Abstract
DNA polymerases (Pols) of the Y family rescue stalled replication forks by promoting replication through DNA lesions. Humans have four Y family Pols, η, ι, κ, and Rev1, of which Pols η, ι, and κ have been shown to physically interact with proliferating cell nuclear antigen (PCNA) and be functionally stimulated by it. However, in sharp contrast to the large increase in processivity that PCNA binding imparts to the replicative Pol, Polδ, the processivity of Y family Pols is not enhanced upon PCNA binding. Instead, PCNA binding improves the efficiency of nucleotide incorporation via a reduction in the apparent Km for the nucleotide. Here we show that Polι interacts with PCNA via only one of its conserved PCNA binding motifs, regardless of whether PCNA is bound to DNA or not. The mode of PCNA binding by Polι is quite unlike that in Polδ, where multisite interactions with PCNA provide for a very tight binding of the replicating Pol with PCNA. We discuss the implications of these observations for the accuracy of DNA synthesis during translesion synthesis and for the process of Pol exchange at the lesion site.
Proliferating cell nuclear antigen (PCNA), a highly conserved, ring-shaped homotrimeric eukaryotic protein, forms a sliding clamp at the template-primer junction. PCNA is loaded onto the primer-template junction in an ATP-dependent manner by a multiprotein clamp loader, replication factor C (RFC). After the loading of PCNA, RFC stays on the DNA via interaction with replication protein A (RPA) bound to single-stranded DNA (1, 18, 37). The binding of the replicative DNA polymerase (Pol), Polδ, to PCNA endows it with a very high processivity (25, 30), and that presumably is the essential function of PCNA in DNA replication.
In Saccharomyces cerevisiae, Polδ is comprised of three subunits of 125, 55, and 40 kDa, encoded by the POL3, POL31, and POL32 genes, respectively (6). While the Pol3 catalytic subunit and the Pol31 subunit are highly conserved among eukaryotes, the Pol32 subunit shows a high degree of divergence. The S. cerevisiae Pol3, Pol31, and Pol32 subunits are the respective homologs of Schizosaccharomyces pombe Polδ subunits Pol3, Cdc1, and Cdc27. Whereas the Pol3 and Pol31 subunits and their counterparts are essential in both S. cerevisiae and S. pombe, the third subunit, Cdc27, is essential for completion of the S phase in S. pombe (22, 27), but its counterpart in S. cerevisiae, Pol32, is not. pol32Δ cells, however, grow poorly and exhibit DNA replication defects (6).
A series of genetic and biochemical observations with S. cerevisiae Polδ have indicated that at least two separate domains on Polδ interact with at least two separate domains on PCNA, and furthermore, it has been suggested that, during replication, Polδ binds to at least two PCNA monomers (15). Overall, the various studies with Polδ from S. cerevisiae, S. pombe, and humans have strongly indicated that several distinct domains on Polδ interact with different regions of PCNA, and these multiple interactions provide the high degree of processivity that PCNA binding imparts to Polδ. Briefly, we review this evidence below.
A consensus PCNA binding motif, QXX(L/I)XXFF, is present at the extreme C terminus of Pol32 in S. cerevisiae and also in its S. pombe and human counterparts Cdc27 and p66, respectively, and mutational inactivation of this domain in PCNA from S. cerevisiae and S. pombe affects the processivity of Polδ (2, 15). In addition, biochemical studies with two mutant PCNAs from S. cerevisiae, pcna-79 and pcna-90, have shown that they both affect the processivity of Polδ (5, 15). In the pcna-79 mutant (I126A/L128A), the hydrophobic pocket in the interdomain connector loop (IDCL) of PCNA is impaired (5), and this mutant PCNA fails to interact with proteins via their consensus QXX(L/I)XXFF PCNA binding motif (8, 15, 32). The pcna-90 (P252A/K253A) mutant has mutational changes in the carboxy-terminal tail of PCNA (5). Since the carboxy-terminal tail of PCNA does not interact with the consensus PCNA binding motif present in Pol32, the adverse effects of mutations in this PCNA region on Polδ processivity (15) must derive from interactions of PCNA with Polδ at a site different from the IDCL interacting domain of Pol32. In keeping with this idea, at least two PCNA binding sites have been identified in the p125 catalytic subunit of human Polδ: one of these is contained in the N2 region toward the amino terminus (38), and the other is in the succeeding N4 region (36). The latter sequence is characterized by the presence of a highly conserved KA motif. The association of Polδ with PCNA thus would be considerably strengthened by these multisite interactions.
The Y family DNA Pols, such as Polη, Polι, and Polκ, promote replication through distorting DNA lesions, but they replicate DNA with a low fidelity and low processivity (24). Although all these Pols interact with PCNA, physically as well as functionally, binding to PCNA does not improve their processivity (9-11, 13). For example, PCNA, when loaded onto DNA by RFC in the presence of RPA, stimulates the DNA synthetic activity of both yeast and human Polη approximately 10- to 15-fold, but the processivity in the presence of these protein factors remains the same as in their absence, at about three or four nucleotides per DNA binding event (9, 11). Instead, the increase in the efficiency of nucleotide incorporation is achieved primarily by a reduction in the apparent Km for the nucleotide (9, 11). PCNA, in the presence of RFC and RPA, also greatly stimulates the DNA synthetic activity of Polι and Polκ, and this again is achieved by a decrease in the apparent Km for the nucleotide, whereas the processivity remains unaffected (10, 13).
Since PCNA binding does not improve the processivity of Y family DNA Pols, these Pols must differ in their mode of PCNA binding from Polδ. As multisite interactions would provide for the strong association of Polδ with PCNA and the ensuing large increase in Polδ processivity, we have examined the possibility that the lack of PCNA stimulation of the processivity of Y family Pols derives from their binding to PCNA rather weakly. The presence of multiple putative PCNA binding motifs in Polι prompted us to determine whether this Pol made multisite contacts with PCNA or whether only one of the sites was involved. Here we show that Polι interacts with PCNA via only one of these sites and discuss the implications of this observation for translesion DNA synthesis (TLS) by Polι as well as other Y family Pols.
MATERIALS AND METHODS
Proteins.
Human PCNA, RFC, and RPA were purified as described previously (3, 7, 20). Six-His-tagged human PCNA used for the interaction studies was overexpressed in Escherichia coli and purified as described previously (19). Wild-type and mutant human Polι proteins in fusion with glutathione S-transferase (GST) were expressed in the yeast strain BJ5464 and bound to a glutathione-Sepharose 4B column as described previously (17). To purify Polι, the GST-Polι-containing beads were incubated overnight at 4°C with PreScission protease, which cleaves the GST-Polι fusion protein at 7 amino acids amino-terminal from the first methionine of Polι, in buffer E containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM dithiothreitol, 0.01% NP-40, and 10% glycerol. The purified Polι was concentrated with a Microcon 30 (Amicon) concentrator, aliquoted, and frozen at −70°C.
Pull-down assay of Polι and PCNA complexes.
To constitute complexes, Polι (2 μg) was mixed with His6-hPCNA (4 μg) in buffer E in 30-μl samples and incubated for 30 min at 4°C followed by 10 min at 25°C. To 20 μl of these samples, 10 μl of nickel-nitrilotriacetic acid (Ni-NTA; Qiagen) beads was added to bind His6-PCNA and His6-PCNA-Polι complex, and the samples were further incubated with constant rocking for 30 min at 4°C followed by the elution of bound proteins with 500 mM imidazole-containing buffer E. The samples containing the protein mixture before the addition of affinity beads, the flowthrough plus washing fractions, and the eluted proteins were precipitated with 5% trichloroacetic acid and separated on a sodium dodecyl sulfate-12% polyacrylamide gel followed by Coomassie blue R-250 staining.
Gel filtration analysis of Polι-PCNA interaction.
Gel filtration of Polι, PCNA, and their complexes was performed at 4°C with a Superdex 200 PC 3.2/30 column (Amersham Pharmacia Biotech, Piscataway, N.J.) equilibrated with buffer I, containing 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 0.01% NP-40, and 10% glycerol. To constitute complexes, Polι (4 μg), His6-PCNA (5 μg), or the mixture of these proteins was incubated in 25 μl of buffer I for 60 min at 4°C followed by incubation for 10 min at 25°C. The protein mixture was then gel filtered at a 20-μl/min flow rate at 4°C. Fractions were collected and analyzed on 10% sodium dodecyl sulfate-polyacrylamide gels stained with Coomassie blue R-250.
DNA Pol assays.
DNA substrate was generated by annealing a 75-nucleotide oligomer template, 5′-biotin-AGCAAGTCACCAATGTCTAAGAGTTCGTATTATGCCTACACTGGAGTACCGGAGCATCGTCGTGACTGGGAAAAC-biotin-3′, which contained one biotin molecule attached at each end, to the 5′ 32P-labeled oligonucleotide primer N4577, 5′-GTTTTCCCAGTCACGACGATGCTCCGGTA-3′. To bind streptavidin to biotin present at the ends of the linear DNA substrates, these primer-templates (2.5 pmol) were preincubated with streptavidin (5 μg) in 25 μl of DNA Pol buffer which contained no MgCl2, for 10 min at 30°C, before their addition to the DNA Pol reaction mixtures. The standard DNA Pol reaction mixture (10 μl) contained 40 mM Tris-HCl (pH 7.5); 8 mM MgCl2; 150 mM NaCl; 1 mM dithiothreitol; 10% glycerol; 100 μg of bovine serum albumin/ml; 500 μM ATP; and 100 μM (each) dGTP, dATP, dTTP, and dCTP. As indicated in the figure legends, mutant and wild-type Polι proteins (1 nM) were mixed with PCNA (50 ng), RFC (5 ng), and RPA (50 ng) and the linear primer-template DNA (10 nM). Assay mixtures were assembled on ice and incubated at 37°C for 10 min, and assays were stopped by the addition of loading buffer (40 μl) containing 20 mM EDTA, 95% formamide, 0.3% bromphenol blue, and 0.3% cyanol blue. The reaction products were resolved on 10% polyacrylamide gels containing 8 M urea. Visualization of the results was done using a Molecular Dynamics Storm PhosphoImager and ImageQuant software.
Two-hybrid analyses.
The HF7c yeast strain was transformed with the Polι and PCNA fusion constructs by the lithium acetate method. Transformants harboring both the GAL4 DNA binding domain (BD) and the GAL4 activation domain (AD) fusion constructs were grown on synthetic complete medium, lacking leucine and tryptophan. β-Galactosidase activity was examined to determine the interaction between Polι and PCNA, as described in the Clontech yeast protocols handbook (PT3024-1, chapter VI). β-Galactosidase activities were quantitated using O-nitrophenyl-β-d-galactopyranoside as substrate. Experiments were performed at least three times in triplicate samples.
RESULTS
Putative PCNA binding motifs in Polι.
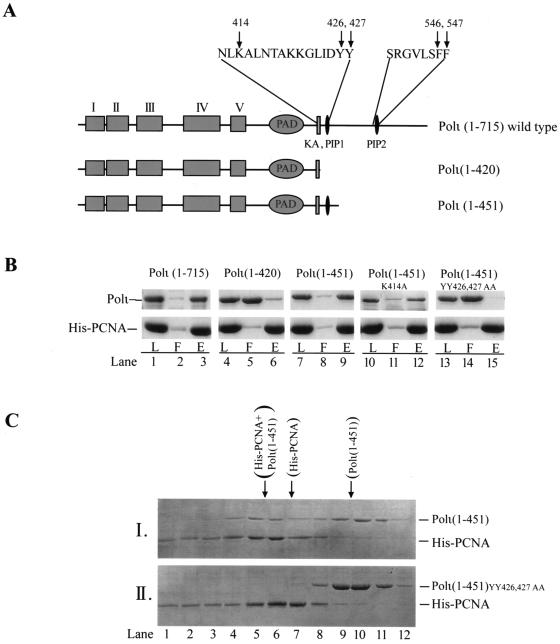
A consensus PCNA binding motif QXX(I, L, or M)XXF(F or Y), also referred to as the PCNA interaction protein box (PIP box), is found in many proteins involved in DNA metabolic processes, such as DNA replication and repair, DNA methylation, and chromatin assembly (4, 21, 28). Structural and mutational studies have indicated the involvement of the conserved hydrophobic residues within this motif in interaction with PCNA (23, 26, 33). Polι has two putative PIP boxes, KKGLIDYY and SRGVLSFF, which are located toward the C terminus after the five conserved Pol domains and the polymerase-associated domain (PAD), and they encompass residues 420 to 427 (PIP1 box) and 540 to 547 (PIP2 box), respectively, of the 715-amino-acid protein (Fig. 1A).
FIG. 1.
Putative PCNA binding motifs of human DNA Polι. (A) Amino acids 418 to 430 and 538 to 550 of human Polι were aligned with the PIP motif identified in various PCNA binding proteins and shown to bind the IDCL of PCNA. The highly conserved residues are indicated in boldface. Hs, Homo sapiens; Sc, S. cerevisiae. (B) Residues 412 to 424 of human Polι were aligned with the PCNA binding KA box of Polδ, which is present also in many other PCNA binding proteins.
Recently, another conserved PCNA binding sequence characterized by a KA motif has been identified in the catalytic subunit of human Polδ (36), and a KA motif is present also in Polι between residues 412 and 424 (Fig. 1B). The presence of three potential PCNA binding motifs in Polι raised the possibility that Polι binds to PCNA at more than one site.
Generation of mutations in the putative PCNA binding motifs of Polι.
To test if the three PCNA binding motifs of Polι have roles in the interaction with PCNA, we generated several Polι mutations (Fig. 2A). In the Polι(1-420) deletion mutant, both the PIP1 and PIP2 motifs are deleted, and only the KA motif is retained. In the Polι(1-451) deletion mutant, the PIP2 motif is deleted, but the KA and PIP1 motifs are retained. In the Polι(1-451) deletion mutant, we also changed the conserved lysine 414 residue of the KA motif to alanine, resulting in the Polι(1-451)K414A mutant, and changed the two tyrosines at positions 426 and 427 in the PIP1 motif to alanine, resulting in Polι(1-451)YY426,427,AA. Additionally, we mutated the YY residues of PIP1 and the FF residues of PIP2 in full-length Polι, generating PolιYY426,427,AA, PolιFF546,547,AA, and PolιYY426,427,AA,FF546,547,AA.
FIG. 2.
Identification of a PCNA binding motif in Polι (A) Mutations made in the putative PCNA binding motifs of human Polι. In the schematic representation of Polι, the boxes indicate the five conserved motifs characteristic of Y family DNA Pols. The location and sequences of the three putative PCNA binding motifs, KA, PIP1, and PIP2, present in Polι are indicated. Arrows indicate the amino acid residues of PCNA binding motifs which were changed to alanine in the various mutant Polι proteins. In the Polι(1-420) deletion mutant protein, the two PIP motifs have been deleted, which leaves only the KA motif, while the Polι(1-451) deletion mutant protein contains the KA box as well as the PIP1 motif. In the Polι(1-451) deletion mutant, the K residue at 414 and the Y residues at 426 and 427 were changed to alanine, resulting in Polι(1-451)K414A and Polι(1-451)YY426,427,AA mutant proteins, respectively. (B) The PIP1 motif mediates complex formation of Polι with PCNA. As indicated on the top, wild-type and mutant Polι proteins (2 μg each) were mixed with His6-PCNA (4 μg). After incubation, samples were bound to Ni-NTA beads followed by washing and elution of the bound proteins with imidazole-containing buffer. Aliquots of each sample before addition to the beads (L), the flowthrough plus wash (F), and the eluted proteins (E) were analyzed on a sodium dodecyl sulfate-12% polyacrylamide gel stained with Coomassie blue. The positions of Polι and His6-PCNA are shown on the left. (C) Polι(1-451) mutant protein is able to form a stable complex with PCNA. The mixture of Polι(1-451) (4 μg) and His6-PCNA (5 μg) (I) and the mixture of Polι(1-451)YY426,427,AA (4 μg) and His6-PCNA (5 μg) (II) were gel filtered after incubation in a 30-μl volume of reaction mixture for 60 min at 4°C followed by 10 min at 25°C. The elution positions of Polι(1-451) in complex with PCNA, PCNA homotrimer, and the free Polι(1-451) are indicated on top. His6-PCNA and Polι mutant proteins are identified on the right.
PIP1 motif of Polι mediates physical interaction with PCNA.
To examine the physical interactions of mutant Polι proteins with PCNA, the Polι proteins were incubated with His6-PCNA and a pull-down assay was carried out using the Ni-NTA affinity beads (Fig. 2B). Because Polι alone cannot bind to Ni-NTA, Polι could be pulled down only if it interacted with PCNA.
When His6-PCNA is bound to the Ni-NTA beads, a large proportion of the wild-type Polι is retained on the beads via PCNA (Fig. 2B, lanes 1 to 3). The Polι(1-420) mutant protein, however, is impaired in interaction with PCNA, indicating that the KA motif alone is not able to mediate the binding of Polι to PCNA (Fig. 2B, lanes 4 to 6). The Polι(1-451) mutant protein, on the other hand, interacts with PCNA as well as the wild-type Polι, which suggests a role for the PIP1 motif in the binding of Polι to PCNA. To provide evidence that the conserved PIP1 motif is, in fact, involved in PCNA binding, we tested the ability of the Polι(1-451)YY426,427,AA mutant protein to bind PCNA. As shown in Fig. 2B, lanes 13 to 15, this mutational alteration of the PIP1 motif in Polι(1-451) inactivated the interaction with PCNA. The Polι(1-451) K414A mutation, however, did not affect PCNA binding (Fig. 2B, lanes 10 to 12). These results indicate that the PIP1 PCNA binding domain in Polι is sufficient to mediate the physical interaction of Polι with PCNA.
To examine further whether the PIP1 domain of Polι alone is able to mediate a stable interaction with PCNA, we analyzed the complexes of mutant Polι proteins and PCNA by gel filtration. Polι(1-451) or Polι(1-451)YY426,427,AA proteins were mixed with PCNA in almost a 1:1 molar ratio, and after incubation, their interaction was examined by gel filtration, a nonequilibrium technique wherein only rather stable protein complexes survive. While PCNA alone eluted mainly in fractions 6 and 7 and Polι(1-451) alone eluted around fractions 9 and 10 (data not shown; also Fig. 2C, panel II), when PCNA was preincubated with Polι(1-451), both of them together eluted earlier around fractions 5 and 6 (Fig. 2C, panel I). This shift in the elution position of both proteins indicates the formation of a complex between Polι(1-451) and PCNA. By contrast, Polι(1-451)YY426,427,AA and PCNA did not elute together after preincubation (Fig. 2C, panel II). These observations support the requirement of the PIP1 motif in Polι(1-451) in PCNA binding.
PIP1 motif of Polι mediates the functional interaction with PCNA.
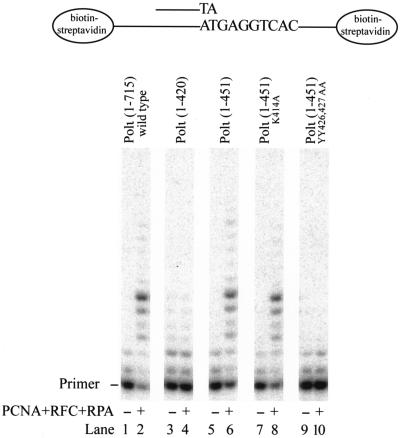
Many PCNA binding proteins interact with PCNA via at least two different domains; one of these is more important when PCNA is in solution, and the other becomes essential when PCNA is loaded onto the DNA (8, 15, 32). To determine if the binding of Polι to PCNA on DNA requires the PIP1 motif or some other motif, we compared the effects of PCNA on the DNA synthetic activity of the wild-type and mutant Polι proteins. First, we loaded PCNA by RFC onto an RPA-coated singly primed 75-nucleotide-long template DNA containing biotin-streptavidin at both ends, which prevented the PCNA from sliding off. DNA synthesis was then initiated by adding the wild-type or mutant Polι protein. The DNA synthetic activity of wild-type Polι is greatly enhanced upon the addition of PCNA, RFC, and RPA (Fig. 3, compare lanes 1 and 2), and PCNA also stimulated the synthetic activity of Polι(1-451) (Fig. 3, compare lanes 5 and 6) and Polι(1-451)K414A (Fig. 3, compare lanes 7 and 8) mutant enzymes, indicating that the KA motif has no role in the functional interaction of Polι with PCNA. By contrast, Polι(1-420) (Fig. 3, compare lanes 3 and 4) and Polι(1-451)YY426,427,AA (Fig. 3, compare lanes 9 and 10) mutant proteins lost their ability to be stimulated by PCNA. The PIP1 motif of Polι thus is sufficient to mediate the physical and functional interactions of Polι with PCNA.
FIG. 3.
The PIP1 motif of Polι is sufficient to mediate the stimulatory effect of PCNA on the DNA synthetic activity of Polι. The reaction mixtures contained wild-type or mutant Polι proteins (1 nM each), along with singly primed 75-nucleotide-long DNA substrate (10 nM) in which the 29-nucleotide primer was 32P labeled at its 5′ end, and the template contained biotin-streptavidin complex at both ends to prevent the PCNA sliding off the DNA and all four deoxynucleotides (100 μM each), in the presence or absence of PCNA (50 ng), RFC (5 ng), and RPA (50 ng). After incubation for 10 min at 37°C, samples were quenched and run on a 10% polyacrylamide gel containing 8 M urea followed by PhosphorImager analysis.
PIP2 motif of Polι has no role in physical or functional interactions with PCNA.
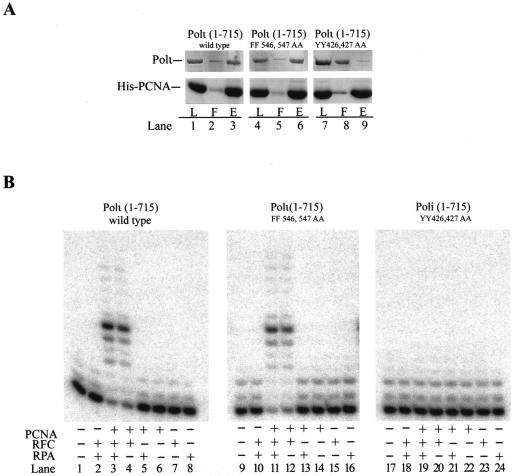
Our observations with Polι(1-451) protein indicating that the PIP1 motif of Polι is required for the physical and functional interactions of Polι with PCNA do not exclude the possibility that the PIP2 motif also influences the PCNA binding of Polι. For this reason, we examined the interactions of full-length Polι proteins carrying mutations in the PIP1 or PIP2 motifs with PCNA. From pull-down analysis with His6-PCNA on Ni-NTA beads, we found that, whereas mutational inactivation of the PIP2 motif of Polι has no effect on the physical interaction of Polι with PCNA (Fig. 4A, lanes 4 to 6), mutational inactivation of the PIP1 motif in full-length Polι impairs the interaction of Polι with PCNA (Fig. 4A, lanes 7 to 9).
FIG. 4.
Only the PIP1 PCNA binding domain of Polι mediates interactions with PCNA. (A) Pull-down analysis of complex formation between Polι proteins and PCNA. Wild-type or mutant Polι (2 μg each) was incubated with His6-PCNA (4 μg) and pulled down on Ni-NTA beads. Aliquots of each sample before addition to the beads (L), the flowthrough plus wash (F) and the eluted proteins (E) were analyzed on a polyacrylamide gel. (B) Wild-type or mutant Polι (1 nM each) was incubated with the DNA substrate (10 nM) in the presence of each of four deoxynucleoside triphosphates under standard reaction conditions. As indicated, the reactions were carried out in the presence or absence of PCNA (50 ng), RFC (5 ng), and RPA (50 ng).
Next, we examined the effect of addition of PCNA, RFC, and RPA on the DNA synthetic activity of full-length Polι proteins carrying mutations in the PIP1 or PIP2 motif (Fig. 4B). While the addition of PCNA, RFC, or RPA alone did not enhance the DNA synthetic activity of the wild-type or mutant Polι proteins (Fig. 4B, compare lanes 1, 9 and 17 to lanes 6 to 8, 14 to 16, and 22 to 24, respectively), robust stimulation of DNA synthesis was observed with the Polι(1-715)FF546,547,AA protein upon the addition of PCNA, RFC, and RPA or only PCNA and RFC (Fig. 4B, compare lanes 9, 11, and 12), and the degree of stimulation was the same as for the wild-type Polι(1-715) protein (Fig. 4B, lanes 1, 3, and 4). DNA synthesis by the Polι(1-715)YY426,427,AA mutant, however, was not stimulated upon the addition of PCNA, RFC, and RPA (Fig. 4B, compare lanes 17, 19, and 20). Thus, the presence of the C-terminal PCNA binding motif, PIP2, in the complete Polι protein with the YY426,427,AA mutation does not enable Polι to bind PCNA or be stimulated by it, and conversely, the mutational inactivation of the PIP2 domain has no adverse effect on the physical or functional interactions of Polι with PCNA. From these results, we conclude that the PIP2 domain has no role in the binding of Polι with PCNA, whereas the PIP1 domain is essential for the physical as well as functional interactions of Polι with PCNA.
Interaction of Polι with PCNA—two-hybrid analysis.
We used the yeast two-hybrid system to examine the interaction of Polι with PCNA in vivo. In one of the plasmids, the GAL4 DNA BD was fused with either the complete wild-type RAD30B Polι-encoding gene or the mutant rad30B A546-A547 and rad30B A426-A427-A546-A547 genes, and in the other plasmid, the GAL4 AD was fused with human PCNA. The HF7c yeast reporter strain harboring the GAL4-AD plasmid was transformed with one of the GAL4-BD plasmids. Interaction of the wild-type and mutant Polι proteins with PCNA in these transformants was analyzed by a β-galactosidase liquid assay, and the results are summarized in Table 1. Compared to the low level of β-galactosidase activity measured with GAL4-BD and the GAL4-AD-PCNA plasmids, the wild-type GAL4-BD Polι protein showed strong interaction with PCNA bound to GAL4-AD, resulting in 38-fold-higher β-galactosidase activity. The A546-A547 point mutations in the PIP2 motif of Polι did not affect the interaction of Polι and PCNA. The additional inactivation of the PIP1 motif, however, as in Polι A426-A427-A546-A547, completely abolished the interaction of Polι with PCNA, yielding β-galactosidase activity similar to that in the control. These results provide further evidence that the interaction of Polι with PCNA occurs via the PIP1 motif of Polι.
TABLE 1.
Interaction of Polι with PCNA in the yeast two-hybrid system
| AD fusion | DNA BD fusion | Mean β-galactosidase activity ± SD | Fold activation |
|---|---|---|---|
| GAL4 AD-PCNA | GAL4 BD | 0.03 ± 0.002 | 1 |
| GAL4 AD-PCNA | GAL4 BD-Polι (WTa) | 1.14 ± 0.05 | 38 |
| GAL4 AD-PCNA | GAL4 BD-Polι (A546A547) | 1.27 ± 0.03 | 42.3 |
| GAL4 AD-PCNA | GAL4 BD-Polι (A426A427A546A547) | 0.029 ± 0.003 | 0.97 |
WT, wild type.
DISCUSSION
Polι contains three potential PCNA binding motifs, the KA motif, and the PIP1 and PIP2 motifs; however, only one of these, PIP1, mediates the physical and functional interactions of Polι with PCNA. We consider it very unlikely that Polι could also interact with PCNA via some other domain, because the PIP1 domain follows right after the PAD. The region of Polι N-terminal to PAD contains the highly conserved motifs I to V, characteristic of Y family Pols, and these are all essential for DNA Pol activity together with the PAD. The PIP1 domain of Polι, KKGLIDYY, resembles the conserved PCNA binding motif that has been identified in a number of proteins and shown to interact with the IDCL region of PCNA.
The observation that the PIP1 domain of Polι is both necessary and sufficient for the interaction of Polι with PCNA in the absence of DNA as well as when PCNA is bound to the DNA substrate at the template-primer junction makes a clear distinction between the mode of PCNA binding by Polι, a TLS Pol, and Polδ, a replicative Pol. Thus, in contrast to Polδ, where distinct domains in different subunits mediate its interactions with PCNA (2, 15, 36, 38), contributing to the tight binding and the concomitant increase in processivity of Polδ, Polι binds PCNA at only one site—the IDCL region. In contrast, and as indicated from biochemical analysis of PCNA mutants of S. cerevisiae, Polδ binds DNA-bound PCNA in at least two regions, the IDCL region and the C-terminal region (15).
The mode of PCNA binding by Polι differs also from that in Fen1, a 5′→3′ nuclease that functions in the removal of RNA primers during Okazaki fragment maturation, and Apn2, a nuclease which functions in base excision repair (8, 32). Whereas in the absence of DNA both these nucleases bind PCNA in the IDCL region, when PCNA is bound to the DNA substrate, they additionally require its C-terminal region for enforcing productive binding. Such an inference is supported from the effects that mutations in the IDCL and C-terminal regions of PCNA have upon the Fen1 and Apn2 binding of PCNA (8, 32): while in the absence of DNA mutations in the IDCL region impair Fen1 and Apn2 binding to PCNA and mutations in the C-terminal region have little effect, in the presence of DNA mutations in both the IDCL and C-terminal regions affect their binding to PCNA, but mutations in the C-terminal region have a more profound effect than do mutations in the IDCL region.
The ability of Y family Pols to replicate through DNA lesions implies that they are not as sensitive to geometric distortions of DNA as are the replicative Pols, which are unable to replicate through DNA lesions. As a consequence, the Y family DNA Pols synthesize DNA with a low fidelity, the fidelity of Polι being particularly poor (24). In contrast to almost all other DNA Pols, Polι incorporates nucleotides opposite the four template bases with very different efficiencies (kcat/Km) and fidelities, and opposite template T, it even misincorporates a G approximately 10-fold better than an A (10, 16, 31, 34, 39). Also, Polι synthesizes DNA with a very low processivity, regardless of whether it is bound to PCNA. Under conditions of DNA synthesis, where Polι was allowed to bind the DNA substrate only once, even with PCNA present Polι incorporated only one nucleotide before dissociation from the DNA (10). Since Polι functions primarily at the nucleotide incorporation step of lesion bypass, wherein it incorporates nucleotides opposite from highly distorting lesions, with the subsequent extension step being performed by another TLS Pol (16, 35), Polι's role in TLS would then mostly be limited to the incorporation of a single nucleotide. Therefore, the very low processivity of PCNA-bound Polι is in accord with its role in lesion bypass; moreover, this attribute would contribute to keeping the incidence of inadvertent nucleotide misincorporations at undamaged sites low.
Our previous studies indicating that the binding of yeast and human Polη to DNA-bound PCNA or to PCNA in the absence of DNA is also mediated by a consensus IDCL binding motif present at the C terminus of these proteins (9, 11) support the idea that all Y family Pols resemble one another in their manner of PCNA binding. Although the mutational studies for the identification of PCNA binding domains in Polη were not as exhaustive as those that we have done for Polι, the fact that mutations in the IDCL binding motif in both yeast and human Polη inactivated their interactions with PCNA in both the absence and presence of DNA implies that this PCNA binding region of Polη makes a paramount contribution to interactions with PCNA in both situations. In summary, then, we suggest that Y family Pols differ strikingly from Polδ and from other PCNA binding proteins, such as Fen1 and Apn2, as they limit their interaction with DNA-bound PCNA only to the IDCL region, and that contributes to their low processivity even with PCNA.
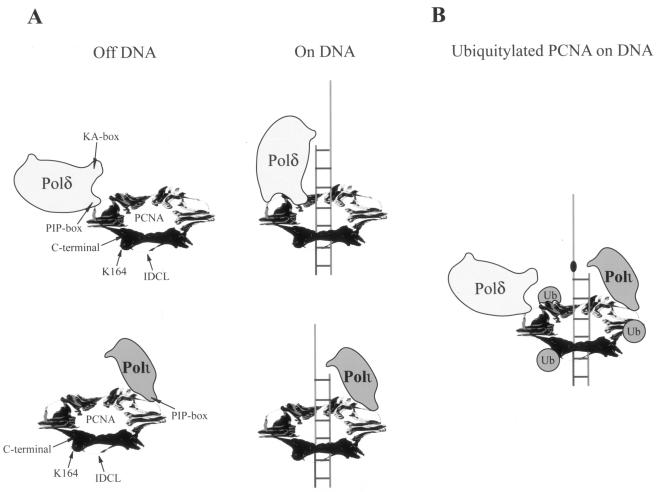
The ability of Polι and other Y family Pols to contact PCNA at only the IDCL site could have important consequences for Pol exchange at the lesion site. By virtue of its ability to contact PCNA at many sites, Polδ would remain stably bound to PCNA when replicating undamaged DNA (Fig. 5A). However, upon encountering a DNA lesion, Polδ would stall, and that presumably activates the Rad6-Rad18-dependent ubiquitylation of PCNA (14), resulting in the displacement of Polδ from the template-primer junction (Fig. 5B). Importantly, however, because of its multisite binding to PCNA, we expect Polδ to still remain bound to PCNA. Furthermore, the ubiquitylation of PCNA at the lysine 164 residue could be important for exposing the IDCL region to allow access of Polι and other Y family Pols to PCNA (12, 14, 29) (Fig. 5B). However, because Polι and other Y family Pols would be loosely anchored to PCNA at only one site, they would dissociate from DNA soon after their role in lesion bypass has been accomplished, whereupon Polδ would regain access to the template-primer junction.
FIG. 5.
Model for the binding of PCNA by Polδ versus Polι and for Pol exchange at the lesion site. (A) Binding of Polδ and Polι on or off DNA. In the absence of DNA, both Pols could bind to PCNA via contacts with the IDCL region. On DNA, Polδ, via its multiple subunits, interacts with PCNA at multiple sites, whereas Polι contacts only the IDCL region of PCNA. Although Polδ could also be bound to different PCNA monomers, Polδ binding to only one monomer is shown. K164 in PCNA is the site of ubiquitin (Ub) attachment by the Rad6-Rad18 enzyme complex. (B) Pol exchange at the lesion site. Rad6-Rad18-dependent PCNA ubiquitylation destabilizes the interactions of Polδ with PCNA, resulting in the displacement of Polδ from the primer end and allowing for the access of Polι to the IDCL region of PCNA. Polδ presumably still remains bound to PCNA through multisite contacts (data not shown) and would regain access to the primer junction soon after the completion of lesion bypass and the concomitant exit of Polι from the replication ensemble.
Acknowledgments
This work was supported by National Institute of Environmental Health Sciences grant ES012411, the Wellcome Trust International Senior Research Fellowship, the Hungarian Science Foundation grant T043354, and the EMBO Restart Fellowship.
REFERENCES
- 1.Bambara, R. A., R. S. Murante, and L. A. Henricksen. 1997. Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem. 272:4647-4650. [DOI] [PubMed] [Google Scholar]
- 2.Bermudez, W. P., S. A. MacNeill, I. Tappin, and J. Hurwitz. 2002. The influence of the Cdc27 subunit on the properties of the Schizosaccharomyces pombe DNA polymerase δ. J. Biol. Chem. 277:36853-36862. [DOI] [PubMed] [Google Scholar]
- 3.Cai, J., E. Gibbs, F. Uhlmann, B. Phillips, N. Yano, M. O'Donnell, and J. Hurwitz. 1997. A complex consisting of human replication factor C, p40, p37, and p36 subunits is a DNA-dependent ATPase and an intermediate in the assembly of the holoenzyme. J. Biol. Chem. 272:18974-18981. [DOI] [PubMed] [Google Scholar]
- 4.Chen, U., S. Chen, P. Saha, and A. Dutta. 1996. p21/Cip1/Waf1 disrupts the recruitment of human Fen1 by proliferating-cell nuclear antigen into the DNA replication complex. Proc. Natl. Acad. Sci. USA 93:11597-11602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eissenberg, J. C., R. Ayyagari, X. V. Gomes, and P. M. J. Burgers. 1997. Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase δ and DNA polymerase ɛ. Mol. Cell. Biol. 17:6367-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerik, K. J., X. Li, A. Pautz, and P. M. J. Burgers. 1998. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 273:19747-19755. [DOI] [PubMed] [Google Scholar]
- 7.Gibbs, E., Z. Kelman, J. M. Gulbis, M. O'Donnell, J. Kuriyan, P. M. Burgers, and J. Hurwitz. 1997. The influence of the proliferating cell nuclear antigen-interacting domain of p21 (CIP1) on DNA synthesis catalyzed by the human and Saccharomyces cerevisiae polymerase delta holoenzymes. J. Biol. Chem. 272:2373-2381. [DOI] [PubMed] [Google Scholar]
- 8.Gomes, X. V., and P. M. J. Burgers. 2000. Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 19:3811-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haracska, L., R. E. Johnson, I. Unk, B. Phillips, J. Hurwitz, L. Prakash, and S. Prakash. 2001. Physical and functional interactions of human DNA polymerase η with PCNA. Mol. Cell. Biol. 21:7199-7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haracska, L., R. E. Johnson, I. Unk, B. B. Phillips, J. Hurwitz, L. Prakash, and S. Prakash. 2001. Targeting of human DNA polymerase ι to the replication machinery via interaction with PCNA. Proc. Natl. Acad. Sci. USA 98:14256-14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haracska, L., C. M. Kondratick, I. Unk, S. Prakash, and L. Prakash. 2001. Interaction with PCNA is essential for yeast DNA polymerase η function. Mol. Cell 8:407-415. [DOI] [PubMed] [Google Scholar]
- 12.Haracska, L., C. A. Torres-Ramos, R. E. Johnson, S. Prakash, and L. Prakash. 2004. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:4267-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haracska, L., I. Unk, R. E. Johnson, B. B. Phillips, J. Hurwitz, L. Prakash, and S. Prakash. 2002. Stimulation of DNA synthesis activity of human DNA polymerase κ by PCNA. Mol. Cell. Biol. 22:784-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoege, C., B. Pfander, G.-L. Moldovan, G. Pyrowolakis, and S. Jentsch. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135-141. [DOI] [PubMed] [Google Scholar]
- 15.Johansson, E., P. Garg, and P. M. J. Burgers. 2004. The Pol32 subunit of DNA polymerase δ contains separable domains for processive replicatin and proliferating cell nuclear antigen (PCNA) binding. J. Biol. Chem. 279:1907-1915. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, R. E., M. T. Washington, L. Haracska, S. Prakash, and L. Prakash. 2000. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 406:1015-1019. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, R. E., M. T. Washington, S. Prakash, and L. Prakash. 2000. Fidelity of human DNA polymerase η. J. Biol. Chem. 275:7447-7450. [DOI] [PubMed] [Google Scholar]
- 18.Kelman, Z., and J. Hurwitz. 1998. Protein-PCNA interactions: a DNA-scanning mechanism? Trends Biol. Sci. 23:236-238. [DOI] [PubMed] [Google Scholar]
- 19.Kelman, Z., N. Yao, and M. O'Donnell. 1995. Escherichia coli expression vectors containing a protein kinase recognition motif, His6-tag and hemagglutinin epitope. Gene 166:177-178. [DOI] [PubMed] [Google Scholar]
- 20.Lee, S. H., T. Eki, and J. Hurwitz. 1989. Synthesis of DNA containing the simian virus 40 origin of replication by the combined action of DNA polymerases alpha and delta. Proc. Natl. Acad. Sci. USA 86:7361-7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, X., J. Li, J. Harrington, M. R. Lieber, and P. M. Burgers. 1995. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by PCNA. J. Biol. Chem. 270:22109-22112. [DOI] [PubMed] [Google Scholar]
- 22.MacNeill, S. A., S. Moreno, N. Reynolds, P. Nurse, and P. A. Fantes. 1996. The fission yeast Cdc1 protein, a homologue of the small subunit of DNA polymerase δ, binds to Pol3 and Cdc27. EMBO J. 15:4613-4628. [PMC free article] [PubMed] [Google Scholar]
- 23.Nakanishi, M., R. S. Robetorye, O. M. Pereira-Smith, and J. R. Smith. 1995. The C-terminal region of p21SD11/WAF1/CIP1 is involved in proliferating cell nuclear antigen binding but does not appear to be required for growth inhibition. J. Biol. Chem. 270:17060-17063. [DOI] [PubMed] [Google Scholar]
- 24.Prakash, S., and L. Prakash. 2002. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 16:1872-1883. [DOI] [PubMed] [Google Scholar]
- 25.Prelich, G., M. Kostura, D. R. Marshak, M. B. Matthews, and B. Stillman. 1987. The cell-cycle regulated proliferating cell nuclear antigen is required for SV40 DNA replication in vitro. Nature 326:471-475. [DOI] [PubMed] [Google Scholar]
- 26.Reynolds, N., E. Warbrick, P. A. Fantes, and S. A. MacNeill. 2000. Essential interaction between the fission yeast DNA polymerase δ subunit Cdc27 and Pcn1 (PCNA) mediated through a C-terminal p21Cip1-like PCNA binding motif. EMBO J. 19:1108-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynolds, N., A. Watt, P. A. Fantes, and S. A. MacNeill. 1998. Cdm1, the smallest subunit of DNA polymerase δ in the fission yeast Schizosaccharomyces pombe, is non-essential for growth and division. Curr. Genet. 34:250-258. [DOI] [PubMed] [Google Scholar]
- 28.Shibahara, K.-I., and B. Stillman. 1999. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96:575-585. [DOI] [PubMed] [Google Scholar]
- 29.Stelter, P., and H. D. Ulrich. 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425:188-191. [DOI] [PubMed] [Google Scholar]
- 30.Tan, C. K., C. Castillo, A. G. So, and K. M. Downey. 1986. An auxiliary protein for DNA polymerase δ from fetal calf thymus. J. Biol. Chem. 261:12310-12316. [PubMed] [Google Scholar]
- 31.Tissier, A., J. P. McDonald, E. G. Frank, and R. Woodgate. 2000. Polι, a remarkably error-prone human DNA polymerase. Genes Dev. 14:1642-1650. [PMC free article] [PubMed] [Google Scholar]
- 32.Unk, I., L. Haracska, X. V. Gomes, P. M. J. Burgers, L. Prakash, and S. Prakash. 2002. Stimulation of 3′→5′ exonuclease and 3′-phosphodiesterase activities of yeast Apn2 by proliferating cell nuclear antigen. Mol. Cell. Biol. 22:6480-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warbrick, E., D. P. Lane, D. M. Glover, and L. S. Cox. 1995. A small peptide inhibitor of DNA replication defines the site of interaction between the cyclin-dependent kinase inhibitor p21WAF1 and proliferating cell nuclear antigen. Curr. Biol. 5:275-282. [DOI] [PubMed] [Google Scholar]
- 34.Washington, M. T., R. E. Johnson, L. Prakash, and S. Prakash. 2004. Human DNA polymerase ι utilizes different nucleotide incorporation mechanisms dependent upon the template base. Mol. Cell. Biol. 24:936-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Washington, M. T., I. G. Minko, R. E. Johnson, W. T. Wolfle, T. M. Harris, R. S. Lloyd, S. Prakash, and L. Prakash. 2004. Efficient and error-free replication past a minor groove DNA adduct by the sequential action of human DNA polymerases ι and κ. Mol. Cell. Biol. 24:5687-5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu, H., P. Zhang, L. Liu, and M. Y. W. T. Lee. 2001. A novel PCNA-binding motif identified by the panning of a random peptide display library. Biochemistry 40:4512-4520. [DOI] [PubMed] [Google Scholar]
- 37.Yuzhakov, A., Z. Kelman, J. Hurwitz, and M. O'Donnell. 1999. Multiple competition reactions for RPA order the assembly of the DNA polymerase δ holoenzyme. EMBO J. 18:6189-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, P., J.-Y. Mo, A. Perez, A. Leon, L. Liu, N. Mazloum, H. Xu, and M. Y. W. T. Lee. 1999. Direct interaction of proliferating cell nuclear antigen with the p125 catalytic subunit of mammalian DNA polymerase δ. J. Biol. Chem. 274:26647-26653. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, Y., F. Yuan, X. Wu, and Z. Wang. 2000. Preferential incorporation of G opposite template T by the low-fidelity human DNA polymerase ι. Mol. Cell. Biol. 20:7099-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]