Abstract
We purified the oncoprotein SnoN and found that it functions as a corepressor of the tumor suppressor p53 in the regulation of the hepatic α-fetoprotein (AFP) tumor marker gene. p53 promotes SnoN and histone deacetylase interaction at an overlapping Smad binding, p53 regulatory element (SBE/p53RE) in AFP. Comparison of wild-type and p53-null mouse liver tissue by using chromatin immunoprecipitation (ChIP) reveals that the absence of p53 protein correlates with the disappearance of SnoN at the SBE/p53RE and loss of AFP developmental repression. Treatment of AFP-expressing hepatoma cells with transforming growth factor-β1 (TGF-β1) induced SnoN transcription and Smad2 activation, concomitant with AFP repression. ChIP assays show that TGF-β1 stimulates p53, Smad4, P-Smad2 binding, and histone H3K9 deacetylation and methylation, at the SBE/p53RE. Depletion, by small interfering RNA, of SnoN and/or p53 in hepatoma cells disrupted repression of AFP transcription. These findings support a model of cooperativity between p53 and TGF-β effectors in chromatin modification and transcription repression of an oncodevelopmental tumor marker gene.
Transforming growth factor beta (TGF-β) signaling and p53 response have pleiotropic, cell type-specific effects on cell cycle progression, differentiation, and tumor suppression, which may converge toward functionally similar outcomes. Transcription activation of genes encoding proteins that mediate cell cycle arrest and/or cell death is shared by TGF-β and p53 induction but by unrelated, diverse mechanisms (54). Loss of function in these tumor suppressor pathways is common among transformed cells, cancer-derived cell lines, and tumors (5, 7; reviewed in references 50 and 83).
TGF-β signaling is mediated by an interwoven series of steps, which are highly regulated and modulated by cross talk with other signaling networks (25, 67). Briefly, the TGF-β signal received by specific serine-threonine kinase receptors is transduced through phosphorylation of ligand-specific intracellular receptor-Smad (R-Smad) proteins, interaction of R-Smads with a common Smad (Smad4), and transport of the R-Smad/common Smad complex to the nucleus. The activated Smad complex can then bind to Smad binding element (5′ AGAC 3′) (SBE)-containing target genes in association with various classes of DNA-binding transcription factors (4, 48, 75, 89). Activation of Smad target genes is autoregulated, since TGF-β stimulation induces expression of corepressors of TGF-β-responsive genes, the oncoproteins SnoN and Ski. Transcription repression is reestablished as SnoN/Ski proteins interact with DNA-bound Smad proteins, forming a heteromeric complex that can include mSin3A, NCoR, and histone deacetylase (HDAC) (1, 45, 47, 55, 73, 74, 81, 82).
SnoN is a member of a small family of proteins, premier of which is Ski, first characterized by an ability to promote oncogenic transformation of chick cells (44; reviewed in reference 46). Ski and SnoN, as well as alternatively spliced SnoN isoforms, have highly conserved domains that are sufficient for transformation and can form homo- and heterodimers (18). Their action as oncoproteins is highly overlapping, but they exhibit different regulatory functions during development. Ski, but not SnoN, is a corepressor of bone morphogenetic protein signaling (85), and mice either mutated or null for each gene have different phenotypes (6, 19, 58). Ski and SnoN also function as tumor suppressor proteins in a cell-specific manner (69, 70).
The essential role that TGF-β signaling plays in normal embryonic development is illustrated through numerous studies with mouse models, Xenopus, and zebra fish embryos (13, 14, 28, 36, 71). These systems, as well as human genetic disorders and studies with cultured cells, reveal how TGF-β functions are influenced directly and indirectly by multiple signaling pathways, including Ras, Stat, NF-κB, Wnt, and others (reviewed in references 25 and 49). More recently, a positive role for p53 function in convergence with TGF-β signaling effectors was found in transcription regulation during embryonic development and patterning in Xenopus laevis (20). Using Xenopus embryos and transfected human cells, the authors showed that p53 and TGF-β effectors cooperated to activate Mix.2 gene expression by binding to widely separated p53 and Smad DNA elements in the Mix.2 promoter.
The p53 protein itself is a nuclear target of multiple stress-activated or differentiation-associated pathways (reviewed in references 3, 5, 41, 43, and 63). Mice null for the TRP53 gene appear developmentally normal; however, further studies show that p53 levels are tightly and critically regulated during embryogenesis (39, 53). Thus, p53 may have cell-specific or developmental functions, which are masked by compensating family members (reviewed in references 17, 33, and 52). As a transcription factor, p53 binds directly to specific DNA elements to regulate expression of genes that are generally involved in maintaining genome stability and tumor suppression. Characterization of p53-regulated genes by differential, genome-wide expression and bioinformatics analyses supports p53 action as either a transcription activator or a repressor of specific sets of genes (40, 84).
Our previous studies of differentiation-associated silencing of the hepatic oncodevelopmental tumor marker gene, AFP, revealed that p53 repressed AFP transcription by sequence-specific binding to a distal regulatory element (42). AFP protein has been reported to act as an immunosuppressor, an antioxidant, and a growth-promoting factor, with aberrantly activated expression levels correlated to size and aggressiveness of liver tumors. Postnatal silencing of AFP expression is disrupted by hepatocellular carcinoma (HCC), liver regeneration, and tissue damage, which promote reentry into the cell cycle (reviewed in references 9 and 16). Reactivation of silenced AFP is also observed during chronic hepatitis B virus infection prior to onset of HCC (reviewed in references 2 and 37).
We describe here a direct, molecular link between nuclear effectors of TGF-β signaling and p53 in regulating tumor marker gene expression. A composite SBE/p53 regulatory element (SBE/p53RE), centered at −850 of the AFP gene, acts as a scaffold where p53, Smads2/4 and SnoN proteins interact to form a complex with histone modifiers. Our findings support a mechanism of SnoN/p53-targeted deacetylation alongside methylation of histone H3K9 at the AFP SBE/p53RE. Neither Smad/SnoN nor p53 binding at the SBE/p53RE is individually sufficient for transcription repression, but together cross talk between these signaling pathways mediates silencing of AFP gene expression.
MATERIALS AND METHODS
Plasmids and transfection assays.
AFP/lacZ and DelA/lacZ have been described (72). For in vitro chromatin transcription reactions, AFP/lacZ was coupled to streptavidin-coated paramagnetic beads as described previously (24). SnoN and Ski expression plasmids were the kind gift of E. Stavnezer. Hepa 1-6 murine hepatoma cells were transfected using Lipofectamine (Invitrogen, Inc.) per the manufacturer's instructions and as previously described (42).
Protein expression and cellular extracts.
Cellular extracts were prepared from HeLa, HepG2 (AFP positive; ATCC catalog number HB-8065) and Hepa 1-6 (AFP positive; ATCC catalog number CRL 1830) and adult mouse liver as previously described (26, 32) with minor modifications (42). Xenopus egg extract high speed supernatant was prepared exactly as described previously (24). Recombinant histidine-tagged p53Δ30 (38) and HBx proteins (34) were prepared as previously described (56).
Solid-phase protein purification and MALDI analysis.
Solid-phase DNA oligomers were generated by annealing the 5′-biotinylated p53 regulatory element (5′-Bio GATCCTTAGCAAACATGTCTGGACCTCTAGAC) or −1007 (5′-BioGATCCAATATCCTCTTGAC) (Invitrogen, Inc.) to its complementary strand prior to coupling to streptavidin-coated paramagnetic beads (Dynal). Single-copy and ligated multicopy oligomers gave similar results (Fig. 1A). Extracts were precleared with 150 ng of nonspecific bead-DNA for 20 min at 4°C prior to a 30-min incubation with specific bead-DNA at 25°C. Approximately 200 ng of solid-phase oligomeric bead DNA was washed one time in 1× phosphate-buffered saline-1% NP-40 and equilibrated in extract buffer prior to addition to binding reactions. Proteins copurified with recombinant p53 protein from adult mouse liver extract on solid-phase oligomers were washed twice in 1× phosphate-buffered saline-1% NP-40 and eluted one time in a solution containing 100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2, 1 mM dithiothreitol, followed by a second elution in a solution containing 5 M urea, 10 mM Tris (pH 8.0), 100 mM NaH2PO4. Proteins were separated by denaturing gel electrophoresis and visualized by silver staining. Following staining, specific protein bands were cut out and digested in gel; peptides were purified as described elsewhere (http://donatello.ucsf.edu/ingel.html). Purified peptides were analyzed using a Voyager DE-PRO matrix-assisted laser desorption ionization (MALDI)-time of flight mass spectrometer (Applied Biosystems) in reflector mode. Spectra were analyzed by using the Protein Prospector MS-Fit algorithm with the NCBInr database (http://prospector.ucsf.edu).
FIG. 1.
A TGF-β effector protein is purified from hepatic cells in the presence of the p53 protein. (A) Purification of SnoN protein. Solid-phase DNA oligomers, either biotinylated, 31-bp p53RE single-copy (lanes 3 to 6), ligated p53RE multimers (lanes 7 to 10), or single-copy, −1007 control (lanes 11 to 14) sequences, were incubated with 100 μg of total protein of adult mouse liver nuclear extract (ML, input extract; lane 2), in the presence of recombinant p53 (p53 alone; lane 1, recombinant protein stabilized by addition of BSA). Following binding, bead-DNA-protein complexes were washed, eluted, and then separated by denaturing gel electrophoresis alongside extract proteins not binding to DNA (unbound, lanes 3, 7, and 11). The gel was silver stained; eluted p53 (dagger, lanes 6 and 9) and a protein enriched in the presence of p53 (asterisk, lanes 5 and 9) were analyzed by MALDI mass spectrometry. (B) In vivo ChIP analysis of wild-type and p53-null mouse liver tissue. PCR analysis of antibody-precipitated genomic fragments was conducted with primers amplifying the region of the SBE/p53RE. Sonicated fragments of formaldehyde-cross-linked chromatin from 2-month-old wild-type (WT) and p53-null mouse liver tissue were antibody precipitated with tetra-acetylated histone H4 antibody (AcH4), Smad4, p53 (Ab-1), SnoN, P-Smad2, or control IgG. (C) Quantifying in vivo ChIP of Smad4 and P-Smad2 binding to the SBE/p53RE and distal genomic sites in WT and p53-null mouse liver tissue. Chromatin lysates were serially diluted (Input) and PCR amplified as described above to determine a linear range of amplification for all lysates. Antibody-precipitated fragments of DNA bound to Smad4 and SnoN were quantified by comparison to this range of values (% Bound). (D) Semiquantitative RT-PCR analysis of AFP RNA. RNA was isolated from 1- and 2-month-old WT and p53-null mouse liver tissue and analyzed in parallel for determination of GAPDH control RNA levels and AFP RNA levels.
TGF-β treatment, RNA isolation, and analysis.
Hepa 1-6 cells were treated with TGF-β1 ligand (2 ng/ml; R&D Systems) or vehicle control (4 mM HCl containing 0.1% bovine serum albumin [BSA]) for 56 h. RNA was isolated at the indicated time points, using the TRIzol reagent (Invitrogen) per the manufacturer's instructions. Northern analysis was performed as described by Carter and colleagues (11). The probe for AFP was generated by reverse transcriptase PCR (RT-PCR) by using the QIAGEN Omniscript RT-KIT. PCR was performed using the following primer set: AFP 3′ (+162), 5′ GGAAAATAGCTCCCAAGTCACAAAGC 3′; and AFP start site (+4), 5′ CCCACTTCCAGCACTGCCTGCGG 3′. The probes for SnoN, actin, and cyclophilin RNAs were generated by random-primed, 32P labeling of their respective cDNA clones (Promega). RNA values were quantified using the IMAGE QUANT software V5.2 (Molecular Dynamics). AFP and SnoN mRNA values were normalized against those of actin or cyclophilin.
For semiquantitative RT-PCR analysis, total RNA from 1-month-old and 2-month-old C57BL/6J wild-type and p53−/− mouse liver tissue was extracted with TRIzol reagent (Invitrogen), and the cDNA was obtained by reverse transcription of 2 μg of RNA (SuperScript First-Strand synthesis system; Invitrogen). Primers for AFP and GAPDH primers were as follows: AFP (forward primer, 5′-CCCACTTCCAGCACTGCCTGCGG-3′; reverse primer, 5′-GGCTGCAGCAGCCTGAGAGTC-3′), GAPDH (forward primer, 5′-TTCACCACCATGGAGAAGGC-3′; reverse primer, 5′-GGCATGGACTGTGGTCATGA-3′).
PCR products were amplified for 35 cycles, separated on 6% polyacrylamide gels, and stained with Sybr Green (Sigma).
Immunoblotting.
Immunoblotting was performed as previously described (42). The primary antibodies used are as follows: p53 (Oncogene Ab1 and Santa Cruz SC-240), SnoN (SC-9141; Santa Cruz), Phospho-Smad2 (07-392; Upstate), Smad4 (06-693; Upstate) (SC-7966; Santa Cruz), AFP (SC C-19; Santa Cruz), and actin (A5316; Sigma).
ChIP.
Hepa 1-6 cells were treated with TGF-β1 ligand at a concentration of 2 ng/ml or vehicle only for 24 h. Following treatment, in vivo chromatin immunoprecipitation (ChIP) assays of cultured cells were performed as described previously (12) with the following modifications: the cell lysate was sonicated eight times with 15-s bursts to yield input DNA enriched for fragments of less than 500 bp in size. The following antibodies were used for the immunoprecipitation reactions: p53 (oncogene Ab1), SnoN (SC9141; Santa Cruz), P-Smad2 (07-392; Upstate), Smad4 (06-693; Upstate), normal sheep immunoglobulin G (IgG) (12-369; Upstate), AcH4 (06-860, ChIP grade; Upstate), AcH3K9 (06-942; Upstate), and DimethylH3K9 (07-212, ChIP grade; Upstate). Primers specific for detection of the SBE/p53RE regions amplified a region from −887 to −762 of the AFP gene, and those specific for the start site of transcription were specific for a region from −82 to +94.
Liver tissue was isolated, cross-linked, and processed for ChIP analyses as described previously (86; also see http://genomecenter.ucdavis.edu/farnham/farnham/protocols/tissues.html). Modifications to this protocol included preclearing of the chromatin lysate with IgG-bound beads and recovery of antibody precipitates with protein A beads (12, 15), as well as digestion with MNase (4 units/μl; Worthington Biochemicals) after sonication to generate fragments less than 400 to 500 bp in length, if needed, as determined empirically by agarose gel electrophoresis of fragmented chromatin samples.
To analyze specific antibody-bound DNA fractions, quantitative PCRs were performed using Taq polymerase (Continental Labs), and primers were generated to detect the AFP SBE/p53RE region of −887 to −762 (forward primer, TAAAAAATAAACTCAACTACATATG; reverse primer, GAAAACTTTTAAAACTTCCC). To ensure that the PCRs were in linear range, several serial dilutions of the input DNA and two dilutions of each of the bound DNA fractions were used. In addition, PCR cycles ranging in number from 20 to 26 (hepatoma) or 23 to 27 (liver) were performed for each bound DNA fraction and input. PCR products were separated on 8% polyacrylamide gels and stained with Sybr Green (Sigma). DNA bands were visualized and quantified as described above. The percent input bound was calculated by dividing the value of the bound fraction by the values of each of the input dilutions in the linear range and multiplying by the respective dilution factor of the input. Quantifications from multiple individual experiments were taken, and the average values were plotted graphically by using XCEL software (V10). Error bars represent the standard errors of the means. In vitro ChIP assays were performed on in vitro chromatin-assembled DNA templates exactly as previously described (57). Results were quantified using ImageQuant analysis (Molecular Dynamic). Values are expressed as percent bound normalized to IgG control.
RNA interference assay.
Small interfering RNA (siRNA) constructs targeting murine SnoN and murine p53 were designed as described at katahdin.cshl.org (9331/RNAi/html/docs/Web_version_of_PCR_strategy1.pdf). Briefly, constructs which encode short-hairpin RNAs (shRNAs) transcribed from the U6 promoter were generated by PCR, using a pGEM U6 promoter template as previously described (35). The amplification reaction was carried by using a 5′ primer corresponding to the SP6 site of the U6 promoter cassette and a long 3′ primer complementary to the 3′ end of the promoter followed by sequences encoding SnoN and p53 shRNA and a polymerase III termination sequence. shRNA sequences encoding inverted repeats of 17 to 19 bp separated by a 9-nucleotide spacer were designed by using the siRNA target finder (http://www.ambion.com/techlib/misc/siRNA_finder.html). The most effective target shRNAs corresponded to nucleotides 1232 to 1250 of the murine p53 cDNA and 761 to 779 of the murine SnoN cDNA. The resulting PCR products were cloned directly into the pGEM-T Easy Vector (Promega) as per the manufacturer's instructions. Transient transfections of the shRNAs were carried out using Lipofectamine 2000 (Invitrogen) as recommended by the manufacturer.
HepG2 extract immunodepletion and chromatin transcription.
HepG2 whole-cell extract was immunodepleted by using SnoN antibody (Upstate) cross-linked to protein A agarose beads (Pierce). Extract was incubated with antibody beads for 1 h at 4°C. The effectiveness of the immunodepletion was judged by immunoblotting 40 μg (input) of control HepG2 extract protein alongside an equal amount of depleted extract protein (supernatant) and the boiled, SnoN antibody and IgG beads (pellet) used in depletions. Solid-phase AFP DNA templates were incubated with control or SnoN-immunodepleted HepG2 extract in the presence or absence of recombinant p53 protein for 20 min prior to chromatin assembly and in vitro transcription (24).
HDAC assay.
HDAC assays were performed exactly as described in the work of Carmen et al. (10).
RESULTS
Purification of SnoN protein.
Our previous studies showed that p53 mediated repression of AFP gene expression by binding specifically to a p53 regulatory element (p53RE) within the 5′ distal regulatory region of AFP (−860 to −830), but transcription repression occurred only in liver-derived cells in vivo or extracts of liver-derived cells in transcription assays in vitro (42, 56, 57). These results led us to isolate potential corepressors that act with p53 to repress transcription. We incubated paramagnetic bead-bound p53RE or control oligomers (−1007 to −977 of AFP) in transcription extracts prepared from adult mouse liver nuclei (32) (Fig. 1A, ML) in the presence of recombinant p53 protein. We analyzed proteins eluted sequentially from the p53/p53RE-protein complexes (E1 and E2) by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and silver staining of the gels. One protein band doublet, which was p53RE specific (compare lanes 5 and 9 to 13), migrated between 68 and 94 kDa (lanes 5 and 9; see also supplemental Fig. S1). This protein was absent from control oligonucleotide incubations, which also did not bind p53 (Fig. 1A, lanes 12 to 14), but interacted with the p53RE oligonucleotides in a p53-dependent and tissue-specific manner (supplemental Fig. S1A and B).
The p53RE-specific protein band was excised, treated with trypsin, and subjected to MALDI- time-of-flight mass spectrophotometry analysis. Spectra were analyzed using the Protein Prospector MS-Fit algorithm against the NCBInr database, which revealed the likely identity of the protein as oncoprotein/corepressor SnoN (four peptides within 0.01 to 0.06 Da of the computed mass). TGF-β signaling is autoregulated by induced expression of the SnoN protein, which is targeted to TGF-β-regulated genes by interaction with DNA-bound Smad proteins (reviewed in reference 45). This identification of SnoN was surprising, since it suggested that the p53 and TGF-β pathways might intersect at a single, overlapping regulatory element to promote transcription repression, which has never been reported previously.
Examination of the AFP p53RE site revealed tandem, consensus SBEs. The SBEs are defined by a minimal core (5′ AGAC 3′) amid varied sequence representation and bind Smad2/3/4 proteins (and subsets thereof) only in the presence of other transcription factors that serve a stabilizing and/or targeting function for DNA binding (reviewed in reference 67). The p53RE, centered at −850 relative to the AFP start site of transcription, is now referred to as an SBE/p53RE to reflect the presence of overlapping p53 and putative Smad protein binding sites (Fig. 2A).
FIG. 2.
SnoN, but not Ski, mediates repression of AFP expression. (A) SnoN-mediated transcription repression is compromised by disruption of p53 DNA binding. (Top) Hepa 1-6 hepatoma cells were transfected with AFP/lacZ (wild-type sequence for p53 binding) or DelA/lacZ (mutated for p53 binding) reporter constructs (500 ng/plate) along with the indicated expression vectors (p53 and SnoN; 500 ng/plate [each]). Each plate was also cotransfected with the pGL2 luciferase vector (50 ng/plate) to standardize and control for transfection efficiency. Expression levels relative to baseline are indicated for AFP/lacZ (gray) and DelA/lacZ (black). Cotransfected proteins exhibited equivalent levels of protein expression by Western blot analysis (data not shown). (Bottom) The p53/HNF-3 (FoxA) regulatory element centered at −850 in the AFP promoter (42) overlaps with consensus SBEs (89). The DelA/lacZ binding element has a 10-bp deletion within the 5′ p53 half site (shaded) and point mutations in the 3′ half site. (B) Ski activates AFP expression in the presence of p53. Identical transient transfection analyses were performed with the AFP/lacZ reporter and coexpressed p53, SnoN, and c-Ski constructs, as indicated. Expressed Ski protein repressed AFP only slightly and activated expression in the presence of p53. β-gal, β-galactosidase; luc, luciferase.
Endogenous p53, Smad, and SnoN bind to chromatin in vivo.
We performed ChIP analyses of normal mouse liver and p53-null mouse liver tissues isolated from 2-month-old mice to assess potential interactions of p53, Smad, and SnoN proteins with AFP chromatin in vivo (Fig. 1B and C). AFP expression is developmentally repressed and undetectable by Northern blot analysis within 2 to 3 weeks after birth (77). Using semiquantitative RT-PCR analysis of RNAs isolated from normal mouse liver tissue at 1 and 2 months of age, AFP expression is detectable at 1 month but fully repressed within 2 months. However, in p53-null mice, AFP expression is maintained even at 2 months of age (Fig. 1D). Antibody-mediated precipitation of fragmented chromatin from formaldehyde-cross-linked mouse liver lysates and PCR amplification (ChIP) showed that AFP repression in normal mouse liver (Fig. 1B, 2-month WT) correlated with the presence of chromatin-bound Smad4, phosphorylated Smad2 (P-Smad2), p53, and SnoN proteins at the SBE/p53RE in vivo. ChIP analyses of endogenous protein binding at the AFP SBE/p53RE in p53−/− mice suggest that recruitment of the SnoN protein to the SBE/p53RE site within the developmental repressor region of AFP is p53 dependent (Fig. 1B, 2-month p53-null). In the p53-null (−/−) mouse liver we see an absence of the SnoN protein concomitant with a lack of p53 binding, while binding of Smad4 and P-Smad2 is maintained.
To quantify Smad4 and SnoN chromatin binding at the SBE/p53RE, antibody-precipitated samples were PCR amplified within a linear range of detection and compared with multiple concentrations of input lysate (Fig. 1C). The percent SnoN protein binding to the SBE/p53RE is increased almost sevenfold in the presence of p53 (2-month wild type) and is equivalent to bound Smad4 protein levels. However, in the absence of p53 (2-month p53-null), very little chromatin-bound SnoN protein is detectable. Smad4 and P-Smad2 binding (Fig. 1B) is observed even in the absence of p53, although the levels of bound Smad4 are decreased twofold. A number of controls, including PCR amplification of AFP regulatory sites distal to the SBE/p53RE (AFP start) of sequence within the same genomic locus (albumin) and at separate chromosomal sites (Brn-3b and GAPDH genes) affirmed the specificity of ChIP analyses (Fig. 1C).
p53 and TGF-β nuclear effectors act through a composite DNA regulatory site.
We performed transient-transfection studies to establish a direct, functional link between SnoN and regulation of AFP expression (Fig. 2A). An AFP reporter construct with approximately 3.8 kb of upstream AFP regulatory sequences fused to a bacterial β-galactosidase lacZ gene (AFP/lacZ, gift of B. Spear) was introduced into mouse hepatoma (Hepa 1-6) cells, which express p53 protein that can be stress activated (42). The baseline expression driven by AFP regulatory sequences was repressed more than fourfold when p53 protein was coexpressed. Transfections with DelA/lacZ, which harbors a deletion and altered bases within the p53-binding site (Fig. 2A, sequence: deletion in shaded area), confirmed our previously published finding that p53 represses AFP transcription by binding to the SBE/p53RE (42).
Overexpression of SnoN protein repressed the AFP-driven lacZ reporter construct more than p53 (10-fold compared with 4-fold, respectively). Introduction of both exogenous p53 and SnoN expression induced slightly more repression (12-fold). The DelA/lacZ construct is mutated in a way that eliminates the 5′SBE while the 3′SBE remains intact. To assess if endogenous p53 contributed to SnoN-mediated repression of AFP, we cotransfected SnoN and the DelA/lacZ reporter, mutated for p53 binding, and saw an 87% loss in SnoN-mediated repression (1.3-fold versus 10-fold).
We compared SnoN and Ski function in regulation of hepatoma-specific AFP expression by transient transfection of each in Hepa 1-6 cells (Fig. 2B). Unlike the case with SnoN, overexpression of Ski had little effect on AFP reporter expression and, surprisingly, reversed p53-mediated repression rather than increasing it. Expression of all three proteins (p53, Ski, and SnoN) to equal levels (data not shown) inhibited the 5-fold activation by p53 and Ski but did not repress AFP expression beyond the minor amount seen with Ski alone. We have not determined the reasons behind opposing Ski and SnoN modulation of p53-regulated transcription. We have focused here on understanding the mechanisms of SnoN-specific roles in p53-mediated transcription repression.
Regulation of endogenous AFP expression by TGF-β1 ligand.
We treated AFP-expressing, Hepa 1-6 hepatoma cells with increasing concentrations of TGF-β1 ligand or its vehicle and determined endogenous AFP, p53, Smad4, and P-Smad2 protein levels by immunoblotting of cell lysates (Fig. 3A). This series of treatments was conducted after serum starvation of Hepa 1-6 cells for 24 h, which induced p53 protein levels without repressing AFP (compare lanes 1 and 2). Increasing amounts of TGF-β1 ligand added to the cell culture medium maintained activated p53 levels and promoted AFP repression, concomitant with the appearance of P-Smad2 protein (lanes 3 to 7). Available antibodies for Smad3 protein cross-reacted with other Smad proteins; therefore, we restricted our analysis to P-Smad2, which could be detected specifically. Addition of vehicle only (lanes 8 to 12) did not induce P-Smad2 and had no effect on AFP protein levels. Although p53 is activated by serum starvation of the cells (lane 2), AFP repression is not observed in the absence of TGF-β ligand (lanes 8 to 12).
FIG. 3.
TGF-β represses endogenous AFP levels in hepatoma cells. (A) TGF-β and p53 repress AFP. Serum-starved Hepa 1-6 cells were treated with increasing concentrations of TGF-β1 (2 to 20 ng/ml of medium, lanes 3 to 7), vehicle (lanes 8 to 12), or no additions (lanes 1 and 2) and harvested at 0 h (lane 1) or 24 h (lanes 2 to 12) posttreatment. Whole-cell lysates were prepared from these cells, and immunoblot analyses were performed to analyze the effects of TGF-β1 treatment on AFP, p53, Smad4, and P-Smad2 protein levels. (B) AFP is a downstream physiological target of TGF-β. Hepa 1-6 cells were treated with 2 ng of TGF-β1/ml or vehicle control and harvested at the indicated time points posttreatment (0 to 56 h). Total RNA was harvested, and Northern blot analysis was performed to examine the levels of AFP and SnoN mRNA. The graph represents AFP (solid line) and SnoN (broken line) RNA levels in cells treated with ligand relative to those that received only the vehicle, each normalized to an actin RNA loading control. (C) TGF-β induces SnoN protein degradation, Smad2 protein phosphorylation, and a decrease in AFP. Hepa 1-6 cell lysates were treated with 4 ng of TGF-β1 ligand/ml or vehicle (V) only (as in panel B) over a 24-h interval and harvested at the indicated times for preparation of nuclear and cytoplasmic extracts. Immunoblot analysis of nuclear extracts was performed to determine SnoN, P-Smad2, p53, and actin protein levels. AFP levels were determined in cytoplasmic extracts of the same cells.
We set up a timed exposure of Hepa 1-6 cells to TGF-β1 ligand and analyzed the levels of stable AFP and SnoN RNA populations by Northern blotting (Fig. 3B). TGF-β1 activated SnoN expression within 2 h, as previously reported (73). Concomitant with this increase in SnoN RNA levels, AFP gene expression was measurably repressed within 2 h and decreased 10-fold within 24 h. Continued treatment beyond 24 h increased AFP repression and SnoN expression.
Corepressors of TGF-β signaling, Ski and SnoN proteins, are found at high levels in a number of tumors and tumor-derived cell lines, such as those for human melanoma, breast cancer, and esophageal cancer (51, 60, 74). Comparison of cell extract proteins isolated from untreated (0 h) and vehicle-treated (24 h) Hepa 1-6 cells by immunoblotting confirmed that SnoN is also abundant in our tumor-derived, cultured hepatoma cells (Fig. 3C). Addition of TGF-β1 ligand for 2 h induced phosphorylation of the Smad2 protein (P-Smad2) and, as previously reported, degradation of the SnoN protein (reviewed in reference 67). Continued TGF-β1 treatment led to a recovery in SnoN protein levels in parallel with SnoN RNA expression (Fig. 3B). Only when the nuclear SnoN, P-Smad2, and p53 proteins were detectable, 24 h after addition of TGF-β1 ligand, was there a noticeable decrease in secreted AFP protein present in cytoplasmic extracts of these cells (Fig. 3C).
p53 and TGF-β effectors bind AFP chromatin at the SBE/p53RE in vivo.
We determined whether endogenous p53, Smad4, P-Smad2, and SnoN proteins were associated with chromatin at the SBE/p53RE when AFP is maximally repressed by TGF-β1 (24-h time point). ChIP assays were performed with mouse Hepa 1-6 cells, which were exposed to ligand or vehicle, cross-linked with formaldehyde, and processed to yield sonicated chromatin fragments of 500 bp or less, in association with bound proteins. A graphic summary of multiple ChIP analyses of three to four independently isolated chromatin lysates, vehicle and ligand treated in parallel, is shown (Fig. 4A). After reversal of cross-links, PCR-amplified, antibody-precipitated DNA was quantified by comparison to input DNA populations in the linear range of analysis, as shown in Fig. 1C. Representative PCR results for all proteins in both vehicle- and ligand-treated cells are shown for two concentrations of bound chromatin fragments and a range of input values (supplemental Fig. S2).
FIG. 4.
Downstream effectors of TGF-β and p53 are recruited to SBE/p53RE in vivo and effect chromatin modifications in response to TGF-β treatment. (A) Chromatin immunoprecipitation of proteins binding to the AFP SBE/p53RE. ChIP assays of SBE/p53RE were performed with Hepa 1-6 cells treated with a 2-ng/ml concentration of TGF-β1 (black bars) or vehicle control (gray bars) for 24 h. Immunoprecipitation of formaldehyde-cross-linked lysate was performed using 1.5 μg of antibody specific for p53, 3 μg of SnoN, 5 μg of Smad4, and 5 μl of P-Smad2 antibodies followed by PCR using oligonucleotides specific for the SBE/p53RE. As negative controls, lysis buffer alone was added to the immunoprecipitation or lysate was incubated with 5 μg of anti-IgG (see supplementary Fig. S2). PCRs using oligonucleotides specific for the AFP start site were also performed as an additional test for antibody specificity. Graphs indicate the average values of percent input bound calculated from multiple experiments and the standard error of the mean. n-fold changes of ligand versus vehicle control are shown below each data set. (B) Deacetylation and concomitant methylation of histone H3 lysine 9 is targeted at SBE/p53RE in response to TGF-β signaling. ChIP assays were performed as described above, except that 5 μg of anti-AcH4, 5 μg of anti-AcH3K9, and 5 μl of anti-diMetH3K9 antibodies were used for ChIP of formaldehyde-cross-linked chromatin lysates. Graphs indicate the average values of percent input bound calculated from multiple experiments and the standard errors of the means. n-fold changes are shown below each data set.
We found that TGF-β ligand treatment of Hepa 1-6 cells induced binding of Smad4, P-Smad2, p53, and SnoN proteins to AFP chromatin at the SBE/p53RE. P-Smad2 protein showed a 5.4-fold-increase in binding to this site, which was roughly equivalent to the increase observed in bound p53 and SnoN proteins (fourfold and sevenfold, respectively) (Fig. 4A). Surprisingly, Smad4 protein showed an even greater level of increased binding than these factors, though its chromatin interaction was marginally p53 dependent in normal liver (Fig. 1C). Both Smad4 and SnoN are readily detectable as soluble proteins in untreated Hepa 1-6 cell lysates (Fig. 3) but bind to chromatin only when Smad2 is activated by phosphorylation.
TGF-β ligand effects chromatin modification at the site of p53, SnoN, and Smad protein binding.
A global view of transcription repression is that it is associated with histone deacetylation, while activation correlates with histone acetylation. More-detailed analyses of multiple genes under various conditions reveal that specific posttranslational marks are present within histone N-terminal tails as well as the histone core region. Unique combinations of these posttranslational modifications establish local and global patterns of chromatin modifications that may dictate downstream functions (62, 79). Analysis of histone modifications present at the SBE/p53RE within cells treated with TGF-β1 ligand or vehicle revealed only little change in histone H4 acetylation with use of an antibody raised against multiple acetylated lysine residues within the amino-terminal tail (AcH4, possible acetylation sites K5, K8, K12 and K16) (Fig. 4B). However, there was a fivefold decrease in histone H3 acetylation at lysine 9 (AcH3K9) with 24 h of TGF-β1 ligand treatment; and, interestingly, a general marker of gene repression, dimethylation of H3K9 (diMetH3K9), increased 2.5-fold in parallel with decreased acetylation at H3K9. Thus, acetylation levels of histone H4 were unaltered but specific repression-associated, individual lysine modifications at histone H3 were observed. These findings compare well with those of in vitro chromatin analyses, which also showed minimal effects on histone H4 acetylation levels and greater consequences for histone H3 modification in the presence of p53 (see below).
Depletion of SnoN or p53 disrupts TGFβ-mediated repression of AFP expression.
We performed siRNA-targeted depletion of SnoN and p53 in Hepa 1-6 cells to test our hypothesis that each protein is essential for repression of AFP. We depleted endogenous SnoN, p53, or both by transfection of plasmids that express short, hairpin double-stranded-RNA targets, which are homologous to coding sequences of each target gene and induce RNA degradation by formation of siRNA and activation of the dicer pathway (reviewed in references 64, 65, and 66). Three target sequences for each gene were tested by transfection into untreated (SnoN) and actinomycin-D-activated (p53) Hepa 1-6 cells (Fig. 5A). A subset of these siRNA constructs for SnoN and p53 was effective in reducing the protein concentration of the specific target: SnoN protein was reduced 7-fold and p53 decreased 2.8-fold, each by their respective target 1 shRNA construct. Immunoblotting with an actin-specific antibody showed that there were no measurable, nonspecific effects on protein expression.
FIG. 5.
Both SnoN and p53 are essential for TGF-β-induced repression of AFP transcription. (A) Depletion of SnoN and p53 by siRNA methodology. Double-stranded oligomers homologous to specific sites within the coding regions of SnoN and p53, respectively, were cloned into U6 promoter-containing vectors and introduced by transfection of Hepa 1-6 cells. Three targets sequences for each gene were tested, each with various degrees of success in knockdown of protein expression. Left panel: transfection was performed without short, hairpin RNA-expressing constructs (shRNA; mock), in duplicate (a and b) with target 1 and separately with targets 2 and 3. Immunoblotting for SnoN revealed that targets 1 and 3 were most effective in knockdown of SnoN expression. Target 2 oligonucleotides contained sequence mismatches for the SnoN target region. Actin was analyzed as a control for nonspecific effects on total protein. Right panel: analysis of p53 knockdown was conducted on lysates of Hepa 1-6 cells treated with actinomycin D to activate p53 protein as previously described (42). Transfections of shRNA constructs were performed as described above. Lysate of actinomycin D-treated cells that were not transfected are analyzed in the lane labeled as “no siRNA. ” (B) Ligand-mediated repression of AFP transcription requires p53 and SnoN. Plasmid constructs capable of inducing siRNA for SnoN or p53 (target 1 constructs for each) were introduced separately or together into Hepa 1-6 cells by transient transfection. A vector specific for siRNA-mediated knockdown of firefly luciferase was used as a nonspecific, target control for RNA depletion. The “no siRNA” cells were not transfected. RNA was isolated from Hepa 1-6 cells, all of which were treated with TGFβ-1 ligand (2 ng/μl) or vehicle alone for 24 h, following a 24-h recovery period after transfection. Levels of AFP RNA were determined by Northern blot analysis and comparison to a cyclophilin RNA loading control. Left panel: a graph summarizing the results of four siRNA experiments; RNA levels were determined by Northern blot as described above. Error bars indicate standard deviation. Right panel: representative Northern blot of AFP and cyclophilin RNA isolated from vehicle only (V) or TGF-β1 ligand-treated (L) cells, which received siRNA constructs as indicated. (C) Immunoblot analysis of siRNA-treated nuclear extracts. SiRNA and ligand or vehicle-only conditions were exactly as shown in panel B and as described above. Actin protein analysis serves as a loading control and a control for nonspecific effects of siRNA treatment.
The effects that knockdown of SnoN, p53 or the two together have on AFP repression were assessed by RNA isolation and Northern blot analysis of AFP RNA levels, as well as a cyclophilin RNA loading control (Fig. 5B). Hepa 1-6 cells were transfected with control and target-specific siRNA constructs (target 1 clones for SnoN and p53, respectively). Twenty-four hours posttransfection, these cells were treated with ligand or vehicle only for 24 h prior to RNA and cell extract isolation. The level of specific knockdown was assessed by immunoblotting of siRNA-treated and control nuclear extracts (Fig. 5C). TGF-β1 ligand effectively repressed AFP transcription levels in the absence of siRNA treatment, as well as in control, luciferase siRNA-targeted cells (Fig. 5B). Knockdown of either p53 or SnoN disrupted repression of AFP expression by TGF-β1. Interestingly, depletion of both SnoN and p53 proteins also disrupted AFP repression but did not increase this loss in an additive manner. This suggests that the two tumor suppressor pathways may act cooperatively in ligand-mediated repression of AFP.
p53 recruits histone deacetylase activity to the SBE/p53RE.
Linear DNA coupled to paramagnetic beads can be assembled into chromatin in vitro by addition of X. laevis egg extract (Fig. 6A) (21, 22). Assembled chromatin can then be analyzed at the structural level or for transcription function in vitro. We showed previously that AFP DNA assembled into chromatin transcribes with tissue and developmental specificity in vitro (21, 23, 24). Addition of protein extracts isolated from AFP-expressing cells (e.g., newborn mouse liver, HepG2, or Hepa 1-6) during chromatin assembly establishes active AFP chromatin. However, extracts of cells that do not express AFP (e.g., HeLa or adult mouse liver) or buffer only, added during chromatin assembly, establish chromatin that is repressed for transcription.
FIG. 6.
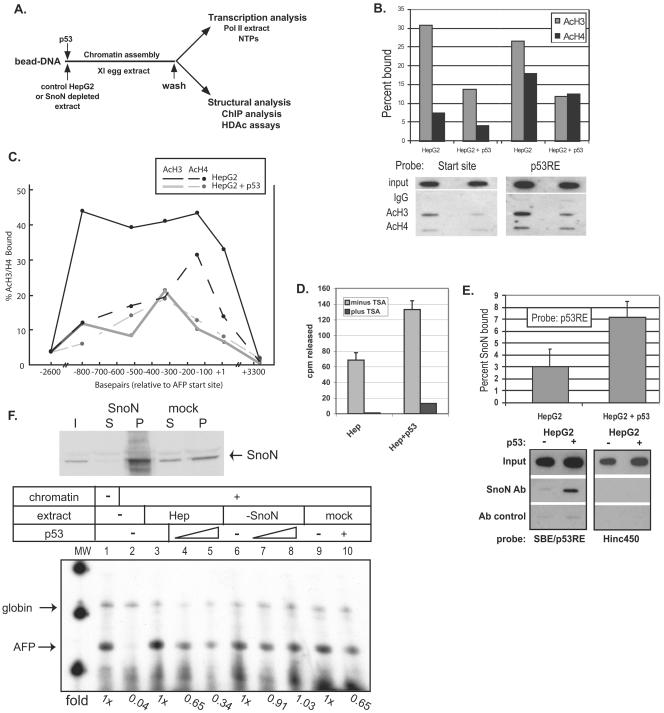
p53 targets histone deacetylation to the SBE/p53RE and mediates SnoN-dependent repression of AFP transcription. (A) Diagram of in vitro chromatin assembly and analysis. AFP bead DNA is preincubated with protein extracts and/or recombinant p53 protein prior to chromatin assembly over a 1-h incubation in X. laevis (Xl) egg extract. Assembled chromatin bead DNA is then washed in an isotonic buffer prior to functional assay by transcription in HeLa extract or structural analysis by ChIP and HDAC assays. (B and C) p53 triggers deacetylation of local and distal chromatin. ChIP assays (two to three independent assays for each data point) were performed in vitro on AFP/lacZ DNA, which was chromatin assembled in the presence of HepG2 whole-cell extract or HepG2 plus 500 ng of activated p53. DNA was purified from protein-chromatin immunoprecipitates and analyzed for percent SBE/p53RE and transcription start site DNA bound to acetylated histone H3 (AcH3) or histone H4 (AcH4) by Southern blotting with labeled (B) SBE/p53RE or start site (+4 to +26) oligonucleotides or (C) across the AFP templates with oligonucleotides specific for +10/+32, −140/−118, −327/−305, −520/−398, −800/−778, −2637/−2615, and +3339/+3361. Representative data are shown below the summary graph (B). (D) p53-dependent HDAC activity at SBE/p53RE. Immobilized SBE/p53RE DNA oligomers were incubated in 100 μg of HepG2 extract, either tryptic soy agar treated (10 μM; black bars) or untreated (gray bars), in the presence or absence of recombinant, activated p53. Protein complexes bound to the oligomers were washed and incubated with in vivo-labeled, 3H-acetylated histones, which were isolated and purified. HDAC enzymatic activity was measured by scintillation counting of released [3H]acetyl groups. The graphrepresents average values from multiple experiments and standard deviations of the means. (E) p53-dependent SnoN association at the SBE/p53RE. A series of ChIP studies (≥3 for each data point) were performed as described above. Values for percentage of SnoN bound relative to input at the SBE/p53RE in the presence and absence of recombinant p53 were normalized to antibody (Ab) control reactions. Probes used for Southern blots of precipitated DNA were labeled SBE/p53RE oligonucleotides or Hinc450 oligonucleotides (+3333 to +3356), which will detect nonspecific binding to AFP/lacZ DNA. (F) SnoN is required for p53-mediated repression of in vitro AFP chromatin transcription. HepG2 whole-cell extract was immunodepleted for SnoN protein (upper panel) and then analyzed by in vitro chromatin transcription (lower panel). Upper panel: 40 μg of protein of starting HepG2 extract, input (I), equal levels of protein from depleted extract, supernatant (S) and pellet (P) derived from boiled antibody-coupled beads used for immunodepletion. Lower panel: immobilized AFP DNA templates were incubated in (i) control (lanes 3 to 5; approximately 200 μg of total protein), (ii) immunodepleted (lanes 6 to 8; approximately 200 μg of total protein), or (iii) mock-depleted (lanes 9 and 10; approximately 200 μg of total protein) extracts or (iv) buffer (lanes 1 and 2) in the presence of recombinant p53 (500 ng, lanes 4 and 7; 1 μg, lanes 5, 8, and 10) or p53 dialysis buffer (lanes 1 to 3, 6 and 9) prior to chromatin assembly. Chromatin-assembled DNA templates were washed in nuclear extract buffer and in vitro transcribed in HeLa nuclear extract. Chick β-globin (50 ng) DNA was added during the transcription period as an RNA recovery control (57). AFP and Globin primer extension products are indicated. n-fold changes of transcribed AFP in vitro RNA signals are shown below each data point.
Recently, p53 and activated Smad2 protein, present in activin-treated, human HepG2 cells, were found to bind their respective regulatory elements, separated by approximately 100 bp of DNA, and cooperatively induce transcription of a specific subset of genes (20). We used extracts of HepG2 cells in our in vitro chromatin system to determine the direct effects of p53 addition on AFP chromatin structure and function in the absence of TGF-β signaling (Fig. 6A). As in other tumor-derived cells, SnoN protein is abundant in the absence of TGF-β ligand addition (data not shown), but unlike many other cultured cell lines, the p53 gene is wild type in HepG2 (20). Use of recombinant p53 protein, which binds DNA constitutively when the C-terminal 30 amino acids are deleted (38), permitted analysis of p53 function in vitro without stress-induced activation.
ChIP assays were performed on in vitro-assembled, AFP chromatin (Fig. 6B), purified by magnetic concentration, washed, and then cross-linked with formaldehyde prior to micrococcal nuclease digestion to mono- and dinucleosomal fragments, as we previously described (57). Antibody-precipitated fragments were heated to reverse the cross-links, purified, and analyzed by Southern blotting for specific regions of AFP DNA associated with acetylated histones H3 and H4. The average values from two to three assays are summarized in the graph, and a representative Southern blot analysis is shown. These assays were repeated for sites across the entire AFP DNA template, from −2000 to +3300 (Fig. 6C). AFP chromatin, incubated with HepG2 extract in the absence of p53 in vitro, was composed of acetylated nucleosomes between the SBE/p53RE (p53RE, −850) and the core promoter (start site, +1) region, with histone H3 more highly acetylated than H4. Addition of p53 during chromatin assembly induced a significant decrease in the level of histone H3 acetylation across the AFP 5′ regulatory region from −850 to +25, while acetylation of histone H4 tails was minimally reduced in response to p53 protein. The specific, p53-mediated effect on H3 acetylation was the same as that observed in vivo after TGF-β1 ligand treatment of Hepa 1-6 cells (Fig. 4). The 5′ enhancer region of AFP (−2000) and plasmid DNA sequences (+3300) exhibited minimal histone acetylation (Fig. 6C).
These effects of p53 on histone modification show an exact parallel with HDAC enzymatic activity (Fig. 6D). Protein pull-down assays, as described for ML extract (Fig. 1A and supplemental Fig. S1) were performed with HepG2 extracts. Purified bead-bound SBE/p53RE protein complexes were incubated with purified 3H-acetylated histones and assayed as described previously (10). Release of [3H]acetyl groups from 3H-acetylated histones occurs as a result of HDAC enzymatic activity and is quantified by scintillation counting. Inclusion of p53 in HepG2-derived protein complexes bound at the SBE/p53RE increased total HDAC activity more than twofold.
SnoN protein is required for p53-mediated repression of AFP transcription in vitro.
In parallel with a p53-targeted increase in HDAC activity, ChIP analysis of AFP chromatin, assembled in vitro in the presence of HepG2 extract, revealed that addition of recombinant p53 protein more than doubled the amount of SnoN protein, present in HepG2 extract, which is associated with cross-linked chromatin fragments at the SBE/p53RE (Fig. 6E). These data, taken together with in vivo ChIP assays of wild-type and p53−/− liver tissue, support a mechanism whereby p53 may recruit SnoN and histone deacetylase complexes and/or stabilize their association with AFP chromatin,
We examined the ability of p53 to repress transcription of activated AFP chromatin assembled in the absence of SnoN protein. To this end, we immunodepleted HepG2 extract of endogenous SnoN protein (immunoblot of control and depleted extract as well as bound SnoN; Fig. 6F, top) and compared the ability of SnoN-depleted, mock-depleted, and control HepG2 extract to establish p53-responsive AFP chromatin in vitro (bottom). We transcribed these chromatin-assembled templates in vitro as previously described (21). Chromatin assembly in Xenopus egg extract in the absence of transactivating proteins is highly repressive for transcription of AFP (∼25-fold repression, lane 2, compared to naked AFP bead-DNA transcribed in lane 1). Chromatin transcription is activated to levels achieved by naked DNA when AFP bead-DNA is assembled into chromatin in the presence of control HepG2 extract (lane 3). Chromatin assembled in the presence of recombinant, constitutively activated p53 protein and control HepG2 extract was repressed for AFP transcription up to 2.9-fold (lanes 4 and 5), as shown previously (56, 57).
The ability to activate AFP chromatin transcription was unaffected by immunodepletion of SnoN protein from HepG2 extract (Fig. 6F, lane 6). However, constitutively activated p53 was unable to repress transcription of AFP chromatin assembled in the presence of SnoN-depleted HepG2 extract (lanes 7 and 8). Control mock depletion with IgG removed some SnoN protein from the HepG2 extract (immunoblot, top) and reduced p53 responsiveness of the assembled chromatin (lane 10). Taken together, these data suggest that p53 cannot repress AFP chromatin transcription in the absence of SnoN protein in human tumor cell extracts and further support a reciprocal link between p53 and the TGF-β pathway.
DISCUSSION
We used affinity chromatography to purify a protein from normal liver and hepatoma cell extracts that interacts specifically with a defined p53 regulatory element, one that functions in transcription repression of the AFP gene during liver development. Once this protein was identified as the SnoN corepressor of the TGF-β pathway, we established that this association occurs in vivo and is p53 dependent by comparing normal and p53-null adult mouse liver tissue. We defined a mechanism for the observed downstream effects on chromatin structure and transcription regulation in vivo and in vitro using hepatoma cells. ChIP analyses revealed that SnoN interaction correlated with p53 binding of chromatin at a regulatory element composed of intercalated p53- and Smad-binding consensus sequences. Taken together, our data support a model of p53- and TGF-β-mediated alterations in protein complexes that interact with a composite SBE/p53RE and effect chromatin modification.
Database scanning for the consensus sequences of p53 and Smad protein binding sites revealed that they overlap to form a potential, composite p53/Smad regulatory element at more than 50 loci within the human genome (S. Zhao, personal communication). Using a compilation of p53 regulatory elements in a human p53 target gene database, Wang et al. found that 16 of 22 TGF-β-regulated genes contained p53 regulatory elements within their regulatory regions (84). Interestingly, in this work, global expression profiling of both p53-mediated activation and repression of gene transcription showed AFP as the most highly p53-repressed gene.
p53 can act as either a transcription activator or a transcription repressor depending on DNA sequence and/or cell specificity. Promotion of transcription repression, versus activation, by TGF-β signaling in concert with p53 may rely on action through an intercalated, composite regulatory element, as shown here, and tissue-specific expression of corepressor or activator complexes. Heteromeric interactions between Smad4, R-Smads, and conserved domains of Ski are sufficient for repression of Smad-dependent transcription of synthetic, SBE reporter constructs (81). The elegant studies of Piccolo and colleagues show that p53 and TGF-β cooperate in transcription activation of a specific subset of genes by binding to regulatory elements separated by more than 100 bp (20). They find that p53 binds its regulatory element in a transfected Mix.2 promoter in the absence of activin ligand, but both p53 and Smad binding are needed for full transcription activation. Activin did not induce activation of the p53 protein, but activated R-Smad (Smad2) exhibited protein-protein interactions with p53, which was purified by affinity for DNA lacking an SBE.
We found that the TGF-β ligand induces endogenous AFP chromatin association of both the p53 and Smad2/4 complexes, although the sequence of this process during embryonic development remains to be determined. HNF-3 (FoxA) is expressed early in development and is an important, endoderm-enriched activator of hepatic gene expression, including that of AFP (88). Winged-helix transcription factors, which include FoxA, can recruit active Smad protein complexes to their binding elements (30). FoxA binds to the 30-bp SBE/p53RE in the absence of p53 and is excluded from the protein-SBE/p53RE complex in the presence of p53 in vitro (42). Thus, Smad proteins interacting with FoxA and SBEs may play a role in AFP activation during embryogenesis prior to p53 activation, which occurs in postnatal mouse liver (42).
The ability of Smad4, P-Smad2, and corepressor SnoN to bind chromatin simultaneously is predicted from the crystal structure of Smad4 and Ski protein domains and in vitro protein-binding assays (87). Chromatin binding of p53 at the histone-acetylated SBE/p53RE region promotes the interaction of corepressor SnoN and HDAC complexes (and likely a histone methyltransferase), altering chromatin structure concomitantly with AFP repression. p53 may promote the binding of SnoN to the DNA-bound, Smad protein complex by direct protein-protein interactions with SnoN and/or by inducing a specific conformation of Smad proteins that exposes interactive surfaces for SnoN binding. Extrapolating from studies of Ski and Smad proteins, SnoN is likely to interact with Smad heteromeric complexes at multiple, potential binding surfaces (59, 81).
There have been previous hints that the p53 and TGF-β pathways could somehow modify each other, although the majority of their downstream effects have been ascribed to independent functions. Mutation of p53 reduces the ability of TGF-β1 to inhibit growth of human bronchial epithelial cells (31) and to stimulate the growth of mesenchymal murine fibroblasts (27). TGF-β1 treatment of rat liver epithelial cells and immortalized human cervical cells induced apoptosis and/or cell cycle arrest, concomitant with p53 activation (61, 76). HBx, the only transactivator protein encoded within the hepatitis B viral genome, can disrupt regulation of a subset of target genes in both TGF-β (68) and p53 signaling pathways (29, 56, 78, 80). These links, which underscore the variety of p53 and TGF-β responses, have now been extended to growth control during development (20).
The biological consequences of the p53 and TGF-β pathways acting in synergy are likely influenced by a number of cis- and trans-acting modifiers. Cellular levels of TGF-β corepressor proteins, SnoN and Ski, are tightly controlled by multiple mechanisms. Overexpression leads to transformation of certain cell types (8, 44; reviewed in reference 46), and decreased expression in mouse models leads to tumor formation (69, 70). Transformation and tumor development in vivo may not only be promoted by dysfunction among direct regulators of TGF-β signaling but also amplified by loss of p53 cooperativity at a subset of target genes. The interplay between these two major tumor suppressor pathways, their function through chromatin modification, and potential cell-specific activities may prove significant in future assessment of transformation, tumor development, and treatment.
Supplementary Material
Acknowledgments
We gratefully acknowledge E. Stavnezer, X.-H. Feng, R. Serra, D. Robbins, K. Luo, D. Edmondson, Y. Evrard, M. Coombes, and S. R. Dent for helpful discussion and/or materials; K. Cho for the RT-PCR analysis performed during a laboratory rotation; K. Wernke-Dollries and A. Lawrence for technical help; and G. Lozano and the National Cell Culture Center for essential materials. D.S.W. especially thanks W. K. Chan and M. Wilkinson for advice, materials, and support.
This work was supported in part by the NCI Cancer Center Support Grant to the UT MD Anderson Cancer Center, NIH grants GM53683 and GM60213 to M.C.B., and predoctoral support from NIH (T32-HD07325 to D.S.W and T32-CA59268 and the Albert J. Ryan Foundation to S.K.O.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akiyoshi, S., H. Inoue, J. Hanai, K. Kusanagi, N. Nemoto, K. Miyazono, and M. Kawabata. 1999. c-Ski acts as a transcriptional co-repressor in transforming growth factor-beta signaling through interaction with smads. J. Biol. Chem. 274:35269-35277. [DOI] [PubMed] [Google Scholar]
- 2.Andrisani, O. M., and S. Barnabas. 1999. The transcriptional function of the hepatitis B virus X protein and its role in hepatocarcinogenesis. Int. J. Oncol. 15:373-379. [DOI] [PubMed] [Google Scholar]
- 3.Ashcroft, M., Y. Taya, and K. H. Vousden. 2000. Stress signals utilize multiple pathways to stabilize p53. Mol. Cell. Biol. 20:3224-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attisano, L. 2000. Smads as transcriptional co-modulators. Curr. Opin. Cell Biol. 12:235-243. [DOI] [PubMed] [Google Scholar]
- 5.Bargonetti, J., and J. J. Manfredi. 2002. Multiple roles of the tumor suppressor p53. Curr. Opin. Oncol. 14:86-91. [DOI] [PubMed] [Google Scholar]
- 6.Berk, M., S. Y. Desai, H. C. Heyman, and C. Colmenares. 1997. Mice lacking the ski proto-oncogene have defects in neurulation, craniofacial, patterning, and skeletal muscle development. Genes Dev. 11:2029-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blain, S. W., and J. Massague. 2000. Different sensitivity of the transforming growth factor-beta cell cycle arrest pathway to c-Myc and MDM-2. J. Biol. Chem. 275:32066-32070. [DOI] [PubMed] [Google Scholar]
- 8.Boyer, P. L., C. Colmenares, E. Stavnezer, and S. H. Hughes. 1993. Sequence and biological activity of chicken snoN cDNA clones. Oncogene 8:457-466. [PubMed] [Google Scholar]
- 9.Camper, S. A., and S. M. Tilghman. 1991. The activation and silencing of gene transcription in the liver. Bio/Technology 16:81-87. [PubMed] [Google Scholar]
- 10.Carmen, A. A., S. E. Rundlett, and M. Grunstein. 1996. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J. Biol. Chem. 271:15837-15844. [DOI] [PubMed] [Google Scholar]
- 11.Carter, M. S., S. Li, and M. F. Wilkinson. 1996. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 15:5965-5975. [PMC free article] [PubMed] [Google Scholar]
- 12.Chadee, D. N., M. J. Hendzel, C. P. Tylipski, C. D. Allis, D. P. Bazett-Jones, J. A. Wright, and J. R. Davie. 1999. Increased Ser-10 phosphorylation of histone H3 in mitogen-stimulated and oncogene-transformed mouse fibroblasts. J. Biol. Chem. 274:24914-24920. [DOI] [PubMed] [Google Scholar]
- 13.Chang, H., C. W. Brown, and M. M. Matzuk. 2002. Genetic analysis of the mammalian transforming growth factor-beta superfamily. Endocr. Rev. 23:787-823. [DOI] [PubMed] [Google Scholar]
- 14.Chang, H., A. L. Lau, and M. M. Matzuk. 2001. Studying TGF-beta superfamily signaling by knockouts and knockins. Mol. Cell. Endocrinol. 180:39-46. [DOI] [PubMed] [Google Scholar]
- 15.Chaya, D., T. Hayamizu, M. Bustin, and K. S. Zaret. 2001. Transcription factor FoxA (HNF3) on a nucleosome at an enhancer complex in liver chromatin. J. Biol. Chem. 276:44385-44389. [DOI] [PubMed] [Google Scholar]
- 16.Chen, H., J. O. Egan, and J. F. Chiu. 1997. Regulation and activities of alpha-fetoprotein. Crit. Rev. Eukaryot. Gene Expr. 7:11-41. [DOI] [PubMed] [Google Scholar]
- 17.Choi, J., and L. A. Donehower. 1999. p53 in embryonic development: maintaining a fine balance. Cell. Mol. Life Sci. 55:38-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen, S. B., G. Zheng, H. C. Heyman, and E. Stavnezer. 1999. Heterodimers of the SnoN and Ski oncoproteins form preferentially over homodimers and are more potent transforming agents. Nucleic Acids Res. 27:1006-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colmenares, C., H. A. Heilstedt, L. G. Shaffer, S. Schwartz, M. Berk, J. C. Murray, and E. Stavnezer. 2002. Loss of the SKI proto-oncogene in individuals affected with 1p36 deletion syndrome is predicted by strain-dependent defects in Ski-/- mice. Nat. Genet. 30:106-109. [DOI] [PubMed] [Google Scholar]
- 20.Cordenonsi, M., S. Dupont, S. Maretto, A. Insinga, C. Imbriano, and S. Piccolo. 2003. Links between tumor suppressors: p53 is required for TGF-beta gene responses by cooperating with Smads. Cell 113:301-314. [DOI] [PubMed] [Google Scholar]
- 21.Crowe, A. J., and M. C. Barton. 1999. Functional analysis of chromatin assembled in synthetic nuclei. Methods 17:173-187. [DOI] [PubMed] [Google Scholar]
- 22.Crowe, A. J., and M. C. Barton. 1999. In vitro reconstitution of nuclei for replication and transcription. Methods Enzymol. 304:63-76. [DOI] [PubMed] [Google Scholar]
- 23.Crowe, A. J., J. L. Piechan, L. Sang, and M. C. Barton. 2000. S-phase progression mediates activation of a silenced gene in synthetic nuclei. Mol. Cell. Biol. 20:4169-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crowe, A. J., L. Sang, K. K. Li, K. C. Lee, B. T. Spear, and M. C. Barton. 1999. Hepatocyte nuclear factor 3 relieves chromatin-mediated repression of the α-fetoprotein gene. J. Biol. Chem. 274:25113-25120. [DOI] [PubMed] [Google Scholar]
- 25.Derynck, R., R. J. Akhurst, and A. Balmain. 2001. TGF-β signaling in tumor suppression and cancer progression. Nat. Genet. 29:117-129. [DOI] [PubMed] [Google Scholar]
- 26.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dkhissi, F., S. Raynal, P. Jullien, and D. A. Lawrence. 1999. Growth stimulation of murine fibroblasts by TGF-beta1 depends on the expression of a functional p53 protein. Oncogene 18:703-711. [DOI] [PubMed] [Google Scholar]
- 28.Dunker, N., and K. Krieglstein. 2000. Targeted mutations of transforming growth factor-beta genes reveal important roles in mouse development and adult homeostasis. Eur. J. Biochem. 267:6982-6988. [DOI] [PubMed] [Google Scholar]
- 29.Feitelson, M. A., M. Zhu, L. X. Duan, and W. T. London. 1993. Hepatitis B x antigen and p53 are associated in vitro and in liver tissues from patients with primary hepatocellular carcinoma. Oncogene 8:1109-1117. [PubMed] [Google Scholar]
- 30.Germain, S., M. Howell, G. M. Esslemont, and C. S. Hill. 2000. Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev. 14:435-451. [PMC free article] [PubMed] [Google Scholar]
- 31.Gerwin, B. I., E. Spillare, K. Forrester, T. A. Lehman, J. Kispert, B. Vogelstein, and C. C. Harris. 1992. Mutant p53 can induce tumorigenic conversion of human bronchial epithelial cells and reduce their responsiveness to a negative growth factor, transforming growth factor β1. Proc. Natl. Acad. Sci. USA 89:2759-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorski, K., M. Carneiro, and U. Schibler. 1986. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell 47:767-776. [DOI] [PubMed] [Google Scholar]
- 33.Hall, P. A., and D. P. Lane. 1997. Tumor suppressors: a developing role for p53? Curr. Biol. 7:R144-R147. [DOI] [PubMed] [Google Scholar]
- 34.Haviv, I., D. Vaizel, and Y. Shaul. 1996. pX, the HBV-encoded coactivator, interacts with components of the transcription machinery and stimulates transcription in a TAF-independent manner. EMBO J. 15:3413-3420. [PMC free article] [PubMed] [Google Scholar]
- 35.Hemann, M. T., J. S. Fridman, J. T. Zilfou, E. Hernando, P. J. Paddison, C. Cordon-Cardo, G. J. Hannon, and S. W. Lowe. 2003. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat. Genet. 33:396-400. [DOI] [PubMed] [Google Scholar]
- 36.Hill, C. S. 2001. TGF-beta signalling pathways in early Xenopus development. Curr. Opin. Genet. Dev. 11:533-540. [DOI] [PubMed] [Google Scholar]
- 37.Hsia, C. C., D. J. Kleiner, C. A. Axiotis, B. A. Di, A. M. Nomura, G. N. Stemmermann, and E. Tabor. 1992. Mutations of p53 gene in hepatocellular carcinoma: roles of hepatitis B virus and aflatoxin contamination in the diet. J. Natl. Cancer Inst. (Bethesda) 84:1638-1641. [DOI] [PubMed] [Google Scholar]
- 38.Hupp, T. R., D. W. Meek, C. A. Midgley, and D. P. Lane. 1992. Regulation of the specific DNA binding function of p53. Cell 71:875-886. [DOI] [PubMed] [Google Scholar]
- 39.Jones, S. N., A. E. Roe, L. A. Donehower, and A. Bradley. 1995. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 8:206-208. [DOI] [PubMed] [Google Scholar]
- 40.Kho, P. S., Z. Wang, L. Zhuang, Y. Li, J. L. Chew, H. H. Ng, E. T. Liu, and Q. Yu. 2004. p53-regulated transcriptional program associated with genotoxic stress-induced apoptosis. J. Biol. Chem. 279:21183-21192. [DOI] [PubMed] [Google Scholar]
- 41.Ko, L. J., and C. Prives. 1996. p53: puzzle and paradigm. Genes Dev. 10:1054-1072. [DOI] [PubMed] [Google Scholar]
- 42.Lee, K. C., A. J. Crowe, and M. C. Barton. 1999. p53-mediated repression of alpha-fetoprotein gene expression by specific DNA binding. Mol. Cell. Biol. 19:1279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levine, A. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 44.Li, Y., C. M. Turck, J. K. Teumer, and E. Stavnezer. 1986. Unique sequence, ski, in Sloan-Kettering avian retroviruses with properties of a new cell-derived oncogene. J. Virol. 57:1065-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu, X., Y. Sun, R. A. Weinberg, and H. F. Lodish. 2001. Ski/Sno and TGF-β signaling. Cytokine Growth Factor Rev. 12:1-8. [DOI] [PubMed] [Google Scholar]
- 46.Luo, K. 2004. Ski and SnoN: negative regulators of TGF-β signaling. Curr. Opin. Genet. Dev. 14:65-70. [DOI] [PubMed] [Google Scholar]
- 47.Luo, K., S. L. Stroschein, W. Wang, D. Chen, E. Martens, S. Zhou, and Q. Zhou. 1999. The Ski oncoprotein interacts with the Smad proteins to repress TGFbeta signaling. Genes Dev. 13:2196-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Massague, J. 1998. TGF-beta signal transduction. Annu. Rev. Biochem. 67:753-791. [DOI] [PubMed] [Google Scholar]
- 49.Massague, J., S. W. Blain, and R. S. Lo. 2000. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell 103:295-309. [DOI] [PubMed] [Google Scholar]
- 50.Massague, J., and D. Wotton. 2000. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 19:1745-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Medrano, E. E. 2003. Repression of TGF-beta signaling by the oncogenic protein SKI in human melanomas: consequences for proliferation, survival, and metastasis. Oncogene 22:3123-3129. [DOI] [PubMed] [Google Scholar]
- 52.Melino, G., X. Lu, M. Gasco, T. Crook, and R. A. Knight. 2003. Functional regulation of p73 and p63: development and cancer. Trends Biochem. Sci. 28:663-670. [DOI] [PubMed] [Google Scholar]
- 53.Montes de Oca Luna, R., D. S. Wagner, and G. Lozano. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378:203-206. [DOI] [PubMed] [Google Scholar]
- 54.Moustakas, A., and D. Kardassis. 1998. Regulation of the human p21/WAF1/Cip1 promoter in hepatic cells by functional interactions between Sp1 and Smad family members. Proc. Natl. Acad. Sci. USA 95:6733-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nomura, T., M. M. Khan, S. C. Kaul, H. D. Dong, R. Wadhwa, C. Colmenares, I. Kohno, and S. Ishii. 1999. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev. 13:412-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogden, S. K., K. C. Lee, and M. C. Barton. 2000. Hepatitis B viral transactivator HBx alleviates p53-mediated repression of α-fetoprotein gene expression. J. Biol. Chem. 275:27806-27814. [DOI] [PubMed] [Google Scholar]
- 57.Ogden, S. K., K. C. Lee, K. Wernke-Dollries, S. A. Stratton, B. Aronow, and M. C. Barton. 2001. p53 targets chromatin structure alteration to repress α-fetoprotein gene expression. J. Biol. Chem. 276:42057-42062. [DOI] [PubMed] [Google Scholar]
- 58.Pearson-White, S., and M. McDuffie. 2003. Defective T-cell activation is associated with augmented transforming growth factor β sensitivity in mice with mutations in the Sno gene. Mol. Cell. Biol. 23:5446-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qin, B. Y., S. S. Lam, J. J. Correia, and K. Lin. 2002. Smad3 allostery links TGF-beta receptor kinase activation to transcriptional control. Genes Dev. 16:1950-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reed, J. A., E. Bales, W. Xu, N. A. Okan, D. Bandyopadhyay, and E. E. Medrano. 2001. Cytoplasmic localization of the oncogenic protein Ski in human cutaneous melanomas in vivo: functional implications for transforming growth factor beta signaling. Cancer Res. 61:8074-8078. [PubMed] [Google Scholar]
- 61.Rorke, E. A., D. Zhang, C. K. Choo, R. L. Eckert, and J. W. Jacobberger. 2000. TGF-beta-mediated cell cycle arrest of HPV16-immortalized human ectocervical cells correlates with decreased E6/E7 mRNA and increased p53 and p21(WAF-1) expression. Exp. Cell Res. 259:149-157. [DOI] [PubMed] [Google Scholar]
- 62.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 63.Ryan, T. M., R. R. Rehringer, N. C. Martin, T. M. Townes, R. D. Palmiter, and R. L. Brinster. 1989. A single erythroid-specific DNase I super-hypersensitive site activates high levels of human β-globin gene expression in transgenic mice. Genes Dev. 3:314-323. [DOI] [PubMed] [Google Scholar]
- 64.Scherr, M., M. A. Morgan, and M. Eder. 2003. Gene silencing mediated by small interfering RNAs in mammalian cells. Curr. Med. Chem. 10:245-256. [DOI] [PubMed] [Google Scholar]
- 65.Schutze, N. 2004. siRNA technology. Mol. Cell. Endocrinol. 213:115-119. [DOI] [PubMed] [Google Scholar]
- 66.Shi, Y. 2003. Mammalian RNAi for the masses. Trends Genet. 19:9-12. [DOI] [PubMed] [Google Scholar]
- 67.Shi, Y., and J. Massague. 2003. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113:685-700. [DOI] [PubMed] [Google Scholar]
- 68.Shih, W.-L., M.-L. Kuo, S.-E. Chauang, A.-L. Cheng, and S.-L. Doong. 2000. Hepatitis B virus X protein inhibits transforming growth factor-beta induced apoptosis through the activation of phosphatidylinositol 3-kinase pathway. J. Biol. Chem. 275:25858-25864. [DOI] [PubMed] [Google Scholar]
- 69.Shinagawa, T., H. D. Dong, M. Xu, T. Maekawa, and S. Ishii. 2000. The sno gene, which encodes a component of the histone deacetylase complex, acts as a tumor suppressor in mice. EMBO J. 19:2280-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shinagawa, T., T. Nomura, C. Colmenares, M. Ohira, A. Nakagawara, and S. Ishii. 2001. Increased susceptibility to tumorigenesis of ski-deficient heterozygous mice. Oncogene 20:8100-8108. [DOI] [PubMed] [Google Scholar]
- 71.Sirotkin, H. I., M. A. Gates, P. D. Kelly, A. F. Schier, and W. S. Talbot. 2000. Fast1 is required for the development of dorsal axial structures in zebrafish. Curr. Biol. 10:1051-1054. [DOI] [PubMed] [Google Scholar]
- 72.Spear, B. T., T. Longley, S. Moulder, S. L. Wang, and M. L. Peterson. 1995. A sensitive lacZ-based expression vector for analyzing transcriptional control elements in eukaryotic cells. DNA Cell Biol. 14:635-642. [DOI] [PubMed] [Google Scholar]
- 73.Stroschein, S. L., W. Wang, S. Zhou, Q. Zhou, and K. Luo. 1999. Negative feedback regulation of TGF-beta signaling by the SnoN oncoprotein. Science 286:771-774. [DOI] [PubMed] [Google Scholar]
- 74.Sun, Y., X. Liu, E. Eaton, W. S. Lane, H. F. Lodish, and R. A. Weinberg. 1999. Interaction of the Ski oncoprotein with Smad3 regulates TGF-beta signaling. Mol. Cell 4:499-509. [DOI] [PubMed] [Google Scholar]
- 75.ten Djike, P., K. Miyazono, and C.-H. Heldin. 2000. Signaling inputs converge on nuclear effectors in TGF-β signaling. Trends Biochem. Sci. 25:64-70. [DOI] [PubMed] [Google Scholar]
- 76.Teramoto, T., A. Kiss, and S. S. Thorgeirsson. 1998. Induction of p53 and Bax during TGF-beta 1 initiated apoptosis in rat liver epithelial cells. Biochem. Biophys. Res. Commun. 251:56-60. [DOI] [PubMed] [Google Scholar]
- 77.Tilghman, S. M. 1985. The structure and regulation of the alpha-fetoprotein and albumin genes. Oxf. Surv. Eukaryotic Genes 2:160-206. [PubMed] [Google Scholar]
- 78.Truant, R., J. Antunovic, J. Greenblatt, C. Prives, and J. A. Cromlish. 1995. Direct interaction of the hepatitis B virus HBx protein with p53 leads to inhibition by HBx of p53 response element-directed transactivation. J. Virol. 69:1851-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Turner, B. M. 2000. Histone acetylation and an epigenetic code. Bioessays 22:836-845. [DOI] [PubMed] [Google Scholar]
- 80.Uchida, T., K. Takahashi, K. Tatsuno, U. Dhingra, and J. F. Eliason. 1996. Inhibition of hepatitis-B-virus core promoter by p53: implications for carcinogenesis in hepatocytes. Int. J. Cancer 67:892-897. [DOI] [PubMed] [Google Scholar]
- 81.Ueki, N., and M. J. Hayman. 2003. Direct interaction of Ski with either Smad3 or Smad4 is necessary and sufficient for Ski-mediated repression of transforming growth factor-beta signaling. J. Biol. Chem. 278:32489-32492. [DOI] [PubMed] [Google Scholar]
- 82.Ueki, N., and M. J. Hayman. 2003. Signal-dependent N-CoR requirement for repression by the Ski oncoprotein. J. Biol. Chem. 278:24858-24864. [DOI] [PubMed] [Google Scholar]
- 83.Wahl, G. M., and A. M. Carr. 2001. The evolution of diverse biological responses to DNA damage: insights from yeast and p53. Nat. Cell Biol. 3:E277-E286. [DOI] [PubMed] [Google Scholar]
- 84.Wang, L., Q. Wu, P. Qiu, A. Mirza, M. McGuirk, P. Kirschmeier, J. R. Greene, Y. Wang, C. B. Pickett, and S. Liu. 2001. Analyses of p53 target genes in the human genome by bioinformatic and microarray approaches. J. Biol. Chem. 276:43604-43610. [DOI] [PubMed] [Google Scholar]
- 85.Wang, W., F. V. Mariani, R. M. Harland, and K. Luo. 2000. Ski represses bone morphogenic protein signaling in Xenopus and mammalian cells. Proc. Natl. Acad. Sci. USA 97:14394-14399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wells, J., C. R. Graveel, S. M. Bartley, S. J. Madore, and P. J. Farnham. 2002. The identification of E2F1-specific target genes. Proc. Natl. Acad. Sci. USA 99:3890-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu, J. W., A. R. Krawitz, J. Chai, W. Li, F. Zhang, K. Luo, and Y. Shi. 2002. Structural mechanism of Smad4 recognition by the nuclear oncoprotein Ski: insights on Ski-mediated repression of TGF-beta signaling. Cell 111:357-367. [DOI] [PubMed] [Google Scholar]
- 88.Zaret, K. S. 1994. Genetic control of hepatocyte differentiation, p. 53-68. In I. M. Arias, J. L. Boyer, N. Fausto, W. B. Jakoby, D. A. Schachter, and D. A. Shafritz (ed.), The liver: biology and pathobiology, 3rd ed. Raven Press, New York, N.Y.
- 89.Zawel, L., J. L. Dai, P. Buckhaults, S. Zhou, K. W. Kinzler, B. Vogelstein, and S. E. Kern. 1998. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol. Cell 1:611-617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.