Abstract
Long-range and local RNA-RNA contacts in viral RNA genomes result in tertiary structures that modulate the function of enhancers, promoters, and silencers during translation, RNA replication, and encapsidation. In the case of flaviviruses, the presence of inverted complementary sequences at the 5′ and 3′ ends of the genome mediate long-range RNA interactions and RNA cyclization. The circular conformation of flavivirus genomes was demonstrated to be essential for RNA amplification. New ideas about the mechanisms by which circular genomes participate in flavivirus replication have emerged in the last few years. Here, we will describe the latest information about cis-acting elements involved in flavivirus genome cyclization, RNA promoter elements required for viral polymerase recognition, and how these elements together coordinate viral RNA synthesis.
Keywords: flavivirus, genome cyclization, viral RNA synthesis, viral cis-acting elements, RNA-RNA interactions, viral UTRs
1. Introduction: Flavivirus replication cycle
Flaviviruses comprise one of the three genera within the Flaviviridae family; the other two are the Pestivirus and the Hepacivirus. The Flavivirus genus is divided into three groups based on their ecological characteristics, mosquito-borne, tick-borne, and no-known vector flaviviruses (Kuno et al., 1998). Among the mosquito-borne flaviviruses there are important human pathogens such as dengue virus (DENV), yellow fever virus (YFV), West Nile virus (WNV), Saint Luis encephalitis virus (SLEV), and Japanese encephalitis virus (JEV).
Flaviviruses are enveloped viruses with a single stranded, ~ 11 kb, positive-sense RNA genome with a type 1 cap (m7GpppAmp) structure at the 5′ end (Cleaves and Dubin, 1979; Wengler and Gross, 1978). The viral RNA encodes a single long open reading frame (ORF) flanked by highly structured 5′ and 3′ untranslated regions (UTR). The 5′UTRs are about 100 nucleotides long, while the 3′UTRs are between 350 to 700 nucleotides.
The virus enters the host cell by receptor mediated endocytosis. Upon internalization and acidification of the endosome, fusion of viral and vesicular membranes allows release of the genomic RNA into the cytoplasm, which serves as mRNA. Translation of the single ORF at the rough ER produces a large polyprotein that is cleaved co- and posttranslationally into the mature proteins. The N-terminal of the polyprotein encodes the three structural proteins (C-prM-E), followed by seven nonstructural (NS) proteins (NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5) (Rice et al., 1985) (Figure 1). The amino termini of prM, E, NS1, and NS4B are generated upon cleavage by the host signal peptidase in the ER lumen, while the processing of most of the NS proteins and the carboxyl terminus of the C protein is carried out by the viral NS3 serine protease with the NS2B cofactor in the cytoplasm of the infected cell.
Figure 1.
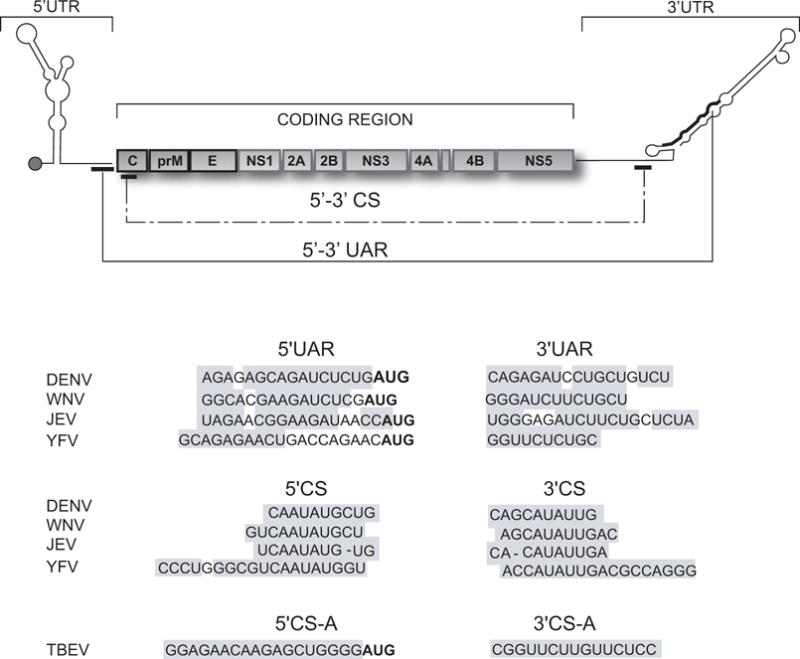
Flavivirus genome. On the top, schematic representation showing the coding and untranslated regions (UTRs) of the viral genome. The three structural proteins C, capsid; prM precursor to membrane protein; E, envelope; and the seven non-structural (NS) proteins NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5 are indicated. The location of the complementary sequences 5′-3′UAR and 5′-3′CS of mosquito-borne flaviviruses is also shown. The bottom panels show the nucleotide sequences of the complementary regions 5′-3′CS and 5′-3′UAR of dengue virus type 2 (DENV2, Genebank number U87412), West Nile virus (WNV, Genebank number M12294), Japanese encephalitis virus (JEV, Genebank number NC001437), and yellow fever virus (YFV, Genebank number NC002031); and the 5′-3′CSA of tick-borne encephalitis virus (TBEV, Genebank number U27495). The grey boxes denote the inverted complementary sequences. In boldface the translation initiator AUG is indicated.
After translation of the viral RNA, virus-induced hypertrophy of intracellular membranes occurs, originating structures known as convoluted membranes and vesicle packets (for review see (Westaway et al., 2003). Viral RNA synthesis occurs in close association with cellular membranes inside the vesicle packets in so called viral replication complexes. The process begins with the synthesis of a negative strand RNA, which serves as template for the amplification of additional positive strand genomic RNA. The enzymatic reaction is catalized by the RdRp activity of the viral NS5 protein, in association with the viral protease/helicase NS3, other viral NS proteins, and presumably host factors. The newly synthesized RNA associates to the C protein by a mechanism still unknown. The RNA-C complex buds into the ER lumen acquiring the lipid bilayer, and the viral E and prM proteins. Furin-mediated proteolysis of prM in the trans-Golgi network (Stadler et al., 1997) triggers rearrangement, homodimerization of E, and formation of new viral particles (Allison et al., 2003).
2. Inverted complementary sequences at the terminal regions of flavivirus genomes
The presence of 5′ and 3′ complementary sequences (CS) in the genome of flaviviruses was first noticed by Hahn et al., who defined an original 8 nucleotide core sequence conserved among mosquito-borne flaviviruses (Hahn et al., 1987). The 5′ CS element was found within the viral ORF, coding for the amino-terminal part of protein C. The complementary 3′CS was identified within the 3′UTR, just upstream of the conserved 3′ terminal stem-loop structure (3′SL) (Figure 1). The requirement of complementarity between 5′ and 3′ CS elements for viral replication was demonstrated using infectious clones and replicon systems of Kunjin virus (KUNV), DENV, and WNV (Alvarez et al., 2005a; Khromykh et al., 2001; Lo et al., 2003). Mutations at the 5′ or 3′CS regions impaired RNA synthesis without altering translation of the input RNA, providing the first evidence for a role of genome cyclization during RNA replication. In these experiments it was demonstrated that reconstitution of base pairings between 5′-3′CS with foreign nucleotide sequences was sufficient to rescue replicon and viral RNA replication, confirming that complementarity rather than the nucleotide sequence per se was essential for RNA synthesis. Using functional assays, the originally recognized 8 nt core CS was extended to 10, 11 and 18 nts for DENV, WNV, and YFV respectively (Alvarez et al., 2005b; Corver et al., 2003).
Additional complementary sequences between the ends of mosquito-borne flavivirus genomes, outside of CS, were noticed using folding prediction algorithms (Hahn et al., 1987; Khromykh et al., 2001; Leyssen et al., 2002; Thurner et al., 2004). A sequence located just upstream of the translation initiator AUG at the 5′UTR was found to be complementary to a region present within the stem of the 3′SL. This pair of complementary sequence was named cyclization sequence 5′-3′UAR, (the name stands for upstream AUGregion) (Alvarez et al., 2005b) (Figure 1). Using a similar approach as the one used to study the requirement of 5′-3′CS, an essential role of 5′-3′ UAR complementarity in viral RNA synthesis was demonstrated in both DENV and WNV (Alvarez et al., 2008; Zhang et al., 2008). Mutations within 5′UAR in the context of infectious DENV RNAs resulted in spontaneous revertions that reconstituted 5′-3′UAR base pairings. Thus, for mosquito-borne flaviviruses two pairs of complementary sequences were found to be necessary for viral replication (Figure 2).
Figure 2.
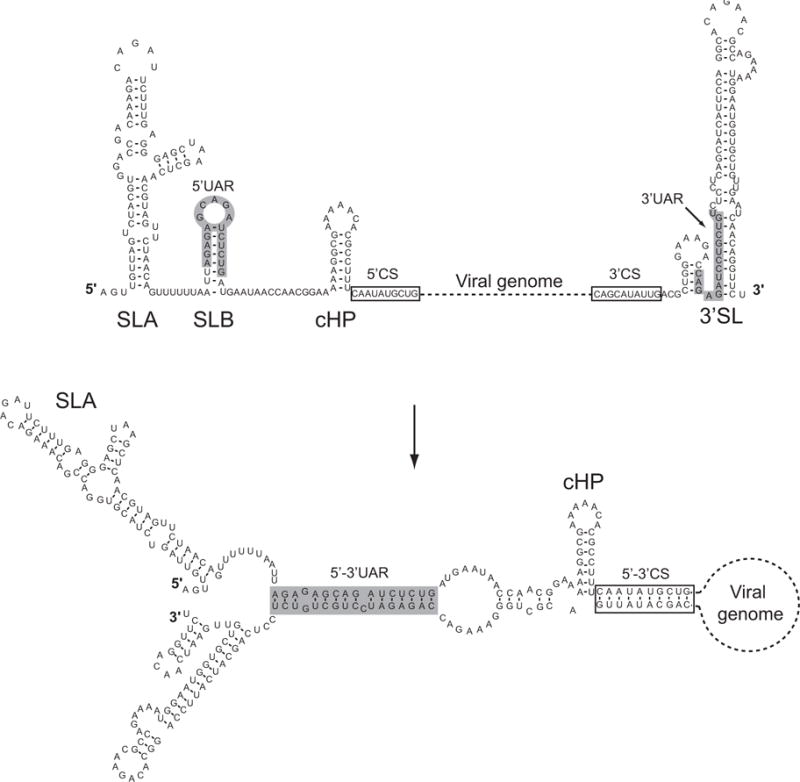
Sequence and predicted secondary structure of 5′ and 3′ terminal sequences of DENV2. On top, t structural elements located at the 5′ end of the genome: stem loop A (SLA), stem loop B (SLB), and capsid region hairpin (cHP) are shown linked by a dash line, representing the viral genome, to the 3′stem loop (3′SL). The bottom structure shows the changes upon hybridization of the complementary sequences, which results in RNA cyclization. Note that the predicted SLA and cHP are maintained while the SLB and the 3′SL changed after 5′-3′UAR and 5′-3′CS interaction.
In the case of tick-borne flaviviruses, two pairs of complementary sequences, 5′-3′ CSA and 5′-3′CSB, were observed by prediction analysis (Khromykh et al., 2001; Mandl et al., 1993). The 5′CSA is located upstream of the initiator AUG and is complementary to the 3′CSA located within the stem of the 3′SL, which is reminiscent of the location of 5′-3′UAR in the mosquito-borne flaviviruses. In addition, the 5′-3′CSB sequences are in similar locations as the 5′-3′CS. Recent studies, using tick-borne flavivirus RNAs, demonstrated that 5′-3′CSA hybridization is essential for viral RNA synthesis, while no crucial function was connected with the CSB elements (Kofler et al., 2006). Sequence complementarity between the ends of the genome of flaviviruses with no-known vector was also reported. For Modoc virus (MODV), two predicted cyclization elements were found. At the 5′ end of the genome, these elements were upstream of the translation initiation codon and in the coding sequence of the capsid protein. At the 3′ end, the complementary sequences were located upstream and within the 3′SL (Leyssen et al., 2002).
3. Long range RNA-RNA contacts mediate flavivirus RNA cyclization
The presence of inverted complementary sequences at the ends of viral RNAs has been observed not only in flaviviruses, but also in the negative stranded RNA bunya-, arena-, and orthomyxoviruses (Flick and Hobom, 1999; Kohl et al., 2004; Mir et al., 2006; Mir and Panganiban, 2005; Perez and de la Torre, 2003); as well as in the double stranded RNA rotaviruses (Patton, 2001). These sequences have been suggested to allow the ends of the genome to associate through base pairing, leading to circular conformations of the RNA. The conformation of the viral RNA, in which the complementary sequences at the ends are hybridized, is known as panhandle-like structures. The significance of genome cyclization during viral replication is now beginning to be uncovered in different systems. For instance, in bunyaviruses, the binding of the nucleocapsid protein to the panhandle structures was shown to regulate viral encapsidation (Mir et al., 2006; Mir and Panganiban, 2005). In the case of influenza virus, the panhandle is part of the promoter for RNA replication (Flick and Hobom, 1999). Thus, in unrelated viruses, RNA cyclization appears to play different roles during the replication cycle.
Direct interaction between two RNA molecules carrying the 5′ and 3′ terminal nucleotide sequences of a flavivirus genome was first observed using psoralen/UV cross-linking assays (You et al., 2001). More recently, an electrophoretic mobility shift assay was employed to characterize these RNA-RNA interactions (Alvarez et al., 2005b). Using this method, the contribution of specific nucleotide sequences on RNA-RNA complex formation was investigated. In these studies, an RNA molecule containing the first 160 nucleotides of DENV2 was incubated with a radiolabeled RNA molecule corresponding to the last 106 nucleotides of the viral RNA. The two RNA molecules were heat denatured, mixed, cooled to room temperature to allow folding, and then used in gel shift assays. Formation of RNA-RNA complexes with slower mobility were observed only in the presence of Mg2+. RNA titration experiments indicated high affinity between the two RNAs with an apparent dissociation constant (Kd) of 8 nM. The requirement of both 5′-3′CS and 5′-3′UAR regions for RNA-RNA complex formation was demonstrated by mutating and reconstituting sequence complementarity within the predicted cyclization elements. Single mismatches within 5′-3′CS or 5′-3′UAR greatly decreased the affinity between the two RNA molecules (Alvarez et al., 2005b).
The first direct evidence of long-range RNA-RNA interactions between the ends of a flavivirus RNA was obtained by visualization of individual molecules using atomic force microscopy (AFM) (Alvarez et al., 2005b). This technique is useful to obtain the topology of single molecules in different environments (Hansma et al., 2004; Henn et al., 2001). Because single-stranded RNA molecules acquire compact tertiary structures that preclude visualization of intramolecular contacts, the RNAs used in this report were hybridized with antisense RNA molecules to generate elongated double stranded segments. This strategy leaves the sequences presumably involved in long range interactions as single stranded overhangs, allowing RNA-RNA contacts mediated by the viral sequences of interest. Using tapping mode AFM in air, a model RNA molecule of 2 kb carrying the 5′ and 3′ terminal sequences of DENV2 were visualized in linear and circular conformations, whereas control molecules with deletions of 3′CS and 3′UAR were only observed in linear forms. According to the concentration of the sample used RNA-RNA head to tail dimers were also observed. A similar strategy was used to demonstrate cyclization of full length DENV genomic RNA. In this case, the 10.7 kb RNA was in vitro transcribed from a cDNA clone. In order to visualize intramolecular interactions, a double stranded RNA segment corresponding to the coding sequence of NS4B and NS5 was used (Figure 3). AFM analysis of these molecules revealed both linear and circular conformations of the RNA, similar to the results obtained with the model molecule.
Figure 3.
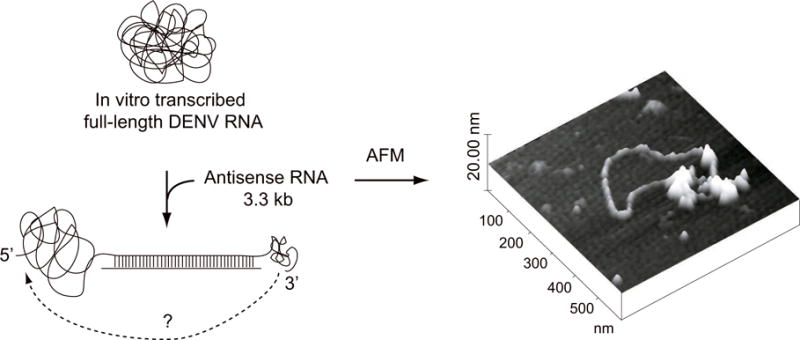
Visualization of the DENV genomic RNA by atomic force microscopy. On the left, a schematic representation of the viral genome in a compact conformation and the strategy used to visualize RNA-RNA contacts. The 10.7 kb RNA molecule was hybridized with an antisense RNA of 3.3 kb resulting in a linear double stranded region with single stranded overhands at the 5′ and 3′ ends. On the right, a representative image of a single molecule of DENV genomic RNA obtained by tapping mode atomic force microscopy in circular conformation is shown.
Although it is possible that binding of cellular or viral proteins to the ends of the RNA could enhance or disrupt flavivirus RNA cyclization, the reported studies were performed in the absence of proteins. Several viral and cellular proteins have been identified to bind the 5′ and/or the 3′ ends of flavivirus genome (Blackwell and Brinton, 1997; De Nova-Ocampo et al., 2002; Garcia-Montalvo et al., 2004; Ta and Vrati, 2000). Thus, it remains to be defined whether these proteins modulate the conformation acquired by flavivirus genomes.
4. Template specificity of flavivirus polymerases
In vitro polymerase activity in cytoplasmic extracts prepared from infected cells has been demonstrated for WNV, KUNV, DENV, and JEV. The RdRp activity was associated with heavy cytoplasmic membrane fractions (Chu and Westaway, 1992; Grun and Brinton, 1988; Takegami and Hotta, 1990; Uchil and Satchidanandam, 2003). The main enzymatic activity observed in the infected extracts with endogenous templates was the elongation of already initiated RNA synthesis. The replication complexes were enriched in NS1, NS2A, NS3, NS4A and contain only about 10% of the NS5 protein (Grun and Brinton, 1987). More recently, data on the composition of the replication complexes in flavivirus-infected cells was obtained by confocal and electron microscopy together with co-immunoprecipitations using specific antibodies to different NS proteins and to dsRNA. The results confirmed that proteins NS1, NS2A, NS3, NS4A, NS5, and for some viruses NS4B co-localize with dsRNA (Mackenzie et al., 1996; Mackenzie et al., 1998; Miller et al., 2006; Westaway et al., 1997). A large fraction of the polymerase was found in the nucleus of DENV and YFV infected cells (Brooks et al., 2002; Buckley et al., 1992; Kapoor et al., 1995; Miller et al., 2006, Uchil et al., 2006)
To evaluate template specificity for a flavivirus RdRp, an in vitro system using DENV infected mosquito cells with exogenous RNAs was developed (You and Padmanabhan, 1999). These studies revealed the requirement of both ends of the viral genome for polymerase activity. A subgenomic RNA of 770 nucleotides, carrying the 5′ and 3′ cyclization sequences, was a template for polymerase activity, while the 3′UTR alone was not active (You et al., 2001; You and Padmanabhan, 1999). Interestingly, addition in trans of RNA molecules bearing the viral 5′ terminal sequences was sufficient to allow RNA synthesis using the 3′UTR molecule as template. This was the first evidence supporting the idea that formation of an RNA-RNA complex between the 5′ and 3′ terminal nucleotides of the viral genome was necessary for polymerase activity. Similar requirements of the cyclization sequences were observed using in vitro assays with recombinant RdRp from DENV and WNV (Ackermann and Padmanabhan, 2001; Nomaguchi et al., 2004).
More recently, using a similar in vitro assay with a recombinant NS5 protein from DENV, the determinants for polymerase recognition were defined (Filomatori et al., 2006). An RNA of 160 nucleotides corresponding to the 5′ end of the genome was sufficient to confer polymerase specificity. The folding prediction of this RNA molecule indicated the presence of a large 5′ stem loop (named SLA), a short stem loop that contained the 5′ cyclization UAR sequence (named SLB, (Alvarez et al., 2008)), a stable stem loop that participates in start codon selection during viral translation (cHP, (Clyde and Harris, 2006)), and the 5′ cyclization CS sequence (Figure 2). Deletion of each of these elements indicated that the SLA was sufficient to promote RNA synthesis even when fused to non-viral sequences (Filomatori et al., 2006).
The requirement of specific structural elements within the SLA for viral RNA synthesis was demonstrated in transfected cells using mutated DENV RNAs (Filomatori et al., 2006) and Lodeiro et al. unpublished). Specific mutation within the SLA, which impaired template activity for the recombinant polymerase in vitro, yielded revertant viruses that restored viral replication. A remarkable correlation between the requirement of DENV SLA sequence/structure for viral replication in transfected cells and the need of this element for in vitro polymerase activity was observed.
The in vivo and in vitro data supported the idea that the SLA, present within the viral 5′UTR, functions as the promoter element for the viral polymerase. Recent studies using full-length chimeric WNV-DENV RNAs showed that the 5′SL structure of these two viruses can be exchangeable (Yu et al., 2008). A WNV carrying the DENV 5′ terminal sequences yielded viable viruses, providing direct evidence for a common role of the 5′SL in different flaviviruses.
5. Conserved structures at the 5′ end of flavivirus genomes
The flavivirus 5′UTRs are between 90 to 130 nucleotides in length. Mosquito borne flaviviruses tend to have shorter 5′UTRs than tick borne flavivirses (Markoff, 2003). In 1988, Brinton and Dispoto made a comparison between the 5′UTRs of WNV and SLEV with those of DENV, YFV, and MVEV (Brinton and Dispoto, 1988). While only short regions were conserved among different viruses, almost complete conservation of nucleotide sequence was observed among different strains of the same virus. Interestingly, the predicted secondary structure of the 5′ end of the genome was similar for all these viruses (Figure 4). The structure consisted of a large stem loop with a side loop. In most of the cases, a second short stem loop could be formed that ended or contained the translation start codon. The predicted structure of 5′ end sequences of tick borne flaviviruses and flaviviruses with no-known vector were later reported to have a similar structure to that observed in the mosquito borne flavivirus genomes (Gritsun et al., 1997; Leyssen et al., 2002; Mandl et al., 1993). A more recent analysis including nucleotide sequences of all flavivirus groups confirmed that the proximal region of the 5′UTR forms a 5′SL, called Y-shaped structure (Gritsun and Gould, 2007).
Figure 4.
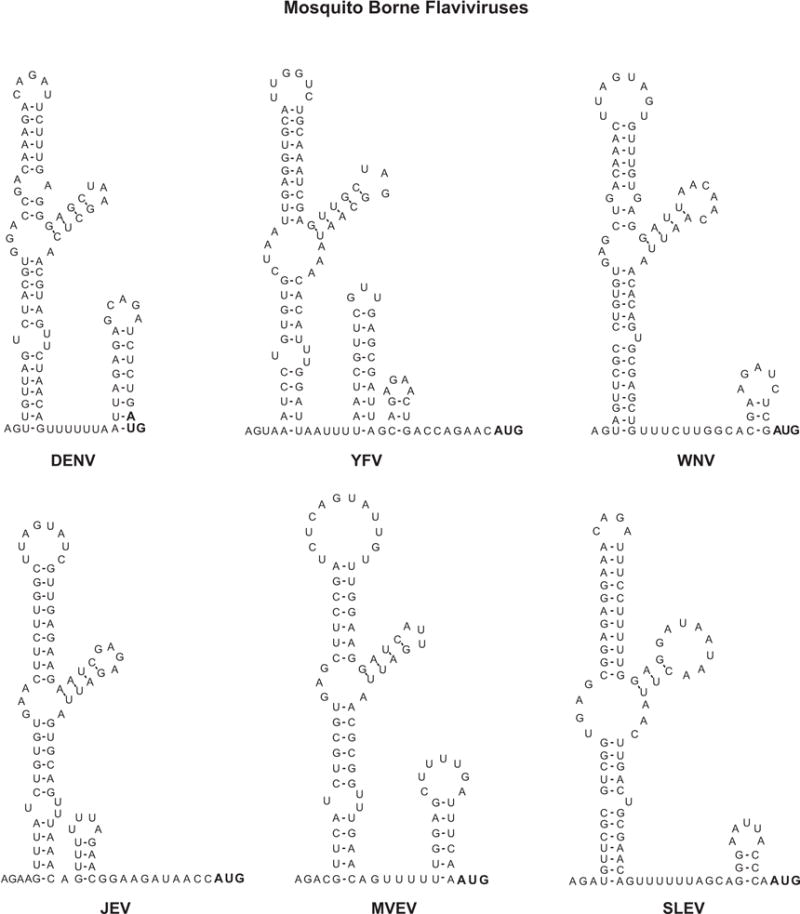
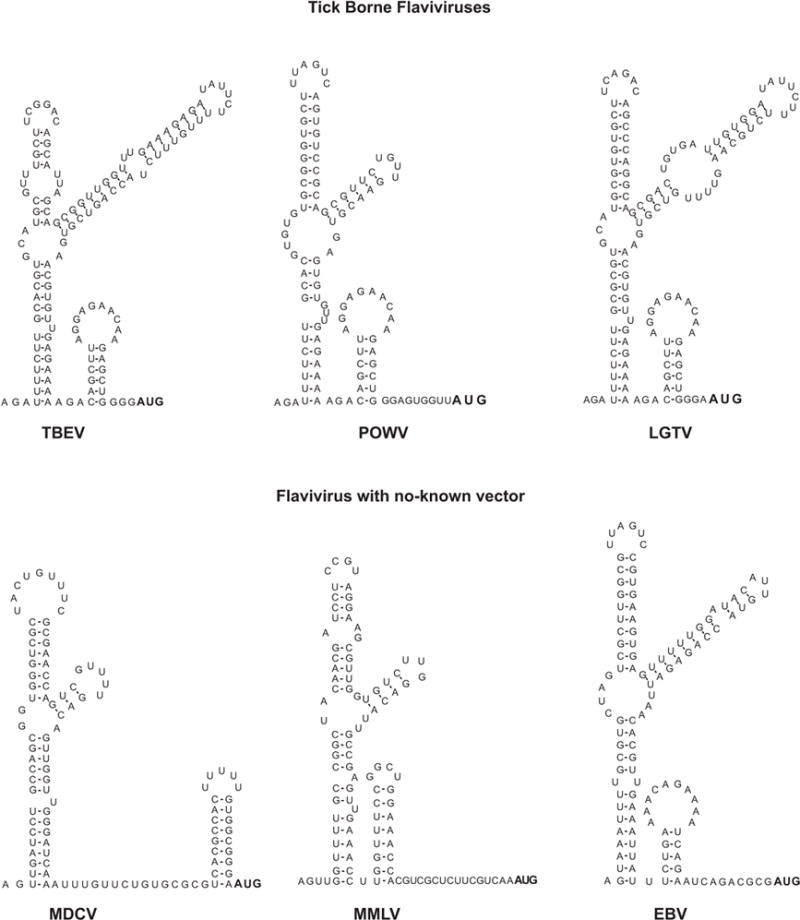
Sequence and predicted secondary structure of 5′UTRs from different members of the flavivirus genus. Mosquito borne flaviviruses: dengue virus 2 (DENV NC001474), yellow fever virus (YFV, NC002031), West Nile virus (WNV, NC001563), Japanese encephalitis virus (JEV, NC001437), Murray Valley encephalitis virus (MVEV, NC000943), and Saint Louis encephalitis virus (SLEV, NC007580). Tick borne flaviviruses: Tick borne encephalitis virus (TBEV, NC001672), Powassan virus (POWV, L06436), and Langat tick-borne virus (LFTV, NC003690). No-known vector flaviviruses: Modoc virus (MDCV, NC003635), Montana Myotis leukoencephalitis virus (MMLV, NC004119), and Entebbe bat virus (EBV, NC008718).
The conservation of the 5′SL in unrelated flavivirus genomes was taken as evidence of its possible role in flavivirus replication. Cahour and co-workers reported the first functional analysis of the role of 5′UTR sequences in flavivirus replication (Cahour et al., 1995). In this study, deletions from 5 to 25 nucleotides were incorporated throughout the 5′UTR by mutagenesis of an infectious DNA copy of DENV4. The dominant effect of the deletions appeared to be at the level of RNA synthesis and many of the mutations were found to be lethal. More recent studies using different flavivirus genomes, indicated that the 5′UTRs contain at least two essential RNA elements with distinct functions during viral RNA synthesis: a) the 5′SL, which was proposed to be the promoter for polymerase recognition and activity; and b) the cyclization sequence located upstream of the translation start AUG codon (Alvarez et al., 2008; Alvarez et al., 2006; Filomatori et al., 2006; Yu et al., 2008; Zhang et al., 2008)
The predicted flavivirus 5′SL contains several common elements (Figure 5). It bears three helical regions (S1, S2, and S3), a top loop, and a side stem loop. Between S1 and S2 there is at least one unpaired U in all flaviviruses (Figure 4). The S1 and S2 regions together are also called S0 and represent one of the most conserved elements within the 5′UTR (Gritsun and Gould, 2007). Co-variations within the double stranded regions S1 and S2 can be found in several positions in different flaviviruses, suggesting a selective pressure to maintain base pairings in those structures. The stem of the side stem loop and the double stranded region of S3 also show significant co-variation. However, the sequence and structure of S3 and the side stem loop show the most variations. Within S3 there are bulges and unpaired nucleotides in different locations, even in different isolates of the same virus. For different tick borne flaviviruses, the length of the side-stem and the size of the side-loop are also different. The side stem-loop structure is formed by a hypervariable stretch of 27 nucleotides (Gritsun et al., 1997). Most of the nucleotide changes observed within this region were compensated by complementary substitutions to support the stem. In the case of Powassan virus (POWV), there is a deletion of the 27 nucleotides (Figure 4). However, this deletion does not affect the predicted folding pattern of the 5′ SL structure (Gritsun et al., 1997).
Figure 5.
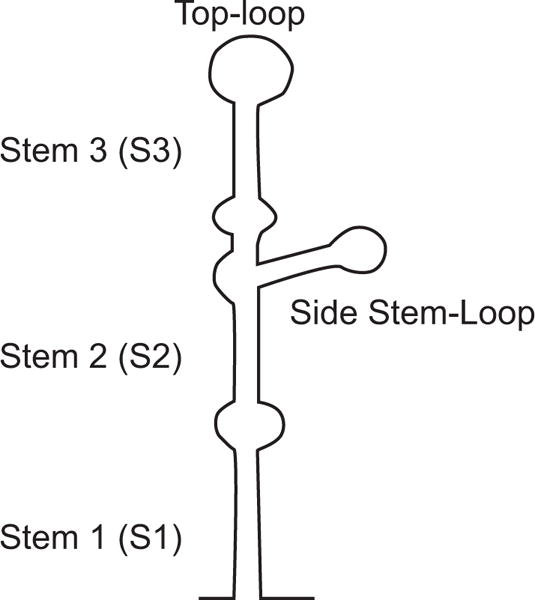
Schematic representation of the 5′ stem loop structure of flavivirus genomes. The regions corresponding to the predicted secondary structures stem 1 (S1), stem 2 (S2), stem 3 (S3), top loop, and side stem loop are indicated.
Functional studies performed with infectious DENV RNAs in transfected cells demonstrated that the Y-shape structure of the 5′SL is necessary for RNA synthesis. Mutations that opened the helical regions S1 or S2 give rise to revertant viruses that spontaneously reconstitute double stranded region (Filomatori et al., 2006); and Lodeiro et al. unpublished). In addition, reconstitution of the helical regions with non viral sequences also re-established viral replication, suggesting that the structure and not the nucleotide sequence per se were necessary for viral replication. In contrast, specific nucleotides at the top loop were found to play a crucial role for both polymerase activity and viral replication in transfected cells (Filomatori et al., 2006).
6. Interaction of NS5 with the viral RNA
NS5 is the largest and the most conserved of the flavivirus proteins. It contains an N-terminal methyl transferase domain (MTase) and a C-terminal RdRp domain. Recently, the structure of the NS5 C-terminal domain of WNV and DENV revealed a classical polymerase fold, bearing palm, thumb, and finger motifs (Malet et al., 2007; Yap et al., 2007). The presence of a priming loop found in these structures is consistent with a primer-independent (de novo) mechanism of initiation of RNA synthesis proposed for flaviviruses. Regarding the MTase domain, using WNV, DENV, and YFV proteins, it was demonstrated both guanine N7 and ribose 2′-O methylation (Dong et al., 2007; Egloff et al., 2002; Ray et al., 2006; Zhou et al., 2007). These activities are involved in formation of the 5′ cap structure required for translation initiation. The crystal structure of WNV and DENV MTase domain showed a single binding site for the methyl donor SAM (Egloff et al., 2002; Zhou et al., 2007). In addition, a positively charged surface adjacent to the SAM binding site was proposed to be the recognition site for the capped-RNA substrate.
The viral protein NS5 has the ability to bind RNA with high affinity. Using mobility shift and filter binding assays, specific interaction of the DENV RdRp with RNA molecules corresponding to the 5′ terminal region of the viral genome was reported (Filomatori et al., 2006). The protein forms complexes with the first 160 nucleotides of the viral genome, with an apparent dissociation constant of 10 nM. The presence of the SLA structure was found to be essential for protein binding to the viral 5′ terminal region, while the presence of SLB, cHP, and 5′CS are dispensable (Figure 2). In addition, using similar gel shift assays, no binding of NS5 to the 3′ end of the DENV RNA was detected. Interaction of the WNV NS5 to the viral RNA was recently reported. Using footprinting experiments, NS5 binding to the 5′ 190 nucleotides of WNV genome was demonstrated, and protection in different regions of the 5′SL was observed (Dong et al., 2007).
Atomic force microscopy was also employed to visualize binding of NS5 to individual RNA molecules carrying the ends of DENV genome in circular conformation (Filomatori et al., 2006). In these studies, model RNA molecules of 2 kb containing the 5′ and 3′ ends of the viral genome were incubated with highly purified recombinant RdRp. The apparent volume of the 5′-3′ interacting regions of circular RNA molecules, in absence and presence of the viral protein, were determined by analysis of a large number of molecules. An increase from 150 to 300 nm3 was reported in the presence of protein, which is consistent with binding of the viral protein to the RNA.
Hybridization of the 5′ and 3′ end sequences of the viral genome mediated by the cyclization elements results in changes around the predicted 3′SL, however, the 5′SL remains with a similar structure as the one predicted for the 5′ RNA molecule alone (Figure 2). Based on these predictions, binding of the viral polymerase to circular RNA molecules, observed in AFM, could be a result of protein binding to the 5′SL. However, it is possible that this binding is modulated by RNA structures formed after 5′-3′ interaction.
7. Model for minus strand RNA synthesis
Important progress has been made in recent years in dissecting the role of cis-acting elements involved in flavivirus RNA replication. Based on information provided by different groups, a model for DENV minus strand RNA synthesis was recently proposed (Figure 6, Filomatori et al., 2006). In this model, the viral polymerase NS5 binds the promoter SLA at the 5′ end of the RNA, ~11 kb away from the 3′ initiation site. Cyclization of the viral genome through long-range RNA-RNA interactions brings the 3′ end of the RNA near the polymerase-SLA complex. This conformation of the RNA allows the polymerase to reach the 3′ end of the genome, which is used as template for initiation of minus strand RNA synthesis.
Figure 6.
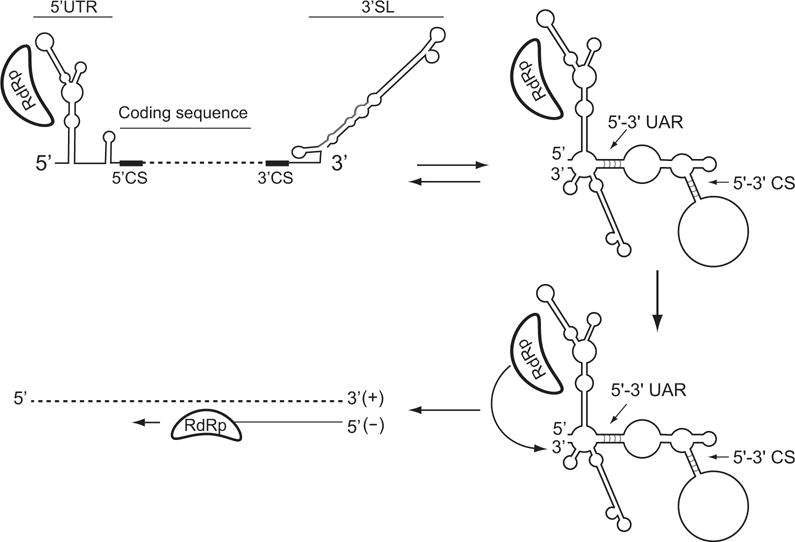
Model proposed for DENV RNA synthesis. The viral RNA-dependent RNA polymerase (RdRp) binds to the promoter SLA at the 5′ end of the genome. Long-range RNA-RNA interactions mediate genome cyclyzation that facilitate polymerase initiation at the 3′ end of the RNA.
A common feature of the resulting structures predicted after RNA-RNA hybridization between the ends of flavivirus genomes is the alteration of the conserved 3′SL element (Figure 2). This element has been shown to be absolutely essential for flavivirus replication (for review see (Markoff, 2003)). Upon base pairings between the 5′ and the 3′ cyclization elements, the bottom half of the 3′SL element is predicted to open. Based on the conserved location of complementary sequences at the bottom of the 3′SL, it has been proposed that cyclization of the viral RNA could have a dual function in the mechanism of RNA replication: i) bring the polymerase-promoter complex near the 3′ initiation site, and ii) provide the correct conformation of the 3′ end of the genome during the initiation process. Structural changes around the 3′ terminal nucleotides were previously reported to be essential for other plus strand RNA viruses. In these cases, melting 3′ end RNA structures was a prerequisite for polymerase initiation of RNA synthesis (Chen and Patton, 1998; Olsthoorn et al., 1999; Pogany et al., 2003; Zhang et al., 2006; Zhang et al., 2004).
According to the crystal structure of flavivirus RdRp, the template channel, which should accommodate the 3′ end of the RNA, has dimensions that would only permit access to a ssRNA chain (Yap et al., 2007). Therefore, it is likely that the 3′SL unwinds before entering the template channel of the enzyme. This process could be aided by the helicase activity of the viral protein NS3 or by cellular trans-acting factors interacting with the viral 3′SL. In addition, base pairings between 5′ and 3′UAR of mosquito borne flaviviruses or between 5′and 3′ CSA of tick borne encephalitis virus are predicted to open the bottom half stem of the 3′SL, releasing the 3′ end nucleotides.
The model proposed for DENV involves three essential cis-acting elements: the 5′SL promoter, the cyclization regions, and the 3′SL structure. The presence of a conserved structure at the 5′ end of flavivirus genomes and the crucial role of genome cyclization reported for mosquito and tick borne flavivirus replication, suggests a common strategy of RNA synthesis in different members of this genus. In this regard, using chimeric viruses, a functional compatibility between the 5′SL and the viral polymerase of WNV and DENV (Yu et al., 2008) was demonstrated, providing functional evidence of conserved cis acting elements in these two viruses. Further studies are needed in order to define whether the 5′SL has a conserved role in flavivirus RNA synthesis.
8. Perspectives
Despite the recent advances in our understanding the role of flavivirus genome cyclization, many questions remain. How does the 5′SL promote RNA synthesis on the authentic 3′ end of the genome? How does the 3′SL function during the initiation process? Does binding of cellular or viral proteins to the 5′ or 3′ ends of the RNA regulate the conformation of the viral genome? Another important question is whether RNA cyclization is necessary for both minus and plus strand RNA synthesis. Tools to discriminate the requirements for these two processes are still lacking, which represent an important challenge to establish the similarities and differences between these two steps of RNA replication. Although it was previously shown that alteration of the long range RNA-RNA interaction did not affect translation of the input viral RNA, it is possible that RNA cyclization plays a role in modulating later rounds of translation. Further studies are necessary to explore whether RNA cyclization also participates in flavivirus translation and/or encapsidation.
Finally, it is important to highlight that panhandle structures have been reported for distantly related RNA viruses. It is still puzzling why different viruses required RNA cyclization for distinct processes during viral infection. Perhaps, communicating the 5′ and the 3′ ends of the RNA provides a general advantage for viral replication. For instance, it could be a control mechanism to amplify only full-length templates or a simple strategy to overlap translation, replication, and encapsidation signals to coordinate these processes, similar to that reported for picornaviruses (Gamarnik and Andino, 1998). In this regard RNA-RNA interactions may serve as molecular switches hiding or exposing signals involved in specific processes. Dissecting the specific conformations of the RNA required in each step of the viral life cycle will be necessary to fully understand these regulatory mechanisms.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackermann M, Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J Biol Chem. 2001;276(43):39926–37. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- Allison SL, Tao YJ, O’Riordain G, Mandl CW, Harrison SC, Heinz FX. Two distinct size classes of immature and mature subviral particles from tick-borne encephalitis virus. J Virol. 2003;77(21):11357–66. doi: 10.1128/JVI.77.21.11357-11366.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez DE, De Lella Ezcurra AL, Fucito S, Gamarnik AV. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005a;339(2):200–12. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Alvarez DE, Filomatori CV, Gamarnik AV. Functional analysis of dengue virus cyclization sequences located at the 5′ and 3′UTRs. Virology. 2008;375(1):223–35. doi: 10.1016/j.virol.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Alvarez DE, Lodeiro MF, Filomatori CV, Fucito S, Mondotte JA, Gamarnik AV. Structural and functional analysis of dengue virus RNA. Novartis Found Symp. 2006;277:120–32. discussion 132–5, 251–3. [PubMed] [Google Scholar]
- Alvarez DE, Lodeiro MF, Luduena SJ, Pietrasanta LI, Gamarnik AV. Long-range RNA-RNA interactions circularize the dengue virus genome. J Virol. 2005b;79(11):6631–43. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell JL, Brinton MA. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J Virol. 1997;71(9):6433–44. doi: 10.1128/jvi.71.9.6433-6444.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinton MA, Dispoto JH. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology. 1988;162(2):290–9. doi: 10.1016/0042-6822(88)90468-0. [DOI] [PubMed] [Google Scholar]
- Brooks AJ, Johansson M, John AV, Xu Y, Jans DA, Vasudevan SG. The interdomain region of dengue NS5 protein that binds to the viral helicase NS3 contains independently functional importin beta 1 and importin alpha/beta-recognized nuclear localization signals. J Biol Chem. 2002;277(39):36399–407. doi: 10.1074/jbc.M204977200. [DOI] [PubMed] [Google Scholar]
- Buckley A, Gaidamovich S, Turchinskaya A, Gould EA. Monoclonal antibodies identify the NS5 yellow fever virus non-structural protein in the nuclei of infected cells. J Gen Virol. 1992;73(Pt 5):1125–30. doi: 10.1099/0022-1317-73-5-1125. [DOI] [PubMed] [Google Scholar]
- Cahour A, Pletnev A, Vazielle-Falcoz M, Rosen L, Lai CJ. Growth-restricted dengue virus mutants containing deletions in the 5′ noncoding region of the RNA genome. Virology. 1995;207(1):68–76. doi: 10.1006/viro.1995.1052. [DOI] [PubMed] [Google Scholar]
- Chen D, Patton JT. Rotavirus RNA replication requires a single-stranded 3′ end for efficient minus-strand synthesis. J Virol. 1998;72(9):7387–96. doi: 10.1128/jvi.72.9.7387-7396.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu PW, Westaway EG. Molecular and ultrastructural analysis of heavy membrane fractions associated with the replication of Kunjin virus RNA. Arch Virol. 1992;125(1–4):177–91. doi: 10.1007/BF01309636. [DOI] [PubMed] [Google Scholar]
- Cleaves GR, Dubin DT. Methylation status of intracellular dengue type 2 40 S RNA. Virology. 1979;96(1):159–65. doi: 10.1016/0042-6822(79)90181-8. [DOI] [PubMed] [Google Scholar]
- Clyde K, Harris E. RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. J Virol. 2006;80(5):2170–82. doi: 10.1128/JVI.80.5.2170-2182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corver J, Lenches E, Smith K, Robison RA, Sando T, Strauss EG, Strauss JH. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J Virol. 2003;77(3):2265–70. doi: 10.1128/JVI.77.3.2265-2270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nova-Ocampo M, Villegas-Sepulveda N, del Angel RM. Translation elongation factor-lalpha, La, and PTB interact with the 3′ untranslated region of dengue 4 virus RNA. Virology. 2002;295(2):337–47. doi: 10.1006/viro.2002.1407. [DOI] [PubMed] [Google Scholar]
- Dong H, Ray D, Ren S, Zhang B, Puig-Basagoiti F, Takagi Y, Ho CK, Li H, Shi PY. Distinct RNA elements confer specificity to flavivirus RNA cap methylation events. J Virol. 2007;81(9):4412–21. doi: 10.1128/JVI.02455-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. Embo J. 2002;21(11):2757–68. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006;20(16):2238–49. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick R, Hobom G. Interaction of influenza virus polymerase with viral RNA in the ’corkscrew’ conformation. J Gen Virol. 1999;80(Pt 10):2565–72. doi: 10.1099/0022-1317-80-10-2565. [DOI] [PubMed] [Google Scholar]
- Gamarnik AV, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12(15):2293–304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Montalvo BM, Medina F, del Angel RM. La protein binds to NS5 and NS3 and to the 5′ and 3′ ends of Dengue 4 virus RNA. Virus Res. 2004;102(2):141–50. doi: 10.1016/j.virusres.2004.01.024. [DOI] [PubMed] [Google Scholar]
- Gritsun TS, Gould EA. Origin and evolution of flavivirus 5′UTRs and panhandles: trans-terminal duplications? Virology. 2007;366(1):8–15. doi: 10.1016/j.virol.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Gritsun TS, Venugopal K, Zanotto PM, Mikhailov MV, Sall AA, Holmes EC, Polkinghorne I, Frolova TV, Pogodina VV, Lashkevich VA, Gould EA. Complete sequence of two tick-borne flaviviruses isolated from Siberia and the UK: analysis and significance of the 5′ and 3′-UTRs. Virus Res. 1997;49(1):27–39. doi: 10.1016/s0168-1702(97)01451-2. [DOI] [PubMed] [Google Scholar]
- Grun JB, Brinton MA. Dissociation of NS5 from cell fractions containing West Nile virus-specific polymerase activity. J Virol. 1987;61(11):3641–4. doi: 10.1128/jvi.61.11.3641-3644.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun JB, Brinton MA. Separation of functional West Nile virus replication complexes from intracellular membrane fragments. J Gen Virol. 1988;69(Pt 12):3121–7. doi: 10.1099/0022-1317-69-12-3121. [DOI] [PubMed] [Google Scholar]
- Hahn CS, Hahn YS, Rice CM, Lee E, Dalgarno L, Strauss EG, Strauss JH. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J Mol Biol. 1987;198(1):33–41. doi: 10.1016/0022-2836(87)90455-4. [DOI] [PubMed] [Google Scholar]
- Hansma HG, Kasuya K, Oroudjev E. Atomic force microscopy imaging and pulling of nucleic acids. Curr Opin Struct Biol. 2004;14(3):380–5. doi: 10.1016/j.sbi.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Henn A, Medalia O, Shi SP, Steinberg M, Franceschi F, Sagi I. Visualization of unwinding activity of duplex RNA by DbpA, a DEAD box helicase, at single-molecule resolution by atomic force microscopy. Proc Natl Acad Sci U S A. 2001;98(9):5007–12. doi: 10.1073/pnas.071372498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner KE, Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995;270(32):19100–6. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- Khromykh AA, Meka H, Guyatt KJ, Westaway EG. Essential role of cyclization sequences in flavivirus RNA replication. J Virol. 2001;75(14):6719–28. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler RM, Hoenninger VM, Thurner C, Mandl CW. Functional analysis of the tick-borne encephalitis virus cyclization elements indicates major differences between mosquito-borne and tick-borne flaviviruses. J Virol. 2006;80(8):4099–113. doi: 10.1128/JVI.80.8.4099-4113.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl A, Dunn EF, Lowen AC, Elliott RM. Complementarity, sequence and structural elements within the 3′ and 5′ non-coding regions of the Bunyamwera orthobunyavirus S segment determine promoter strength. J Gen Virol. 2004;85(Pt 11):3269–78. doi: 10.1099/vir.0.80407-0. [DOI] [PubMed] [Google Scholar]
- Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB. Phylogeny of the genus Flavivirus. J Virol. 1998;72(1):73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyssen P, Charlier N, Lemey P, Billoir F, Vandamme AM, De Clercq E, de Lamballerie X, Neyts J. Complete genome sequence, taxonomic assignment, and comparative analysis of the untranslated regions of the Modoc virus, a flavivirus with no known vector. Virology. 2002;293(1):125–40. doi: 10.1006/viro.2001.1241. [DOI] [PubMed] [Google Scholar]
- Lo MK, Tilgner M, Bernard KA, Shi PY. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J Virol. 2003;77(18):10004–14. doi: 10.1128/JVI.77.18.10004-10014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JM, Jones MK, Young PR. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology. 1996;220(1):232–40. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- Mackenzie JM, Khromykh AA, Jones MK, Westaway EG. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology. 1998;245(2):203–15. doi: 10.1006/viro.1998.9156. [DOI] [PubMed] [Google Scholar]
- Malet H, Egloff MP, Selisko B, Butcher RE, Wright PJ, Roberts M, Gruez A, Sulzenbacher G, Vonrhein C, Bricogne G, Mackenzie JM, Khromykh AA, Davidson AD, Canard B. Crystal Structure of the RNA Polymerase Domain of the West Nile Virus Non-structural Protein 5. J Biol Chem. 2007;282(14):10678–89. doi: 10.1074/jbc.M607273200. [DOI] [PubMed] [Google Scholar]
- Mandl CW, Holzmann H, Kunz C, Heinz FX. Complete genomic sequence of Powassan virus: evaluation of genetic elements in tick-borne versus mosquito-borne flaviviruses. Virology. 1993;194(1):173–84. doi: 10.1006/viro.1993.1247. [DOI] [PubMed] [Google Scholar]
- Markoff L. 5′ and 3′ NCRs in Flavivirus RNA. In: Chambers TJ, Monath TP, editors. Flaviviruses, Adv Virus Res. Vol. 60. Elsevier Academic Press; 2003. pp. 177–223. [Google Scholar]
- Miller S, Sparacio S, Bartenschlager R. Subcellular localization and membrane topology of the Dengue virus type 2 Non-structural protein 4B. J Biol Chem. 2006;281(13):8854–63. doi: 10.1074/jbc.M512697200. [DOI] [PubMed] [Google Scholar]
- Mir MA, Brown B, Hjelle B, Duran WA, Panganiban AT. Hantavirus N protein exhibits genus-specific recognition of the viral RNA panhandle. J Virol. 2006;80(22):11283–92. doi: 10.1128/JVI.00820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir MA, Panganiban AT. The hantavirus nucleocapsid protein recognizes specific features of the viral RNA panhandle and is altered in conformation upon RNA binding. J Virol. 2005;79(3):1824–35. doi: 10.1128/JVI.79.3.1824-1835.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomaguchi M, Teramoto T, Yu L, Markoff L, Padmanabhan R. Requirements for West Nile virus (−)- and (+)-strand subgenomic RNA synthesis in vitro by the viral RNA-dependent RNA polymerase expressed in Escherichia coli. J Biol Chem. 2004;279(13):12141–51. doi: 10.1074/jbc.M310839200. [DOI] [PubMed] [Google Scholar]
- Olsthoorn RC, Mertens S, Brederode FT, Bol JF. A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. Embo J. 1999;18(17):4856–64. doi: 10.1093/emboj/18.17.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton JT. Rotavirus RNA replication and gene expression. Novartis Found Symp. 2001;238:64–77. doi: 10.1002/0470846534.ch5. discussion 77–81. [DOI] [PubMed] [Google Scholar]
- Perez M, de la Torre JC. Characterization of the genomic promoter of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol. 2003;77(2):1184–94. doi: 10.1128/JVI.77.2.1184-1194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J, Fabian MR, White KA, Nagy PD. A replication silencer element in a plus-strand RNA virus. Embo J. 2003;22(20):5602–11. doi: 10.1093/emboj/cdg523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J Virol. 2006;80(17):8362–70. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice CM, Lenches EM, Eddy SR, Shin SJ, Sheets RL, Strauss JH. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985;229(4715):726–33. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- Stadler K, Allison SL, Schalich J, Heinz FX. Proteolytic activation of tick-borne encephalitis virus by furin. J Virol. 1997;71(11):8475–81. doi: 10.1128/jvi.71.11.8475-8481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta M, Vrati S. Mov34 protein from mouse brain interacts with the 3′ noncoding region of Japanese encephalitis virus. J Virol. 2000;74(11):5108–15. doi: 10.1128/jvi.74.11.5108-5115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takegami T, Hotta S. Synthesis and localization of Japanese encephalitis virus RNAs in the infected cells. Microbiol Immunol. 1990;34(10):849–57. doi: 10.1111/j.1348-0421.1990.tb01063.x. [DOI] [PubMed] [Google Scholar]
- Thurner C, Witwer C, Hofacker IL, Stadler PF. Conserved RNA secondary structures in Flaviviridae genomes. J Gen Virol. 2004;85(Pt 5):1113–24. doi: 10.1099/vir.0.19462-0. [DOI] [PubMed] [Google Scholar]
- Uchil PD, Kumar AV, Satchidanandam V. Nuclear localization of flavivirus RNA synthesis in infected cells. J Virol. 2006;80(11):5451–64. doi: 10.1128/JVI.01982-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchil PD, Satchidanandam V. Architecture of the flaviviral replication complex. Protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J Biol Chem. 2003;278(27):24388–98. doi: 10.1074/jbc.M301717200. [DOI] [PubMed] [Google Scholar]
- Wengler G, Gross HJ. Studies on virus-specific nucleic acids synthesized in vertebrate and mosquito cells infected with flaviviruses. Virology. 1978;89(2):423–37. doi: 10.1016/0042-6822(78)90185-x. [DOI] [PubMed] [Google Scholar]
- Westaway EG, Mackenzie JM, Kenney MT, Jones MK, Khromykh AA. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J Virol. 1997;71(9):6650–61. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westaway EG, Mackenzie JM, Khromykh AA. Kunjin RNA replication and applications of Kunjin replicons. Adv Virus Res. 2003;59:99–140. doi: 10.1016/s0065-3527(03)59004-2. [DOI] [PubMed] [Google Scholar]
- Yap TL, Xu T, Chen YL, Malet H, Egloff MP, Canard B, Vasudevan SG, Lescar J. Crystal Structure of the Dengue Virus RNA-Dependent RNA Polymerase Catalytic Domain at 1.85-Angstrom Resolution. J Virol. 2007;81(9):4753–65. doi: 10.1128/JVI.02283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You S, Falgout B, Markoff L, Padmanabhan R. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J Biol Chem. 2001;276(19):15581–91. doi: 10.1074/jbc.M010923200. [DOI] [PubMed] [Google Scholar]
- You S, Padmanabhan R. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J Biol Chem. 1999;274(47):33714–22. doi: 10.1074/jbc.274.47.33714. [DOI] [PubMed] [Google Scholar]
- Yu L, Nomaguchi M, Padmanabhan R, Markoff L. Specific requirements for elements of the 5′ and 3′ terminal regions in flavivirus RNA synthesis and viral replication. Virology. 2008 doi: 10.1016/j.virol.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Dong H, Stein DA, Iversen PL, Shi PY. West Nile virus genome cyclization and RNA replication require two pairs of long-distance RNA interactions. Virology. 2008;373(1):1–13. doi: 10.1016/j.virol.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Zhang G, Zhang J, George AT, Baumstark T, Simon AE. Conformational changes involved in initiation of minus-strand synthesis of a virus-associated RNA. RNA. 2006;12(1):147–62. doi: 10.1261/rna.2166706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Zhang J, Simon AE. Repression and derepression of minusstrand synthesis in a plus-strand RNA virus replicon. J Virol. 2004;78(14):7619–33. doi: 10.1128/JVI.78.14.7619-7633.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. Structure and function of flavivirus NS5 methyltransferase. J Virol. 2007;81(8):3891–903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


