G, Xiang A, Zhang Q, Li W, Yang G, et al. (2015). Over-expression of miR-125a-5p inhibits proliferation in C2C12 myoblasts by targeting E2F3. Acta Biochim Biophys Sin (Shanghai), 47: 244-249 [DOI] [PubMed] [Google Scholar]
- [46].Wang G, Mao W, Zheng S, Ye J (2009). Epidermal growth factor receptor-regulated miR-125a-5p--a metastatic inhibitor of lung cancer. FEBS J, 276: 5571-5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Graff JW, Dickson AM, Clay G, McCaffrey AP, Wilson ME (2012). Identifying functional microRNAs in macrophages with polarized phenotypes. J Biol Chem, 287: 21816-21825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kim SW, Ramasamy K, Bouamar H, Lin AP, Jiang D, Aguiar RC (2012). MicroRNAs miR-125a and miR-125b constitutively activate the NF-kappaB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20). Proc Natl Acad Sci U S A, 109: 7865-7870 [DOI] [PMC free article] [PubMed] [Google Scholar]