Abstract
Molecular portraits of numerous tumors have flooded oncologists with vast amounts of data. In parallel, effective inhibitors of central pathways have shown great clinical benefit. Together, this promises potential clinical benefits to otherwise end-stage cancer patients. Here, we report a clinical service offering mutation detection of archived samples using the ion Ampliseq cancer panel coupled with clinical consultation.
A multidisciplinary think tank consisting of oncologists, molecular-biologists, genetic counselors, and pathologists discussed 67 heavily pretreated, advanced cancer patient cases, taking into account mutations identified using ion Ampliseq cancer panel, medical history, and relevant literature.
The team generated a treatment plan, targeting specific mutations, for 41 out of 64 cases. Three patients died before results were available. For 32 patients, the treating oncologists chose not to include the panel recommendation in the treatment plan for various reasons. Nine patients were treated as recommended by the panel, 5 with clinical benefit, and 4 with disease progression.
This study suggests that routine use of massive parallel tumor sequencing is feasible and can judiciously affect treatment decisions when coupled with multidisciplinary team-based decision making. Administration of personalized based therapies at an earlier stage of disease, expansion of genetic alterations examined, and increased availability of targeted therapies may lead to further improvement in the clinical outcome of metastatic cancer patients.
Keywords: DNA, high-throughput nucleotide sequencing, mutation, neoplasms, precision medicine
1. Introduction
In recent years, molecular profiles of tumors such as breast,[1,2] prostate,[3] colon,[4] lung,[5] ovary,[6] and glioblastoma[7] have been reported. In parallel, inhibitors of molecular pathways are commonly used in oncological practice including inhibitors of ABL1,[8] Adenylyl cyclase,[9] ALK,[10] BRAF,[11,12] CDK4/6,[13] DNMT,[14] EGFR,[15–18] HER2,[19–22] JAK,[23] KIT,[24] MEK,[25] mTOR,[26] RET,[5] ROS,[27] SMO,[28] VEGF,[29,30] and VEGFR.[31,32] Some of these inhibitors have shown clinical activity in diverse organs—HER2 inhibition in HER2-positive breast[22] and gastric tumors;[33] CKIT inhibition in gastrointestinal stroma tumor[24] and melanoma,[34] and mTOR inhibition in renal cell carcinoma,[35] Astrocytoma,[26] pancreatic neuroendocrine tumors,[36] and ER-positive breast cancer.[37] These reports, in conjunction with phase II,[38] phase I[39] and case reports[40] where patients derived clinical benefit from pathway inhibition, provide the clinical rationale for testing mutations in tumor samples and utilizing mutation analysis to choose a pathway inhibitor to treat patients. Several academic institutions[41,42] and commercial companies[43,44] offer a molecular profiling service[41,42] that hundreds of cancer patients in Israel have chosen to utilize, indicating an unmet need.
In this report, we describe a comprehensive molecular service based in an academic hospital setting. We detail the validation of the molecular technique, patient population and mutations found, as well as the decision-making process, clinical decisions taken by the molecular oncology forum and clinical outcome.
2. Methods
2.1. Patient population
Patients were referred by their treating physician, at their discretion after a detailed discussion with the patient where the possible benefits and expected limitations were carefully reviewed prior to ordering this service. The clinical service included mutation detection, data analysis, and panel treatment recommendation. Patients receiving off-label treatment signed an informed consent (29c) that was approved by the head of the Hadassah Medical Center ethics (Helsinki) committee prior to treatment.
2.2. Molecular profiling
Formalin fixed paraffin embedded (FFPE) tissue was examined by a pathologist to identify the region for sampling and percentage of tumor cells in the analyzed region. DNA was extracted using QIamp DNA FFPE Tissue Kit and the Ion Ampliseq cancer panel was applied. Up to 4 samples were loaded on a 314 chip (10 million bases (Mb)capacity) or up to 8 samples were loaded on a 316 chip (100 Mb capacity) and run on an Ion Torrent Personal Genome Machine (PGM) System. Mutations were identified by the Ion Variant caller as previously described.[43] The V1 panel amplifies 13,311 bp in ABL1, AKT1, ALK, APC, ATM, BRAF, CDH1, CDKN2A, CSF1R, CTNNB1, EGFR, ERBB2/4, FBXW7, FGFR1/2/3, FLT3, GNAS, HNF1A, HRAS, IDH1, JAK2/3, KDR, KIT, KRAS, MET, MLH1, MPL, NOTCH1, NMP1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RB11, RET, SMAD4, SMARCB1, SMO, SRC, STK11, TP53 and VHL. The V2 panel amplifies 22,027 bp in the same genes and also in EZH2, GNA11, GNAQ, and IDH2. Sanger sequencing was performed as previously described.
2.3. Data interpretation
All variants were (manually) visualized using the integrative genome viewer.[44] Noncoding and synonymous variants were not investigated further. All variants with allelic fraction of 100% ± 3% or 50% ± 3% were perceived as potential germ line changes. If a variant was previously identified, in the study population, as a known germline variant, it was appraised as such. All others were perceived as somatic changes. Nonsynonymous somatic variants were examined using the COSMIC database,[45] and variants not identified in the database were not further evaluated. The variants identified in COSMIC were investigated by a literature review initiated by references found in the COSMIC database. A report including a summary of the case, the variant caller report, and review of the literature was sent to the treating physician. Based on the treating physician's remarks, a revised report was sent to the molecular oncology forum members including molecular-biologists, genetic counselors, oncologists, and pathologists. Each case was presented, reviewed, and discussed to reach a consensus recommendation.
2.4. FFPE-based somatic panel validation
To validate the test, we sequenced 20 samples, 19 of the samples tested positive for KRAS, BRAF, or EGFR and 1 sample was positive for several mutations. In 19 out of 20 samples we succeeded in generating amplified DNA amenable for massive parallel sequencing. The average number of bases read was 229 Mb per chip which resulted in average coverage of 3503X. Sanger sequencing was performed on previously unknown mutations for further validation. The previously known mutations in all samples were identified.
Reproducibility was tested using duplicates prepared separately from the same DNA sample. There was full concordance between variants called, a total of 14 pairs. The average difference of variant allelic fraction (i.e., the percentage of the DNA reads that are mutated) in the duplicates was 1.6% with a median of 0.5%. A sample of normal tissue was analyzed and the variations found were either 50% ± 3% or 100% ± 3%, all perceived as germline. Based on these results, a clinical service was established where each tumor sample is tested twice, and certain mutations are regarded as germline.
3. Results
3.1. Patient population
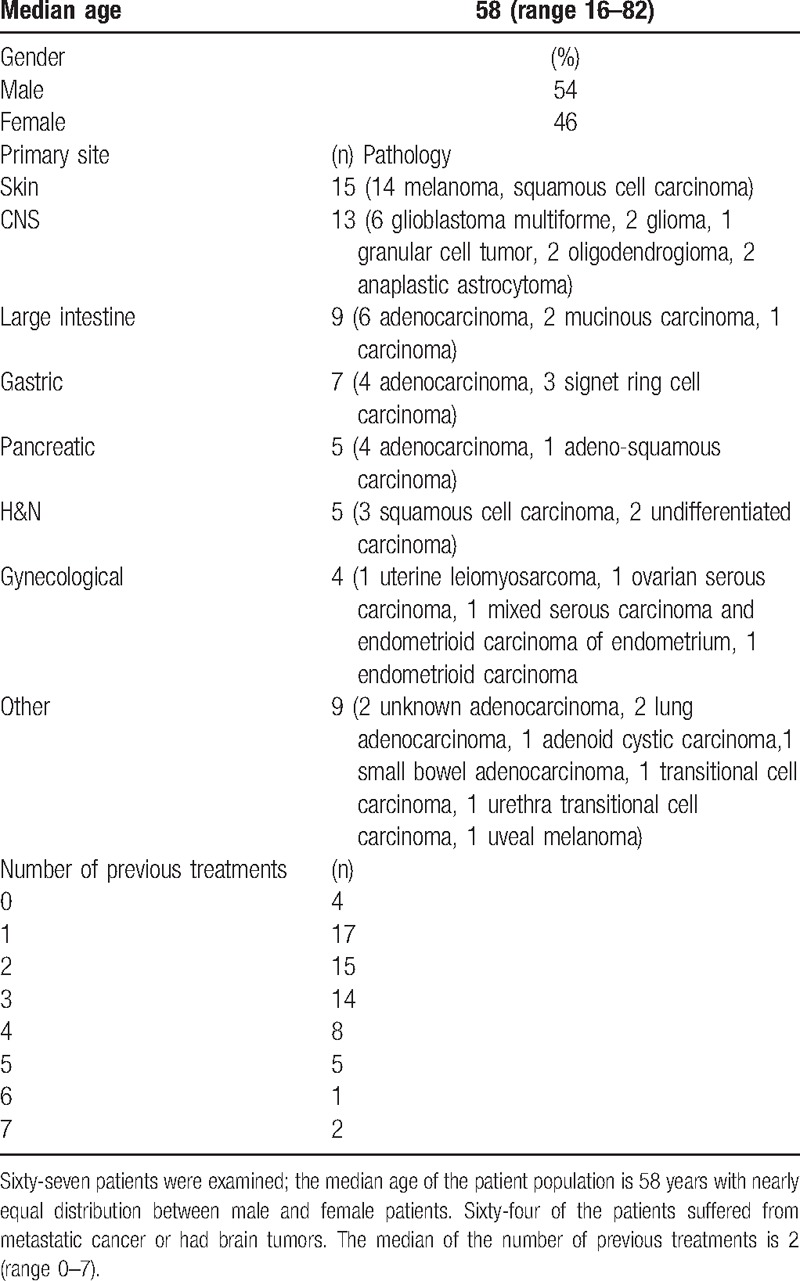
Table 1 reports the patients’ characteristics. The median number of previous treatments is 2. The advanced stage of disease in this population is demonstrated by the fact that 3 patients died while the test was processed, in a span of weeks (Table 1). In 64 cases the test was performed on existing FFPE samples. In 3 cases where no tissue was available for testing, test-designated biopsies were performed, 2 from lung metastases, and 1 from the primary gastric tumor. DNA was extracted from the tumor primary site (n = 33), local recurrences or distant metastasis (n = 29). The tumors were either naive to chemotherapy (n = 47), or previously treated (n = 16).
Table 1.
Clinical characterization of patients examined.
3.2. Molecular profile
Some samples tested harbored known mutations; 3 BRAF V600E, 3 KRAS G12D, 1 KRAS G13C, and 1 IDH1 R132H were reidentified. A KRAS G12D positive case was reclassified as KRAS wild type, a KRAS negative case was reclassified as KRAS A146T positive, and a BRAF V600E negative case was reclassified as BRAF V600E positive.
3.3. Clinical outcome
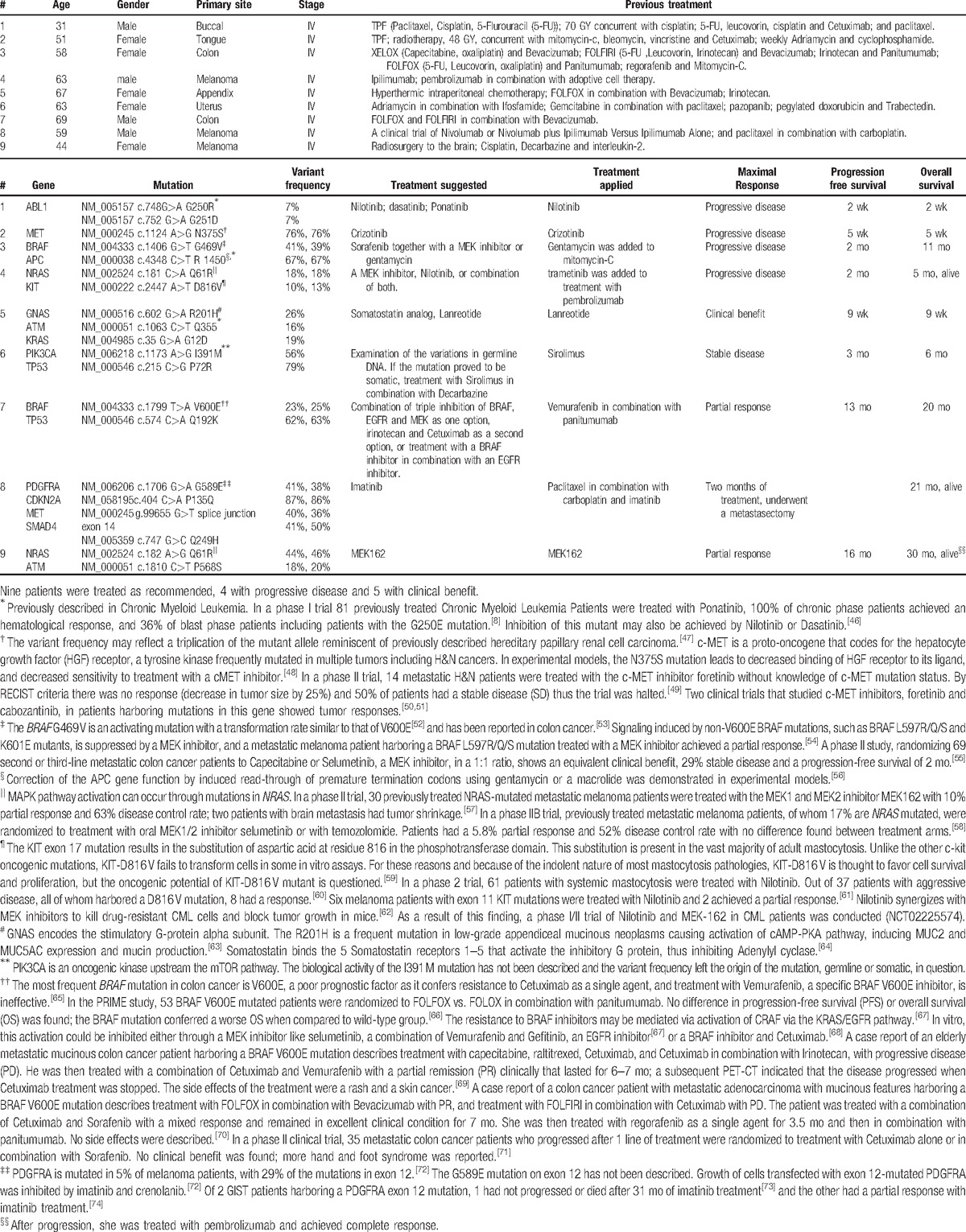
Of 67 patients assessed, 3 died before results were processed. Of the remaining patients, for 23 patients no novel perceived actionable somatic mutations were detected; in 41 patients, 75 novel actionable somatic mutations were detected with a median of 1 mutation per sample (range 1–3). One sample harboring hundreds of somatic mutations is not described. Actionable mutations are listed in Table 2. Of the 41 patients with actionable mutations that led to treatment recommendations, 9 patients received the treatment recommended by the forum. In 4 patients the disease progressed, however in 5, following the recommended treatment a clinical benefit, stable disease for more than 2 months or partial response was achieved (Table 3).[46–74] In 32 patients, treatment was deferred due to a combination of reasons including availability of pathway inhibitors in clinical trials outside the country, poor clinical condition, and other available treatment options. In cases where germline mutations were suspected, genetic consultation was recommended.
Table 2.
Actionable mutations identified in 67 cancer patients.
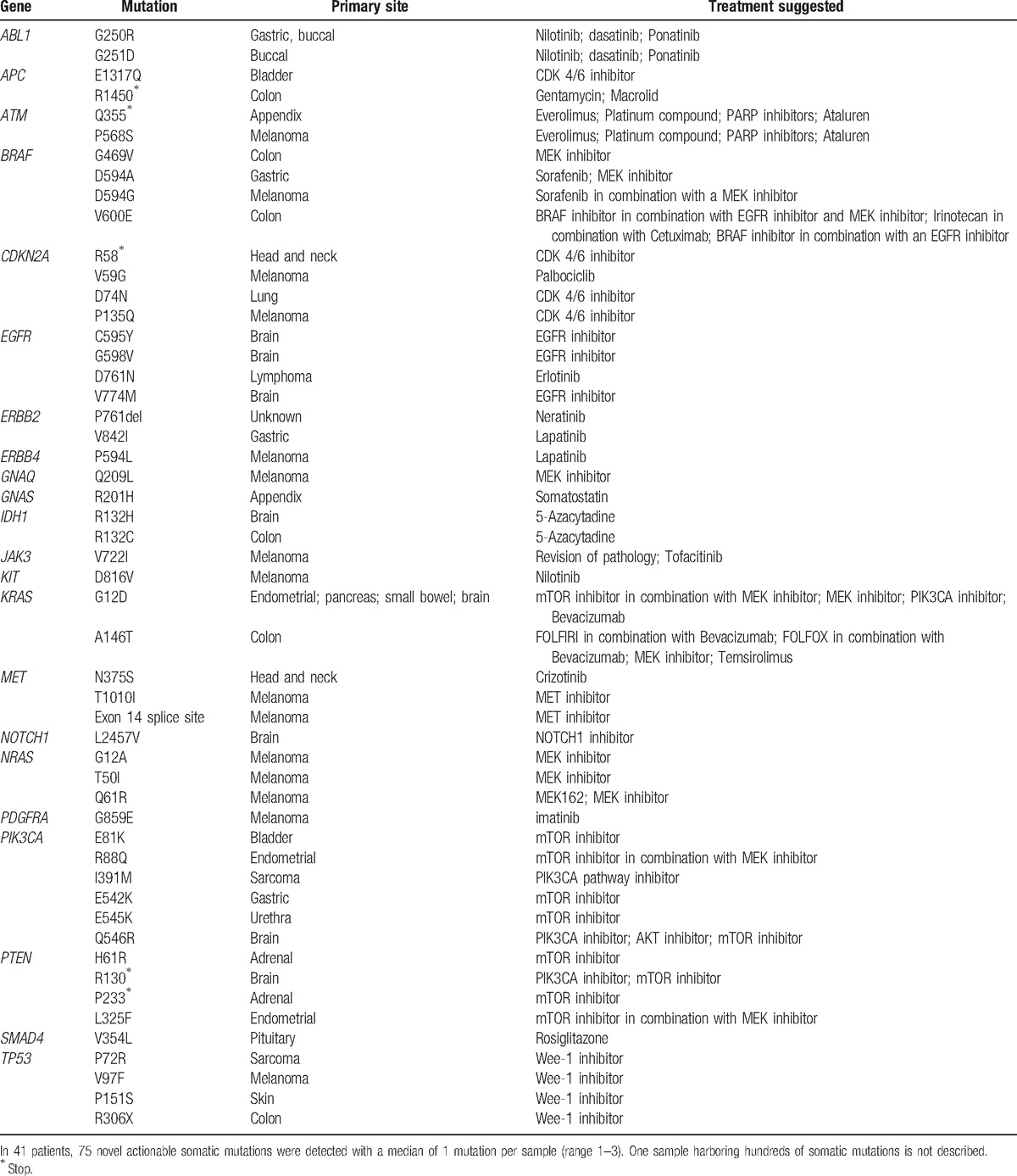
Table 3.
Clinical outcome of patients treated as recommended.
4. Discussion
This series of 67 metastatic cancer or brain tumor patients whose tumors were tested for actionable mutations demonstrates that in the majority of patients, actionable mutations can be identified. When the recommended treatment was applied, clinical benefit was achieved in a significant portion of the patients. This work has several limitations, including being conducted in a single institution, retrospective study, limited accessibility to pathway inhibitors, a small heterogeneous population, and lack of clear indication.
A proof that such a service prolongs the life of patients in a randomized prospective study was not found.[75] The extendibility of such a proof will be hard to come by, as the paradigm in oncology is shifting from large randomized trials to highly tailored small trials,[76] and following the perception that each patient's cancer is unique and genomic characterization of the tumor can have clinical significance in treating cancer patients[76] in a patient-centered research approach.[77,78] An impetus to establishing and applying this test clinically was the ever-increasing utilization of genomic tests performed by private companies. It was felt that a service that includes a validated test followed by a discussion by a multidisciplinary forum should be established.
There is limited availability to pathway inhibitors recommended by the forum, as phase I/II trials targeting molecular pathways are currently sparsely available in Israel.[79] The recommended treatment options often include treatments that may not be covered by the health insurance, and when purchased privately, may cost thousands of dollars a month.
The small patient population described is very heterogonous as to cancer site, number of treatments, and clinical statuses. It also does not represent the general patient population as these patients were able to pay for the service and were selected at the treating physician's discretion. Patient selection could have led to deference of treatments proposed as some patients were on one hand too ill to receive treatment, or on the other hand had other treatment options. The limitations of this work mirror the realty of implementing tumor biology into day-to-day clinical practice. These include, among other things, complicated issues involving ethics, drug accessibly, and clinical indication.[80]
This study highlights the growing ethical dilemmas a treating oncologist is faced with daily,[78] and questions such as whether it is ethical to offer an unproven test or treatment to patients that are suffering from end stage cancer? Is it ethical to deny a test that may decrease suffering and prolong life? As with others,[81] in our experience, it is essential to conduct a detailed discussion with the patient where the possible benefits and expected limitations are carefully reviewed prior to ordering this service.
For all patients with an actionable mutation, a clinical trial outside of Israel could be found using www.clinicaltrials.gov.[79] This option is considered not relevant by the molecular oncology forum due to the effort and suffering of advanced stage cancer patients traveling to a foreign country and living there, the very high costs and the inherently unknown clinical benefit. As another option, the concept of suitable off-trial possibilities was opted.[81] It is clear that this treatment concept is inferior to including patients in clinical trials. As molecular characterization of tumors has been democratized, increasing access to molecular inhibitors should be the next challenge of the pharmaceutical, research, and clinical community. This approach may help solve poor accrual as once uniform clinical entities are fragmented to an assortment of rare tumors with hundreds of compounds and thousands of combinations waiting to be tested in phase I/II trials.[82]
Other groups have recently published the clinical results of harnessing molecular profiling to metastatic cancer patients. A study of 1283 advanced metastatic cancer patients tested FFPE tumor tissue using targeted sequencing of hotspot regions in PIK3CA, BRAF, KRAS, NRAS, PTEN, EGFR, KIT, GNAQ and MET. Using these tests, clinical targets were found in 40% of the patients. Sixteen percent received targeted treatment with 4% of the total population achieving a clinical response. A similar group of patients who received nontargeted therapy had an inferior response rate, time to treatment failure, and overall survival.[83] In 2 studies including 109 and 423 metastatic breast cancer patients, fresh tissue biopsies were tested for amplifications and deletions using comparative genome hybridization and hot spot sequencing of AKT1 and PIK3CA. Using these tests, clinical targets were found in 50% and 46% of the patients respectively. Sixteen percent and 13% received targeted treatment; the treatment was outside a clinical trial protocol in 40% of the patients, with a total of 8% and 3% respectively achieving clinical benefit.[84,85] Another study of 11 advanced metastatic cancer patients tested fresh tissue biopsies using whole genome sequencing and whole transcriptome sequencing. Using these tests, clinical targets were found in 89% of the patients. One patient was treated according to the targets identified with a short-lived partial response.[86] Initiatives such as the AURORA trial where hundreds of metastatic breast cancer patients will be subjected to molecular characterization and treated per mutation with a pathway inhibitor[87] and the NCI-MATCH trial that aims at recruiting 2400 metastatic cancer patients who will be treated in 24 different arms based on somatic mutations identified in the tumor sample will better quantify the benefit of this approach. Molecular profiling in the NCI-MATCH is based on the Oncomine Cancer Panel assay, using AmpliSeq chemistry and the PGM sequencer. Using this assay achieved an overall sensitivity of 96.98% and 99.99% specificity in detecting mutations. High reproducibility in detecting all reportable variants was observed, with a 99.99% mean interoperator pairwise concordance.[88]
Our experience is in line with these studies, putative targets are identified in most patients, and clinical benefit is achieved in modest numbers. This study suggests that routine use of massive parallel tumor sequencing is feasible and can judiciously affect treatment decisions when coupled with multidisciplinary team based decision making. Administration of personalized therapies at earlier stages of therapy, expansion of genetic alterations examined, and availability of targeted therapies may lead to further improvement in the clinical outcome of patients.
Footnotes
Abbreviations: FFPE = formalin fixed paraffin embedded, Mb = million bases, PGM = Personal Genome Machine.
AZ and TP equality contributed to this work.
This work is supported by THE ISRAEL SCIENCE FOUNDATION (Grant No. 1985/13) and the Sharett Fund.
The authors have no conflicts of interest to disclose.
References
- [1].Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Grasso CS, Wu YM, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012;487:239–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 2012;18:382–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cortes JE, Kantarjian H, Shah NP, et al. Ponatinib in refractory Philadelphia chromosome-positive leukemias. N Engl J Med 2012;367:2075–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lin JD, Lee ST, Weng HF. An open, phase III study of lanreotide (Somatuline PR) in the treatment of acromegaly. Endocrine J 1999;46:193–8. [DOI] [PubMed] [Google Scholar]
- [10].Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385–94. [DOI] [PubMed] [Google Scholar]
- [11].Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011;364:2507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Flaherty KT, Infante JR, Daud A, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 2012;367:1694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Turner NC, Ro J, Andre F, et al. Palbociclib in hormone-receptor-positive advanced breast cancer. N Engl J Med 2015;373:209–19. [DOI] [PubMed] [Google Scholar]
- [14].Kantarjian HM, Thomas XG, Dmoszynska A, et al. Multicenter, randomized, open-label, phase III trial of decitabine versus patient choice, with physician advice, of either supportive care or low-dose cytarabine for the treatment of older patients with newly diagnosed acute myeloid leukemia. J Clin Oncol 2012;30:2670–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008;359:1757–65. [DOI] [PubMed] [Google Scholar]
- [16].Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947–57. [DOI] [PubMed] [Google Scholar]
- [17].Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med 2005;353:123–32. [DOI] [PubMed] [Google Scholar]
- [18].Douillard JY, Siena S, Cassidy J, et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol 2010;28:4697–705. [DOI] [PubMed] [Google Scholar]
- [19].Geyer CE, Forster J, Lindquist D, et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med 2006;355:2733–43. [DOI] [PubMed] [Google Scholar]
- [20].Baselga J, Cortes J, Kim SB, et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med 2012;366:109–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Verma S, Miles D, Gianni L, et al. Trastuzumab emtansine for HER2-positive advanced breast cancer. N Engl J Med 2012;367:1783–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001;344:783–92. [DOI] [PubMed] [Google Scholar]
- [23].Verstovsek S, Kantarjian H, Mesa RA, et al. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. N Engl J Med 2010;363:1117–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Demetri GD, von Mehren M, Blanke CD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med 2002;347:472–80. [DOI] [PubMed] [Google Scholar]
- [25].Flaherty KT, Robert C, Hersey P, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med 2012;367:107–14. [DOI] [PubMed] [Google Scholar]
- [26].Krueger DA, Care MM, Holland K, et al. Everolimus for subependymal giant-cell astrocytomas in tuberous sclerosis. N Engl J Med 2010;363:1801–11. [DOI] [PubMed] [Google Scholar]
- [27].Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 2012;30:863–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Von Hoff DD, LoRusso PM, Rudin CM, et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N Engl J Med 2009;361:1164–72. [DOI] [PubMed] [Google Scholar]
- [29].Hurwitz H, Fehrenbacher L, Novotny W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med 2004;350:2335–42. [DOI] [PubMed] [Google Scholar]
- [30].Van Cutsem E, Tabernero J, Lakomy R, et al. Addition of aflibercept to fluorouracil, leucovorin, and irinotecan improves survival in a phase III randomized trial in patients with metastatic colorectal cancer previously treated with an oxaliplatin-based regimen. J Clin Oncol 2012;30:3499–506. [DOI] [PubMed] [Google Scholar]
- [31].Motzer RJ, Hutson TE, Tomczak P, et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med 2007;356:115–24. [DOI] [PubMed] [Google Scholar]
- [32].Escudier B, Eisen T, Stadler WM, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med 2007;356:125–34. [DOI] [PubMed] [Google Scholar]
- [33].Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010;376:687–97. [DOI] [PubMed] [Google Scholar]
- [34].Hodi FS, Corless CL, Giobbie-Hurder A, et al. Imatinib for melanomas harboring mutationally activated or amplified KIT arising on mucosal, acral, and chronically sun-damaged skin. J Clin Oncol 2013;31:3182–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Motzer RJ, Escudier B, Oudard S, et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet 2008;372:449–56. [DOI] [PubMed] [Google Scholar]
- [36].Yao JC, Shah MH, Ito T, et al. Everolimus for advanced pancreatic neuroendocrine tumors. N Engl J Med 2011;364:514–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Baselga J, Campone M, Piccart M, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med 2012;366:520–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yoon DH, Ryu MH, Park YS, et al. Phase II study of everolimus with biomarker exploration in patients with advanced gastric cancer refractory to chemotherapy including fluoropyrimidine and platinum. Br J Cancer 2012;106:1039–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Soria JC, Baselga J, Hanna N, et al. Phase I-IIa study of BMS-690514, an EGFR, HER-2 and -4 and VEGFR-1 to -3 oral tyrosine kinase inhibitor, in patients with advanced or metastatic solid tumours. Eur J Cancer 2013;49:1815–24. [DOI] [PubMed] [Google Scholar]
- [40].Butrynski JE, D’Adamo DR, Hornick JL, et al. Crizotinib in ALK-rearranged inflammatory myofibroblastic tumor. N Engl J Med 2010;363:1727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ross JS, Ali SM, Wang K, et al. Comprehensive genomic profiling of epithelial ovarian cancer by next generation sequencing-based diagnostic assay reveals new routes to targeted therapies. Gynecol Oncol 2013;130:554–9. [DOI] [PubMed] [Google Scholar]
- [42].Von Hoff DD, Stephenson JJ, Jr, Rosen P, et al. Pilot study using molecular profiling of patients’ tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol 2010;28:4877–83. [DOI] [PubMed] [Google Scholar]
- [43].Singh RR, Patel KP, Routbort MJ, et al. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn 2013;15:607–22. [DOI] [PubMed] [Google Scholar]
- [44].Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings Bioinformatics 2013;14:178–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 2011;39:D945–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Reddy EP, Aggarwal AK. The ins and outs of bcr-abl inhibition. Genes Cancer 2012;3:447–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Schmidt L, Duh FM, Chen F, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet 1997;16:68–73. [DOI] [PubMed] [Google Scholar]
- [48].Krishnaswamy S, Kanteti R, Duke-Cohan JS, et al. Ethnic differences and functional analysis of MET mutations in lung cancer. Clin Cancer Res 2009;15:5714–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Seiwert T, Sarantopoulos J, Kallender H, et al. Phase II trial of single-agent foretinib (GSK1363089) in patients with recurrent or metastatic squamous cell carcinoma of the head and neck. Investig New Drugs 2013;31:417–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Choueiri TK, Vaishampayan U, Rosenberg JE, et al. Phase II and biomarker study of the dual MET/VEGFR2 inhibitor foretinib in patients with papillary renal cell carcinoma. J Clin Oncol 2013;31:181–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hart CD, De Boer RH. Profile of cabozantinib and its potential in the treatment of advanced medullary thyroid cancer. OncoTargets Ther 2013;6:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949–54. [DOI] [PubMed] [Google Scholar]
- [53].Suehiro Y, Wong CW, Chirieac LR, et al. Epigenetic-genetic interactions in the APC/WNT, RAS/RAF, and P53 pathways in colorectal carcinoma. Clin Cancer Res 2008;14:2560–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Dahlman KB, Xia J, Hutchinson K, et al. BRAF(L597) mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer Discovery 2012;2:791–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Bennouna J, Lang I, Valladares-Ayerbes M, et al. A Phase II, open-label, randomised study to assess the efficacy and safety of the MEK1/2 inhibitor AZD6244 (ARRY-142886) versus capecitabine monotherapy in patients with colorectal cancer who have failed one or two prior chemotherapeutic regimens. Investig New Drugs 2011;29:1021–8. [DOI] [PubMed] [Google Scholar]
- [56].Zilberberg A, Lahav L, Rosin-Arbesfeld R. Restoration of APC gene function in colorectal cancer cells by aminoglycoside- and macrolide-induced read-through of premature termination codons. Gut 2010;59:496–507. [DOI] [PubMed] [Google Scholar]
- [57].Ascierto PA, Schadendorf D, Berking C, et al. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. Lancet Oncol 2013;14:249–56. [DOI] [PubMed] [Google Scholar]
- [58].Kirkwood JM, Bastholt L, Robert C, et al. Phase II, open-label, randomized trial of the MEK1/2 inhibitor selumetinib as monotherapy versus temozolomide in patients with advanced melanoma. Clin Cancer Res 2012;18:555–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chaix A, Arcangeli ML, Lopez S, et al. KIT-D816 V oncogenic activity is controlled by the juxtamembrane docking site Y568-Y570. Oncogene 2014;33:872–81. [DOI] [PubMed] [Google Scholar]
- [60].Hochhaus A, Baccarani M, Giles FJ, et al. Nilotinib in patients with systemic mastocytosis: analysis of the phase 2, open-label, single-arm nilotinib registration study. J Cancer Res Clin Oncol 2015;141:2047–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Cho JH, Kim KM, Kwon M, et al. Nilotinib in patients with metastatic melanoma harboring KIT gene aberration. Investig New Drugs 2012;30:2008–14. [DOI] [PubMed] [Google Scholar]
- [62].Packer LM, Rana S, Hayward R, et al. Nilotinib and MEK inhibitors induce synthetic lethality through paradoxical activation of RAF in drug-resistant chronic myeloid leukemia. Cancer Cell 2011;20:715–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Nishikawa G, Sekine S, Ogawa R, et al. Frequent GNAS mutations in low-grade appendiceal mucinous neoplasms. Br J Cancer 2013;108:951–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lamberts SW, van der Lely AJ, de Herder WW, et al. Octreotide. N Engl J Med 1996;334:246–54. [DOI] [PubMed] [Google Scholar]
- [65].Yaeger R, Saltz L. BRAF mutations in colorectal cancer: clinical relevance and role in targeted therapy. J Natl Comprehensive Cancer Netw 2012;10:1456–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Douillard JY, Oliner KS, Siena S, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013;369:1023–34. [DOI] [PubMed] [Google Scholar]
- [67].Corcoran RB, Ebi H, Turke AB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discovery 2012;2:227–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012;483:100–3. [DOI] [PubMed] [Google Scholar]
- [69].Connolly K, Brungs D, Szeto E, et al. Anticancer activity of combination targeted therapy using cetuximab plus vemurafenib for refractory BRAF (V600E)-mutant metastatic colorectal carcinoma. Current Oncol 2014;21:e151–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Al-Marrawi MY, Saroya BS, Brennan MC, et al. Off-label use of cetuximab plus sorafenib and panitumumab plus regorafenib to personalize therapy for a patient with V600E BRAF-mutant metastatic colon cancer. Cancer Biol Ther 2013;14:703–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Galal KM, Khaled Z, Mourad AM. Role of cetuximab and sorafenib in treatment of metastatic colorectal cancer. Indian J Cancer 2011;48:47–54. [DOI] [PubMed] [Google Scholar]
- [72].Dai J, Kong Y, Si L, et al. Large-scale analysis of PDGFRA mutations in melanomas and evaluation of their sensitivity to tyrosine kinase inhibitors imatinib and crenolanib. Clin Cancer Res 2013;19:6935–42. [DOI] [PubMed] [Google Scholar]
- [73].Heinrich MC, Owzar K, Corless CL, et al. Correlation of kinase genotype and clinical outcome in the North American Intergroup Phase III Trial of imatinib mesylate for treatment of advanced gastrointestinal stromal tumor: CALGB 150105 Study by Cancer and Leukemia Group B and Southwest Oncology Group. J Clin Oncol 2008;26:5360–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Dileo P, Pricl S, Tamborini E, et al. Imatinib response in two GIST patients carrying two hitherto functionally uncharacterized PDGFRA mutations: an imaging, biochemical and molecular modeling study. Int J Cancer J 2011;128:983–90. [DOI] [PubMed] [Google Scholar]
- [75].Meric-Bernstam F, Farhangfar C, Mendelsohn J, et al. Building a personalized medicine infrastructure at a major cancer center. J Clin Oncol 2013;31:1849–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Garraway LA. Genomics-driven oncology: framework for an emerging paradigm. J Clin Oncol 2013;31:1806–14. [DOI] [PubMed] [Google Scholar]
- [77].Van Allen EM, Wagle N, Levy MA. Clinical analysis and interpretation of cancer genome data. J Clin Oncol 2013;31:1825–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Mendelsohn J. Personalizing oncology: perspectives and prospects. J Clin Oncol 2013;31:1904–11. [DOI] [PubMed] [Google Scholar]
- [79].Zarin DA, Tse T, Williams RJ, et al. The ClinicalTrials.gov results database—update and key issues. N Engl J Med 2011;364:852–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Gingras I, Sonnenblick A, de Azambuja E, et al. The current use and attitudes towards tumor genome sequencing in breast cancer. Sci Rep 2016;6:22517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Dienstmann R, Rodon J, Barretina J, et al. Genomic medicine frontier in human solid tumors: prospects and challenges. J Clin Oncol 2013;31:1874–84. [DOI] [PubMed] [Google Scholar]
- [82].Sleijfer S, Bogaerts J, Siu LL. Designing transformative clinical trials in the cancer genome era. J Clin Oncol 2013;31:1834–41. [DOI] [PubMed] [Google Scholar]
- [83].Tsimberidou AM, Iskander NG, Hong DS, et al. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res 2012;18:6373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Arnedos M, Scott V, Job B, et al. Array CGH and PIK3CA/AKT1 mutations to drive patients to specific targeted agents: a clinical experience in 108 patients with metastatic breast cancer. Eur J Cancer 2012;48:2293–9. [DOI] [PubMed] [Google Scholar]
- [85].Andre F, Bachelot T, Commo F, et al. Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: a multicentre, prospective trial (SAFIR01/UNICANCER). Lancet Oncol 2014;15:267–74. [DOI] [PubMed] [Google Scholar]
- [86].Weiss GJ, Liang WS, Demeure MJ, et al. A pilot study using next-generation sequencing in advanced cancers: feasibility and challenges. PLoS One 2013;8:e76438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Zardavas D, Maetens M, Irrthum A, et al. The AURORA initiative for metastatic breast cancer. Br J Cancer 2014;111:1881–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Lih CJ, Harrington RD, Sims DJ, et al. Analytical validation of the next-generation sequencing assay for a nationwide signal-finding clinical trial: molecular analysis for therapy choice clinical trial. J Mol Diagn 2017;19:313–27. [DOI] [PMC free article] [PubMed] [Google Scholar]


