Abstract
Paxillin is a prominent focal adhesion docking protein that regulates cell adhesion and migration. Although numerous paxillin-binding proteins have been identified and paxillin is required for normal embryogenesis, the precise mechanism by which paxillin functions in vivo has not yet been determined. We identified an ortholog of mammalian paxillin in Drosophila (Dpax) and have undertaken a genetic analysis of paxillin function during development. Overexpression of Dpax disrupted leg and wing development, suggesting a role for paxillin in imaginal disc morphogenesis. These defects may reflect a function for paxillin in regulation of Rho family GTPase signaling as paxillin interacts genetically with Rac and Rho in the developing eye. Moreover, a gain-of-function suppressor screen identified a genetic interaction between Dpax and cdi in wing development. cdi belongs to the cofilin kinase family, which includes the downstream Rho target, LIM kinase (LIMK). Significantly, strong genetic interactions were detected between Dpax and Dlimk, as well as downstream effectors of Dlimk. Supporting these genetic data, biochemical studies indicate that paxillin regulates Rac and Rho activity, positively regulating Rac and negatively regulating Rho. Taken together, these data indicate the importance of paxillin modulation of Rho family GTPases during development and identify the LIMK pathway as a critical target of paxillin-mediated Rho regulation.
Regulation of cell adhesion to the surrounding extracellular matrix (ECM) plays an essential role in organizing tissues and organs during development, and dysregulation of cell adhesion has been implicated in tumor invasion and metastasis (15). The adhesion process is mediated predominantly through a class of heterodimeric transmembrane receptors called integrins (17). Engagement of integrin receptors transduces a cascade of signaling events that regulate cell migration, proliferation, and survival. The cytoplasmic domains of integrins associate with a group of dynamic effectors, which includes several actin-binding proteins, focal adhesion kinase (FAK), Src family kinases, p130CAS, and paxillin (17). Focal adhesions play an essential role in linking the ECM to the actin cytoskeleton.
Paxillin was originally identified as a tyrosine phosphorylated protein in v-src-transformed cells and subsequently as a target of cellular Src family kinases (33, 34). Tyrosine phosphorylation of paxillin is also observed upon integrin-mediated cell adhesion and upon growth factor stimulation (5). Molecular analysis revealed that paxillin is a multidomain protein which contains five leucine-rich motifs, known as LD repeats, and four tandemly arranged LIM domains that are conserved among mammalian, avian, and Drosophila paxillin (30, 39, 42). The LD repeats allow paxillin to interact with actin-binding proteins, kinases, and the ARF family GTPase-activating proteins (ARF-GAPs). While a limited number of binding partners have been identified for the LIM domains, this region is crucial for mediating paxillin localization to focal adhesions (3, 30). The array of docking sites in paxillin mediate the recruitment and assembly of multiprotein complexes to focal adhesions and allow paxillin to function as an adapter to coordinate various signaling pathways (30, 37).
Recently, it was demonstrated that the LD4 repeat of paxillin associates with several of the ARF-GAPs, which are known for their role in regulating membrane trafficking and organelle structure (22, 38). The ARF-GAP protein p95PKL could link paxillin to Rho family GTPase signaling by binding to a protein complex containing the Rac-specific guanine nucleotide exchange factor PIX/COOL, the Rac effector target p21-activated serine/threonine kinase PAK, and the SH2-SH3 adaptor protein Nck (20, 38).
The Rho family GTPases, including Rho, Rac, and Cdc42, are known for their function in regulating actin cytoskeletal structure, cell motility, and morphogenesis (27). Activation of Rho induces focal adhesion and stress fiber formation, and activation of Rac and Cdc42 results in the formation of lamellipodia and filopodia, respectively. The recruitment of the PKL-PIX-PAK complex to focal adhesions by paxillin may stimulate the transition from Rho-mediated focal adhesions to Rac-mediated focal contacts to facilitate membrane protrusion and cell migration (40). Thus, these various features of the paxillin protein indicate that it can potentially play an important role in regulating signals from Rho GTPases to the actin cytoskeleton.
The in vivo function of paxillin has recently been addressed using a gene-targeting strategy in mice (12). Targeted disruption of the paxillin gene in the mouse embryo results in multiple defects in the development of mesodermally derived tissues such as heart and somites. Analysis of focal adhesions and lamellipodia in embryo-derived cells lacking paxillin indicated that paxillin is not required for the formation of focal adhesions or lamellipodia; however, paxillin is likely to be important for maintenance of lamellipodia and is required for focal adhesion turnover (41). Consistent with these functions, paxillin-deficient cells also exhibit delayed cell spreading and reduced motility (12).
To further explore the role of paxillin in regulating signaling pathways that influence tissue morphogenesis during development, we have utilized a genetic approach in Drosophila studies. Genetic analysis revealed that a Drosophila ortholog of mammalian paxillin modulates Rho and Rac GTPase signaling during Drosophila development. Significantly, paxillin antagonizes Rho signals to the actin cytoskeleton but promotes signals from Rac. This regulation may be crucial for development of imaginal disc structures such as the wing and leg, as overexpression of paxillin results in leg defects and wing blisters. In support of this hypothesis, a gain-of-function suppressor screening for Dpax-induced wing blisters identified a genetic interaction between Dpax and cdi, the Drosophila homolog of TESK, the cofilin kinase family member. Cofilin kinases include LIMKs, and LIMKs are important mediators of Rho signaling that promote actin polymerization by phosphorylating and inhibiting the actin-severing protein cofilin (18). Consistent with the results from the suppressor screening, Dlimk and other components in this pathway also interacted genetically with Dpax. One mechanism by which paxillin may control Rho signaling is at the level of Rac and Rho activation, as Rac and Rho are misregulated in paxillin-deficient mouse embryo fibroblasts (MEFs). Taken together, these data suggest that a conserved role for paxillin in regulating signals from Rho GTPases to the actin cytoskeleton is critical for normal tissue morphogenesis during development.
MATERIALS AND METHODS
Isolation of the paxillin cDNA and DNA manipulation.
A DNA fragment containing sequences encoding the Drosophila homolog of mammalian paxillin was isolated by degenerate PCR of fly genomic DNA. The PCR product was then used as a probe to screen a Drosophila embryonic cDNA library (Clontech). Phage clones that gave positive results were purified, cloned into the PBSKSII vector, and sequenced using an ABI automated DNA sequencer. The Dpax antibody is a polyclonal antibody generated against a His-tagged fusion protein containing the N-terminal half of Dpax. For generation of the Dpax RNAi, a ∼800-bp fragment of the Dpax N-terminal region was amplified by PCR using two pairs of primers. One PCR used 5′GACTGAATTCGATGCTCTATTAGCCGACCTG-3′ and 5′GAATGCGGCCGCCTGGGTGAGATGTGCCTGCTG-3′ primers and resulted in EcoRI and NotI sites at either end of the fragment. The other reaction used two related primers but with BamHI and XbaI sites at either end. Each of these fragments was then sequentially inserted into the corresponding restriction sites in the pCaSpeR-hs. These inserts resulted in a head-to-head inverted repeat (IR) formation. The hs-Dpax-IR fly was generated by transformation of Drosophila embryo as described below.
Drosophila stocks and genetics.
Fly stocks were maintained on cornmeal-molasses medium at 25°C. For P-element transformation, the Dpax cDNA was cloned into the pGMR and/or pUAST vector and coinjected with the delta2.3 transposase plasmid into W1118 embryos prior to cellularization. More than four independent insertion lines of each construct were established. To drive expression of these upstream activation sequence (UAS) constructs, several GAL4 enhancer lines were used, including engrailed (en)-GAL4, 32B-GAL4, Act5C-GAL4, and hs-GAL4. To generate the 32B>UAS-Dpax allele, we have recombined 32B-GAL4 onto a UAS-Dpax chromosome. The EP collection was obtained from the Berkeley Drosophila Genome Project (28). Individual EP lines were crossed to the 32B>UAS-Dpax at 25°C. UAS-DSRF was a gift from M. Affolter. The following genetic strains were used for the genetic interaction assays. The fly strains tsrK05633 and ssh01207 were provided by the Bloomington Stock Center. Genetic interactions were tested at 25°C unless noted otherwise.
Electron microscopy.
Scanning electron micrographs of Drosophila eyes were generated as described previously (16).
Immunostaining.
Imaginal discs from wandering third instar larvae were dissected, collected, and rinsed in phosphate-buffered saline (PBS) prior to fixing in 4% paraformaldehyde for 30 min. Imaginal discs were incubated overnight at 4°C with anti-Dpax antibody (1:50), washed three times for 5 min each time in PBS-0.1% Triton X-100 followed by 4 h of incubation in secondary antibody, and washed five times in PBS-0.1% Triton X-100. Imaginal discs were then mounted prior to visualization by confocal microscopy. F-actin was stained using tetramethyl rhodamine isocyanate-labeled phalloidin (Sigma).
Western blot analysis.
Flies from different developmental stages were lysed in RIPA buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% deoxycholate, 1% Triton X-100, 5 mM EDTA, 0.1% sodium dodecyl sulfate [SDS]) containing 1 mM sodium orthovanadate, 50 units of aprotinin/ml, 2 μg of leupeptin/ml, 1 mM phenylmethylsulfonyl fluoride, and 10 mM NaF. Insoluble materials were removed by centrifugation. The lysates were separated by SDS-10% polyacrylamide gel electrophoresis (PAGE) and transferred to a polyvinylidene difluoride membrane (Minipore). Membrane was probed with Dpax antibody followed by horseradish peroxidase-conjugated secondary, and signals were detected by enhanced chemiluminescence (Perkin-Elmer Life Sciences). For phosphocofilin analysis, cells were serum starved for 36 h and plated on fibronectin for various times. Lysates were prepared and normalized for total protein. Following SDS-PAGE and immobilization of proteins on nitrocellulose, filters were probed with antibodies to phosphorylated cofilin (Ser3; Cell Signaling) and then reprobed with a cofilin antibody (Cytoskeleton) to assure equal loading. For immunoprecipitations, lysates from Cos7 cells transfected with Flag-LIMK1, hemagglutinin-LIMK2 (HA-LIMK2), HA-p190N, or HA-p190GAP (W. Jiang and J. Settleman, unpublished data) were immunoprecipitated using either 1 μg of paxillin (Transduction Laboratories) antibody or immunoglobulin G. The LIMK1 antibody (UBI) was used at a 1:500 dilution, and the HA antibody (Santa Cruz) was used at a 1:1,000 dilution.
Transcriptional reporter assays.
Mouse embryo-derived fibroblasts from mice containing a targeted disruption of the paxillin gene and the rescued cells were previously described (12). Paxillin-deficient or -rescued cells were cotransfected with a serum response factor (SRF)-luciferase reporter plasmid and a herpes simplex virus-thymidine kinase Renilla plasmid for normalization. After 40 h, cells were lysed and luciferase activity was measured using the luciferase assay system (Promega). To measure the effects of overexpressing paxillin on LIMK-induced SRF activity, COS7 cells were transfected with either the SRF-luciferase reporter plasmid alone or together with a LIMK1 expression vector and, where indicated, a paxillin expression vector (2 μg of each). Luciferase activity was measured after 40 h.
GTPase activation assays.
To measure the activation of Rho and Rac GTPases, paxillin-deficient MEFs or their rescued counterparts were serum starved for 36 h and then plated on fibronectin (Fn) for various times. Assays were done as previously described (26, 29). Briefly, for Rho assays, cells were lysed in buffer A (50 mM Tris [pH 7.6], 500 mM NaCl, 0.1% SDS, 0.5% deoxycholate, 1% Triton X-100, 0.5 mM MgCl2) plus protease inhibitors. Lysates were incubated for 60 min on ice with bacterially produced glutathione S-transferase (GST)-Rhotekin to capture GTP-bound Rho. Beads were then washed several times with buffer B (50 mM Tris [pH 7.6], 150 mM NaCl, 1% Triton X-100, 0.5 mM MgCl2) and then resuspended in Laemmli buffer. To measure Rac activation, cells were lysed in buffer B plus inhibitors and lysates were incubated for 20 min on ice with bacterially produced GST-PAK (RBD) to capture GTP-bound Rac. Beads were washed several times with buffer B and then resuspended in Laemmli buffer. Bound GTPases were analyzed by SDS-PAGE, and resolved proteins were transferred to nitrocellulose (Osmonics). Filters were probed with anti-Rho or anti-Rac antibodies (Santa Cruz Biotechnology and BD Biosciences), followed by fluorescence-conjugated secondary antibody (Molecular Probes). Signals were detected and quantitated using a Li-Cor Odyssey Infrared Imager and Odyssey software. Total lysates were analyzed in parallel for normalization purposes.
RESULTS
Cloning and expression of Drosophila paxillin.
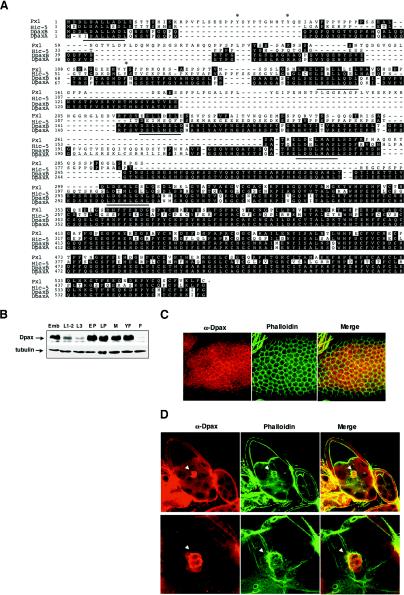
A closely related Drosophila ortholog of mammalian paxillin, designated DpaxA, was isolated by hybridization screening of a Drosophila embryonic cDNA library. DpaxA is highly homologous to mammalian paxillin, including the presence of five LD repeats and four LIM motifs in the C-terminal half. Another group has previously isolated similar Dpax cDNA clones (42). This clone, however, differs slightly at the 5′ end from our clone and also diverges between the third and fourth LD repeats (DpaxB) (Fig. 1A). A second group has also isolated a Dpax clone (called DPxn37) which is identical to the clone that we have isolated (44). It is possible that these differences reflect alternative splicing of the paxillin gene. In addition, while these clones have been designated as orthologs of mammalian paxillin, comparison to Hic-5, a second paxillin family member, also reveals a high (43%) degree of homology (Fig. 1A).
FIG. 1.
Molecular characterization and expression of Drosophila paxillin. (A) Alignment of mammalian paxillin, Hic-5, and the two Dpax isoforms, DpaxA and DpaxB. The DpaxA sequence is similar to that reported by Wheeler et al. (42). Asterisks denote known tyrosine phosphorylation sites in avian and mammalian paxillin. The dotted line represents the region of sequence divergence between DpaxA and -B. LD repeats are underlined. (B) Lysates from the embryonic stage (Emb) (0 to 24 h), larval stages (L1-2, first and second instar; L3, third instar), and pupal stages (EP, early pupae; LP, late pupae) and from male (M) and female (YF, newly enclosed; F, 24-h-old) adult flies were subjected to immunoblotting with anti-Dpax antibody. (C and D) Dpax is expressed in follicle cells in the developing ovary (C) and in border cells (arrowheads) (D). Ovaries were costained with Dpax antibody and phalloidin. In panel D, the lower panel shows higher magnification. Merge, merged images of the left and center panels.
In situ hybridization analysis of Dpax mRNA expression during embryogenesis revealed that Dpax expression is enriched in the peripheral nervous system, leading edge cells, and muscle attachment sites (reference 42 and data not shown). To determine the expression of Drosophila paxillin later in development, total protein extracts from embryonic, first and second instar, third instar, early pupal, late pupal, and adult developmental stages were analyzed by immunoblotting for Dpax. Dpax expression is high during embryonic development, is decreased during larval development, and is relatively high again during pupal stages (Fig. 1B). In the developing egg chamber, Dpax is found in the follicle cells. Although Dpax shows diffuse cytoplasmic staining, it also colocalizes with F-actin at the cell periphery (Fig. 1C). It is also highly expressed in the border cells (Fig. 1D). Intriguingly, although Dpax protein is expressed in both border cells and follicle cells, Western analysis indicates that it is much more highly expressed in males than females (Fig. 1B).
Overexpression of Dpax during development disrupts wing and leg morphogenesis.
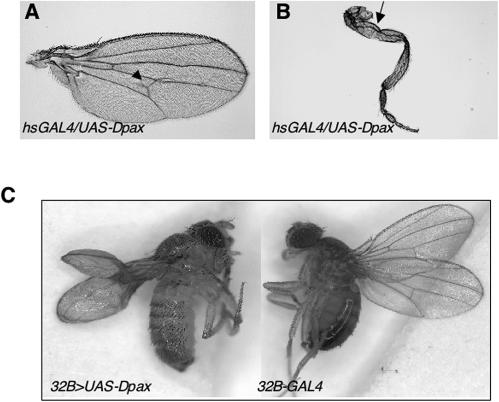
To begin to address the function of Dpax in vivo, we generated transgenic flies that express a wild-type Dpax under the control of the GAL4-responsive UAS element (UAS-Dpax). Expression of wild-type Dpax at the first instar larval stage using an hs-GAL4 driver (heat shock driven) results in lethality. These larvae die within 48 h after heat shock and exhibit an aberrant dentical epidermis (data not shown). Furthermore, overexpression of Dpax at the late larval-pupal stages results in a pupal lethal phenotype with a few escapers. These escaper flies survive to adulthood and exhibit extra-wing-vein or blistered-wing and malformed-leg phenotypes (Fig. 2A and B), suggesting a role for Dpax during imaginal disc morphogenesis. We therefore further examined the role of Dpax during wing morphogenesis by use of the wing-specific drivers 69B-GAL4 and 32B-GAL4. Overexpressing Dpax with these drivers results in a highly penetrant, poorly expanded, and blistered-wing phenotype (Fig. 2C and data not shown), characteristic of altered cell adhesion. This phenotype was also observed using the ubiquitously expressed Actin5C driver (data not shown). The Dpax-induced blistered-wing phenotype was rescued by expression of Dpax double-stranded RNA (hs-Dpax-IR), thus suggesting that the phenotype is Dpax specific (Table 1).
FIG. 2.
Overexpression of Dpax results in malformed wing and leg phenotypes. (A and B) Overexpressing Dpax results in mostly pupa lethal phenotypes, but a few escapers carry malformed-wing (A) and -leg (B) phenotypes. The hsGAL4/+;UAS-Dpax/+ larvae were heat shocked at 96 h after egg deposition (AED) daily for 40 min at 37°C through eclosion. Extra wing veins (arrowhead) and bent and/or twisted legs (arrow) are observed in flies expressing Dpax. (C) Overexpression of UAS-Dpax driven by 32B-GAL4 induces wing blisters.
TABLE 1.
Quantification of the genetic interactions with 32B>UAS-Dpax
| Genotype for suppression of 32B>UAS-Dpax | % of wings with normal pattern (n)a |
|---|---|
| Control 1 (+/+) | 0 (81) |
| tsrk05633/+ | 56 (55) |
| ssh01207/+ | 48 (67) |
| EP3319 (cdi) | 40 (75) |
| UAS-Dlimk/+ | 94 (71) |
| UAS-DSRF/+ | 95 (80) |
| Control2 (+/+)b | 0 (62) |
| hs-Dpax-IR/+b | 74 (82) |
The percentages of wings with normal pattern for flies carrying the genotypes heterozygous for the indicated alleles and containing one copy of the 32B>UAS-Dpax transgene are shown.
Heat shock experiments involved heat treatment at 72 h after egg deposition daily for 30 min at 37°C through eclosion.
Dpax interacts genetically with Rho and Rac GTPases.
A similar malformed-leg phenotype has been previously identified in zipper (zip) and spaghetti squash (sqh) mutant flies (8). zip and sqh encode Drosophila nonmuscle myosin heavy chain and nonmuscle myosin regulatory light chain, respectively. Biochemical and genetic analysis have previously shown that zip and sqh function downstream of the Rho GTPase (43). Significantly, flies carrying both Rho1 and zip mutations exhibit severely malformed wings and legs, raising the possibility that Dpax also functions in a Rho GTPase-mediated signaling pathway required for imaginal disc morphogenesis.
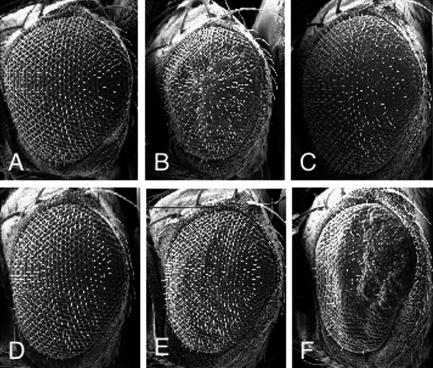
To examine further a potential genetic interaction between Dpax and the Rho GTPases, we took advantage of transgenic flies that we had previously generated that express various Rho family GTPase specifically in the developing eye (13, 24). We have previously shown that overexpression of Drosophila Rho GTPases in the developing eye causes a rough-eye phenotype (Fig. 3B and C). Overexpression of Dpax in the eye, using the eye-specific GMR promoter, results in flies that lack any obvious eye defects (Fig. 3D). Interestingly, in flies overexpressing Dpax together with Rho1, there is a substantial reduction in Rho1-induced eye roughness (Fig. 3E) whereas Dpax expression significantly enhances the Rac1-induced rough-eye phenotype (Fig. 3F). These results suggest that Dpax negatively regulates Rho1-mediated signaling but activates Rac1-mediated signaling in developing tissues in vivo.
FIG. 3.
Dpax genetically interacts with Rho and Rac in eye morphogenesis. Scanning electron micrographs of the compound eye of wild-type (A), GMR-Rho11Rho13/+ (B), GMR-Rac1/+ (C), GMR-Dpax/+ (D), GMR-Rho11Rho13/GMR-Dpax (E), and GMR-Rac1/GMR-Dpax (F) flies are shown. Dpax expression alleviates Rho1 but enhances Rac1-induced rough-eye phenotypes (E and F).
Cofilin kinases influence Dpax signaling.
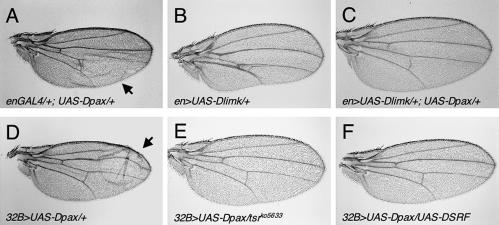
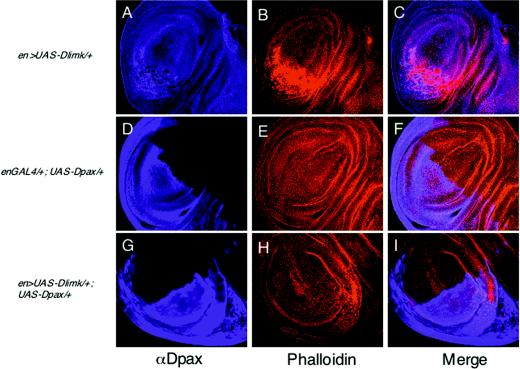
To identify additional components involved in the paxillin-mediated signaling pathway, we conducted a gain-of-function genetic interaction screening for genes that can rescue the Dpax-induced wing blister. We utilized a collection of previously generated fly lines that harbor various P-element insertions (EP lines) in which genes localized adjacent to the insertion site can be specifically overexpressed. In a screening of approximately 1,500 independent EP insertion lines on the second and third chromosome, we identified three lines which, when crossed to the Dpax transgenic flies, can modify the Dpax-induced blistered-wing phenotype (Table 1 and data not shown). Here, we focus on one of these lines, EP3319, in which the insertion drives expression of the center divider gene (cdi), which encodes the Drosophila homolog of human testicular protein kinase (TESK) (21). TESK phosphorylates and inhibits cofilin, the actin-depolymerizing factor, and flies expressing excessive cdi exhibit a substantial reduction in Dpax-induced wing blistering (Table 1). Significantly, the kinase domain of cdi is highly homologous (42% identity) to LIMK, the cofilin-phosphorylating protein. We have recently identified the Drosophila ortholog of LIMK (Dlimk) (6). Consistent with results from the gain-of-function screening, overexpression of Dlimk was able to suppress the Dpax-induced wing blisters (94%, n = 71; compare Fig. 4A, B, and C). In addition, Dpax expression substantially decreases Dlimk-induced F-actin accumulation in wing imaginal disks when the two proteins are coexpressed (Fig. 5; compare 5B and H). Taken together, these results suggest that cofilin kinases and paxillin perform antagonistic functions in cell adhesion that may be mediated by cofilin phosphorylation and actin reorganization.
FIG. 4.
Dpax-induced wing blisters can be rescued by components in the Rho pathway, including Dlimk and its downstream targets. (A and D) Overexpression of UAS-Dpax driven by en-GAL4 or 32B-GAL4 results in blistered wings (arrows). (B) Overexpression of UAS-Dlimk driven by en-GAL4 results in multiple defects in the posterior part of the wing (6). (C and F) Overexpression of Dlimk (C) or DSRF (F) rescues the Dpax-induced blistered-wing phenotype. (E) A mutation in cofilin (tsrko5633), the downstream target of LIMK, rescues the wing-blistering defect. Flies were maintained at 18°C.
FIG. 5.
Dpax rescues Dlimk-induced F-actin accumulation. Third-instar wing imaginal disks were dissected from en >UAS-Dlimk/+ (A to C), enGAL4/+;UAS-Dpax/+ (D to F), or en >UAS-Dlimk/+;UAS-Dpax/+ (G to I) genotypes and stained with anti-Dpax antibody (A, D, and G) and tetramethyl rhodamine isocyanate-labeled phalloidin (B, E, and H). Merged images of the left and center panels are shown in panels C, F, and I.
In mammalian cells, LIMK functions downstream of both Rac and Rho; however, our recent studies on Dlimk indicate that in Drosophila this kinase may be specific to the Rho-mediated pathway during disc morphogenesis (6). The ability of Dlimk to suppress the Dpax-induced wing blisters is consistent with the role of this kinase specifically downstream of Rho. To further examine this relationship, we looked for genetic interactions between Dpax and downstream components of the LIMK-mediated pathway in the developing wing. Dlimk stabilizes F-actin by inactivating cofilin, which functions as an actin-depolymerizing factor (18). Recently, a Drosophila phosphatase called slingshot (ssh) that dephosphorylates cofilin was identified (23). Loss of ssh function causes a dramatic increase in the level of F-actin in developing tissues. We reasoned that loss of either cofilin or ssh function would be similar to increasing the Rho and LIMK activity and therefore would be able to modify the Dpax-induced wing blistering. Indeed, we found that mutations in either twinstar, which encodes Drosophila cofilin, or slingshot are each able to dominantly suppress the blistered-wing phenotype caused by Dpax misexpression (Fig. 4E and Table 1).
In mammals, it has been shown that LIMK is important in regulating SRF via regulation of actin dynamics (31, 32). Blistered (bs) encodes the Drosophila ortholog of SRF (DSRF). A blistered-wing phenotype has also been observed in the blistered mutant flies (10). Therefore, we examined a potential genetic interaction between Dpax and bs/DSRF. Overexpression of DSRF strongly suppresses the Dpax-induced wing blistering phenotype (95%, n = 80; Fig. 4F).
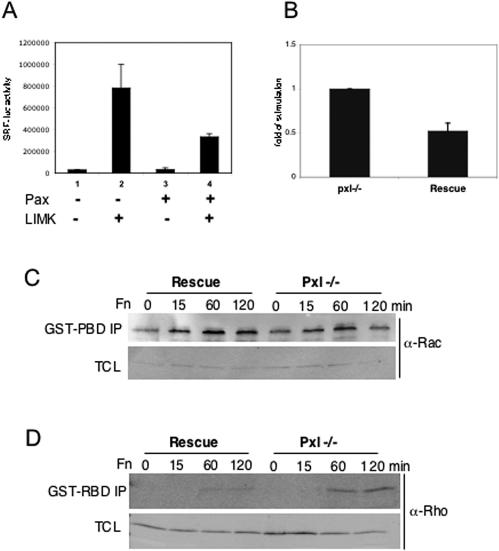
Thus, taken together these data indicate that Dpax may antagonize pathways leading to SRF activation. Indeed, in a cell culture experiment, an SRF transcriptional reporter assay revealed that paxillin overexpression inhibited LIMK-induced SRF activation (Fig. 6A).
FIG. 6.
Paxillin antagonizes the Rho-LIMK-mediated signaling pathway. (A) Paxillin suppresses LIMK1-induced SRF transcriptional activity in Cos7 cells. (B) Paxillin−/− cells exhibit approximately twofold-increased SRF transcriptional activity compared with rescued cells. (C) Paxillin−/− cells exhibit decreased Rac activity, as measured in pulldown assays with GST-PAK (PBD). The lower panel shows levels of rac in total cell lysate (TCL). (D) Paxillin−/− cells have increased Rho activity. The lower panel shows that levels of rho in different lysates are similar.
Cells lacking paxillin exhibit increased Rho and SRF activity.
Since the in vivo studies described above relied largely on overexpression, we wanted to confirm the observed relationship between paxillin and Rho GTPase signaling in a loss-of-function setting. We therefore examined SRF activation in paxillin-deficient (Pxl−/−) MEFs and Pxl−/− MEFs rescued with wild-type paxillin (rescued) by use of an SRF reporter assay. We observed a significant increase in reporter activity in Pxl−/− cells compared to that seen in rescued cells (Fig. 6B). Thus, a normal function of paxillin appears to be the negative regulation of SRF activation.
The SRF reporter studies and the reported defects in lamellipodium formation and increased focal adhesions in Pxl−/− MEFs, as well as the genetic studies described above, strongly suggest that paxillin can regulate Rho family GTPase pathways. Paxillin could be acting upstream or downstream of Rho GTPases to regulate their activation or the activation and localization of their effectors. To determine where in the pathway paxillin regulates Rho GTPase signaling, we examined Rac and Rho activation in Pxl−/− MEFs (Fig. 6C and D). Although little difference in Rac activation was detected at early time points (15 min), at later time points an approximately twofold decrease in Rac activation was observed in Pxl−/− MEFs, consistent with a positive role for paxillin in regulation of Rac. The difference in activation at later time points suggests that paxillin may be more important for maintaining Rac activation. In contrast to what is observed for Rac, Pxl−/− MEFs have enhanced Rho activation compared to the results seen with the Rescued cells, suggesting that paxillin negatively regulates Rho activation (Fig. 6D). While there was little difference in rho activation between paxillin-rescued and null cells at 15 min, a two- to threefold increase in rho activation was observed 60 min after plating and was still detected at 2 h. Taken together, these results provide both genetic and biochemical evidence for a role for paxillin in the regulation of Rac and Rho activity.
Regulation of Rho pathway.
Previous studies have suggested that one mechanism by which paxillin might modulate Rho is through indirect regulation of the RhoGAP, p190. In one model, p190 can bind to the SH2 domain of p120Ras GAP which keeps p190 inactive and release from p120RasGAP results in active p190. Active p190 has been shown to localize to the leading edge. Recent work has shown that paxillin can bind to p120RasGAP which releases p190 and thus would result in Rho inactivation. To determine whether the enhanced Rho activation could be due to loss of p190 regulation, p190 localization was examined in paxillin-deficient MEFs. Loss of paxillin resulted in only a minor decrease in p190 localization to the leading edge (data not shown), suggesting that in this system, regulation of p190 may not be the major mechanism by which paxillin regulates Rho.
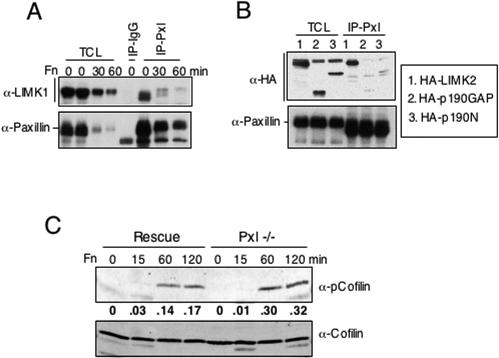
Given that paxillin is a scaffolding protein and that it can genetically interact with LIMK, we examined whether paxillin could bind to LIMK and/or regulate its activity. Because of the low levels of LIMK in fibroblasts, Cos7 cells were transfected with a LIMK1 or LIMK2 cDNA. The tagged LIMK was precipitated from randomly growing transfectants, transfectants in suspension, or transfectants that had been plated on fibronectin. Interestingly, paxillin was able to bind LIMK1 and LIMK2 under each of these conditions (Fig. 7A and B and data not shown). When normalization to the amount of paxillin immunoprecipitated was performed, there was no clear difference in the amounts of LIMK associated with paxillin under these different conditions. Thus, the interaction does not seem to be regulated by adhesion.
FIG. 7.
Paxillin interacts with and regulates LIMK. (A and B) Lysates were prepared from randomly growing or Fn-plated Cos7 cells that had been transfected with LIMK1 or LIMK2 (see Materials and Methods). Immunoblots were probed with antibody to paxillin, LIMK1, or HA. (C) Rescued or Pxl−/− MEFs were subjected to a fibronectin-plating assay as described above. Filters were probed with antibody to phosphocofilin (upper blot) or reprobed with antibody to cofilin (lower blot). Quantitation (shown below the phosphocofilin blot) was done on a Li-Cor infrared imaging system.
To determine whether paxillin may regulate LIMK activity, cofilin phosphorylation was examined in paxillin−/− MEFs. Loss of paxillin resulted in elevated LIMK activity, as measured by increased cofilin phosphorylation at serine 3 (Fig. 7C). While both rescued and Pxl−/− cells had elevated levels of cofilin phosphorylation at 60 min, Pxl−/− cells plated on fibronectin for 60 min showed a twofold increase in phosphocofilin levels at 60 min, and this difference was still detected at 2 h. This was not due to differences in the amount of cofilin. Taken together these studies suggest that paxillin might regulate the Rho pathway by modulating Rho activation as well as by regulating a downstream effector, LIMK.
DISCUSSION
Paxillin is a scaffolding protein found in focal adhesions (30, 37). Targeted disruption of paxillin in mice results in an early embryonic lethal phenotype with defects in multiple mesodermally derived structures (12). The recent completion of the Drosophila genome revealed the evolutionary conservation of many of the key molecules found in focal adhesions, including integrins, paxillin, vinculin, FAK, p130CAS, and ILK (14). We have identified the Drosophila paxillin, which is predominantly expressed in embryos, pupae, and male adults. In situ analysis of staged embryos revealed a restricted expression pattern of Dpax (42). In particular, Dpax is highly expressed in tissues undergoing cell shape changes or cell migration. Overexpression of Dpax in late larval stages results in a pupal lethal phenotype with few escapers bearing malformed phenotypes, suggesting that Dpax also plays an important role during later stages of development.
A loss-of-function mutant of Drosophila paxillin has not yet been reported. Therefore, we employed the UAS/GAL4 system to investigate the function of Dpax in the later stages of development. As has been reported for Drosophila FAK, overexpressing Dpax results in a blistered-wing phenotype. In mammals, paxillin is a substrate of FAK in transducing signals from integrins. FAK regulates focal adhesion disassembly and has been shown to be involved in Drosophila Wnt4-mediated cell movement during ovarian morphogenesis and is also required for border cell migration during oogenesis (7, 9). The function of Dpax in oogenesis is not clear; however, we found that Dpax is also highly expressed in the border cells (Fig. 1D).
The blistered-wing phenotype is also found in integrin mutant flies (2). In the prepupal stage, the wing is a single epithelial sheet; integrins have been suggested to play a regulatory role. As development progresses this sheet folds into a dorsal and ventral side, and the integrins play an adhesive role at these later stages. Using drivers that are expressed at different stages of development, the studies suggest that paxillin could be important for both the regulatory and adhesive functions of the integrins. Such functions would be consistent with studies of mammalian systems in which paxillin functions downstream of multiple integrins and can regulate both inside out and outside in signaling (12, 19, 45). In addition, both paxillin and FAK are important for focal adhesion turnover (41). Thus, too much paxillin or FAK may increase the turnover of focal complexes and perturb the stable adhesion between two epithelia, thereby resulting in the blistering phenotype.
Using a gain-of-function screen for modifiers that can rescue the Dpax-induced wing blistering, we identified Cdi/TESK. Like LIMK, Cdi/TESK phosphorylates the actin-depolymerizing factor cofilin and stabilizes F-actin. Cdi/TESK is highly homologous to LIMK in the kinase domain; however, a recent study demonstrated that Cdi/TESK functions downstream of Rac1 during spermatogenesis (25). Drosophila LIMK functions downstream of Rho1 in regulating disk morphogenesis (6). Dlimk and components in the Rho-LIMK pathway, including ssh, tsr, and bs/DSRF, also rescue the blistering phenotype. In addition, another regulator of SRF and actin, diaphanous, also showed genetic interactions with Dpax (data not shown). Diaphanous is a direct effector of Rho which cooperates with LIMK to regulate SRF activation (11, 35). All of these components play important roles in regulating F-actin synthesis. Taken together, these data indicate that it is possible that an increase in actin levels can prevent the increase in focal adhesion turnover caused by the excess level of paxillin, therefore suppressing the blistering phenotype. It is possible that simply overexpressing actin might be sufficient to rescue the blistering phenotype, although our results shown in Fig. 5 suggest that paxillin itself does not affect F-actin synthesis or actin organization. The ability of paxillin, however, to coimmunoprecipitate with LIMK and the increased cofilin phosphorylation in Pxl−/− MEFs suggests that paxillin can modulate LIMK function. These data, combined with the genetic and biochemical evidence that paxillin can regulate Rho, suggest that paxillin could act at multiple points to regulate the Rho pathway.
Interestingly, while modulation of some components downstream of Rho was able to suppress the blistering phenotype, overexpression of other components such as ROK did not alter this phenotype (data not shown). While this could reflect insufficient expression levels or more complex regulation of ROK, the data suggest that paxillin's regulation of the Rho pathway may involve either modulation of only certain downstream components or a lack of function for these components in the paxillin-induced phenotypes.
Rho GTPases play an important role in regulating actin cytoskeleton organization. Genetic and biochemical analysis revealed that paxillin activates Rac signaling but inactivates Rho signaling. Previous binding and localization studies suggest that paxillin may regulate Rac through its indirect association with at least two Rac exchange factors (30, 37). Pix/Cool is linked to paxillin via PKL/Git2, the ARF-GAP, and overexpression studies with mutants of paxillin and other members of this complex have led to the suggestion that paxillin may be important for recruiting this complex to focal contacts (1, 20). A second binding partner, Crk, can also link paxillin to Rac activation via a nontraditional exchange factor, Dock180. Mislocalization of one or both complexes in Pxl−/− MEFs could therefore lead to defects in Rac activation and subsequent defects in lamellipodium dynamics and migration. We examined both Pix/Cool and Crk localization in Rescued and Pxl−/− MEFs and detected only a minor decrease in Cool and Crk positive peripheral adhesions in Pxl−/− cells (data not shown). In MEFs, therefore, paxillin is not required for localization of these proteins to peripheral adhesions. This may be due to functional redundancy, as the paxillin family member Hic-5 can also bind the PKL-Pix complex and Crk can bind to other focal adhesion proteins, including p130Cas. In any case, mislocalization of these complexes is unlikely to account for the differences in Rac activation. In contrast, genetic studies of Drosophila have shown that deletion of a region encompassing the Drosophila homolog of Cool was able to suppress the Dpax-induced blistering (data not shown). Thus, one potential mechanism by which paxillin may control Rac activation in Drosophila is through regulation of Pix/Cool. As Rac and Rho have been shown to antagonize each other, it remains possible that in higher eukaryotes, paxillin could indirectly regulate Rac via regulation of Rho (4).
It is not clear how paxillin down-regulates Rho activity. Paxillin might be important for spatial regulation of Rho activity and/or controlling the activity or localization of a Rho GAP or GEF. Two Rho GAPs have been linked to paxillin. Graf is a Rho GAP that was originally identified as a Fak-binding partner, and a homolog of this protein has been identified in Drosophila studies. As paxillin can interact with Fak, it is possible that loss of paxillin may somehow affect Graf localization or activation. While Fak localization to focal adhesions is less efficient in Pxl−/− MEFs, the effects are minimal and thus this is unlikely to account for the enhanced Rho activity. It is worth noting that another group has recently reported that mammalian paxillin binds to the p120 RasGAP and competes with p120 RasGAP for binding to p190 RhoGAP (36). These investigators suggest that paxillin inhibits Rho by promoting the formation of free p190 RhoGAP. We have found that the Drosophila ortholog of p190 RhoGAP does not bind to the Drosophila p120 RasGAP. In addition, only minor changes in p190 localization to the leading edge were detected in Pxl−/− MEFs. Thus, paxillin may antagonize Rho function through multiple distinct regulatory mechanisms.
Taken together, our data suggest that while paxillin has the ability to interact with multiple proteins involved in diverse signaling pathways, a major function of this scaffolding protein in vivo is to regulate Rho family GTPases. Thus, misregulation of these GTPases is likely to account for the adhesion defects observed during development in mouse and Drosophila studies.
Acknowledgments
We thank O. Bernard, M. Affolter, T. Nakamura, C. Thummel, and the Bloomington Stock Center for providing various Drosophila stocks and reagents and S. Bagrodia and R. Cerione for Cool antibody. We are grateful to Bill Fowle for expert assistance with electron microscopy. We thank members of the Settleman and Thomas laboratories for helpful discussions.
G.-C.C. was supported by a postdoctoral fellowship from the Charles A. King Trust. B.T. was supported by a postdoctoral fellowship from the Massachusetts Public Health Breast Cancer Program. P.J.R. was supported by a postdoctoral National Institutes of Health Cancer Biology Training grant fellowship. M.H. was supported by National Institutes of Health award RO1CA75621 to S.M.T. This work was supported by National Institutes of Health award RO1GM60466 to J.S. and RO1CA75621 and a Leukemia Society Scholar award to S.M.T.
REFERENCES
- 1.Bagrodia, S., S. J. Taylor, K. A. Jordon, L. Van Aelst, and R. A. Cerione. 1998. A novel regulator of p21-activated kinases J. Biol. Chem. 273:23633-23636. [DOI] [PubMed] [Google Scholar]
- 2.Brabant, M. C., and D. L. Brower. 1993. PS2 integrin requirements in Drosophila embryo and wing morphogenesis. Dev. Biol. 157:49-59. [DOI] [PubMed] [Google Scholar]
- 3.Brown, M. C., J. A. Perrotta, C. E. Turner, and J. T. Miller. 1996. Identification of LIM3 as the principal determinant of paxillin focal adhesion localization and characterization of a novel motif on paxillin directing vinculin and focal adhesion kinase binding. J. Cell Biol. 135:1109-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burridge, K. 1999. Crosstalk between Rac and Rho. Science 283:2028-2029. [DOI] [PubMed] [Google Scholar]
- 5.Burridge, K., C. E. Turner, and L. H. Romer. 1992. Tyrosine phosphorylation of paxillin and pp125FAK accompanies cell adhesion to extracellular matrix: a role in cytoskeletal assembly. J. Cell Biol. 119:893-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, G. C., P. Gajowniczek, and J. Settleman. 2004. Rho-LIM kinase signaling regulates ecdysone-induced gene expression and morphogenesis during Drosophila metamorphosis. Curr. Biol. 14:309-313. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, E. D., M. C. Mariol, R. M. Wallace, J. Weyers, Y. G. Kamberov, J. Pradel, and E. L. Wilder. 2002. DWnt4 regulates cell movement and focal adhesion kinase during Drosophila ovarian morphogenesis. Dev. Cell 2:437-448. [DOI] [PubMed] [Google Scholar]
- 8.Edwards, K. A., and D. P. Kiehart. 1996. Drosophila nonmuscle myosin II has multiple essential roles in imaginal disc and egg chamber morphogenesis. Development 122:1499-1511. [DOI] [PubMed] [Google Scholar]
- 9.Fox, G. L., I. Rebay, and R. O. Hynes. 1999. Expression of DFak56, a Drosophila homolog of vertebrate focal adhesion kinase, supports a role in cell migration in vivo. Proc. Natl. Acad. Sci. USA 96:14978-14983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fristrom, D., P. Gotwals, S. Eaton, T. B. Kornberg, M. Sturtevant, E. Bier, and J. W. Fristrom. 1994. Blistered: a gene required for vein/intervein formation in wings of Drosophila. Development 120:2661-2671. [DOI] [PubMed] [Google Scholar]
- 11.Geneste, O., J. W. Copeland, and R. Treisman. 2002. LIM kinase and Diaphanous cooperate to regulate serum response factor and actin dynamics. J. Cell Biol. 157:831-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagel, M., E. L. George, A. Kim, R. Tamimi, S. L. Opitz, C. E. Turner, A. Imamoto, and S. M. Thomas. 2002. The adaptor protein paxillin is essential for normal development in the mouse and is a critical transducer of fibronectin signaling. Mol. Cell. Biol. 22:901-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hariharan, I. K., K. Q. Hu, H. Asha, A. Quintanilla, R. M. Ezzell, and J. Settleman. 1995. Characterization of rho GTPase family homologues in Drosophila melanogaster: overexpressing Rho1 in retinal cells causes a late developmental defect. EMBO J. 14:292-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hynes, R. O., and Q. Zhao. 2000. The evolution of cell adhesion. J. Cell Biol. 150:F89-F96. [DOI] [PubMed] [Google Scholar]
- 15.Keely, P., L. Parise, and R. Juliano. 1998. Integrins and GTPases in tumour cell growth, motility and invasion. Trends Cell Biol. 8:101-106. [DOI] [PubMed] [Google Scholar]
- 16.Kimmel, B. E., U. Heberlein, and G. M. Rubin. 1990. The homeo domain protein rough is expressed in a subset of cells in the developing Drosophila eye where it can specify photoreceptor cell subtype. Genes Dev. 4:712-727. [DOI] [PubMed] [Google Scholar]
- 17.Kumar, C. C. 1998. Signaling by integrin receptors. Oncogene 17:1365-1373. [DOI] [PubMed] [Google Scholar]
- 18.Lawler, S. 1999. Regulation of actin dynamics: the LIM kinase connection. Curr. Biol. 9:R800-R802. [DOI] [PubMed] [Google Scholar]
- 19.Liu, S., S. M. Thomas, D. G. Woodside, D. M. Rose, W. B. Kiosses, M. Pfaff, and M. H. Ginsberg. 1999. Binding of paxillin to alpha4 integrins modifies integrin-dependent biological responses. Nature 402:676-681. [DOI] [PubMed] [Google Scholar]
- 20.Manser, E., T. H. Loo, C. G. Koh, Z. S. Zhao, X. Q. Chen, L. Tan, I. Tan, T. Leung, and L. Lim. 1998. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol. Cell 1:183-192. [DOI] [PubMed] [Google Scholar]
- 21.Matthews, B. B., and S. T. Crews. 1999. Drosophila center divider gene is expressed in CNS midline cells and encodes a developmentally regulated protein kinase orthologous to human TESK1 DNA. Cell Biol. 18:435-448. [DOI] [PubMed] [Google Scholar]
- 22.Mazaki, Y., S. Hashimoto, K. Okawa, A. Tsubouchi, K. Nakamura, R. Yagi, H. Yano, A. Kondo, A. Iwamatsu, A. Mizoguchi, and H. Sabe. 2001. An ADP-ribosylation factor GTPase-activating protein Git2-short/KIAA0148 is involved in subcellular localization of paxillin and actin cytoskeletal organization. Mol. Biol. Cell 12:645-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niwa, R., K. Nagata-Ohashi, M. Takeichi, K. Mizuno, and T. Uemura. 2002. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell 108:233-246. [DOI] [PubMed] [Google Scholar]
- 24.Nolan, K. M., K. Barrett, Y. Lu, K. Q. Hu, S. Vincent, and J. Settleman. 1998. Myoblast city, the Drosophila homolog of DOCK180/CED-5, is required in a Rac signaling pathway utilized for multiple developmental processes. Genes Dev. 12:3337-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raymond, K., E. Bergeret, A. Avet-Rochex, R. Griffin-Shea, and M. O. Fauvarque. 2004. A screen for modifiers of RacGAP(84C) gain-of-function in the Drosophila eye revealed the LIM kinase Cdi/TESK1 as a downstream effector of Rac1 during spermatogenesis. J. Cell Sci. 117(Pt. 13):2777-2789. [DOI] [PubMed] [Google Scholar]
- 26.Ren, X. D., and M. A. Schwartz. 2000. Determination of GTP loading on Rho. Methods Enzymol. 325:264-272. [DOI] [PubMed] [Google Scholar]
- 27.Ridley, A. J. 1997. Signalling by Rho family proteins. Biochem. Soc. Trans. 25:1005-1010. [DOI] [PubMed] [Google Scholar]
- 28.Rorth, P., K. Szabo, A. Bailey, T. Laverty, J. Rehm, G. M. Rubin, K. Weigmann, M. Milan, V. Benes, W. Ansorge, and S. M. Cohen. 1998. System gain-of-function genetics in Drosophila. Development 125:1049-1057. [DOI] [PubMed] [Google Scholar]
- 29.Sander, E. E., S. van Delft, J. P. ten Klooster, T. Reid, R. A. van der Kammen, F. Michiels, and J. G. Collard. 1998. Matrix-dependent Tiam1/Rac signaling in epithelial cells promotes either cell-cell adhesion or cell migration and is regulated by phosphatidylinositol 3-kinase. J. Cell Biol. 143:1385-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaller, M. D. 2001. Paxillin: a focal adhesion-associated adaptor protein. Oncogene 20:6459-6472. [DOI] [PubMed] [Google Scholar]
- 31.Schratt, G., U. Philippar, J. Berger, H. Schwarz, O. Heidenreich, and A. Nordheim. 2002. Serum response factor is crucial for actin cytoskeletal organization and focal adhesion assembly in embryonic stem cells. J. Cell Biol. 156:737-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sotiropoulos, A., D. Gineitis, J. Copeland, and R. Treisman. 1999. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell 98:159-169. [DOI] [PubMed] [Google Scholar]
- 33.Thomas, S. M., and J. S. Brugge. 1997. Cellular functions regulated by Src family kinases. Annu. Rev. Cell Dev. Biol. 13:513-609. [DOI] [PubMed] [Google Scholar]
- 34.Thomas, S. M., P. Soriano, and A. Imamoto. 1995. Specific and redundant roles of Src and Fyn in organizing the cytoskeleton. Nature 376:267-271. [DOI] [PubMed] [Google Scholar]
- 35.Treisman, R., A. S. Alberts, and E. Sahai. 1998. Regulation of SRF activity by Rho family GTPases. Cold Spring Harbor Symp. Quant. Biol. 63:643-651. [DOI] [PubMed] [Google Scholar]
- 36.Tsubouchi, A., J. Sakakura, R. Yagi, Y. Mazaki, E. Schaefer, H. Yano, and H. Sabe. 2002. Localized suppression of RhoA activity by Tyr31/118-phosphorylated paxillin in cell adhesion and migration. J. Cell Biol. 159:673-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turner, C. E. 2000. Paxillin interactions. J. Cell Sci. 113:4139-4140. [DOI] [PubMed] [Google Scholar]
- 38.Turner, C. E., M. C. Brown, J. A. Perrotta, M. C. Riedy, S. N. Nikolopoulos, A. R. McDonald, S. Bagrodia, S. Thomas, and P. S. Leventhal. 1999. Paxillin LD4 motif binds PAK and PIX through a novel 95-kD ankyrin repeat, ARF-GAP protein: a role in cytoskeletal remodeling. J. Cell Biol. 145:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner, C. E., and J. T. Miller. 1994. Primary sequence of paxillin contains putative SH2 and SH3 domain binding motifs and multiple LIM domains: identification of a vinculin and pp125Fak-binding region. J. Cell Sci. 107:1583-1591. [DOI] [PubMed] [Google Scholar]
- 40.Turner, C. E., K. A. West, and M. C. Brown. 2001. Paxillin-ARF GAP signaling and the cytoskeleton. Curr. Opin. Cell Biol. 13:593-599. [DOI] [PubMed] [Google Scholar]
- 41.Webb, D. J., K. Donais, L. A. Whitmore, S. M. Thomas, C. E. Turner, J. T. Parsons, and A. F. Horwitz. 2004. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 6:154-161. [Online.]. [DOI] [PubMed] [Google Scholar]
- 42.Wheeler, G. N., and R. O. Hynes. 2001. The cloning, genomic organization and expression of the focal contact protein paxillin in Drosophila. Gene. 262:291-299. [DOI] [PubMed] [Google Scholar]
- 43.Winter, C. G., B. Wang, A. Ballew, A. Royou, R. Karess, J. D. Axelrod, and L. Luo. 2001. Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell 105:81-91. [DOI] [PubMed] [Google Scholar]
- 44.Yagi, R., S. Ishimaru, H. Yano, U. Gaul, H. Hanafusa, and H. Sabe. 2001. A novel muscle LIM-only protein is generated from the paxillin gene locus in Drosophila. EMBO Rep. 2:814-820. [Online.]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young, B. A., Y. Taooka, S. Liu, K. J. Askins, Y. Yokosaki, S. M. Thomas, and D. Sheppard. 2001. The cytoplasmic domain of the integrin α9 subunit requires the adaptor protein paxillin to inhibit cell spreading but promotes cell migration in a paxillin-independent manner. Mol. Biol. Cell 12:3214-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]