Abstract
Diploid Saccharomyces cells experiencing a double-strand break (DSB) on one homologous chromosome repair the break by RAD51-mediated gene conversion >98% of the time. However, when extensive homologous sequences are restricted to one side of the DSB, repair can occur by both RAD51-dependent and RAD51-independent break-induced replication (BIR) mechanisms. Here we characterize the kinetics and checkpoint dependence of RAD51-dependent BIR when the DSB is created within a chromosome. Gene conversion products appear within 2 h, and there is little, if any, induction of the DNA damage checkpoint; however, RAD51-dependent BIR occurs with a further delay of 2 to 4 h and cells arrest in response to the G2/M DNA damage checkpoint. RAD51-dependent BIR does not require special facilitating sequences that are required for a less efficient RAD51-independent process. RAD51-dependent BIR occurs efficiently in G2-arrested cells. Once repair is initiated, the rate of repair replication during BIR is comparable to that of normal DNA replication, as copying of >100 kb is completed less than 30 min after repair DNA synthesis is detected close to the DSB.
Eukaryotic cells have evolved several mechanisms to repair broken chromosomes (reviewed in references 28 and 34). In Saccharomyces cerevisiae, gene conversion (GC) is the dominant pathway for double-strand break (DSB) repair. GC requires both ends of a DSB to share homology with sequences located on a sister chromatid, on a homologous chromosome, or in some ectopic location. This homology is used as a template for DSB repair, resulting in a limited patch of new DNA synthesis. In mitotic cells of budding yeast, GC generally occurs without an exchange of flanking chromosome arms, although conversions are accompanied by crossing over 5 to 10% of the time (10, 12). GC depends on the Rad51 strand exchange protein, as well as the recombination proteins Rad52, Rad54, Rad55, and Rad57 (11, 34).
In diploids, a DSB is repaired by GC >98% of the time, making other DSB repair mechanisms difficult to study. When GC is eliminated, by deleting the RAD51 gene, a DSB in the middle of a chromosome can be repaired by RAD52-dependent break-induced replication (BIR), in which sequences centromere-proximal to the DSB invade the unbroken homologous chromosome, establish a replication fork, and copy the template chromosome to the chromosome end (22). At least 100 kb can be copied, resulting in loss of heterozygosity of all markers distal to the point of strand invasion. The RAD51-independent BIR process requires Rad59, the Rad54 homolog Tid1, and the Mre11-Rad50-Xrs2 (MRX) complex (11, 32). A striking feature of RAD51-independent BIR initiated at the MAT locus on chromosome III is that the site where repair apparently initiates is far from the DSB site, predominantly making use of a facilitator of BIR (FBI) sequence located more than 30 kb closer to the centromere (23). When RAD51-independent BIR occurs on a plasmid or when a linear DNA fragment lacking a telomere is transformed into budding yeast, there may not be need for such facilitating sequences (5, 11).
There is also a RAD51-dependent BIR process, which has been studied when only the centromere-proximal end of a DNA molecule is able to invade homologous sequences on a template chromosome (1, 5, 6, 11, 15, 22, 25, 27). In early studies, whether the BIR events were RAD51 dependent was not established. Direct evidence for the existence of a RAD52-dependent, RAD51-dependent BIR mechanism first came from studying how telomeres are maintained in the absence of telomerase (17). Erosion of telomeres presents the cells with unprotected chromosome ends, each of which shares homology to other sequences in the genome only on the centromere-proximal side. There are two RAD52-dependent BIR processes, leading to different outcomes (17, 21). A RAD51-independent BIR process requires Rad59 and the MRX complex, resulting in the elongation of the TG1-3 telomere sequences themselves (35, 36). This process seems analogous to the RAD51-independent repair of a single DSB in the middle of a chromosome. But there is also a RAD51-dependent BIR process to maintain telomeres which leads to the proliferation of long, subtelomeric Y′ and other subtelomeric sequences at nearly all chromosome ends (21). The different outcomes in the RAD51-dependent and RAD51-independent BIR processes at telomeres seem to reflect different requirements for homology between the DSB end and its template, as seen in analogous events on plasmids in which a DSB is introduced (11). Recently, Davis and Symington (5) provided direct evidence for RAD51-dependent BIR following the transformation of a linearized DNA fragment that acquired a telomere by BIR.
Although genetic studies have clearly demonstrated that BIR occurs, there has been no molecular characterization of these events or direct comparison with the process of GC. In this paper, we present a detailed characterization of RAD51-dependent BIR resulting from a single DSB created in a chromosome. To study BIR we eliminated most GC by using diploids in which only the centromere-adjacent end of a DSB shares substantial homology with the intact, template chromosome. We show that RAD51-dependent BIR is more efficient than RAD51-independent BIR and does not require an FBI sequence to initiate repair. We confirm that RAD51-dependent BIR is strongly out-competed by GC and show that this is largely due to the inherently slow kinetics of initiating BIR, which also triggers the DNA damage checkpoint. Although steps leading to the initiation of BIR are slow, the subsequent rate of DNA synthesis along the 100-kb template is comparable to that of normal DNA replication, even when the entire repair process is carried out in G2-arrested cells.
MATERIALS AND METHODS
Diploid strain MLN134 (Fig. 1A) has the genotype MATa/MATα-inc ade1/ade1 met13/MET13 ura3/ura3 leu2/leu2 THR4/thr4 LYS5/lys5 trp1/TRP1 pho87::URA3/pho87::URA3 hmlΔ::ADE1/HML hmrΔ::ADE1/HMR ade3::GAL::HO/ADE3. This strain is isogenic to strains published by Malkova et al. (22), resulting from a cross between EI515 transformed to pho87::URA3 and AM133. Plasmid pMN1 was created by inserting a PCR fragment containing the MAT-Ya sequence and 46 bp of MAT-Z1 sequence into MluI- and SalI-digested plasmid pJH1202. The resultant plasmid contained the Ya sequence and HO recognition site adjacent to the LEU2 gene and Tetrahymena telomere repeats. Diploid strain MLN141 was constructed by transformation of MLN134 by a BamHI-MluI fragment of pMN1. Transformants were selected as Leu+ colonies that also became thr4 and proved to contain a terminal truncation of the MATa-containing chromosome III. Strain MY006 was constructed by transforming strain MLN141 with a BsaI restriction fragment from plasmid pEC4, resulting in the replacement of HMR on the MATα-inc-containing chromosome III by the NAT (nourseothricin [Clonat]-resistance) gene. Strain AM792 is isogenic to MY006, but MATα-inc is replaced by a MATa-inc sequence by insertion and subsequent excision of the URA3-containing plasmid pJH32. Additional isogenic strains include MY012 (rad50Δ::HPH/rad50Δ::KAN), MY014 (rad51Δ::HPH/rad51Δ::KAN), and AM803 (rad9Δ::HPH/rad9Δ::KAN). Centromeric plasmid pJH1086, containing MATa, the LEU2 marker, and a yeast origin of replication (ARS1), was used to transform MATα-inc/MATα-inc BIR survivors to permit sporulation and tetrad analysis.
FIG. 1.
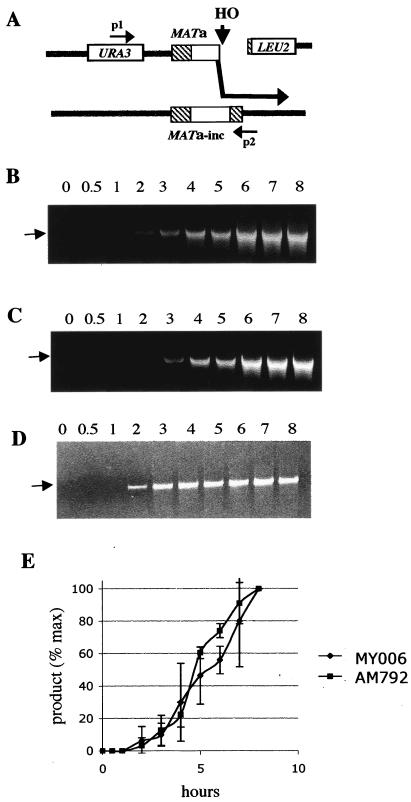
Experimental system to study BIR and GC. (A) Chromosome III in diploid strain MLN134, used to study kinetics of GC. Positions of restriction endonuclease recognition sites are indicated as follows: S, StuI; Bg, BglII; K, KpnI; B, BamHI. p1, p2, and p3 indicate positions of primers that were used for PCR analyses of intermediates of GC and BIR. (B) Chromosome III in diploid strain MY006, used to study BIR. The MATa-containing copy of chromosome III is truncated by insertion of a LEU2 gene fused to telomere sequences. All abbreviations are similar to those in panel A. (C) Illustration of GC repair of MY006. (D) BIR repair of MY006. (E) Chromosome loss when the HO cut in MY006 is not repaired. In all cases, repair occurring in just one chromatid is illustrated for simplicity. In a case of DSB repair in G2, the colony phenotype depends on independent repair of two chromatids.
Media and growth conditions.
Rich medium (yeast extract-peptone-dextrose [YEPD]), synthetic complete medium with bases and amino acids omitted as specified, and sporulation media were as described previously (14). YEP-lactate and YEP-galactose (YEP-Gal) consisted of 1% yeast extract-2% Bacto Peptone medium supplemented with 3.7% lactic acid (pH 5.5) and 2% (wt/vol) galactose, respectively. Cultures were incubated at 30°C.
Analysis of DNA repair.
Logarithmically growing cells grown in YEP-glycerol were plated on YEP-Gal and grown into colonies. The colonies were then replica plated onto nutritional dropout medium to determine the fate of the ADE1, THR4, LEU2, and URA3 markers. Cell viability following the HO induction was derived by dividing the number of CFU on YEP-Gal by that on YEPD.
HO induction and measurement of the kinetics of DSB repair.
Time course experiments were conducted as described previously (29). YEP-lactate (500 to 1,000 ml) was inoculated with 1 × 106 to 3 × 106 cells/ml. Cultures were grown at 30°C overnight to reach 5 × 106 cells/ml. HO endonuclease was induced by the addition of galactose to achieve a final concentration of 2%. For DNA extraction, 50-ml aliquots were removed, and sodium azide was added to achieve a concentration of 0.1% to stop DNA repair processes. For PCR and Southern blot analyses, DNA was immediately extracted by a glass bead-phenol-sodium dodecyl sulfate protocol (9). For CHEF gel electrophoresis, aliquots were processed immediately using CHEF genomic DNA plug kits (Bio-Rad). For fluorescence-activated cell sorter (FACS) analyses, 4-ml aliquots were removed, fixed by addition of 11 ml of 96% ethanol, and stored at 4°C.
To synchronize cells at the G2 stage before HO induction, nocodazole (USB) (final concentration, 0.015 mg/ml) was added to cultures (5 × 106 cells/ml) grown in YEP-lactate, and cultures were incubated with nocodazole for 3 h at 30°C, until more than 97% of the cells showed characteristic arrest in microscopic analysis.
DNA analysis.
Purified DNA was digested with appropriate restriction enzymes and separated on a 0.8% native gel. To analyze kinetics of BIR, one of the following digests were used: StuI and BglII, StuI and BamHI, or KpnI. Southern blotting was carried out as described previously (2). The blots were probed with appropriate DNA fragments labeled with 32P. A 320-bp PstI-EcoRI fragment from pJH454 containing a MAT-proximal region was used to analyze BIR. Blots were analyzed by using a Molecular Dynamics PhosphorImager. Pulse-field (CHEF) gel electrophoresis was performed by running genomic DNA embedded in plugs of 1% agarose, at 200 V, for 40 h (initial time = 10 s; final time = 35 s), followed by Southern blotting and hybridization, using as a probe a DNA fragment containing URA3 sequence (a BamHI fragment from pJH632) or ADE1 sequence (SalI fragment from pJH879).
For PCR analysis, equal amounts of purified genomic DNA were diluted, and PCR was carried out using buffer E from Epicentre and Taq polymerase (Roche). The following primers were used to identify early BIR and GC products: p1 (specific to URA3; 5′-ACC CGG GAA TCT CGG TCG TAA TGA-3′), p3 (specific to the Yα-inc region of MAT; 5′-GAA ATC AGC TTA GAA GTG GGC AAG-3′), and p2 (a sequence distal to the Z1 region of MAT; 5′-ATC CGT CAC CAC GTA CTT CAG C-3′). PCR products were subjected to gel electrophoresis in 1% agarose, stained with ethidium bromide, and quantified using Bio-Rad Quantity One software.
FACS analysis.
Flow cytometry analysis was done with a Becton Dickinson fluorescence-activated cell analyzer, as described previously (18). The DNA content reflects an average of about 15,000 cells.
RESULTS
GC predominates when both sides of a DSB share extensive homology with a homologous chromosome.
We have studied the repair of a single DSB on chromosome III in diploid MLN134, in which MATa is cleaved by a galactose-inducible HO endonuclease (Fig. 1A). The chromosome III that suffers the DSB is lacking HML and HMR, the mating-type switching donors, so repair nearly always occurs by GC by use of the uncleaveable MATα-inc allele on the homologous chromosome as a template for repair (22, 32). This strain is heterozygous for THR4+/thr4− distal to the MAT locus and carries a URA3 marker 3 kb centromere-proximal to the DSB. The initial phenotype of this strain is Ade+ Ura+ Thr+ and is nonmating, because both MATa and MATα-inc are expressed. Following the induction of GAL::HO, different repair outcomes can be monitored genetically (Table 1) (23). Complete loss of the HO-cleaved chromosome III is seen by the absence of ADE1 that marks deletions of both HML and HMR (Fig. 1E). Ade+ cells, having repaired the DSB and retained at least the ADE1 marker on the opposite side of the centromere, are homozygous for MATα-inc. Retention of URA3 indicates that strand invasion occurred close to the DSB. Loss of heterozygosity at THR4 can reflect chromosome truncation, repair by BIR, or one alternative outcome of GC associated with crossover (Table 1).
TABLE 1.
Repair of HO-induced DSBs in diploids containing full-length chromosome III
| Strain | No. of colonies tested | Classes of events (%)a
|
|||||
|---|---|---|---|---|---|---|---|
| GC | GC + crossover (or + BIR) | BIR | Chrom. loss + GC | Chrom. loss + BIR | Others | ||
| MLN134 MATa/MATα-inc | 408 | 86.3 | 10.7 | 1.7 | 1.0 | 0 | 0.2 |
| MY017 MATa/mat::KAN | 300 | 48.9 | 26.3 | 10.3 | 5.0 | 8.0 | 1.3 |
Some colonies are sectored for one or more nutritional marker (A+ = Ade+, T+ = Thr+, U+ = Ura+). A colony that is half Ade+ and half Ade− is indicated as A+/−. To determine the proportion of each event, each colony is treated as two half sectors even when the colony was uniform. For MLN 134, GC represents A+T+U+ (62.3%) plus A+T+U+/− (20.6%) plus A+ T+ U− (3.4%); GC + crossover represents A+T+/− U+/− U+ (3.9%) plus A+T+/−U+/− (6.6%) plus A+/−T+/−U− (0.2%); BIR represents A+T−U+ (1.0%) plus A+T−U− (0.7%); Chrom. loss + GC represents A+T+/−U+/− (1.0%); others represents A−T+U+ (0.2%). For MY017, GC represents A+T+U+ (33.3%) plus A+T+U+/− (11.3%) plus A+T+U− (4.3%); GC + crossover represents A+T+/−U+ (8.3%) plus A+T+/−U+/− (14%) plus A+T+/−U− (4%); BIR represents A+T−U+ (1.3%) plus A+T−U− (6.3%); Chrom.loss + GC represents A+/− T+/−U+/− (3.0%) plus A+/−T+/− U+/− (0.7%) plus A+T+/−U−/+ (1.3%); Chrom. loss + BIR represents A+/−T−U− (7.7%) and A+/−T+U+/− (0.3%); others represents A−T+U+ (0.2%) plus A+T+/−U+/− (0.75%).
As evaluated by plating cells on YEP-Gal to induce HO, DSB repair is highly efficient and occurs almost always by GC. Only 1.7% of the total events were distinguished as BIR events, with Ade+ Thr− colonies. About 86% of the plated colonies were Ade+ diploids homozygous for MATα-inc and still heterozygous for THR4/thr4 (Table 1) (22). Approximately 11% of the initially plated cells gave rise to Ade+ colonies that were sectored, half Thr+ and half Thr−. These colonies result from DSB repair in G2 as a consequence of crossover between two nonsister chromatids or as a result of two independent repair events in G2 (one GC and one BIR) (7). Thus, as expected from earlier studies (22), the majority of DSBs were repaired by GC.
BIR occurs in RAD51 cells when only the centromere-proximal end of the DSB shares extensive homology with the template chromosome.
To study RAD51-dependent BIR events we created a modified diploid isogenic to MLN134 from which all but 46 bp of homology distal to the HO cleavage site was deleted by creating a LEU2-marked truncation ending in an artificial telomere (Fig. 1B). Strain MY006 is hemizygous for thr4 and also lacks HMRa on the “bottom” chromosome. When these diploid cells were plated on YEP-Gal to induce HO, only about 11% of the colonies were Ade+ Ura+ and α-mating and either completely (49%) or partially (51%) Leu+, the phenotype expected if repair had proceeded by GC (Fig. 1C and Table 2). Those colonies that were Leu+/− represent cases where two independent repair events occurred in G2 cells, with one being repaired by GC and the other by BIR (Fig. 1C and 1D and see below).
TABLE 2.
Repair of HO-induced DSBs in strain MY006 and its derivatives
| Strain | Viability on YEP-GAL | No. of colonies tested | Phenotype of colonies (%)a
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| GC | BIR
|
BIR + chromosome loss
|
Chromosome loss | ||||||
| Ura+ | Ura+/− | Ura− | Ura+/− | Ura− | |||||
| MY006 | 97 | 1,119 | 10.7 | 33.2 | 20.5 | 8.4 | 12.8 | 14.0 | 0.4 |
| AM792 | 93 | 344 | 12.5 | 22.7 | 9.3 | 0 | 11.3 | 43.6 | 0.6 |
| MY006 G2 arrested | 5 | 117 | 16.2 | 24.8 | 0.9 | 30.8 | 5.1 | 11.1 | 11.1 |
| rad50Δ rad50Δ | 94 | 574 | 6.6 | 9.2 | 8.5 | 7.3 | 25.4 | 38.2 | 4.8 |
| rad51Δ rad51Δ | 73 | 155 | 5.2 | 78.7 | 16.1 | ||||
| rad9Δ rad9Δ | 86 | 832 | 6.1 | 0.7 | 3.5 | 5.5 | 9.8 | 73.9 | 0.5 |
| tid1Δ tid1Δ | 89 | 909 | 16.0 | 44.5 | 13.6 | 15.4 | 3.0 | 6.3 | 1.2 |
Some colonies were sectored for one or more nutritional marker. A colony that was half Ade+ and half Ade− is indicated as Ade+/−. To determine the proportion of each event, each colony is treated as two half sectors, even when the colony was uniform. GC, Ade+ Leu+ Ura+ and Ade+ Leu+/− Ura+; BIR, Ade+ Leu− and either Ura+, Ura+/−, or Ura−; BIR + chromosome loss, Ade+/− Leu− and either Ura+ or Ura−; Chromosome loss, Ade− Leu− Ura−.
Despite the reduced level of GC, only 0.4% of diploids experienced a loss of the broken chromosome, i.e., Ade− Ura− Leu− colonies (Table 2). Thus, alternative repair mechanisms efficiently substitute for GC. Approximately 62% of the colonies were α-mating Ade+ Leu−, characteristic of BIR. Thus, with limited homology on the right side of the DSB, BIR becomes the dominant repair pathway. Because the majority of DSB repair by BIR occurred in G2 (see below), the repair phenotypes most often should have resulted from two independent repair events in two broken sister chromatids. Therefore, to determine the proportion of each event, each colony was treated as two half sectors even when the colony was uniform (Table 2). In particular, the uniform Ade+ Ura+ Leu− colonies (comprising about a half of all Ade+ Leu−) correspond, most likely, to the repair in G2, with both chromatids repaired similarly by BIR initiated within 3 kb of the DSB.
In 13% of the Ade+ Leu− cases, colonies were uniformly Ura−, indicating that repair occurred more than 3 kb from the DSB in both broken sister chromatids. In the remaining one-third of BIR events, colonies were sectored for URA3; most likely BIR occurred also in G2 cells, with two HO-cleaved chromatids undergoing independent repair by BIR initiated between URA3 and MAT in one chromatid and proximal to URA3 in another chromatid (Table 2).
The remaining 27% of the colonies in Table 2 were sectored Ade+/Ade− but were fully Leu−. About half of these ADE1-sectored colonies were similarly sectored for URA3 (Ade− Ura−/Ade+ Ura+). These sectored colonies likely represent cases where one of two sister chromatids completed repair whereas the second chromatid was left unrepaired and lost in the next cell division.
In strain MY006, the centromere proximal (left) side of the DSB does not begin to share extensive homology with the template chromosome until after the removal of about 650 bp of Ya sequences that differ from the 700-bp Yα sequences on the donor chromosome. To examine the situation where the left side of the DSB is more readily available for DSB repair, we created the isogenic diploid strain AM792, in which the bottom chromosome carries MATa-inc rather than MATα-inc. After HO induction, about 12.5% of the DSB repair events proved to be GC and nearly 90% were apparently BIR (Ade+ Thr− Leu−) (Table 2); these results are quite similar to those obtained with MY006. The efficiency of BIR repair, judging by the proportion of unsectored colonies, is statistically significantly lower in AM792 than in MY006, despite the improvement in the homology at the DSB end. We believe that this difference is explained by the fact that MY006 is heterozygous MATa/MATα whereas AM792 is homozygous for MATa; cells expressing both Mata1 and Matα2 are known to have increased recombination proficiency compared to diploids expressing only one mating type (3). Thus, BIR results in an efficient outcome when there is insufficient homology to ensure GC repair of the DSB; moreover, the presence of 650 bp of nonhomology to the left of the DSB is not responsible for the absence of efficient GC.
We confirmed that Ade+ Ura+ Leu− colonies arose by BIR and not from de novo telomere formation, as the repaired (ADE1-containing) chromosome was the size of a full-length chromosome III (data not shown, but see Fig. 2). Tetrad analysis of eight Ade+ Ura+ Leu− products of MY006 showed that the repaired diploids contained two complete copies of chromosome III (see Materials and Methods) and also that the repair products were heterozygous for hml::ADE1/HML.
FIG. 2.
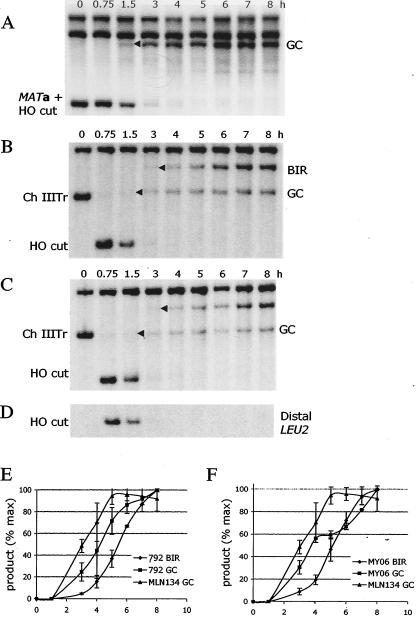
BIR and GC analyzed by pulse-field gel electrophoresis. DNA was prepared for pulse-field gel (CHEF) electrophoresis at intervals after induction of a DSB at MAT. Southern blots were probed with ADE1, which hybridizes to chromosome I (Ch I) and to hml::ADE1 on chromosome III. (A) GC in strain MLN134. The initial appearance of product is indicated with an arrowhead. (B and C) A diploid carrying a truncated chromosome III (Ch IIITr) yields both GC and BIR in strain MY006 (B) and strain AM792 (C). The first significant increase in product formation is indicated with an arrowhead. (D) Kinetics of accumulation of GC in strain MLN134 (□) and BIR in strain MY006 (Δ) and strain AM792 (•).
We also performed a pedigree analysis of strain MY006 undergoing DSB repair. Unbudded G1 cells were micromanipulated on YEP-Gal plates. After the first mitotic division, mother and daughter cells were separated, grown into colonies, and scored. If the repair in MY006 resulted from a half-crossover between the broken and intact chromosomes, then only one cell out of each mother-daughter pair would be viable, the other having two truncated chromosomes, or, depending on chromosome segregation, both cells would be monosomic for chromosome III. Out of 59 mother-daughter pairs dissected, 54 gave two viable colonies. Among these pairs, 17 consisted of two Ade+ Ura+ Leu− colonies, suggesting that BIR occurred with strand invasion between URA3 and the HO-cut MAT locus. Nine pairs were Ade+ Ura− Leu−, where strand invasion occurred to the left of URA3. Another 17 Leu− pairs had one Ade+ Ura+ colony and one Ade+ Ura− colony, suggesting independent repair of broken chromatids in G2. In six cases, one or both colonies of the mother-daughter pair were sectored for ADE1 and probably represent cases in which BIR took place in subsequent cell generations. Five cases were consistent with repair by GC in the mother or daughter cell or both. We also performed a PCR analyses of the HML chromosomal regions in the 11 mother-daughter pairs and demonstrated that in each case the repaired cells were indeed heterozygous (HML/hml::ADE1) (data not shown). These findings suggest that all repair outcomes contained two full copies of the chromosome III. These data support earlier conclusions (1) that repair occurred by BIR and not by half-crossovers.
Thus, in a strain where homology is severely limiting on one side of the DSB, the broken chromosome is repaired efficiently by BIR, most of the time beginning within a few kilobases of the DSB. If we consider each cell to have repaired the DNA in the G2 stage of the cell cycle, after DNA replication, then 8.2% of these RAD51 cells repaired the DSB by GC, 78% of the cells repaired the DSB by BIR, and 13.8% fail to repair the DSB, leading to chromosome loss.
RAD51-dependent BIR is more efficient than RAD51-independent BIR.
To confirm that strain MY006 predominantly used a RAD51-dependent BIR mechanism, we constructed a rad51Δ homozygous derivative. As shown previously in diploids with two full-length chromosome IIIs (22, 23, 32), DSB repair in rad51Δ homozygotes was inefficient (Table 2), as reflected by the large increase in cells that lost the broken chromosome. There was no GC, as expected. Cells that did repair by BIR were invariably Ura−, as expected if RAD51-independent repair requires FBI sequences (23). Moreover, only 5.2% of the colonies were fully Ade+; nearly 80% of the time, repair only occurred on one of two sister chromatids or in later generations (Table 2). There were often multiple white (Ade+) sectors against a red (Ade−) background, indicative of a high amount of chromosome loss, with several independent repair events in subsequent cell cycles (22). We conclude that the majority of the repair events in Rad51+ MY006 cells occur by a RAD51-dependent BIR pathway.
RAD50 plays an important role in RAD51-dependent BIR, but TID1 does not.
RAD50 and TID1 are both essential for RAD51- independent BIR (32). Here we show that RAD50 is also important in RAD51-dependent BIR. A rad50Δ derivative of MY006 did not prevent either GC or BIR, but there was an overall decrease in repair efficiency, as seen by an increase in chromosome loss (Table 2). About 26% of repair events were fully Ade+ Leu− compared with 62% in an isogenic RAD50 strain. Still, only 4.8% of colonies showed complete chromosome loss, indicating that after initially suffering a DSB, some descendants of a cell with a DSB were capable of repairing the DSB in a RAD51-dependent, RAD50-independent fashion. A similar analysis of a tid1Δ derivative of MY006 revealed that repair was more efficient than in the wild-type strain, as determined on the basis of a lower frequency of chromosome loss in tid1 mutants (Table 2), although it is difficult to say whether the efficiency of BIR per se is increased. Nevertheless, we conclude that the efficiency of BIR in tid1Δ mutants is the same as or even higher than that which occurs in the wild type. As tid1Δ prevents cells that have not completed repair from exiting the DNA damage checkpoint by adaptation (19), it is possible that the increased repair efficiency represents cells that would otherwise have been compromised by mitosis before repair was complete.
BIR and GC occur with different kinetics.
The kinetics of DNA repair can be monitored by pulse-field gel electrophoresis to see formation of GC and BIR products by examining whole-chromosome sizes. We compared strain MLN134 with two full-length chromosome IIIs, where 98% of repair was by GC, to both strain MY006 and strain AM792, where >75% of repair was by BIR. We examined events that retained hmlΔ::ADE1 on the left arm of chromosome III. The product of DSB repair by GC in strain MLN134 accumulates early (between 1.5 and 2 h after HO cleavage) (Fig. 2A and D). DSB repair by BIR (yielding a full-length chromosome III) appears approximately 2 h later in both strain MY006 and strain AM792, indicating that repair by BIR is slower than by GC (Fig. 2B, C, and D). On these same gels we could detect apparent GC repairing the truncated chromosome III in both strain MY006 and strain AM792 (Fig. 2B and C). Conversions appeared significantly earlier than BIR.
The slowness of BIR could reflect either slow initiation or slow replication of the 100 kb of the template chromosome. To distinguish between these possibilities we analyzed the kinetics of initiation by Southern blotting on the basis of restriction site polymorphisms between the donor and recipient chromosomes (Fig. 1 and 3). As shown in Fig. 3A, GC in strain MLN134 yields a novel BglII restriction fragment that appears 1.5 to 2 h after HO cleavage; these kinetics are similar to those of other HO-induced GC events (12, 33). In strains MY006 (Fig. 3B) and AM792 (Fig. 3C) one sees both GC and BIR, with GC appearing significantly earlier than BIR. The combined data from three separate experiments show a nearly 2-h delay in the 50% level of BIR relative to that of GC (Fig. 3E and F). Although it appears that GC may occur somewhat later in these strains than in MLN134, this is largely caused by the fact that cells repairing the DSB by GC do not exhibit cell cycle arrest whereas cells repairing by BIR have a substantial DNA damage checkpoint-mediated arrest (see below). Hence, continued proliferation of cells that have completed GC leads to an overestimation of GC in later time points, thus resulting in underestimation of the percentage of GC formed at early times. In any case, the delay of BIR relative to GC is evident.
FIG. 3.
Southern blot analysis of GC and BIR. (A) DNA digested with BglII to analyze GC in strain MLN134. The initial product appearance is shown by an arrowhead. (B and C) DNA digested with KpnI was analyzed for GC and BIR in strain MY006 (B) and strain AM792 (C). Blots were probed with a 320-bp fragment proximal to MAT locus. (D) The same blot as shown in panel C but probed with a LEU2-specific probe. (E) The appearance of GC and BIR products for AM792 is compared with GC results for MLN134. The rate of appearance of GC in AM792 is apparently retarded compared to that of MLN134 because the maximum level of GC is overestimated, as cells that repair DSBs by GC resume division whereas repair by cells using BIR is delayed. (F) Appearance of GC and BIR in MY006 compared with that of GC in MLN134.
We also note that the distal side of the DSB, containing LEU2 adjacent to telomere sequences, is degraded within the first 2 h (Fig. 3D), indicating that proximity to the telomere did not prevent its degradation.
BIR initiation is also slower than GC initiation.
It is possible that extension of BIR by even several kilobases might be delayed relative to earlier steps of strand invasion. We used a PCR assay (39) to detect intermediates of recombination in which new DNA synthesis has extended the invading strand by new DNA synthesis. In strain AM792, where the HO-cut end invades close to the cut site, PCR primers can detect the initiation of new synthesis that extends no more than 200 bp, as this primer extension creates a covalent single strand to which both PCR primers can anneal (Fig. 4A). The p1-p3 PCR product indicative of GC in strain MLN134 appeared 2 h after DSB induction (Fig. 1 and Fig. 4D). In contrast, the p1-p2 PCR product corresponding to the strand invasion intermediate in strains AM792 and MY006 appeared in significant amounts only 4 h after DSB induction, again showing a 2-h delay compared to GC results (Fig. 4B and C). A faint band observed at 3 h after induction corresponds to the formation of a very small amount of BIR product (5 to 10% of maximum). Thus, the kinetics of GC and BIR are different, even in the earliest steps. The absence of significant amounts of a PCR-detected intermediate for the BIR strain at 2 or 3 h also suggests another important conclusion: BIR is unlikely to result from a simple modification of the strand invasion and primer extension that occurs in GC. If this were the case, we should expect BIR events as well as GC events all to start at the same time, but it is clear that PCR also reveals a 2-h delay in the initiation of BIR compared to GC initiation. Thus, establishment of a repair replication fork for BIR occurs by a separate, slower process. Again, the fact that strains AM792 and MY006 yield similar results (Fig. 4E) argues that the removal of the Ya nonhomology at the left end of the DSB in MY006 is not responsible for the delay in BIR.
FIG. 4.
Timing of strand invasion during BIR versus GC. (A) PCR detection of a strand invasion and extension intermediate in DSB repair. The example shown is for strain AM792, where the left end of the DSB is perfectly homologous to the donor template. In strain MY006, the first 650 bp of the left end of the DSB are not homologous to the MATα-inc donor sequence. (B) PCR analysis of strand invasion and later steps of repair in diploid strain MY006 by use of primers p1 and p2 (Fig. 1B). The arrow indicates a position of the 5-kb PCR product corresponding to the strand invasion product formed by BIR. (C) PCR analyses of samples of strain AM792 by use of primers p1 and p2 (Fig. 1B) reveals a 5-kb product corresponding to the strand invasion product formed by BIR. (D) Timing of strand invasion during GC as determined by PCR analysis of samples from MLN134 (Fig. 1A). Use of primers p1 and p3 (Fig. 1A) revealed a 4-kb product corresponding to the strand invasion product formed by GC. (E) Accumulation of BIR strand invasion products in MY006 and AM792. The PCR data presented in panels A and B are quantitative for the initial time points up to 5 h. For the quantitation of the amounts of products at the later time points we used PCR performed on diluted DNAsamples. The results presented in panel C are quantitative only for the 2-h time point, as the amount of product was saturated for all later time points.
BIR occurs efficiently in G2/M-arrested cells.
BIR in strains MY006 and AM792 demands the copying of >100 kb from the homologous chromosome; moreover, the delay in initiating BIR means that nearly all cells became checkpoint arrested in G2/M before DNA replication could be completed (see below). Some components of normal DNA replication, including the Mcm helicase proteins, are excluded from the nucleus in nocodazole-arrested cells (16, 26). To confirm that BIR could occur outside of normal S phase, we induced HO in MY006 cells after arrest with nocodazole. FACS analysis confirmed that the cells remained with a 2C DNA content during the experiment (data not shown). By probing for URA3 sequences located just proximal to the HO-cut MATa locus, we could detect BIR both by Southern blot analysis, assaying the elongation of the DSB end for at least a few kilobases, and by CHEF gel electrophoresis to see the appearance of a full-length chromosome III (data not shown). Both Southern blots and CHEF gel analysis showed less BIR product detected with the URA3 probe than when HO was induced in log-phase cells. When these same CHEF gels were probed with sequences homologous to hml::ADE1 on the left end of the truncated chromosome, however, we found that repair was highly efficient in nocodazole-arrested cells (Fig. 5), as 100% of broken molecules were repaired at 10 h after the DSB (data not shown). Cell viability after long incubation in nocodazole was reduced to about 9%, a result also seen in cells that cannot experience a DSB and that were treated in the same way (8% viable). Among survivors 44% of BIR events were Ura+ compared to 70% in logarithmically growing cells (Table 2). This could have arisen if there had been more extensive resection of DSB ends prior to strand invasion.
FIG. 5.
BIR in strain MY006 cells arrested in G2 by nocodazole. A Southern blot of the appearance of BIR products on a CHEF gel probed with ADE1, which hybridizes to chromosome I and to the HO-cut truncated chromosome III, is shown. A BIR product that is the same size as an intact chromosome III and hybridizes also to a THR4 probe (not shown) is indicated as Ch III BIR. A later-appearing translocation that does not hybridize with THR4 is indicated as Tx.
In addition to the BIR-sized product that also hybridizes with the URA3 probe, we observed an additional, larger band that does not hybridize to URA3 (Fig. 5; indicated as Tx). This band is not seen when BIR is induced in growing cells, even though they arrest for several hours in G2/M because of the damage checkpoint; this exceptional product is seen only when G2/M arrest is established prior to the HO induction by nocodazole. The kinetics of appearance of this second band is slower than that of the URA3-containing BIR product. Preliminary analysis suggests that this product arises by recombination between Ty1 sequences 30 kb proximal to the DSB with Ty1 sequences on another chromosome. Characterization of this product will be presented elsewhere.
The rate of DNA synthesis in BIR is comparable to that seen in S phase.
To estimate the time it takes for BIR to traverse >100 kb of the template chromosome, we used physical analysis to detect the beginning and completion of the process. Initial strand invasion and replication of a few kilobases can be seen by Southern blotting or PCR (Fig. 3 and 4). The completion of BIR can be seen by the appearance of full-length chromosomes on pulse-field gels (Fig. 2). Comparisons of times of appearance of the initial BIR product with times of appearance of the full-length chromosome for strain MY006 for three different experiments are shown in Fig. 6. The kinetics of the two processes are quite similar, suggesting that once BIR is initiated, the rate of DNA replication to the chromosome end in these G2/M-arrested cells is sufficiently rapid that we do not see a difference, given the resolution of 30-min time points. Thus, once BIR begins, the rate of replication is about 100 kb/30 min, which is comparable to estimates of normal S-phase elongation of between 2 and 4 kb/min (31). Previous studies of the kinetics of DNA replication often use cells grown at temperatures as low as 18°C to see the progression of the replication fork. We also attempted to use cells grown at 18°C to study BIR; a previous study of MAT switching at such low temperatures showed that even GC was not completed for 7 h (9). With strain MY006, BIR was so delayed (>12 h) that this proved not to be informative.
FIG. 6.
Rate of replication associated with BIR. Accumulation of products corresponding to the beginning and to the end of BIR in strain MY006 is shown. Panels A, B, and C show the results of three independent experiments. In experiments shown in panels A and B, samples were taken every 30 min from initially exponential cultures of strain MY006 undergoing BIR. DNA samples were analyzed by CHEF similarly to the analysis described for Fig. 2B (finish) or were digested by BamHI-StuI and analyzed by Southern blotting (start). In the experiment represented by panel C, samples were taken at 1-h intervals and Southern analyses were performed on DNA samples digested by KpnI.
Nonhomology at DSB ends increases the proportion of BIR events.
In isogenic diploid MY017, MATα-inc is replaced by a KAN-MX-marked deletion event that removes all of MAT-X, Yα, and Z1 sequences. Consequently, HO-induced DSB ends at MATa have about 230 bp of nonhomology to the right and >1 kb of nonhomology to the left before homology is encountered with the donor chromosome; nonhomology must be removed before the 3′ end of an invading strand can be used to prime new DNA synthesis necessary for repair. Whereas GC is highly efficient when only one DSB end is nonhomologous, as in strain MLN134, nonhomology at both ends provoked a significant increase in repair events that apparently use BIR (Table 1). The proportion of Ade+ Thr+ colonies, repaired by GC, dropped from 86 to 49%, whereas the proportion of Ade+ Thr− colonies, repaired by BIR, increased from 1.7 to 10.3%. There was also an increased level of chromosome loss. A significant increase in colonies sectored for THR4/thr4 is most likely a result of instances in which one MATa chromatid was repaired by GC and the other by BIR. Thus, BIR may be an important outcome even in wild-type cells when both DSB ends lack immediate homology to the template.
BIR and GC exhibit different DNA damage checkpoint responses.
The different kinetics of GC and BIR were reflected in different DNA damage checkpoint responses. In parent strain MLN134, where GC predominates, the distribution characteristics of G1-, S-, and G2-phase cells, as determined by FACS analysis, did not significantly differ following HO induction, suggesting that there is no or little checkpoint delay associated with GC (Fig. 7). This result is similar to that seen for intrachromosomal recombination (MAT switching) in haploid cells, where there was no induction of the DNA damage checkpoint even though the repair process takes more than 1 h (30). In contrast, HO induction in strain MY006 (MATa/MATα-inc) resulted in nearly all cells becoming arrested at the G2/M stage of the cell cycle and resuming cell cycle progression only at the time that Southern blot analysis showed that BIR had occurred (Fig. 7). We note that the apparent increase in DNA content above 2C in cells that remain arrested for long periods of time is an artifact of FACS scanning of cells that grow to very large size (38).
FIG. 7.
FACS analysis of strains undergoing DSB repair after HO cleavage. Repair in strain MLN134 occurs predominantly by GC, whereas repair in strain MY006 occurs predominantly by BIR. Delayed cell cycle progression in strain AM792, in which sequences on both sides of the cleavage site are fully homologous, is shown.
The behavior of a small proportion of cells that completed GC did not significantly contribute to the distribution of cells. Heterology is not the cause of this delay in the cell cycle, as eliminating any heterology around the DSB, as in strain AM792 (MATa/MATa-inc), does not eliminate the cell cycle arrest (Fig. 7). This result suggests that nearly all the repair of the DSB by BIR occurs in G2/M-arrested cells, when the cell contains two HO-cleaved chromatids, although the outcomes appear to be somewhat different from those seen when cells are first arrested in G2 by nocodazole. Overall, we conclude that BIR and GC exhibit different DNA damage checkpoint responses. This result is consistent with our observation that deletion of the SRS2 gene that is required for recovery from checkpoint arrest (38) leads to low (14%) viability associated with DSB in a strain MY006 derivative and has no effect on viability when introduced in strain MLN134 (data not shown).
Effect of deleting the RAD9 checkpoint gene on BIR.
The discovery that cells engaged in BIR activate the G2/M checkpoint led us to examine the requirement for checkpoint activation in completing BIR. Rad9 is required for Mec1-dependent cell cycle arrest (37). A rad9Δ derivative of MY006 failed to arrest after HO induction (data not shown). The viability of these cells was near that of the wild type, but the pattern of repair events was different (Table 2). The proportion of GC which occurs rapidly and may not engage the checkpoint was similar to wild-type results (6%), but the proportion of colonies in which repair was efficient enough so that all cells retained the left arm of the broken chromosome (Ade+) dropped from 62% to less than 10%. Most colonies (83.7%) were sectored for ADE1, indicating that half or more of the cells had lost the broken chromosome. Moreover, the site of successful repair events was shifted so that nearly all of the products did not retain the URA3 marker. Still, only a few cells gave rise to colonies with no indication of repair at all, as only 0.5% of the colonies were fully Ade−. These results suggest that repair by BIR is decreased in rad9Δ cells due to the inability of these cells to arrest, although the change in the point of strand invasion might reflect an increased rate of resection in rad9Δ strains (13).
Southern blot analysis of CHEF gels showed formation of only 5% of BIR product compared to the truncated HO-cut chromosome results, consistent with checkpoint-defective cells continuing to divide before repair (data not shown). To demonstrate that rad9Δ cells are repair competent, we arrested them in G2 with nocodazole before inducing the DSB. Southern blot analysis of CHEF gels, probed with ADE1, showed the formation of a repair band similar to the pattern seen with nocodazole-arrested MY006 cells. Overall, our data suggest that even though deletion of RAD9 does not prevent BIR per se, the establishment of RAD9-mediated arrest is crucial to ensure that there is enough time to repair a DSB by BIR before mitosis.
As mentioned above, the adaptation-defective tid1Δ reduced the proportion of colonies in which there were sectors with one part exhibiting BIR and the other exhibiting chromosome loss (Table 2), but there was no apparent change in the kinetics of either rare GCs or BIR on pulse field gels (data not shown). It is possible that the improvement in recovering full BIR colonies reflects tid1Δ's restraint on adaptation, allowing more time for repair to be accomplished. This result reinforces our finding that adaptation need not precede the repair of a broken chromosome end by BIR, although adaptation may lead to loss, by missegregation, of acentric chromosome fragments, leading to an increase in BIR in adapting cells (8).
DISCUSSION
We initially described a RAD51-independent BIR process that could repair a chromosomal DSB (22). However, we suggested that there was almost certainly a RAD51-dependent BIR process as well, but this was obscured by the very high efficiency of GC. The existence of a RAD51-dependent BIR process was confirmed first by analysis of telomere maintenance in the absence of telomerase that showed the existence of both RAD51-dependent and -independent pathways (17). In this paper we describe in detail the properties of RAD51-dependent BIR. RAD51-dependent BIR is much more efficient that the RAD51-independent mechanism; it is not eliminated by deletion of either RAD50 or TID1 and does not depend on a special facilitating sequence, as does RAD51-independent BIR.
Recently Davis and Symington (5) used a transformation-based BIR assay to study both RAD51-dependent and RAD51-independent BIR. RAD51-dependent BIR proved to be much more efficient, as we find with chromosomal DSBs. In their system, in which one cannot judge the kinetics of the process or whether repair occurs in only a fraction of the progeny of a transformed cell, it appears that rad50Δ improves RAD51-dependent repair, possibly by preventing rapid degradation of transforming DNA ends, whereas RAD50 was not required in RAD51-independent events. In our chromosomal assay, RAD50 plays an important but not essential role in RAD51-mediated BIR but is strongly required for RAD51-independent events, which also could not use homology close to the DSB (23, 32). These contrasting results may reflect the differences in chromatin structure between broken preexisting chromosomes and transforming DNA.
The hierarchy of RAD51-dependent repair is intriguing. When there is little or no homology to one side of the DSB, BIR is efficient and there is almost no loss of the broken chromosome; but BIR loses out to GC when there is substantial homology on both sides of the DSB. When the distal side of the DSB has only 46 bp of homology, about 10% of the events occur by GC, and these events are kinetically distinct from the slower, checkpoint-triggering BIR process, even when the proximal side of the DSB is fully homologous to the donor template.
How the recombination machinery determines whether there is homology on both sides of the DSB remains a mystery, and how this assessment is tied to the absence of triggering the DNA damage checkpoint is also not understood. We note that the LEU2-containing terminal fragment of the truncated chromosome disappears on Southern blots as rapidly as the proximal HO-cut fragment (Fig. 2D). When both ends of the DSB become productively engaged in Rad52-mediated recombination, there is apparently very little activation of the DNA damage signal, even though repair takes more than 1 h. An absence of checkpoint activation is also seen during MAT switching, an intrachromosomal GC event (30); however, checkpoint activation is seen when a DSB in MAT sequences is repaired by interchromosomal, ectopic GC, when the coordinated action of two ends may be more difficult (38).
When one end of the DSB is unable to engage in recombination, continuing resection of this end and the lack of strand invasion apparently trigger the checkpoint. In spite of the activation of the DNA damage checkpoint prior to BIR, this checkpoint is not, in and of itself, required to promote BIR; nocodazole-arrested rad9Δ cells proved to be proficient in repair.
We also find that when a DSB is created in a small island of nonhomology between generally homologous chromosomes, the presence of nonhomology at both ends of the DSB leads to a decrease in GC and a marked increase in BIR events. Removal of nonhomology from the end that can then initiate GC is an inherently slow step (4, 9). This situation could readily occur at spontaneous DSBs arising within short interspersed repeated sequences that are at polymorphic positions in two homologous chromosomes.
What is the slow step in BIR?
Why should BIR be so slow to initiate, especially if the sequences to the left of the DSB are exactly the same in cells initiating GC and in those engaged in BIR? We have ruled out the possibility that the delay in BIR is caused by a region of nonhomology to the left of the DSB, as diploid AM792 exhibits the same slow kinetics as does strain MY006 and both exhibit a checkpoint-mediated delay at G2/M. These results suggest that there could be an initial phase of homology searching that leads to a coordination of both ends of the DSB.
We have considered an alternative explanation for the slow establishment of BIR, namely, that the W, X, and Ya sequences to the left of the HO cleavage site at MAT are inherently less active in establishing strand invasion than the Z region sequences on the right. We have established a haploid strain in which a DSB is created within a 117-bp HO cleavage site derived from MATa and inserted in another locus such that repair cannot occur by GC (38). The end that carries out strand invasion contains the opposite end of the DSB from that used in the BIR events initiated at MAT. In this case, too, BIR is not seen until 4 to 6 h after HO cutting (M. B. Vaze, N. Sugawara, and J. E. Haber, unpublished data).
Once strand invasion has initiated, DNA repair replication appears to progress at a rate that is not significantly different from that of normal S-phase replication. We note that BIR occurs with approximately the same kinetics in both checkpoint-arrested and nocodazole-blocked G2 cells, when the Mcm helicase proteins are apparently excluded from the nucleus. The Mcm complex is believed to be important both in the initiation of DNA replication at origins and in the processive movement of the replication fork (16). Given that BIR is initiated by an origin-independent recombination process, the absence of a role for Mcm proteins in starting BIR is not surprising; but then, what helicases are responsible for the ability of the replication fork to progress >100 kb down the template chromosome?
What appears to be the slow step in BIR is the establishment of a repair replication fork. In bacteria, for which recombination-dependent DNA replication has also been well documented, the normal initiation role of the DnaA and DnaC proteins in loading DnaB helicase at origins is replaced by the PriA complex which promotes assembly of a repair replication fork (20, 24). There is no obvious PriA homologue in eukaryotes, but we speculate that there must be such a protein. Even so, the initiation step appears to be highly rate limiting. In the future it should be possible to dissect the initial steps of strand invasion and replication fork establishment in BIR by chromatin immunoprecipitation and by searches for additional genes required for BIR.
Acknowledgments
We thank Moreshwar B. Vaze and present members of the Haber lab for their suggestions throughout this work and about the manuscript. This work was supported by National Institutes of Health grants GM20056 and GM61766 as well as National Science Foundation grant MCB0077257.
REFERENCES
- 1.Bosco, G., and J. E. Haber. 1998. Chromosome break-induced DNA replication leads to non-reciprocal translocations and telomere capture. Genetics 150:1037-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clikeman, J., G. Khalsa, S. Barton, and J. Nickoloff. 2001. Homologous recombinational repair of double-strand breaks in yeast is enhanced by MAT heterozygosity through yKU-dependent and -independent mechanisms. Genetics 157:579-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colaiácovo, M. P., F. Pāques, and J. E. Haber. 1999. Removal of one nonhomologous DNA end during gene conversion by a RAD1- and MSH2-independent pathway. Genetics 151:1409-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis, A. P., and L. S. Symington. 2004. RAD51-dependent break-induced replication in yeast. Mol. Cell. Biol. 24:2344-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunn, B., P. Szauter, M. L. Pardue, and J. W. Szostak. 1984. Transfer of yeast telomeres to linear plasmids by recombination. Cell 39:191-201. [DOI] [PubMed] [Google Scholar]
- 7.Esposito, M. S. 1978. Evidence that spontaneous mitotic recombination occurs at the two-strand stage. Proc. Natl. Acad. Sci. USA 75:4436-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galgoczy, D. J., and D. P. Toczyski. 2001. Checkpoint adaptation precedes spontaneous and damage-induced genomic instability in yeast. Mol. Cell. Biol. 21:1710-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holmes, A., and J. E. Haber. 1999. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell 96:415-424. [DOI] [PubMed] [Google Scholar]
- 10.Inbar, O., B. Liefshitz, G. Bitan, and M. Kupiec. 2000. The relationship between homology length and crossing-over during the repair of a broken chromosome. J. Biol. Chem. 275:30833-30838. [DOI] [PubMed] [Google Scholar]
- 11.Ira, G., and J. E. Haber. 2002. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 22:6384-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ira, G., A. Malkova, G. Liberi, M. Foiani, and J. E. Haber. 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115:401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia, X., T. Weinert, and D. Lydall. 2004. Mec1 and Rad53 inhibit formation of single-stranded DNA at telomeres of Saccharomyces cerevisiae cdc13-1 mutants. Genetics 166:753-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Kraus, E., W. Y. Leung, and J. E. Haber. 2001. Break-induced replication: a review and an example in budding yeast. Proc. Natl. Acad. Sci. USA 98:8255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Labib, K., S. E. Kearsey, and J. F. Diffley. 2001. MCM2-7 proteins are essential components of prereplicative complexes that accumulate cooperatively in the nucleus during G1-phase and are required to establish, but not maintain, the S-phase checkpoint. Mol. Biol. Cell 12:3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le, S., J. K. Moore, J. E. Haber, and C. Greider. 1999. RAD50 and RAD51 define two different pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest DNA damage. Cell 94:399-409. [DOI] [PubMed] [Google Scholar]
- 19.Lee, S. E., A. Pellicioli, A. Malkova, M. Foiani, and J. E. Haber. 2001. The Saccharomyces recombination protein Tid1p is required for adaptation from G2/M arrest induced by a double-strand break. Curr. Biol. 11:1053-1057. [DOI] [PubMed] [Google Scholar]
- 20.Lovett, S. T. 2003. Connecting replication and recombination. Mol. Cell 11:554-556. [DOI] [PubMed] [Google Scholar]
- 21.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell 73:347-360. [DOI] [PubMed] [Google Scholar]
- 22.Malkova, A., E. L. Ivanov, and J. E. Haber. 1996. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 93:7131-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malkova, A., L. Signon, C. B. Schaefer, M. Naylor, J. F. Theis, C. S. Newlon, and J. E. Haber. 2001. RAD51-independent break-induced replication to repair a broken chromosome depends on a distant enhancer site. Genes Dev. 15:1055-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marians, K. J. 2000. PriA-directed replication fork restart in Escherichia coli. Trends Biochem. Sci. 25:185-189. [DOI] [PubMed] [Google Scholar]
- 25.Morrow, D. M., C. Connelly, and P. Hieter. 1997. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics 147:371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen, V. Q., C. Co, K. Irie, and J. J. Li. 2000. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr. Biol. 10:195-205. [DOI] [PubMed] [Google Scholar]
- 27.Nickoloff, J. A., D. B. Sweetser, G. J. Khalsa, and S. L. Wheeler. 1999. Multiple heterologies increase the length of double-strand break-induced allelic gene conversion tracts. Genetics 153:665-679. [DOI] [PMC free article] [PubMed]
- 28.Pāques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pāques, F., W. Y. Leung, and J. E. Haber. 1998. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol. Cell. Biol. 18:2045-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pellicioli, A., L. S. E., C. Lucca, M. Foiani, and J. E. Haber. 2001. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from G2/M arrest. Mol. Cell 7:293-300. [DOI] [PubMed] [Google Scholar]
- 31.Raghuraman, M. K., E. A. Winzeler, D. Collingwood, S. Hunt, L. Wodicka, A. Conway, D. J. Lockhart, R. W. Davis, B. J. Brewer, and W. L. Fangman. 2001. Replication dynamics of the yeast genome. Science 294:115-121. [DOI] [PubMed] [Google Scholar]
- 32.Signon, L., A. Malkova, M. Naylor, and J. E. Haber. 2001. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol. 21:2048-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugawara, N., X. Wang, and J. E. Haber. 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell 12:209-219. [DOI] [PubMed] [Google Scholar]
- 34.Symington, L. S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66:630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teng, S., J. Chang, B. McCowan, and V. A. Zakian. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell 6:947-952. [DOI] [PubMed] [Google Scholar]
- 36.Teng, S. C., and V. A. Zakian. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toh, G. W., and N. F. Lowndes. 2003. Role of the Saccharomyces cerevisiae Rad9 protein in sensing and responding to DNA damage. Biochem. Soc. Trans. 31:242-246. [DOI] [PubMed] [Google Scholar]
- 38.Vaze, M., A. Pellicioli, S. Lee, G. Ira, G. Liberi, A. Arbel-Eden, M. Foiani, and J. Haber. 2002. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires srs2 helicase. Mol. Cell 10:373. [DOI] [PubMed] [Google Scholar]
- 39.White, C. I., and J. E. Haber. 1990. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 9:663-673. [DOI] [PMC free article] [PubMed] [Google Scholar]