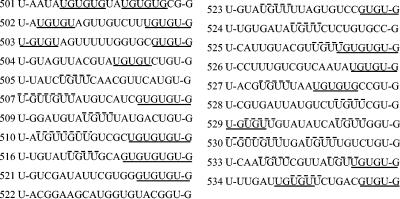Abstract
ETR-3 (also know as BRUNOL3, NAPOR, and CUGBP2) is one of six members of the CELF (CUG-BP1- and ETR-3-like factor) family of splicing regulators. ETR-3 regulates splicing by direct binding to the pre-mRNA. We performed systematic evolution of ligands by exponential enrichment (SELEX) to identify the preferred binding sequence of ETR-3. After five rounds of SELEX, ETR-3 selected UG-rich sequences, in particular UG repeats and UGUU motifs. Either of these selected motifs was able to restore ETR-3 binding and responsiveness to a nonresponsive splicing reporter in vivo. Moreover, this effect was not specific to ETR-3 since minigenes containing either of the two motifs were responsive to two other CELF proteins (CUG-BP1 and CELF4), indicating that different members of the CELF family can mediate their effects via a common binding site. Using the SELEX-identified motifs to search the human genome, we identified several possible new ETR-3 targets. We created minigenes for two of these genes, the CFTR and MTMR1 genes, and confirmed that ETR-3 regulates their splicing patterns. For the CFTR minigene this regulation was demonstrated to be dependent on the presence of the putative binding site identified in our screen. These results validate this approach to search for new targets for RNA processing proteins.
The fact that the large number of proteins that comprise the human proteome are generated from a much smaller number of genes has highlighted the role of posttranscriptional events such as alternative splicing and editing in the regulation of gene expression. These regulatory events allow the production of multiple mRNA species from a single gene, thus increasing the coding potential of the human genome (25, 36, 41).
CUG-binding protein 1 (CUG-BP1) and ELAV-type RNA binding protein 3 (ETR-3, also known as BRUNOL3, NAPOR,and CUGBP2)-like factor (CELF) proteins (also known as bruno-like or BRUNOL) are a family of related RNA binding proteins (see http://www.bcm.edu/pathology/cooper/files/celf_brunol_napor.htm for a description of nomenclature). They have been implicated in the regulation of splicing, editing, and translation and therefore are likely to play a major role in the generation of proteomic diversity (1, 19, 37). The human genome encodes six CELF proteins (CUG-BP1, ETR-3, and CELF3 through CELF6), all having similar architectures: two RNA recognition motifs (RRMs) near the N terminus, a third RRM near the C terminus, and a 160- to 230-amino-acid divergent domain between RRM2 and RRM3 (15, 23, 24). This divergent domain is unique to the CELF proteins and in ETR-3 and CELF4 has been shown to contain one or more activation modules necessary for splicing activity (45).
The CELF proteins have been demonstrated to regulate alternative splicing events for several variable regions (cardiac troponin T [cTNT] exon 5, insulin receptor [IR] exon 11, chloride channel 1 [ClC1] intron 2, NMDAR-1 exons 5 and 21, and the α-actinin muscle-specific exon) (16, 23, 40, 42, 50). The mechanism by which the CELF proteins regulate splicing is still largely unknown; however, it is clear that regulation requires direct binding to the pre-mRNA (7, 8, 16, 40, 42).
Splicing regulation by the CELF proteins has also been implicated in the pathogenesis of myotonic dystrophy (DM), an autosomal dominant multisystemic disorder caused by expansion of a tri- or tetranucleotide repeat (17, 31). Pathogenesis is thought to result from a unique RNA gain-of-function mechanism in which nuclear accumulation of repeat-containing RNA transcribed from the expanded allele creates an unusual toxicity (31, 33). One of the effects of the repeat-containing RNA is to disrupt specific alternative splicing events. These splicing defects can be recapitulated in tissue culture by transiently expressing CUG-containing RNA. Three of the known CELF protein targets (cTNT exon 5, IR exon 11, and ClC1 intron 2) are misspliced in DM patients in a way that is consistent with increased CELF protein splicing activity (8, 40, 42). A second family of splicing regulators, called muscleblind-like (MBNL) proteins, also regulates the splicing of at least two of these exons (18). The effect of MBNL proteins on the splicing of these exons is the opposite of that of the CELF proteins, and it has been proposed that the active form of these proteins is reduced in DM patients via sequestration of the protein on the expanded RNA (34, 35). A MBNL isoform-specific knockout mouse was shown to display several symptoms of DM patients, highlighting the relevance of this family of splicing regulators in DM (21).
ETR-3, one of the best-characterized CELF proteins, has several roles in mRNA processing, such as alternative splicing, editing, translation, and regulation of mRNA stability (1, 23, 37). Splicing regulation by ETR-3 is thought to be particularly relevant in muscle, heart, and brain since ETR-3 mRNA is abundant in these tissues (10, 29, 30). The splicing pattern of the known targets of ETR-3 changes during muscle and heart development and in different regions of the brain (23, 50). Additionally, the level of ETR-3 protein decreases during heart development, suggesting that ETR-3 might be part of a regulated developmental program (A. N. Ladd, M. G. Stenberg, M. S. Swanson, and T. A. Cooper, submitted for publication).
Since the CELF proteins require binding to the pre-mRNA to promote splicing effects, we performed systematic evolution of ligands by exponential enrichment (SELEX) using ETR-3, in order to identify its preferred RNA binding sequence(s). The results obtained in this study demonstrate that ETR-3 binds preferentially to sequences with a high content of U's and G's, in particular to sequences with UG repeats and UGUU motifs. We show that recombinant ETR-3 binds the SELEX-identified sequences in vitro and that introduction of either of the two different enriched motifs into a nonresponsive cTNT minigene restores regulation by ETR-3 in vivo. Interestingly, the two motifs that restored ETR-3 regulation also restored responsiveness to two other CELF proteins (CUG-BP1 and CELF4). In addition, the restoration of ETR-3 binding and splicing activity to the cTNT minigene also restored its responsiveness to the expression of RNA containing CUG repeats. This result strengthens previous observations that the trans-dominant effects of CUG repeat RNA depend on the responsiveness to CELF regulation.
To discover new targets of ETR-3, we used the SELEX-identified motifs to search the human genome for putative ETR-3 binding sites near alternative exons. This strategy allowed us to identify eighteen potential novel ETR-3 targets. We created minigenes containing genomic fragments for two of these genes and confirmed that ETR-3 is able to regulate their splicing patterns. These results demonstrate the usefulness of this approach to identify novel targets for RNA binding proteins.
MATERIALS AND METHODS
Preparation of recombinant protein.
The pET-28a ETR-3 clone was previously described (7), and the protein was purified according to the manufacturer's procedure (Novagen, Madison, Wis.). After purification, the protein was dialyzed against DG buffer (20 mM HEPES [pH 7.9], 84 mM l-glutamic acid, 0.2 mM EDTA, 0.5 mM dithiothreitol, 20% glycerol), aliquoted at 500 ng/μl, and stored at −80°C. Purified recombinant protein was at least 90% full length based on analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie staining.
SELEX.
SELEX procedures were performed as described previously (48). The protein was incubated with the RNA in binding buffer (3-μg/μl heparin, 1.5 mM l-glutamate, 0.3-μg/μl yeast RNA [Ambion, Austin, Tex.], 0.3 mM ATP, 10% DG buffer) at 30°C for 30 min in order to allow the binding of ETR-3 to the RNAs. To increase the specificity of binding, the concentration of ETR-3 was reduced from 0.8 μM in the first three rounds to 0.5 μM in the fourth and fifth rounds of SELEX. To enforce the selection of the full-length RNA, the DNA pool was gel purified after each round of SELEX.
In vitro transcription and EMSA.
RNAs used in the electrophoretic mobility shift assays (EMSAs) were synthesized by IDT, Inc. (Coralville, Iowa) and 5′ end labeled with [α-32P]ATP (Perkin-Elmer, Wellesley, Mass.). The generic sequence of the RNAs used in Fig. 2 was UCCGCAUN20GGAAGCU. For Fig. 3C, the generic sequence of the RNAs used was CCGCAUN26GGAAGC, with N26 being the sequences shown in Fig. 3A.
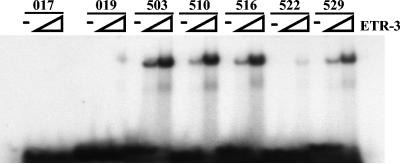
FIG. 2.
EMSA performed with bacterially expressed recombinant human His-ETR-3 and synthetic 5′-end-labeled RNA from the clones indicated. Clones 017 and 019 are from round 0; all others are from round 5. Clone 522 was the only clone from round 5 without a UG or UGUU motif. Zero, 100, or 1,000 ng of ETR-3 was added to each reaction mixture.
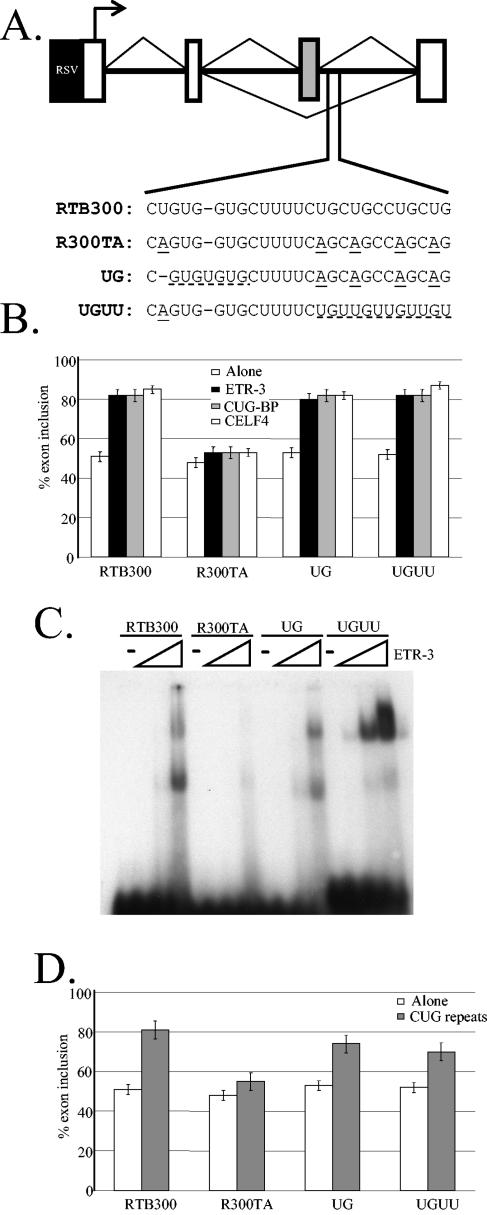
FIG. 3.
ETR-3 SELEX-identified sequences that bind to ETR-3 in vitro restore splicing activity of ETR-3, CUG-BP1, CELF4, and CUG-expanded RNA in vivo. (A) Schematic representation of the minigenes used to analyze the responsiveness of the SELEX-identified motifs in vivo. RTB300 contains a wild-type genomic sequence for human cTNT, while R300TA contains five point mutations (underlined with long dashes) that prevent the binding of CUG-BP1 (40) and ETR-3(Fig. 3C). The 26-nucleotide sequence initially replaced by the SELEX-identified sequences is shown. The UG and UGUU motifs identified in the SELEX-identified sequences were cloned into R300TA, and the minigenes were called the UG and UGUU minigenes, respectively. The SELEX motifs in the new minigenes are underlined with short dashes. (B) ETR-3 SELEX-identified motifs restore regulation of cTNT exon 5 splicing in vivo by ETR-3, CUG-BP1, and CELF4. Derivatives of the cTNT minigene (A) were cotransfected with the expression plasmid for ETR-3, CUG-BP1, or CELF4. The extent of exon inclusion was measured by RT-PCR. Percent exon inclusion is calculated as [(amount of mRNA containing the exon)/(amount of mRNA lacking the exon + amount of mRNA containing the exon)] × 100. The means ± standard deviations from at least three independent experiments are shown. Protein expression was confirmed by Western blotting in a parallel transfection. (C) EMSA performed with recombinant ETR-3 and synthetic 5′ end 32P-labeled RNAs containing the regulatory regions of each of the minigenes tested in vivo. The RNAs used for EMSA contained the sequences indicated in panel A plus the flanking sequence present in the SELEX constructs (see Materials and Methods). Zero, 100, 1,000, or 4,000 ng of ETR-3 was added to the reaction mixture. (D) Only the minigenes that respond to CELF protein coexpression respond to the expression of CUG repeat RNA. All minigenes were expressed with or without RNA containing 960 CUG repeats. The inclusion of the exon was measured by RT-PCR, and the results presented are the averages of at least three independent experiments. Percent exon inclusion is calculated as for panel B.
EMSA binding reactions were performed in binding buffer plus 200 ng of bovine serum albumin, 1 μg of RNA, and His-ETR-3 (amounts indicated in figures). The reaction mixtures were incubated at 30°C for 30 min and immediately analyzed by electrophoresis at 140 V for 2 h with a 6% (acrylamide-bisacrylamide, 37.5:1) native gel in 1× Tris-acetate-EDTA at room temperature. Gels were prerun at 140 V overnight prior to loading.
Plasmids.
ETR-3, CUG-BP1, and CELF4 pcDNA3.1 His expression plasmids were previously described (23). RTB300 and R300TA minigenes were previously described (40). The UG and UGUU mutant minigenes were generated from R300TA by inverse PCR. All clones were confirmed by sequencing.
Myotubularin-related 1 (MTMR1) and cystic fibrosis transmembrane conductance regulator (CFTR) genomic regions were amplified with the following primers: MTMR1C (TGAGTCGACTGCTCTGGGTCCAAGGTGGGGATTTCT), MTMR1B (ACTCTAGACCCCAGTCAGTCATGAATATCCATTTGG), CFTR2 (TAATAAGTCGACCAGTGTAATGGATCATGGG), and CFTR3 (AAATAATTCCCCAGGATCCTGTTAAAA). The mutant CFTR genomic region was amplified with the following primers: CFTR2 and CFTR6mut (TTCCCCGGATCCTGTTAAAAAGAGGAGGGAGGAAAGGGAAGGATCAAAAATAAAAGATGAGTTTGTCAG).
For the MTMR1 minigene, the MTMR1 genomic fragment replaced the human cTNT genomic fragment in R300TA. The CFTR minigene contains an artificial alternative exon flanked upstream by the CFTR genomic sequence indicated in Fig. 4A and downstream by 50 nucleotides of human cTNT containing the R300TA mutation. The remaining portions of the minigene are identical to RTB300 except for deletion of an 1,128-nucleotide fragment including the second exon of the RTB300 minigene. Plasmid sequences are available upon request.
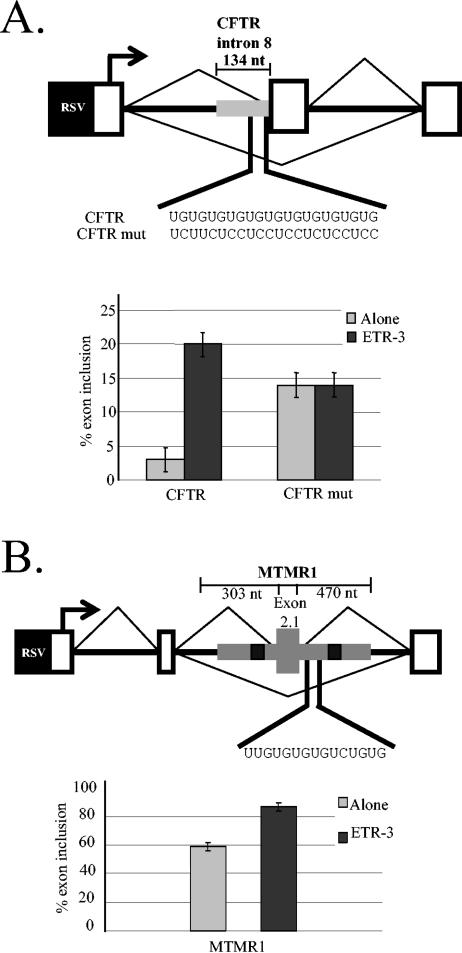
FIG. 4.
CFTR exon 9 and MTMR1 exon 2.1 are responsive to ETR-3. (A) Schematic representation of the minigenes used to analyze the effects of ETR-3 coexpression on the splicing of CFTR. Thick gray bar, CFTR genomic fragment. The sequences indicated represent the wild type (CFTR) and the mutant minigene used in this study (CFTRmut). ETR-3 regulation of minigene splicing is dependent on the putative binding site identified in the genomic screen. The extent of inclusion of the exon was measured by RT-PCR, and the results presented are the averages of at least three independent experiments. Percent exon inclusion is calculated as for Fig. 3B. (B) Schematic representation of the minigene used to analyze the effects of ETR-3 coexpression in the splicing of MTMR exon 2.1. Gray segment, MTMR1 genomic fragment. The sequences indicated represent the putative ETR-3 binding site identified in the genomic screen. Black boxes, locations of other ETR-3 putative binding sites in the MTMR1 genomic fragment. The inclusion of the exon was measured by RT-PCR, and the results presented are the averages of at least three independent experiments. Percent exon inclusion is calculated as indicated for Fig. 3B. Protein expression was confirmed by Western blotting in a parallel transfection.
Transient transfection.
COS cells were plated at 200,000 cells per 60-mm-diameter plate in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, glutamine, and penicillin-streptomycin (Gibco). Twenty-four hours after plating, cells were transfected with 300 ng of the minigene, 300 ng of the expression plasmid, and sp72 as a carrier to a total amount of 2 μg of DNA. Fugene6 (Roche, Indianapolis, Ind.) was added according to the manufacturer's directions. Protein and RNA were harvested 36 to 48 h after transfection.
RT-PCR and Western blot analysis.
RNA isolation and reverse transcription-PCR (RT-PCR) for all minigenes were performed as described previously (40). Protein expression was confirmed by Western blotting as described previously (23).
RESULTS
ETR-3 binding sites enriched by SELEX are UG rich.
SELEX was used to identify the preferred RNA binding sites of recombinant human ETR-3 protein purified from Escherichia coli. The high-affinity targets were selected from a pool of RNAs 54 nucleotides long containing a central region of 20 randomized nucleotides (see Materials and Methods). To identify ETR-3 binding sites, five rounds of SELEX were performed and 20 individual clones from the unselected pool (round 0) and 21 clones from the final round (round 5) were randomly picked and sequenced (Fig. 1). Comparison of the sequences obtained in rounds 0 and 5 revealed a clear increase in the content of U's and G's as well as GU and UG dinucleotides (Table 1). From round 0 to round 5, the combined percentage of U and G nucleotides increased from 43 to 75% and the percentage of UG and GU dinucleotides increased from 10 to 47%.
FIG. 1.
Sequences selected by ETR-3 after five rounds of SELEX. The UG repeat motifs are underlined (UGUGU), and the UGUU motifs are overlined (UGUU). The first U and the last G of each sequence are part of the constant region and therefore were not subjected to selection. All sequenced clones are shown. Numbers indicate the clone number.
TABLE 1.
UG-rich motifs are enriched after five rounds of SELEX with ETR-3
| Motif | % Occurrencea or no. of occurrencesb in sequences from round:
|
|
|---|---|---|
| 0 | 5 | |
| A | 25.3 | 12.6 |
| C | 31.3 | 10.5 |
| G | 20.0 | 30.2 |
| U | 23.4 | 46.7 |
| GU | 5.8 | 25.6 |
| UG | 4.7 | 21.6 |
| UU | 5.3 | 11.5 |
| AU | 5.0 | 8.3 |
| UA | 5.8 | 6.0 |
| UC | 8.0 | 5.3 |
| CG | 6.4 | 4.8 |
| CU | 8.3 | 2.8 |
| CA | 5.8 | 2.3 |
| GA | 6.6 | 2.3 |
| AC | 9.1 | 2.0 |
| GG | 3.9 | 2.0 |
| GC | 3.3 | 1.8 |
| AA | 6.9 | 1.5 |
| AG | 4.7 | 1.5 |
| CC | 10.2 | 1.0 |
| UGUGU | 0 | 18 |
| UAUGU | 0 | 10 |
| GUGUG | 0 | 9 |
| GUUGU | 1 | 9 |
| AUGUU | 1 | 8 |
| UGUUG | 1 | 7 |
| AUGUG | 0 | 6 |
| UUGUU | 0 | 6 |
| CGUGU | 1 | 5 |
| UGUUC | 1 | 5 |
| UGUUU | 0 | 5 |
| CACGA | 3 | 0 |
| CAUCG | 3 | 1 |
| CGUCC | 3 | 0 |
| GCCUA | 3 | 0 |
| GUCCU | 3 | 0 |
| GUGUGU | 0 | 7 |
| AUGUGU | 0 | 6 |
| UGUGUG | 0 | 6 |
| UGUUGU | 0 | 5 |
| GUAUGU | 0 | 4 |
| GUCUGU | 1 | 4 |
| GUUGUU | 0 | 4 |
| UAUGUG | 0 | 4 |
| UAUGUU | 0 | 4 |
| UUGUGU | 0 | 4 |
| CGUCCU | 3 | 0 |
| AAGUUC | 2 | 0 |
| AAUCUA | 2 | 0 |
| CACGAC | 2 | 0 |
| CAUCGC | 2 | 0 |
| GAAUCU | 2 | 0 |
| GUCCUC | 2 | 0 |
| UAAUCC | 2 | 0 |
| UCGUCC | 2 | 0 |
For single nucleotides and dinucleotides.
For pentamers and hexamers.
Binding sites for other RNA binding proteins involved in splicing are relatively small, usually between 5 and 9 nucleotides (20, 32, 49). We therefore performed an analysis of the frequency of pentameric and hexameric motifs within the SELEX-identified sequences (Table 1). In round 0 five pentamers and one hexamer were represented three times. In a sample of 20 20-nucleotide sequences, we would expect that each pentanucleotide and hexanucleotide would be represented 0.31 and 0.07 times, respectively. The relatively high frequencies of some pentamers and hexamers in round 0 might be due to a slight bias toward C's in the unselected pool (Table 1). In fact, all five pentamers and the hexamer most represented in round 0 are composed of at least 40% C's. In contrast, one pentamer (UGUGU) is represented 18 times and one hexamer (GUGUGU) is represented 7 times in round 5, clearly demonstrating enrichment of some motifs during SELEX (Table 1). The enriched pentamers and hexamers found in round 5 have an especially high content of U's and G's. The nucleotide composition of the top 11 pentamers and the top 10 hexamers is approximately 91% U's and G's, 6% A's, and only 3% C's, which further indicates ETR-3's preference for UG-rich motifs. The most enriched motif in round 5 is repeats composed of alternating U's and G's. At least one UGUGU or GUGUG motif is found in 15 out of the 21 sequences from round 5. A second enriched motif, also present in 15 sequences, is UGUU. This motif is present in 4 of the 11 most represented pentamers and in 3 of the top 10 hexamers found in round 5. More importantly, this motif is present in five of the six sequences that lack a repetitive UG motif (Fig. 1). In addition to these predominating motifs, motifs similar to the UG repetitions but with an A replacing one of the G's were also enriched. Since these motifs are less frequent than those containing only UG repetitions and since the A's were not enriched during the SELEX procedure (Table 1), these motifs might be secondary ETR-3 binding sites.
ETR-3 binds to the SELEX-identified sequences in vitro.
To confirm that ETR-3 binds to the SELEX-identified sequences, two sequences from round 0 and four from round 5 were randomly selected to perform an EMSA (Fig. 2). One additional sequence from round 5 was selected (clone 522) because it contained the lowest UG content and was the only selected sequence that lacked UG repetition or UGUU motifs. Labeled synthetic RNAs were incubated with or without bacterially expressed recombinant ETR-3, and bound and unbound RNAs were separated by electrophoresis on a nondenaturing gel. Neither of the sequences from round 0 showed significant affinity for binding to ETR-3, even with large amounts of recombinant ETR-3 (1 μg; Fig. 2A). In contrast, four of the five sequences from round 5 bound to ETR-3 when incubated with 1/10 of that amount of ETR-3 (100 ng). The sequence from round 5 with the poorest binding affinity to ETR-3 was clone 522, the only sequence that lacks a UG or UGUU motif. None of the seven RNAs bound to a recombinant polypyrimidine tract binding protein, an unrelated RRM-containing protein (data not shown).
ETR-3 SELEX-identified motifs function as CELF response elements in vivo.
Next we tested the ability of the selected UG and UGUU motifs to mediate ETR-3 splicing activity in vivo. Alternative splicing of human cTNT exon 5 was previously shown to be regulated by CUG-BP1, which is 78% identical to ETR-3 (40). The CUG-BP1 binding site is located 20 nucleotides downstream of the alternative exon and a 5-nucleotide substitution (Fig. 3A) that disrupts CUG-BP1 binding also prevents regulation by CUG-BP1 in vivo (40). Similarly to what is observed with CUG-BP1, ETR-3 coexpression with the wild-type minigene also leads to an increase of exon 5 inclusion (Fig. 3B). Moreover, the 5-nucleotide mutation that affects CUG-BP1 binding and regulation also disrupts ETR-3's binding and splicing activity (Fig. 3B and C).
We replaced a 26-nucleotide region in R300TA that contains the mutated CUG-BP1 binding sites (Fig. 3A) with two SELEX-identified sequences from round 5 (clones 503 and 510 in Fig. 1) and one from round 0 (UCGAAUUACACAUCGCCUAG). Unexpectedly, all three sequences led to greater-than-90% exon 5 inclusion in COSM6 cells even in the absence of ETR-3 coexpression (data not shown). Because of these high basal levels of exon inclusion, we were unable to evaluate the effects of ETR-3 expression with these minigenes. The elevated levels of exon inclusion might be due to a disruption of an uncharacterized negative element present in the replaced region downstream of the exon.
To avoid disrupting uncharacterized regulatory elements, we made smaller changes in the minigene by inserting short enriched motifs into R300TA. An advantage of inserting smaller ETR-3 binding motifs is that it allows us to determine more precisely the pre-mRNA sequence required for ETR-3 splicing activity. We chose to separately test the two motifs that were most represented in the fifth round of SELEX: UG repeats and UGUU. Each motif was inserted into R300TA by either mutating only 2 nucleotides (UG minigene) or replacing 12 nucleotides (UGUU minigene) (Fig. 3A). Both UG and UGUU minigenes were transiently expressed in COSM6 cells with or without ETR-3. Coexpression of ETR-3 increased exon 5 inclusion for both the UG and UGUU minigenes to levels comparable to the levels seen for the wild-type cTNT minigene (from 50 to 80%), while it had nearly no effect on the parental minigene, R300TA (increase from 50 to 55%) (Fig. 3B). These results indicate that ETR-3, like CUG-BP1, activates human cTNT exon 5 inclusion. It also indicated that mutations inserting the well-represented motifs identified by SELEX into the regulatory region restored responsiveness to ETR-3 splicing activity in vivo to wild-type levels.
We next tested whether other members of the CELF family could regulate splicing via the ETR-3 SELEX-identified motifs. The CELF protein family can be divided into two subgroups based on sequence identity and target specificity (23, 24). One group is composed of ETR-3 and CUG-BP1, while the second group includes CELF3 through CELF6. We transfected one protein from each of these groups, CUG-BP1 and CELF4, with each of the four minigenes. As observed with CUG-BP1 and ETR-3, CELF4 was able to induce exon 5 inclusion when coexpressed with RTB300 but not with R300TA. This indicates that all three CELF proteins act via the previously identified CUG-BP1 binding site (40). Interestingly, coexpression of CUG-BP1 and CELF4 with the UG and UGUU minigenes also led to higher levels of exon 5 inclusion. These results indicate that all three CELF proteins are able to regulate exon 5 inclusion via the motifs identified by SELEX with ETR-3, suggesting that both subgroups can use similar binding sites to regulate alternative splicing in vivo.
To evaluate whether regulation by ETR-3 in vivo correlated with ETR-3 binding, we replaced the randomized region in the sequences from round 0 with the regulatory regions from the RTB300, R300TA, UG, and UGUU minigenes shown in Fig. 3A and tested their abilities to bind ETR-3 by EMSA. The results in Fig. 3C show that ETR-3 binds with higher affinity to RTB300, UG, and UGUU minigene sequences than to R300TA, which correlates directly with activation of splicing regulation by ETR-3 in vivo.
ETR-3 binding sites restore responsiveness to CUG repeat RNA.
Coexpression of expanded CUG RNA with RTB300 induces exon 5 inclusion and is thought to reproduce the effects of expression of the expanded CUG repeat allele in DM (40) (Fig. 3D). This response requires an intact CUG-BP1 binding site, since coexpression of expanded CUG RNA has little effect on the splicing of R300TA (40) (Fig. 3D). When UG and UGUU minigenes were coexpressed with expanded CUG RNA, both showed a greater increase in exon 5 inclusion than R300TA (Fig. 3D). The result obtained with the UG minigene is particularly striking since mutation of only 2 nucleotides in R300TA was enough to restore response to the expanded CUG RNA. These results, together with the results obtained with coexpression of the minigenes and the CELF proteins, demonstrate that the presence of an CELF binding site restores the trans-dominant effect of the CUG-expanded RNA in the splicing of cTNT exon 5, supporting the proposal that the effects of the expanded RNA are mediated, at least in part, by the CELF proteins.
Discovery of new splicing targets regulated by ETR-3.
The preferred ETR-3 binding sites provide a tool to identify potential new targets of ETR-3 splicing regulation. Using the BLAT engine in the University of California, Santa Cruz (UCSC), genome browser (http://genome.ucsc.edu) and the SELEX-identified sequences and motifs, we searched the human genome for potential ETR-3 binding sites located within 500 bp of an exon identified as alternatively spliced based on expressed sequence tag or mRNA sequences available in the UCSC genome browser. Since the SELEX motifs identified are relatively small, we used several combinations of UG-rich elements as probes for our search. We also used strict conditions to consider a match in the human genome as an ETR-3 putative binding site. Besides being near an alternative exon, a putative ETR-3 binding site would have to be at least 10 nucleotides long or have two smaller motifs on each side of the exon. Of the more than 2,500 potential ETR-3 binding sites found in our search, 19 are located near an alternatively spliced exon. Of these, 18 are represented in Table 2 and one is ClC1 intron 2, which was previously shown to be regulated by CUG-BP1 (8). This intron was the only known CELF protein target that could be identified using the criteria of this screen. For all other targets, either the binding sites are smaller or the regulatory elements are located further away from the exon (8, 16, 40, 42, 50). Nevertheless, we wanted to have a relatively small list of genes with strong ETR-3 putative binding sites.
TABLE 2.
List of genes with putative ETR-3 binding sites in introns within 500 nucleotides of an alternatively spliced exon
| Gene product | Exonic sequence | ETR-3 putative binding site and location(s)a |
|---|---|---|
| Ros1 | TTAGGGCCTTTA...CTACAACATCAG | +43, GUGUGU...GUGUGU |
| BITE | ATCTCATCATTT...TCAACAGCTTAG | +29, UUGUUUGUAUG |
| MEF2C | GCTTGCACTAGC...TTCTCCTCCTAG | −55, UGAUGU...UGAGUU |
| MTMR1 | GTTTTCAGGAGGCCTGATCTAAGG | +37, UUGUGUGUGUCUGUG |
| MYH7 | GTGAAAAACCTG...CAAGTGGATGAT | +60, GUGUGU...AUGUUU |
| FXRP1 | GATGATAGTGAA...GATAGACAGCCA | −32, GUUUUU...UUUUGU |
| AGA | GCATGGTTGTAA...CAAAATACATGG | −305, GUUGUG...GUGUGU |
| MBNL | CCAATGTTTTCA...GACAGACTTGAG | +49, GUGUGG...GUGUUU |
| NCX1 | TAGGTTGTGACA...TGATGAAATTGT | +19, UGUGUG...GUGGUU |
| CFTR | GGATTTGGGGAA...GGAGCAGGCAAG | −10, UGUGU...GUGUGU |
| FREQ | TTACCGAGAAGGAGGTCCAGCAGTG | +209, GUGUUUGUUG |
| TPM1 | CTCGAGGAGGAC...GCCGCCGCCAAG | −53, GUGUGUUGUGUGUGU |
| FLJ12871 | ATTCCAAGGTTG...GAGGAGAACAAG | +136, UUUGUGUUGUUGUUGUUG |
| GRID2 | GAAATAAACGAT...CAGAATATGGAG | −44, GUUUGUGUUUUGUG |
| FOXM1 | CCACTGGACCCA...CACTTGGAATCA | −75 and +111, GUGUUUUU and UGUUUUGU |
| VCL | GGCAATGACATC...AACCTCTTACAG | +18, UGCGUGUGU...GUGUUG |
| RASGRF1 | GTTCCCTCATTC...CACTTGACCAAG | +297, UGUGUGUG...AUGUGUUU |
| ATP2A2 | CTCATCTTCCAG...ACCTGGAACCTG | −3 and +7, UGUGUGU and GUGUGUG |
Location(s) of the putative binding site in relation to the alternatively spliced exon (+, upstream; −, downstream).
Minigenes containing two of these exons were created and tested for responsiveness to ETR-3: exon 9 from the CFTR gene and MTMR1 exon 2.1. These exons were chosen because they are involved in human disease, the binding sites are located very close to the exon, and, for CFTR, exon 9 skipping is known to produce a nonfunctional protein.
CFTR is mutated in all cystic fibrosis patients, and less-severe forms of the disease, such as congenital bilateral absence of vas deferens (CBAVD), pancreatitis, nasal polyposis, and disseminated bronchiectases, are associated with an allelic variation located 7 nucleotides upstream of CFTR exon 9 (9, 11, 12, 28). This variable region is composed of from 9 to 13 UG repeats followed by a variable U repeat (from 5 to 9). The UG repeats were identified in our screen as a potential ETR-3 binding site. It is thought that the U5 allele results in disease variation with incomplete penetrance modulated by other mutations and/or polymorphisms (reviewed in reference 13). Interestingly, the pathological effect of the U5 allele has been associated with changes in the alternative splicing of CFTR exon 9. This exon encodes part of a nucleotide-binding domain, and its skipping produces a nonfunctional CFTR protein. In CBAVD patients and unaffected individuals, there is a correlation between the number of UG repeats, and particularly U repeats, in the polymorphic locus and the amount of CFTR mRNA that lacks exon 9 (38, 39). A low number of U repeats favors the exclusion of exon 9 in the mRNA, and the level of exclusion is modulated by the length of the UG repeats. In the presence of a short U tract, a longer UG repeat generally favors exon 9 skipping while shorter UG tracts tend to favor exon inclusion (38).
To evaluate the influence of ETR-3 on the regulation of this exon, we replaced a segment of intron upstream of an artificial alternative exon that is not responsive to ETR-3 with the intronic region (containing the UG repeats and the U tract) upstream of CFTR exon 9 (Fig. 4A). A second minigene containing a mutation that mutates the UG repeats was also constructed (CFTRmut; Fig. 4A). When ETR-3 was coexpressed with the minigene containing the wild-type CFTR intronic segment, an increase in exon inclusion was observed (from 3 to 20%). The substitutions in the CFTR intronic segment in CFTRmut increased the basal level of exon inclusion, presumably because of the increased pyrimidine content of the 3′ splice site. The key result is that enhancement of exon inclusion by ETR-3 was completely abolished when the putative ETR-3 binding site was mutated (Fig. 4A). These results suggest that ETR-3 acts directly on CFTR pre-mRNA via the binding sites identified in the genomic screen.
MTMR1 is a member of a large family of phosphatases thought to have an important role in regulating vesicle trafficking and autophagy (2, 14, 26, 44, 47). Several members of this protein family are affected in human diseases such as X-linked myotubular myopathy (MTM1), Charcot-Marie-Tooth type 4B (MTMR2), and congenital DM1 (MTMR1) (3, 6, 27). In congenital DM1 patients, the splicing of exons 2.1 and 2.2 is disrupted, leading to the formation of abnormal isoforms. Interestingly, the putative ETR-3 binding site identified in our screen (UUGUGUGUGUCUGUG) is located 37 nucleotides downstream of MTMR1 alternative exon 2.1, between this exon and exon 2.2 (Fig. 4B). A 797-nucleotide genomic region that contains exon 2.1 was cloned into R300TA, completely replacing the genomic sequence of cTNT. When ETR-3 was coexpressed with the minigene-containing MTMR1 genomic segment, exon 2.1 inclusion was induced from 60 to 87%. We mutated the region identified in our genomic screen as a putative ETR-3 binding site, but the new minigenes were still responsive to coexpression of ETR-3 (data not shown). The region identified in our genomic screen is the best match to the SELEX motifs, but if we analyze the sequence flanking MTMR1 exon 2.1, we find other good ETR-3 potential binding sites within 200 nucleotides upstream and downstream from the exon. We do not know if these other potential binding sites are the endogenous ETR-3 binding sites or if they just compensate for the mutations made to the principal binding site. Nevertheless, we conclude that ETR-3 is able to modulate MTMR1 exon 2.1 splicing.
DISCUSSION
Here we report the identification of the preferred binding motifs for ETR-3 and demonstrate that these elements bind ETR-3 in vitro and mediate responsiveness to ETR-3 in vivo. The sequences selected by ETR-3 during SELEX were highly enriched in U's and G's, and in particular UG and UGUU motifs. These motifs are similar to those found in some known CELF binding sites in pre-mRNAs regulated by CELF proteins (7, 8). The UG motif is also similar to the sequences obtained with a yeast three-hybrid screen and SELEX performed with CUG-BP1 (4, 22, 46). Two groups also described a motif composed of UAUG repeats obtained by analysis of zebra fish and Xenopus laevis CELF proteins (4, 46). Even though the frequency of this motif is not as high as the frequency of UG repeats or UGUU motifs, it is increased in our SELEX sequences for ETR-3, and it is part of three of the most common pentanucleotides and four common hexanucleotides (Table 1). However, we noticed that one copy of this sequence is present in one clone from round 5 that bound poorly to ETR-3 (clone 522), suggesting that efficient binding to human ETR-3 under our conditions requires more than one copy and/or a different context.
We demonstrated that the SELEX-identified sequences bind ETR-3 in vitro and that mutations to insert either the UG or UGUU motif into a nonresponsive minigene restored responsiveness to ETR-3. Therefore, the responsiveness of those minigenes is wholly consistent with the SELEX data. In addition, both motifs restored responsiveness to two other CELF proteins, CUG-BP1 and CELF4. Of all the CELF proteins, CELF4 has the lowest identity to ETR-3 (43%). Despite this divergence, CELF4 is able to activate splicing via the motifs selected for ETR-3, indicating that different CELF proteins can act through similar motifs to activate splicing in vivo. These results show that ETR-3 optimal binding is functionally significant not only for ETR-3 but also for other CELF proteins. Nevertheless, we cannot assume that all natural ETR-3 binding sites will also be relevant for the other CELF proteins. In fact, it was previously shown that CELF proteins differ in their abilities to regulate alternative splicing of some pre-mRNAs (16, 23, 24). While the optimal binding motifs identified in this work are able to induce splicing regulation by the tested CELF proteins, it is likely that there are specific pre-mRNA sequences that bind only a subset of CELF proteins. Although the mechanism by which the CELF proteins regulate splicing is not known, our working model is that the CELF proteins are part of regulatory complexes that assemble near the alternatively spliced exon. The splicing specificity of each CELF protein might also be explained by differences in the auxiliary factors that bind to each CELF protein directly or that bind to additional pre-mRNA regulatory elements and facilitate splicing regulation by the CELF proteins. Since there is evidence that some CELF proteins regulate other steps of mRNA processing, such as editing and translation, and RNA stability (1, 19, 37), it is likely that there is some degree of overlapping target specificity in these processes as well.
The CELF proteins have been implicated in the pathology of DM. The splicing of several known CELF protein targets is misregulated in DM patients (8, 40, 42, 43). To test the relationship between misregulated splicing induced by CUG-expanded RNA and ETR-3 regulation, we tested the effect of CUG-expanded RNA on the splicing of the minigenes containing the SELEX-identified motifs. The results showed that the three minigenes containing an ETR-3 binding site (the RTB300, UG, and UGUU minigenes) respond to the coexpression of expanded CUG RNA while the minigene without ETR-3 binding sites (R300TA) shows minimal splicing changes. The result obtained with the UG minigene is striking since a mutation in only 2 nucleotides restores binding and splicing regulation by ETR-3 and the splicing effect of the CUG repeat-containing RNA. These results indicate that responsiveness to the trans-dominant effects of expanded CUG RNA on cTNT splicing is dependent on the ability of CELF proteins to regulate the splicing event, rather than the particular sequence of the pre-mRNA.
With a simple genomic screen, we identified 18 new alternatively spliced exons with potential ETR-3 binding sites within 500 nucleotides upstream or downstream of the exon. Interestingly, none of the 2,500 hits that we obtained were located in an exon, which is consistent with the current evidence that the CELF proteins act via intronic elements. We created minigenes for two of these potential targets to analyze the effects of ETR-3 on their splicing patterns. In both cases, ETR-3 coexpression induced exon inclusion. For CFTR, mutation of the ETR-3 binding site demonstrated that this effect was dependent on the presence of the motif identified in our genomic screen. These results indicate that ETR-3 is able to modulate the splicing of MTMR1 exon 2.1 and CFTR exon 9 and that for CFTR exon 9 this regulation is dependent on the region identified in our screen.
The strategy to use the SELEX sequences to search for endogenous targets was first described by Buckanovich and Darnell (5). In that study, the authors searched a database of neuron-specific alternative splicing exons for Nova-1 putative binding sites. This report shows that this strategy can be applied to any database without having previous knowledge of the tissue or developmental stage in which the protein activity will be relevant. The fact that, for most of the RNA binding proteins, little is known about their binding site preferences and characteristics and the binding sites known are usually very small and degenerate makes the search and discovery of new targets for nucleic acid binding proteins very difficult. Even though ETR-3 is an RNA binding protein with a relatively degenerate and small binding site, the approach used was able to reveal new putative targets. This strategy can easily be applied to other DNA and RNA binding proteins involved in different processes, in particular, if the proteins have a very specific binding site and act in a specific location of the DNA or RNA. These two last factors will help reduce the number of false-positive putative targets, thereby increasing the efficiency of the strategy.
Acknowledgments
We thank Donnie Bundman, Jin Han, Saadi Imam, Yousif Shamoo, Gopal Singh, Miles Wilkinson, and Gang Wu for their technical assistance and Young-Hwa Goo, Jin Han, Thai Ho, and Andrea Ladd for helpful discussions on the manuscript.
This work was supported by grants to T.A.C. from NIH (RO1HL45565 and RO1AR45653) and to N.A.F. from the Portuguese Foundation for Science and Technology (BD2744/00).
REFERENCES
- 1.Anant, S., J. O. Henderson, D. Mukhopadhyay, N. Navaratnam, S. Kennedy, J. Min, and N. O. Davidson. 2001. Novel role for RNA-binding protein CUGBP2 in mammalian RNA editing. CUGBP2 modulates C to U editing of apolipoprotein B mRNA by interacting with apobec-1 and ACF, the apobec-1 complementation factor. J. Biol. Chem. 276:47338-47351. [DOI] [PubMed] [Google Scholar]
- 2.Blondeau, F., J. Laporte, S. Bodin, G. Superti-Furga, B. Payrastre, and J. L. Mandel. 2000. Myotubularin, a phosphatase deficient in myotubular myopathy, acts on phosphatidylinositol 3-kinase and phosphatidylinositol 3-phosphate pathway. Hum. Mol. Genet. 9:2223-2229. [DOI] [PubMed] [Google Scholar]
- 3.Bolino, A., M. Muglia, F. L. Conforti, E. LeGuern, M. A. Salih, D. M. Georgiou, K. Christodoulou, I. Hausmanowa-Petrusewicz, P. Mandich, A. Schenone, A. Gambardella, F. Bono, A. Quattrone, M. Devoto, and A. P. Monaco. 2000. Charcot-Marie-Tooth type 4B is caused by mutations in the gene encoding myotubularin-related protein-2. Nat. Genet. 25:17-19. [DOI] [PubMed] [Google Scholar]
- 4.Bonnet-Corven, S., Y. Audic, F. Omilli, and H. B. Osborne. 2002. An analysis of the sequence requirements of EDEN-BP for specific RNA binding. Nucleic Acids Res. 30:4667-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckanovich, R. J., and R. B. Darnell. 1997. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol. Cell. Biol. 17:3194-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buj-Bello, A., D. Furling, H. Tronchere, J. Laporte, T. Lerouge, G. S. Butler-Browne, and J. L. Mandel. 2002. Muscle-specific alternative splicing of myotubularin-related 1 gene is impaired in DM1 muscle cells. Hum. Mol. Genet. 11:2297-2307. [DOI] [PubMed] [Google Scholar]
- 7.Charlet-B, N., P. Logan, G. Singh, and T. A. Cooper. 2002. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell 9:649-658. [DOI] [PubMed] [Google Scholar]
- 8.Charlet-B, N., R. S. Savkur, G. Singh, A. V. Philips, E. A. Grice, and T. A. Cooper. 2002. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell 10:45-53. [DOI] [PubMed] [Google Scholar]
- 9.Chillon, M., T. Casals, B. Mercier, L. Bassas, W. Lissens, S. Silber, M. C. Romey, J. Ruiz-Romero, C. Verlingue, M. Claustres, M., et al. 1995. Mutations in the cystic fibrosis gene in patients with congenital absence of the vas deferens. N. Engl. J. Med. 332:1475-1480. [DOI] [PubMed] [Google Scholar]
- 10.Choi, D. K., K. W. Yoo, S. K. Hong, M. Rhee, Y. Sakaki, and C. H. Kim. 2003. Isolation and expression of Napor/CUG-BP2 in embryo development. Biochem. Biophys. Res. Commun. 305:448-454. [DOI] [PubMed] [Google Scholar]
- 11.Chu, C. S., B. C. Trapnell, S. Curristin, G. R. Cutting, and R. G. Crystal. 1993. Genetic basis of variable exon-9 skipping in cystic fibrosis transmembrane conductance regulator messenger RNA. Nat. Genet. 3:151-156. [DOI] [PubMed] [Google Scholar]
- 12.Cuppens, H., W. Lin, M. Jaspers, B. Costes, H. Teng, A. Vankeerberghen, M. Jorissen, G. Droogmans, I. Reynaert, M. Goossens, B. Nilius, and J. J. Cassiman. 1998. Polyvariant mutant cystic fibrosis transmembrane conductance regulator genes. The polymorphic (Tg)m locus explains the partial penetrance of the T5 polymorphism as a disease mutation. J. Clin. Investig. 101:487-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faustino, N. A., and T. A. Cooper. 2003. Pre-mRNA splicing and human disease. Genes Dev. 17:419-437. [DOI] [PubMed] [Google Scholar]
- 14.Gillooly, D. J., I. C. Morrow, M. Lindsay, R. Gould, N. J. Bryant, J. M. Gaullier, R. G. Parton, and H. Stenmark. 2000. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19:4577-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Good, P. J., Q. Chen, S. J. Warner, and D. C. Herring. 2000. A family of human RNA-binding proteins related to the Drosophila Bruno translational regulator. J. Biol. Chem. 275:28583-28592. [DOI] [PubMed] [Google Scholar]
- 16.Gromak, N., A. J. Matlin, T. A. Cooper, and C. W. Smith. 2003. Antagonistic regulation of alpha-actinin alternative splicing by CELF proteins and polypyrimidine tract binding protein. RNA 9:443-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harper, P. S. 2001. Myotonic dystrophy, 3rd ed., vol. 37. W. B. Saunders, London, United Kingdom.
- 18.Ho, T. H., N. Charlet-B, M. G. Poulos, G. Singh, M. S. Swanson, and T. A. Cooper. 2004. Muscleblind proteins regulate alternative splicing. EMBO J. 23:3103-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iakova, P., G. L. Wang, L. Timchenko, M. Michalak, O. M. Pereira-Smith, J. R. Smith, and N. A. Timchenko. 2004. Competition of CUGBP1 and calreticulin for the regulation of p21 translation determines cell fate. EMBO J. 23:406-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen, K. B., K. Musunuru, H. A. Lewis, S. K. Burley, and R. B. Darnell. 2000. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc. Natl. Acad. Sci. USA 97:5740-5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanadia, R. N., K. A. Johnstone, A. Mankodi, C. Lungu, C. A. Thornton, D. Esson, A. M. Timmers, W. W. Hauswirth, and M. S. Swanson. 2003. A muscleblind knockout model for myotonic dystrophy. Science 302:1978-1980. [DOI] [PubMed] [Google Scholar]
- 22.Kino, Y., D. Mori, Y. Oma, Y. Takeshita, N. Sasagawa, and S. Ishiura. 2004. Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum. Mol. Genet. 13:495-507. [DOI] [PubMed] [Google Scholar]
- 23.Ladd, A. N., N. Charlet-B, and T. A. Cooper. 2001. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell. Biol. 21:1285-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ladd, A. N., N. H. Nguyen, K. Malhotra, and T. A. Cooper. 2004. CELF6, a member of the CELF family of RNA binding proteins, regulates MSE-dependent alternative splicing. J. Biol. Chem. 279:17756-17764. [DOI] [PubMed] [Google Scholar]
- 25.Lander, E. S., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 26.Laporte, J., F. Blondeau, A. Buj-Bello, and J. L. Mandel. 2001. The myotubularin family: from genetic disease to phosphoinositide metabolism. Trends Genet. 17:221-228. [DOI] [PubMed] [Google Scholar]
- 27.Laporte, J., L. J. Hu, C. Kretz, J. L. Mandel, P. Kioschis, J. F. Coy, S. M. Klauck, A. Poustka, and N. Dahl. 1996. A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat. Genet. 13:175-182. [DOI] [PubMed] [Google Scholar]
- 28.Larriba, S., L. Bassas, J. Gimenez, M. D. Ramos, A. Segura, V. Nunes, X. Estivill, and T. Casals. 1998. Testicular CFTR splice variants in patients with congenital absence of the vas deferens. Hum. Mol. Genet. 7:1739-1743. [DOI] [PubMed] [Google Scholar]
- 29.Levers, T. E., S. Tait, M. C. Birling, P. J. Brophy, and D. J. Price. 2002. Etr-r3/mNapor, encoding an ELAV-type RNA binding protein, is expressed in differentiating cells in the developing rodent forebrain. Mech. Dev. 112:191-193. [DOI] [PubMed] [Google Scholar]
- 30.Li, D., L. L. Bachinski, and R. Roberts. 2001. Genomic organization and isoform-specific tissue expression of human NAPOR (CUG-BP2) as a candidate gene for familial arrhythmogenic right ventricular dysplasia. Genomics 74:396-401. [DOI] [PubMed] [Google Scholar]
- 31.Liquori, C. L., K. Ricker, M. L. Moseley, J. F. Jacobsen, W. Kress, S. L. Naylor, J. W. Day, and L. P. Ranum. 2001. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science 293:864-867. [DOI] [PubMed] [Google Scholar]
- 32.Liu, H. X., M. Zhang, and A. R. Krainer. 1998. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 12:1998-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mankodi, A., E. Logigian, L. Callahan, C. McClain, R. White, D. Henderson, M. Krym, and C. A. Thornton. 2000. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science 289:1769-1773. [DOI] [PubMed] [Google Scholar]
- 34.Mankodi, A., C. R. Urbinati, Q. P. Yuan, R. T. Moxley, V. Sansone, M. Krym, D. Henderson, M. Schalling, M. S. Swanson, and C. A. Thornton. 2001. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet. 10:2165-2170. [DOI] [PubMed] [Google Scholar]
- 35.Miller, J. W., C. R. Urbinati, P. Teng-Umnuay, M. G. Stenberg, B. J. Byrne, C. A. Thornton, and M. S. Swanson. 2000. Recruitment of human muscleblind proteins to (CUG)n expansions associated with myotonic dystrophy. EMBO J. 19:4439-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Modrek, B., and C. Lee. 2002. A genomic view of alternative splicing. Nat. Genet. 30:13-19.11753382 [Google Scholar]
- 37.Mukhopadhyay, D., C. W. Houchen, S. Kennedy, B. K. Dieckgraefe, and S. Anant. 2003. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol. Cell 11:113-126. [DOI] [PubMed] [Google Scholar]
- 38.Niksic, M., M. Romano, E. Buratti, F. Pagani, and F. E. Baralle. 1999. Functional analysis of cis-acting elements regulating the alternative splicing of human CFTR exon 9. Hum. Mol. Genet. 8:2339-2349. [DOI] [PubMed] [Google Scholar]
- 39.Pagani, F., E. Buratti, C. Stuani, M. Romano, E. Zuccato, M. Niksic, L. Giglio, D. Faraguna, and F. E. Baralle. 2000. Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a nonevolutionary conserved intronic element. J. Biol. Chem. 275:21041-21047. [DOI] [PubMed] [Google Scholar]
- 40.Philips, A. V., L. T. Timchenko, and T. A. Cooper. 1998. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science 280:737-741. [DOI] [PubMed] [Google Scholar]
- 41.Resch, A., Y. Xing, B. Modrek, M. Gorlick, R. Riley, and C. Lee. 2004. Assessing the impact of alternative splicing on domain interactions in the human proteome. J. Proteome Res. 3:76-83. [DOI] [PubMed] [Google Scholar]
- 42.Savkur, R. S., A. V. Philips, and T. A. Cooper. 2001. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Gen. 29:40-47. [DOI] [PubMed] [Google Scholar]
- 43.Savkur, R. S., A. V. Philips, T. A. Cooper, J. C. Dalton, M. L. Moseley, L. P. Ranum, and J. W. Day. 2004. Insulin receptor splicing alteration in myotonic dystrophy type 2. Am. J. Hum. Genet. 74:1309-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simonsen, A., A. E. Wurmser, S. D. Emr, and H. Stenmark. 2001. The role of phosphoinositides in membrane transport. Curr. Opin. Cell Biol. 13:485-492. [DOI] [PubMed] [Google Scholar]
- 45.Singh, G., N. Charlet-B, J. Han, and T. A. Cooper. 2004. ETR-3 and CELF4 protein domains required for RNA binding and splicing activity in vivo. Nucleic Acids Res. 32:1232-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki, H., Y. Jin, H. Otani, K. Yasuda, and K. Inoue. 2002. Regulation of alternative splicing of alpha-actinin transcript by Bruno-like proteins. Genes Cells 7:133-141. [DOI] [PubMed] [Google Scholar]
- 47.Taylor, G. S., T. Maehama, and J. E. Dixon. 2000. Inaugural article: myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc. Natl. Acad. Sci. USA 97:8910-8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White, E. K., T. Moore-Jarrett, and H. E. Ruley. 2001. PUM2, a novel murine puf protein, and its consensus RNA-binding site. RNA 7:1855-1866. [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan, X., N. Davydova, M. R. Conte, S. Curry, and S. Matthews. 2002. Chemical shift mapping of RNA interactions with the polypyrimidine tract binding protein. Nucleic Acids Res. 30:456-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, W., H. Liu, K. Han, and P. J. Grabowski. 2002. Region-specific alternative splicing in the nervous system: implications for regulation by the RNA-binding protein NAPOR. RNA 8:671-685. [DOI] [PMC free article] [PubMed] [Google Scholar]