Abstract
Purpose
Deregulation or mutation of the EZH2 gene causes various tumors, including ccRCC. Although several splice variants of EZH2 have been identified, little is known about how EZH2 splicing is regulated or the contribution of alternative splicing to its pro-tumorigenic functions.
Experimental Design
We conducted RT-PCR, western blot and immunohistochemistry techniques, to examine EZH2 and its alternative splicing transcript expression in renal cancer tissue and renal cancer cell lines. Proliferation, migration, clonogenicity and tumorigenicity of renal cancer cells either exhibiting knock-down of EZH2 or its splicing factor SF3B3 were assessed by CCK8, Transwell, and murine xenograft experiments.
Results
We found that inclusion of alternative EZH2 exon 14 was significantly increased in ccRCC samples and renal cancer cell lines. In ccRCC lines, enforced expression of EZH2Δ14 inhibited, and EZH2 promoted, cell growth, migration, proliferation and tumorigenicity in a xenograft model. Mechanistic studies demonstrated that EZH2Δ14 isoform functions as a dominant-negative inhibitor of full-length EZH2. Coexpression of EZH2Δ14 variant with full-length EZH2 not only abrogated DAB2IP and HOXA9 suppression but also inhibited EZH2-driven tumorigenesis. Strikingly, the splicing factor SF3B3 stimulates inclusion of exon14 and has pro-proliferative activity. Importantly, the upregulation of SF3B3 expression observed in clinical ccRCC samples parallels the increased inclusion of EZH2 exon14, and the SF3B3 level is associated with higher tumor stage and poor overall survival.
Conclusions
These results suggest that SF3B3 as a key regulator of EZH2 pre-mRNA splicing and SF3B3 may represent a novel prognostic factor and potential therapeutic target in ccRCC.
Keywords: Alternative splicing, EZH2, SF3B3
Introduction
PRC2 catalyses the mono-, di- and trimethylation of lysine 27 of histone H3, which plays an important role in regulation of tumor transformation and progression (1). EZH2, the catalytic subunit of PRC2, is recurrently mutated in several forms of cancer and is highly expressed in numerous others, including renal cell carcinoma (2–6). Mechanismly, EZH2, and by inference H3K27 methylation, targets many tumor suppressor genes such as DAB2IP and HOXA9 (7). Besides, EZH2 also has other HMT-independent functions, including DNA methylation, STAT3 methylation, GATA4 methylation, RORalpha methylation, NOTCH1 activation, and androgen receptor activation (8–13). Numerous studies showed that PRC2 can also have a tumour-suppressive function (reviewed in (14,15)). In numerous experimental settings, it has been established that EZH2 expression and activity are highly regulated at the transcriptional (16–19), posttranscriptional (20–23), and post-translational levels (24–28). Targeting EZH2 is believed to be a promising strategy for cancer therapy (29,30). However, an incomplete understanding of the molecular mechanisms that regulate EZH2 remains a barrier to designing effective therapeutics against EZH2.
Almost all human genes are subject to alternative splicing, a pivotal mechanism for generating protein diversity; indeed different splice isoforms from the same gene may have differing and even opposing functions. Increasing evidence suggests that tumorigenesis often involves large-scale alterations in alternative splicing (reviewed in (31)). For example, SF3B1 is a crucial component of the splicing machinery, which is frequently mutated in refractory anemia with ring sideroblasts (RARS) (32), RARS associated with thrombocytosis (RARS-T) (33), and chronic lymphocytic leukemia (34,35). Thus, the deregulation of alternative RNA splicing plays a critical role in tumor development and progression. However, the roles of spliced variants and splicing factors that control splicing dysregualtion in tumorigenesis need more investigation.
In this study, we observe that inclusion of EZH2 exon 14 is significantly increased in renal cancer cell lines and renal cancer samples. EZH2Δ14 inhibits, and EZH2 promotes, renal tumor tumorigenesis. Mechanistically, EZH2Δ14 competes with EZH2 for other PRC2 components and lncRNA binding, but had little or no histone methyltransferase (HMT) activity, and EZH2Δ14 isoform functions as a dominant-negative inhibitor of full-length EZH2. We also demonstrated that the splicing factor SF3B3 promotes the inclusion of exon14. Knockdown of SF3B3 inhibits cell proliferation, colony formation, and cell migration in vitro and suppresses carcinogenesis in vivo. Importantly, SF3B3 is upregulated in ccRCC and highly correlates with EZH2 exon14 inclusion. Strikingly, the SF3B3 level correlates with higher tumor stage and poor overall survival in renal cancer. Overall, the results of this study suggest that the functions of SF3B3 in modulating alternative splicing of EZH2 exon14 contribute greatly to its role in tumor promotion.
Materials and Methods
Human samples
24 pairs of ccRCC and their corresponding adjacent normal tissues were obtained from ccRCC patients treated at Department of Urology at Tongji Hospital (Wuhan, China) after their written informed consent. All the samples were kept in liquid nitrogen before RNA extraction.
Antibodies
Following antibodies were used in the experiments: anti-Flag (F3165) from Sigma Aldrich; anti-GFP (11814460001), anti-Myc (11667149001) and anti-HA antibody (11583816001) from Roche Applied Science; anti-EZH2 (#3747), anti-Ecadherin (#3195), anti-N-cadherin (#13116) and anti-Vimentin (#5741) from Cell Signaling Technology; anti-DAB2IP (ab87811), anti-Histone H3 (total, ab82454), anti-H3K27me3 (ab6002) and anti-SF3B3 (ab96683) purchased from Abcam; anti-GAPDH (CW0100) purchased from Beijing CWBio; Goat anti-mouse IgG horseradish peroxidase (HRP)-linked whole antibody (31430) and goat anti-rabbit IgG horseradish peroxidase (HRP)-linked whole antibody (31460) purchased from Thermo scientific.
Plasmids
Human EZH2 cDNA was PCR amplified using primers EZH2-5′ and -3′, digested by BamHI and XhoI, and ligated into psi-HA, psi-Myc, psi-Flag and EGFP-myc-C1, respectively. The SUZ12 cDNA was PCR amplified using primers SUZ12-5′ and SUZ12-3′, digested by XhoI and EcoRI, and ligated into psi-Flag cut with XhoI and EcoRI to create the Flag-SUZ12. The EED cDNA was PCR amplified using primers EED-5′ and EED-3′, digested by BamHI and XhoI, and ligated into psi-Flag cut with BamHI and XhoI to create the Flag-EED. The RNAi-resistant EZH2 (Flag EZH2 RM) constructs, which the RNAi-targeted nucleotide sequence was partially substituted without affecting the aa residues (5′-GGACGGCTCCTCTAACCATGT-3′), was generated by a two-step PCR-based mutagenesis procedure using Flag-EZH2 as the template. psi-Rluc-MCS-Fluc recombinant was constructed by cloning the coding region of renilla luciferase and firefly luciferase into psi-EGFP-N1. Splicing reporter Rluc-miniEZH2-Fluc construct, which contains a point change sequence introduced the stop codon in the EZH2 exon 14, was generated by a two-step PCR-based mutagenesis procedure. The primers for making these constructs are showed in Supplementary Table S1. All plasmids were verified by sequencing.
Gene-specific shRNA target sequence was synthesized and cloned into the HpaI and XhoI sites of the pSicoR plasmid (Addgene, #11597). The primers for making these constructs were provided in Supplementary Table S1. The paired primers were annealed and ligated into pSicoR cut with HpaI and XhoI to create shRNA plasmids. For application of Cas9 for site-specific genome editing in human cells, EZH2-specific sgRNA was synthesised and cloned into lentiCRISPRv2 containing two expression cassettes, hSpCas9 and the chimeric guide RNA (Addgene, #52961). The paired primers were annealed and ligated into lentiCRISPRv2 cut with BsmBI to create sgEZH2 plasmids. For activation of endogenous HOTAIR genes by dCas9-SunTag-VP64, sgHOTAIRs targeting HOTAIR promoter were transfected into dCas9-SunTag-VP64-expressing cells. The sgHOTAIRs expression plasmids were cloned by inserting annealed oligos into the lentiGuide-Puro expression vector (Addgene, #52963) that was digested by BsmBI. The primers for making these constructs are showed in Supplementary Table S1. All plasmids were verified by sequencing.
Co-immunoprecipitation and RNA immunoprecipitation (RIP) analysis
To analyze protein interactions, co-immunoprecipitation experiments were performed using HEK293T cells after 48 hours transfection according to previously published protocols (36). To analyze RNA molecules (HOTAIR) associated with EZH2, RIP experiments were performed using Magna RIP Kit (Millipore, Catalog No.17-701) according to the manufacturer’s instructions and previously published RIP-Chip protocol (37). The RIP RNA fraction was extracted using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. The following steps were the same as in RT-PCR and Real-time PCR analysis described in this section.
RT-PCR, Real-time PCR and Chip-qPCR analysis
Total RNAs were extracted by trizol (Invitrogen) and cDNAs were synthetised using Rever Ace qPCR RT Kit (TOYOBO). Real-time PCR was performed using SYBR Green Realtime PCR Master Mix (Roche) and the ABI ViiA7 QPCR System (Applied Biosystems). Chromatin immunoprecipitation (ChIP) was performed to investigate whether EZH2 or/and EZH2 binding to HOXA9 promoter. ChIP assays were performed as described previously (38). Final analysis was performed using qPCR and shown as fold enrichment of the HOXA9 gene promoter.
Colony formation, cell proliferation, cell migration and invasion assays
Colony formation were measured two weeks after seeding 1000 cells per well in 6-well plates. Cell proliferation was estimated using the CCK-8 (Dojindo Laboratories, Japan) according to manufacturer instructions. Migration and invasion assays were performed using uncoated and Matrigel™ coated Transwell® inserts according to manufacturer instructions. All experiments were performed in triplicate.
Animal experiments and metastasis assay
Tumorgenesis in nude mice was determined as described previously (38). Five mice each were injected subcutaneously with prepared cells at a single site. Tumor onset measured with calipers at the site of injection weekly at different times on the same day. The pro-metastatic activity of SF3B3 was tested in the mouse ACHN lung metastasis model as described previously (38). All experiments were approved by the Animal Care and Use Committee of Tongji Medical College of Huazhong University of Science and Technology.
Statistical analysis
The data are presented as the mean ± SD. Differences among groups were determined by a two-way ANOVA followed by a post hoc-Tukey’s test. Comparisons between two groups were performed using an unpaired Student’s t test. Survival curve was plotted using the Kaplan-Meier method and compared by the log-rank test. A value of P < 0.05 was considered significant.
Additional methods are listed in Supplementary Materials and Methods.
Results
Dysregulated EZH2 exon 14 splicing in clinical renal cancer samples
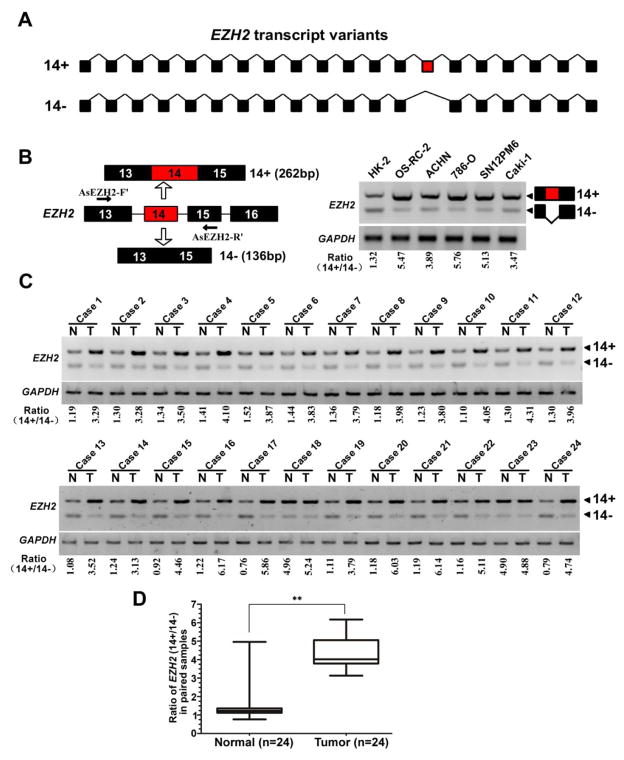
The EZH2 gene can give rise to over 30 different EZH2 mRNAs (39). In pilot experiments of cloning the full-length coding sequence (CDS) of EZH2, we determined that EZH2 gene can generate at least three transcripts by alternative mRNA splicing (data not shown). Human EZH2 spans ~76 kb and contains 20 exons; exon 14 is subject to alternative splicing regulation, which generates variants dependent on inclusion or skipping of the exon 14 (Fig. 1A). To explore whether the expression level of EZH2 isoforms with or without of exon 14 is differently expressed in various renal cell lines, RT-PCR analysis revealed that the EZH2-(14+) isoform was high expressed in cancerous cell lines as compared with immortalized human proximal renal tubule epithelial cell line HK-2 cells (Fig. 1B). Remarkably, the same expression pattern of EZH2 isoforms exists in clinical samples (Fig. 1C, Supplementary Table S2). The ratio of exon 14 inclusion versus skipping (EZH2 14+/14− mRNA) in normal tissues was 1.51 ± 1.07 (n=24), whereas inclusion of exon 14 in tumor samples was increased by a mean value of 2.9 folds to 4.37 ± 0.94(n=24) (Fig. 1D). These data suggested that the dysregulation of EZH2 exon 14 alternative splicing may play an important role in renal cancer development.
Fig. 1. EZH2 exon 14 inclusion is upregulated in renal cancers.
(A) Schematic representation of the EZH2 exon structure to highlight the alternative splicing between exons 13 and 15 generating EZH2-14+ (EZH2) and EZH2-14− (EZH2Δ14) variants. (B) Left: diagrams for detection of EZH2-14+ and EZH2-14− mRNA. Primer pairs and product sizes for the two variants are shown. Right: Expression of EZH2-14+ and EZH2-14− mRNA in one normal kidney proximal tubular epithelial cell line (HK2) and five cancer cell lines (OS-RC-2, ACHN, 786-O, SN12PM6, Caki-1) by RT PCR. Ratio for 14+/14− is listed below the panel. (C) Expression of EZH2-14+ and EZH2-14− mRNA in 24 paired renal cancer tissues and adjacent non-tumor tissues by RT-PCR. GAPDH transcript level was used as the load control. (D) Quantification of data from c for exon14 inclusion/exclusion ratio. ** indicates significant differences, P < 0.01.
EZH2Δ14 has little or no HMT activity and inhibits the tumorigenic features of renal cancer cells
Full length EZH2 is a large protein of 751 amino acids, which is classically associated with PRC2 complex function. In contrast, the EZH2 splice isoform lacking exon 14 (from here on referred to as EZH2Δ14) creates a truncated protein that differs by 43 amino acids located in the CXC domain of EZH2, while the EED-interaction, I, II, and SET domain are all unaltered (Supplementary Fig. S1A). Consistent with this notion, both EZH2 and EZH2Δ14 localize in the cell nucleus (Supplementary Fig. S1B) and interact with EED and SUZ12 of the PRC2 complex, as indicated by the co-immunoprecipitation assays (Supplementary Fig. S1C). Together, these results demonstrated that EZH2Δ14 retains EZH2’s ability to complex with EED and SUZ12 in the nucleus.
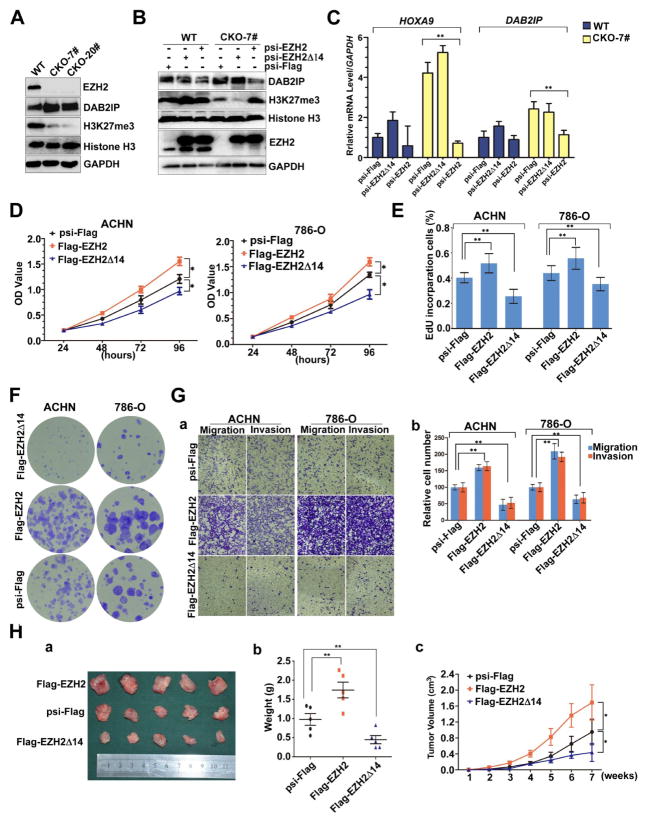
We next assessed the histone methyltransferase activity of EZH2Δ14 in comparison to EZH2 in their abilities to rescue the depletion of H3K27me3 and to induce EZH2 target genes in EZH2-knockout cells. We applied the efficient CRISPR/Cas genome editing system (reviewed in (40–42)) targeting exon 2 of EZH2 to knockout this gene in 786-O cells. We first infected 786-O cells with lentivirus encoding a sgRNA targeting exon 2 of EZH2 (Supplementary Fig. S2A) and Cas9 to the introduction of small insertions or deletions (indels) in EZH2 loci. We then isolated single cell clones and performed PCR, western blot and sequencing analysis (Supplementary Fig. S2B, C, D) to screen the EZH2-knockout clones. Consistent with previous reports, the EZH2-null clones exhibited a significant decrease in H3K27me3 and an increase in DAB2IP level comparing to controls (Fig. 2A). Restoration of EZH2 expression in the EZH2-null clones by the introduction of a sgRNA-resistant EZH2 rescued the EZH2 histone methyltransferase activity as indicated by the elevated H3K27 methylation and suppressed DAB2IP expression, compared to control (Fig. 2B, lane 4 and 6). In contrast, reintroduction of EZH2Δ14 was unable to restore this methyltransferase activity (Fig. 2B, lane 2 and 5). Furthermore, overexpression of EZH2 can, but EZH2Δ14 can not, result in inhibition of HOXA9 and DAB2IP transcription (Fig. 2C). Collectively, these findings suggested that the histone methylation activity of EZH2Δ14 is deficient compared to full length EZH2. This result is in agreement with previous reports that the CXC domain is required for EZH2 histone methyltransferase (HMT) activity (43,44).
Fig. 2. EZH2Δ14 and full-length EZH2 have opposite roles in RCC development, and EZH2Δ14 lacks H3K27 methyltransferase activity.
(A) Western blot analysis shows upregulation of DAB2IP (a known downstream gene of EZH2) and almost loss of histone H3K27 trimethylation in EZH2-knockout 786-O cell lines. Total histone H3 and GAPDH were shown as control. (B) Reintroduction of EZH2 into the KO cells can, but EZH2Δ14 can not, restore the histone H3K27 trimethylation and DAB2IP levels. Rescue experiments were carried out by transfecting EZH2-mut or EZH2Δ14-mut (sgRNA target sequence was partially substituted without affecting the amino acid residues) plasmids. (C) RT qPCR analysis of endogenous EZH2 target genes (HOXA9 and DAB2IP) in EZH2 WT and KO cells transduced with EZH2-mut and EZH2Δ14-mut. Expression was normalized to cells transduced with the control vector in wt cells. Data are plotted as the mean ± SD of 3 independent experiments. **indicates significant differences, P < 0.01. (D) The cell viability of ACHN and 786-O cells expressing Flag-EZH2 or Flag-EZH2Δ14 was determined by CCK8 assays at indicated time points. Data presented are means ± SD from three independent experiments. (E) Quantification of EdU incorporated-cells in indicated engineered cell lines. ** indicates significant differences, P < 0.01. (F) Relative colony formation units of Flag-EZH2- or Flag-EZH2Δ14-transfected stable ACHN and 786-O cells. (G) ACHN and 786-O cells expressing Flag-EZH2 or Flag-EZH2Δ14 were subjected to migration and invasion assay. a. Representative photographs were taken at ×200 magnification. b,c. The number of migrated and invaded cells was quantified in 4 random images from each group. ** indicates significant differences, P < 0.01. (H) a. Photographs of tumors excised 7 weeks after inoculation of stably transfected ACHN cells into nude mice. b. The tumor volume of Flag-EZH2-/Flag-EZH2Δ14-treated ACHN cells in nude mice at the end of 7 weeks after transplantation. c. Mean tumor volume measured by caliper on the indicated weeks. * P < 0.05. **indicates significant differences, P < 0.01.
Given the importance of EZH2 in directing tumor growth and metastasis, we next assessed the tumorigenicity of EZH2 isoforms. Over-expression of full-length EZH2 accelerated proliferation, increased DNA synthesis and colony formation while stable EZH2Δ14 over-expression had the opposite effect of decreasing these 3 activities (Fig. 2D–F, Supplementary Fig. S3A). The EZH2 isoforms displayed similar impact on cancer cell migration and invasion. As shown in Fig. 2G, overexpression of the full-length EZH2 increased, and EZH2Δ14 inhibited cell migration and invasion capability. Consistent with the transwell results, expression of N-cadherin and Vimentin (mesenchymal marker) were reduced after EZH2Δ14 over-expression, whereas E-cadherin (epithelial marker) was upregulated in both ACHN and 786-O cells (Supplementary Fig. S3B), demonstrating that EZH2Δ14 inhibits EMT. Furthermore, in vivo tumorigenic analyses revealed that induced expression of EZH2Δ14 or full-length EZH2 in ACHN cells resulted in decrease or increase, tumor growth rate and tumor size, respectively (Fig. 2H). These results indicate that EZH2Δ14 inhibits, while EZH2 promotes, tumorigenicity of renal tumor cells in both in vitro and in vivo assays.
EZH2Δ14 isoform acts as a dominant-negative inhibitor of full-length EZH2
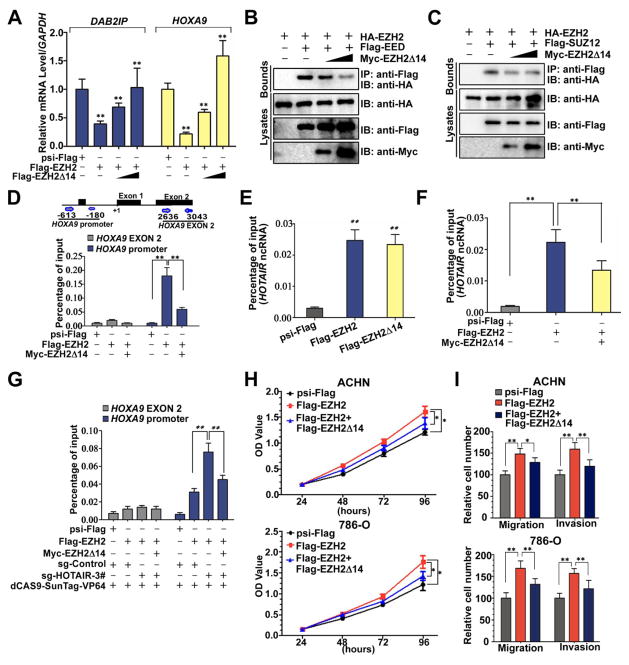
The results above suggested that EZH2Δ14 could function as a dominant-negative inhibitor and interfere with the function of full-length EZH2. As expected, RT-qPCR assays showed that co-expression of EZH2Δ14 along with full-length EZH2 diminished the inhibitory effect of DAB2IP and HOXA9 expression in a dose-dependent manner (Fig. 3A), indicating EZH2Δ14 inhibits full-length EZH2-mediated repression of DAB2IP and HOXA9. Increased expression of EZH2Δ14 led to a dose-dependent decrease in EZH2 EED and EZH2 SUZ12 interaction (Fig. 3B, C), suggesting that EZH2Δ14 competes with full-length EZH2 for EED and SUZ12 binding. Furthermore, EZH2Δ14 also competes with EZH2 for the HOXA9 promoter and HOTAIR lncRNA binding (Fig. 3D, E, F). In addition, the binding of full-length EZH2 to the HOXA9 promoter was significantly increased upon transcriptional activation of HOTAIR by the dCas9-SunTag system, but this effect was blocked by co-expression of EZH2Δ14 (Fig. 3G, Supplementary Fig. S4). Moreover, over-expression of EZH2Δ14 together with full-length EZH2 lowered the growth, migration and invasion promoting potential of full-length EZH2 (Fig. 3H, I Supplementary Fig. S5). Collectively, these results strongly indicate that the EZH2Δ14 splice isoform acts as a dominant-negative inhibitor of full-length EZH2.
Fig. 3. EZH2Δ14 isoform acts as a dominant-negative inhibitor of full-length EZH2.
(A) qPCR analysis of expression of PRC2 target genes HOXA9 and DAB2IP in ACHN cells transfected with psi-Flag (control), or Flag-EZH2 (0.4μg) and Flag-EZH2Δ14 (0, 0.2, or 0.6μg) plasmids. Means ± standard deviations for n = 3 are shown. **p < 0.01 vs. Ctrl. (B) EZH2Δ14 binds to EED and competes with the full-length EZH2. (C) EZH2Δ14 binds to SUZ12 and competes with the full-length EZH2. (D) Effects of EZH2Δ14 overexpression on the full-length EZH2 recruitment to the HOXA9 gene promoter in ACHN cells. At 24 h after transfection, cells were subjected to ChIP qPCR. **indicates significant differences, P < 0.01. (E) RNA immunoprecipitation (RIP) show that EZH2Δ14 and the full-length EZH2 can bind to the HOTAIR in ACHN cells. Transfected ACHN cells were harvested for western blots and RIP with anti-Flag. Retrieved HOTAIR ncRNA was analysed by RT qPCR. Means ± standard deviations for n = 3 are shown. ** P < 0.01 vs. Ctrl. (F) Effect of EZH2Δ14 overexpression on the binding of HOTAIR ncRNA to the full-length EZH2. **indicates significant differences, P < 0.01. (G) EZH2Δ14 inhibits HOTAIR-enhanced binding of EZH2 to the HOXA9 promoter. **indicates significant differences, P < 0.01. (H, I) EZH2Δ14 abrogates cell growth, migration and invasion induced by full-length EZH2. ACHN and 786-O cells were infected with different combinations of lentivirus as indicated. At 72 h after infection, CCK8 assays and transwell assays were performed to determine the cell growth (H), migration and invasion (I). * P < 0.05 vs. Ctrl. ** P < 0.01 vs. Ctrl.
SF3B3 regulates EZH2 alternative splicing and its expression correlates with levels of the full-length EZH2
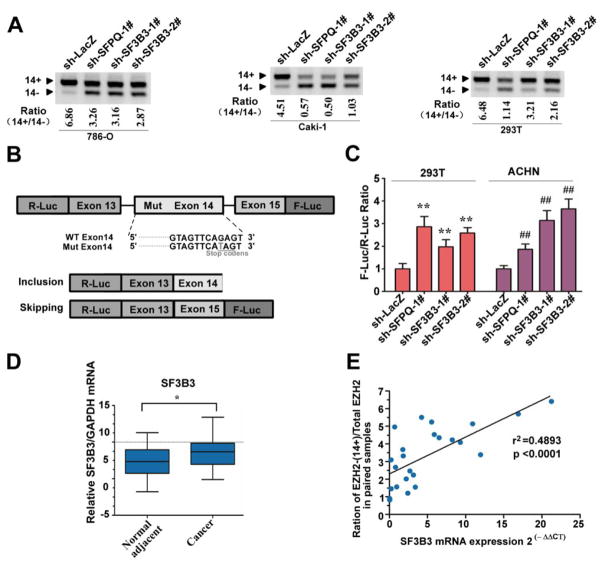
The mechanism controlling exon 14 inclusion/skipping is currently unknown. Aberrant splicing can result from mutations in cis-acting splicing elements and changes in the activity of trans-acting splicing factors (31,45,46). We had previously observed that many splicing regulators were differentially expressed in renal cancer tissues by microarray (Supplementary Table S3) (47). We reasoned that some of these differentially expressed splicing factors may play potential role in EZH2 splicing regulation. We evaluated this possibility by constructing a series of ACHN cell lines stably expressing lentiviral shRNA constructs against the following individual splicing factors: hnRNPA1, hnRNPA2, SRSF1, PRPF3, SFPQ, SF3A3, and SF3B3 (Supplementary Fig. S6A). Based on reproducibility (4 of 4 pilot experiments) and the magnitude of change (>5 fold) in the exon 14+/14− mRNA ratio upon knockdown of indicated factor via shRNAs, SFPQ and SF3B3 were selected for further investigation. We also found that knockdown of SFPQ or SF3B3 promoted EZH2 exon 14 skipping in 786-O, Caki-1, and 293T cells (Fig. 4A), suggesting that the effect of SFPQ or SF3B3 on EZH2 exon 14 splicing was not cell-type specific. To further confirm the effect of SFPQ and SF3B3 on the splicing of EZH2 exon 14, we generated a mini-EZH2 dual luciferase splice reporter gene (Fig. 4B). Transient transfection of EZH2 dual luciferase splice reporter into ACHN and 293T cells revealed that relative firefly luciferase activity (F-Luc/R-Luc ratio) was significantly elevated when SFPQ or SF3B3 expression was knocked down (Fig. 4C). Besides EZH2 and EZH2Δ14, the EZH2 gene generates many other different products (39). It is therefore difficult to distinguish EZH2Δ14 from other isoforms by western blot at protein level. To further confirm the effect of SFPQ and SF3B3 on the protein level of EZH2Δ14 isoforms, we established a 293T cell line stably expressing the mini-EZH2 gene constructs by blasticidin selection (Supplementary Fig. S6B, C). The BSD-EGFP level was significantly increased when SFPQ or SF3B3 was knocked down (Supplementary Fig. S6D), suggesting that the relative expression level of EZH2 and EZH2Δ14 protein isoforms could be changed by SFPQ or SF3B3 depletion. Together, these findings illustrate that down-regulation of SFPQ and SF3B3 promoted the skipping of exon 14 of EZH2.
Fig. 4. SF3B3 regulates EZH2 alternative splicing and its expression correlates with levels of the full-length EZH2.
(A) SF3B3 regulated EZH2 exon 14 inclusion is cell-type independently. EZH2 exon 14 splicing was measured by RT PCR in various cell lines (786-O, Caki-1, 293T) stably expressing sh-LacZ, sh-SFPQ, or sh-SF3B3. RT PCR was performed as described in Fig. 1B. (B) Schematic representation of splicing dual-reporter assay that mimics endogenous splicing by insertion of EZH2 genome fragment (from exon 13 to 15) harbors a point mutation in exon 14 to create stop codons. (C) The bar graph depicts the skipping of EZH2 exon 14 as measured by the firefly (F-Luc) to Renilla (R-Luc) luciferase ratio. 293T and ACHN cells were infected with lentivirus expressing sh-LacZ, sh-SFPQ, sh-SF3B3-1#, or sh-SF3B3-2#. After 3 d, these stable knockdown cell lines were transiently transfected with the splicing reporter. After transfection for 24 h, luciferase activities were measured. Data are mean ± s.d., n = 3 independent experiments, **P < 0.01. (D) SF3B3 is upregulated in renal clear cell carcinomas. Total RNA isolated from paired ccRCC tumors and adjacent normal tissues were assayed by real-time RT-PCR. * P < 0.05. (E) Positive correlation between EZH2-(14+)/EZH2 total mRNA ratio and expression levels of SF3B3 was observed in RCC samples. Relative mRNA levels of SF3B3 and the corresponding levels of EZH2 (ratio of EZH2-(14+)/EZH2 total mRNA) was plotted in each patient sample (P < 0.05, R2 = 0.4319).
SF3B3 is a subunit of the U2 small nuclear ribonucleoprotein (U2 snRNP), which recognizes and binds to branch sites within the intron (Supplementary Fig. S7A). Our microarray experiments suggested that SF3B3 and SF3A3 are highly expressed in ccRCC (Supplementary Table S3). However, the expression patterns of other U2-snRNP components in ccRCC are unclear. We analyzed in 24 paired ccRCC and adjacent non-tumor renal tissues by qPCR. Unexpectedly, our results demonstrated that U2-snRNP protein components were not broadly increased in expression in ccRCC tissues; in fact, of the U2-snRNP components, only SF3B3 was upregulated in RCCC with statistical significance (P<0.05) (Fig. 4D, Supplementary Fig. S7B). Like SF3A3 and SF3B3, knockdown of the U2-snRNP proteins SF3A1, SF3A2, or SF3B1 in ACHN cells also enhanced production of EZH2-(14−) isoform (Supplementary Fig. S7C), implying an important role for U2-snRNP activity in EZH2 exon 14 splicing.
We next wondered whether expression of SF3B3 is associated with the EZH2-(14+)/EZH2 total ratio. We therefore chose to monitor EZH2 mRNA splicing by qPCR using primers that can distinguish between the EZH2-(14+) isoform and the total EZH2 (Supplementary Fig. S7D). Our qPCR analysis revealed consistently higher levels of the EZH2-(14+) isoform and EZH2 total mRNA in the 24 paired renal cancer tissue and tumor adjacent renal tissue specimens (Supplementary Fig. S7E, F). Significantly, a positive correlation was observed between SF3B3 expression levels and the ratio of the EZH2-(14+) splice variants/EZH2 total mRNA in clinical samples (Fig. 4E), corroborating our observation that SF3B3 promotes the production of EZH2-(14+) isoform. Together, these results indicate that SF3B3 is involved in ccRCC development, probably reflecting its ability to regulate the alterative splicing of EZH2 pre-mRNA.
SF3B3 is a renal cancer oncogene
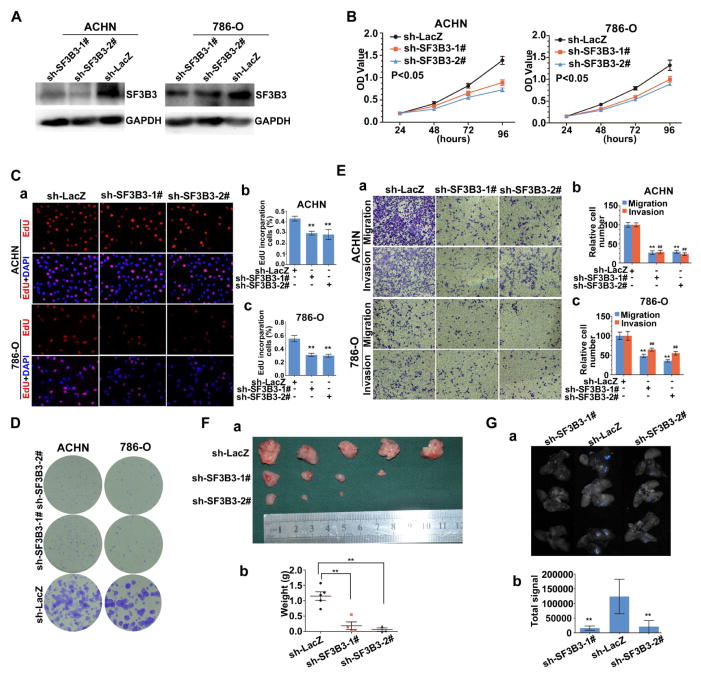
To examine the possible role of SF3B3 in renal cancer progression, we stably knocked down SF3B3 expression with two distinct shRNAs in ACHN and 786-O cells (Fig. 5A), and examined the effect of SF3B3 loss on cell proliferation and colony formation. Our data showed that two independent SF3B3 shRNAs significantly decreased cell viability, DNA synthesis, and reduced the ability of colony formation of these cells in vitro (Fig. 5B–D). Moreover, compared with the negative control, knockdown of SF3B3 inhibited cell migration by 71% and 65% in migratory ACHN and 786-O cells, respectively (Fig. 5E). SF3B3 knockdown also significantly reduced invasion capability of ACHN and 786-O cells. As shown in Fig. 5E, approximately 77% and 45% reduction of invading ACHN and 786-O cells were observed in the sh-SF3B3 infected cells when compared with the cells infected with sh-LacZ control. In addition, SF3B3 knockdown in ACHN and 786-O cells resulted in downregulation of mesenchymal markers expression and upregulation of epithelial marker, demonstrating that SF3B3 knockdown inhibits EMT (Supplementary Fig. S8). Moreover, we also found that knockdown of SF3B3 inhibited in vivo tumor growth in xenograft models with statistical significance (Fig. 5F), and decreased the rates of lung colonization in tail vein xenograft models (Fig. 5G). Taken together, these results suggest that SF3B3 acts as a pro-oncogene to promote cell growth, migration and invasion in renal cancer.
Fig. 5. SF3B3 is required for tumorigenesis of renal cancer cells.
(A) Generation of SF3B3-knockdown ACHN and 786-O cells. ACHN and 786-O cells were infected with lentivirus with control sh-LacZ (Control) and sh-SF3B3s, respectively. Western blot analysis of the selected clones (sh-LacZ, sh-SF3B3-1#, and sh-SF3B3-2#) was performed to evaluate the expression of SF3B3. GAPDH is an internal control. * P < 0.05 vs. sh-LacZ. (B) The cell viability of ACHN and 786-O cells expressing sh-LacZ or sh-SF3B3 was determined by CCK8 assays at indicated time points. Data presented are means ± SD from three independent experiments. (C) Representative micrographs (a) and quantification (b,c) of EdU incorporated-cells in indicated engineered cell lines. ** P < 0.01 vs. sh-LacZ. (D) Relative colony formation units of sh-SF3B3- or sh-LacZ-transfected stable ACHN and 786-O cells. (E) ACHN and 786-O cells expressing sh-SF3B3 or sh-LacZ were subjected to migration and invasion assay. a. Representative photographs were taken at ×200 magnification. b,c. The number of migrated and invaded cells was quantified in 4 random images from each group. ** P < 0.01 vs. sh-LacZ. ## P < 0.01 vs. sh-LacZ. (F) ACHN cells expressing sh-SF3B3 or sh-LacZ were transplanted into mice. a. Representative images of the isolated tumors from injected mice. b. Tumor weight of each nude mouse at the end of 7 weeks. ** P < 0.01 vs. sh-LacZ. (G) a. Representative bioluminescent images of lungs of nude mice at the 30th days after IV. injection of renal cancer cell. b. Quantification analysis of fluorescence signal from captured bioluminescence images. ** P < 0.01 vs. sh-LacZ.
Next, we tested whether exogenous EZH2-(14+) isoform expression could compensate for SF3B3 knockdown. We found that the effect of SF3B3 silencing on proliferation, migration and invasion was partially reversed by concomitant EZH2 over-expression, suggesting that the effect of SF3B3 on cell migration and invasion is mediated, at least in part, through EZH2 (Supplemental Fig. S9).
SF3B3 is upregulated in RCC and is associated with poor clinical outcome
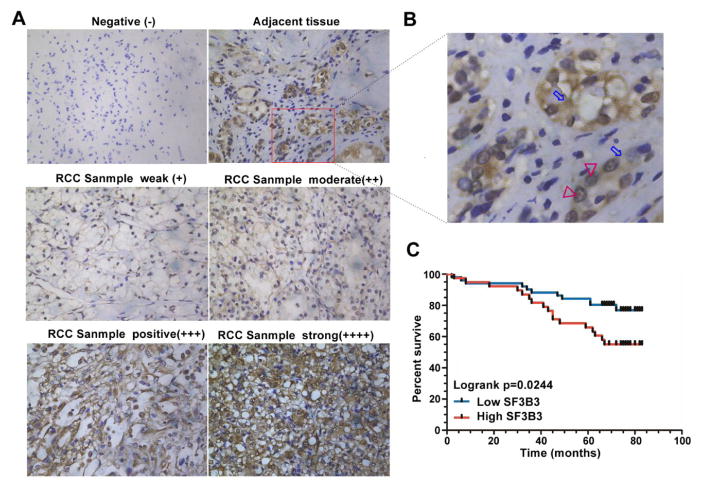
To further study the role of SF3B3 as an oncogene, we examined its clinical relevance in cancer patients. We analyzed SF3B3 expression in 90 paired clinical samples by immunohistochemistry (Fig. 6A, Supplemental Table S4). In normal renal tissue, SF3B3 protein was found mainly in epithelial cells of the renal tubule and the protein was localized in the nucleus and cytoplasm (Fig. 6B). We found that, in the primary renal cancer tissue specimens, the level of SF3B3 expression can be divided into two categories: negative and positive. A higher expression of SF3B3 is associated with higher tumor stage (p=0.032; Supplementary Table S5). Moreover, Kaplan-Meier analysis showed that the SF3B3-positive group showed significantly poorer overall survival than the negative group (Fig. 6C), indicating that SF3B3 may be a potentially valuable biomarker for the prognosis of ccRCC. Together, these clinical data suggest that SF3B3 plays a role in the development of renal cancer.
Fig. 6. High expression of SF3B3 is associated with poor prognosis in patients with renal cancer.
(A) Normal renal tissues and ccRCC samples were collected and subjected to immunohistochemical staining with a SF3B3 antibody. (B) Higher magnification images of the square regions in red line in A. Red triangles indicate the nucleus. Blue arrows indicate the cytoplasm. (C) Kaplan-Meier curve showing overall survival of kidney cancer patients with high or low SF3B3 expression (p = 0.0244 by log-rank test).
Discussion
This study provides the first direct evidence that EZH2 is alternatively spliced at exon 14, and demonstrates that EZH2 exon 14 inclusion is a frequent event in renal cancers. We further observed that knockdown of SF3B3 promoted expression of exon14-lacking EZH2 transcripts through modulation of EZH2 pre-mRNA splicing. Importantly, SF3B3 mRNA level is positively correlated with exon14-containing EZH2 transcripts in clinical ccRCC samples. Together, these data demonstrate that SF3B3 can regulate alternative splicing of EZH2 at exon 14. Splicing dysregulation is one of the molecular hallmarks of cancer (48). However, the underlying molecular mechanisms remain poorly defined. Our finding that SF3B3 regulates EZH2 alternative splicing not only provides a mechanism for the regulation of EZH2 but also suggests that upregulation of SF3B3, a common event in renal cancer, is an important mechanism contributing to the frequently increased expression of exon14-containing EZH2 transcripts in tumors.
Like most of other human genes, the alternative splicing of EZH2 locus can give rise to different EZH2 mRNAs (39). Here we show that the exon14-lacking EZH2Δ14 isoform is downregulated, and exon14-containing EZH2 isoform is upregulated, in renal cancer cell lines and renal cancer tissues, suggesting an important role for the splicing variants of EZH2 in the tumorigenesis of human renal cancer. Studies on human tumors show that EZH2 is frequently over-expressed in a wide variety of cancerous tissue types (49). It remains an open question whether dysregulation of alternative splicing of EZH2 is also involved in other cancer types. Of note, RT-PCR analysis demonstrated that 4 of 4 hepatocellular carcinomas specimens and 6 of 6 bladder carcinomas specimens expressed increased EZH2 exon 14 inclusion when compared with normal tissues (Supplementary Fig. S10). This raises the possibility that the differential inclusion of EZH2 exon 14 may be a frequent event in many human cancers, and EZH2 splice variants may be novel cancer markers.
We also provide functional evidence that full length EZH2 promotes, and EZH2Δ14, inhibits, cancer cell growth, migration, and colony formation in vitro and tumorigenesis in vivo. A large number of studies have revealed that many cancer-related genes exhibit alternative splicing and the alternatively spliced isoforms can drive malignant phenotypes (45,46,48). Our study suggests that EZH2Δ14 lacks histone methyltransferase (HMT) activity and has a distinct function in the tumorigenesis of human renal cancer, and this result supports the notion that the CXC domain of EZH2 is required for histone methyl transferase (HMT) activity (43,44). We propose that EZH2Δ14 functions mainly by disabling full-length EZH2. Apart from the canonical role of EZH2 as a PRC2 component, emerging data suggest that EZH2 promotes cancer via PRC2-independent functions (8–15). Paradoxically, several observations showed that EZH2 also have tumour-suppressive functions. For example, EZH2 depletion accelerates lymphomagenesis in Eμ-Myc transgenic mice (50), which acts as a tumor suppressor for myeloid malignancies (14,44). Previous observations support the notion that EZH2 is a context-dependent oncogene and tumour-suppressor gene (14,15,51). Currently, it remains unclear whether EZH2Δ14 has other functions independent of PRC2 and EZH2. Further studies should be directed to dissect the pathways that are regulated by the dysregulation of EZH2 exon 14 splicing.
Previous studies have revealed that EZH2 can be regulated in different types of human cancers at the transcriptional, post-transcriptional and post-translational levels (49). This study provides the first evidence that EZH2 is also regulated by alternative splicing, and that such alternative splicing is associated with carcinogenesis. Our findings are of particular interest because EZH2 may be a potential therapeutic target in many tumor (52–59), and significant efforts have been made to the tumor-promoting function of EZH2, such as EZH2 inhibitors are currently in clinical trials (Supplementary Table S6) (14,15,29,30,58,60–66). Specifically, our results suggest that modulating EZH2 splicing may be an effective way to influence EZH2 activity in cancer. This approach may provide a potential new strategy for development of anti-cancer compounds that can shift of EZH2 splicing, and may lead to accelerated development of EZH2 targeting agents.
SF3B3 encodes subunit 3 of splicing factor 3b protein complex, a member of the U2 snRNP complex, which is responsible for 3′ splice site recognition (67). Previously, SF3b mutations were identified in multiple cancer types, especially in myelodysplastic syndromes (MDSs) (32–35,68–74). These mutations appear to promote use of many cryptic 3′ splice site that induces global splicing changes and generates aberrant transcripts (75–78); in many cases, inappropriate exon skipping is observed (67,79,80). Here, we found that knockdown of the SF3a/SF3b complexes by shRNA induced the production of exon14-lacking spliced isoform, consistent with previous studies; EZH2Δ14 is produced if the 3′ splice site at the end of intron 13 is skipped and the 3′ splice site at the end of intron 14 is used instead (Supplementary Fig. S11). It is intriguing to speculate that the choice of 3′ splice site around EZH2 exon 14 evolved to be exquisitely sensitive to SF3a/SF3b activity or/and expression levels because of the functional significance of EZH2 exon 14-encoded domain, and may be a key point of regulating EZH2’s function (Supplementary Fig. S11). However, the molecular mechanisms by which the SF3a or SF3b complexes regulate the alternate splicing of EZH2 remain to be defined.
Because we found that EZH2 was not sufficient to rescue the growth defects following SF3B3 inhibition, the effects of SF3B3 knockdown on cell growth likely involves a number of other genes. This hypothesis is supported by previous studies that SF3a/SF3b complexes are required for proper pre-mRNA splicing of many genes, including p27 (67), FIR (81), MyD88 (79), CRNDE, ABCC5 and UQCC (71). Furthermore, SF3b also have other biological functions, such as SF3B3 have been shown to play critical roles in the regulation of chromatin modification and transcription through direct physical interactions with STAGA (82). Thus, the other candidate SF3B3-regulated oncogenes in tumorigenesis remain to be defined.
Interestingly, we show that the expression of multiple components of the SF3a/SF3b complex (SF3A1, SF3A2, SF3A3, SF3B1, SF3B2, SF3B3, and SF3B4) were not coordinately increased in ccRCC, suggesting these proteins may have specific functions in the SF3a/SF3b complex, or that activity level of these proteins is regulated by some other mechanism. Furthermore, we also found that both SF3A1 and SF3B3 were localized in the nucleus, whereas the majority of SF3B3 was localized in the cytoplasm (data not shown), suggesting that the subcellular location of SF3B3 may be critical in regulating the U2 snRNP complex activity. It is intriguing to speculate that the expression levels and subcellular location of SF3B3 may be a key point of regulation to limit the U2 snRNP complex’s activity. We also provide evidence that SF3B3 is upregulated in renal clear cell carcinomas and highly correlates with EZH2-(14+)/-(14−) ratio, reinforcing SF3B3 as an important mRNA splicing factor implicated in regulating EZH2 exon 14 splicing.
A number of studies have revealed that splicing dysregulation can drive malignant phenotypes. Here, we provide evidence for abundant SF3B3 transcripts in renal cancer tissues in comparison with matching adjacent normal renal tissues. We also confirmed that knockdown of SF3B3 can inhibit cancer cell growth, migration and invasion in vitro and tumorigenicity and metastasis in vivo. Together, this suggests that SF3B3 functions as a proto-oncogene in renal cancer development and progression, and may serve as a potential prognostic marker for ccRCC patients. Recently, Gokmen-Polar et al. demonstrated that SF3B1 and SF3B3 levels were significantly increased in breast cancer, and that SF3B3 expression correlates with prognosis and endocrine resistance in estrogen receptor-positive breast cancer (83). We also verified a high abundance of SF3B3 transcripts in renal cancer tissues in comparison with matching adjacent normal renal tissues. Most importantly, we analyzed the data from 90 Chinese ccRCC patients, and found that the expression level of SF3B3 is clearly correlated with the stage of renal cancer and that SF3B3 overexpression is associated with poor prognosis of ccRCC patients. Our findings suggest that SF3B3 may contribute to renal cancer, and raise the possibility that it may be essential for the maintenance of malignant phenotypes of renal cancers.
In summary, our study reveals tumorigenic roles for dysregulated EZH2 exon14 splicing, and identifies SF3B3 as regulator of the splicing of EZH2 pre-mRNA in a manner that contributes to tumorigenesis of renal cancer cells. These findings suggest that a rational drug combination for the treatment of ccRCC could be an EZH2 inhibitor (e.g., EPZ005687) together with a SF3b inhibitor (e.g., Spliceostatin A).
Supplementary Material
Translational Relevance.
Dysregulated expression or mutation of EZH2 is being recognized as a critical event in several genetic disorders and cancer, but the regulation of EZH2 splicing, and the functional and clinical significance of the variants in ccRCC remains unexplored. Herein, we found that inclusion of alternative EZH2 exon 14 was significantly increased in ccRCC samples. Functional assays identified SF3B3 promotes inclusion of EZH2 exon14 and has pro-proliferative activity. Importantly, the upregulation of SF3B3 expression observed in clinical ccRCC samples parallels the increased inclusion of EZH2 exon14, and the SF3B3 level is associated with higher tumor stage and poor overall survival. Collectively, our findings clarified how upregulation of SF3B3 exerts an oncogenic function in ccRCC that may offer a novel prognostic factor and potential therapeutic target in this disease.
Acknowledgments
Grant Support
This work was supported by the National Natural Science Foundation of China (31372562, 81170650, 81270788, 81470935, 81402098, 81402105, 81402087), the National Major Scientific and Technological Special Project for Significant New Drugs Development (2012ZX09303018), the Chenguang Program of Wuhan Science and Technology Bereau (2013072304010833, 2015070404010199), and The National High Technology Research and Development Program 863 (2014AA020607).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
References
- 1.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–9. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 2.Wagener N, Holland D, Bulkescher J, Crnkovic-Mertens I, Hoppe-Seyler K, Zentgraf H, et al. The enhancer of zeste homolog 2 gene contributes to cell proliferation and apoptosis resistance in renal cell carcinoma cells. Int J Cancer. 2008;123:1545–1550. doi: 10.1002/ijc.23683. [DOI] [PubMed] [Google Scholar]
- 3.Wagener N, Macher-Goeppinger S, Pritsch M, Husing J, Hoppe-Seyler K, Schirmacher P, et al. Enhancer of zeste homolog 2 (EZH2) expression is an independent prognostic factor in renal cell carcinoma. BMC cancer. 2010;10:524. doi: 10.1186/1471-2407-10-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu B, Abourbih S, Sircar K, Kassouf W, Mansure JJ, Aprikian A, et al. Enhancer of zeste homolog 2 expression is associated with metastasis and adverse clinical outcome in clear cell renal cell carcinoma: a comparative study and review of the literature. Arch Pathol Lab Med. 2013;137:1326–1336. doi: 10.5858/arpa.2012-0525-OA. [DOI] [PubMed] [Google Scholar]
- 5.Hinz S, Weikert S, Magheli A, Hoffmann M, Engers R, Miller K, et al. Expression profile of the polycomb group protein enhancer of Zeste homologue 2 and its prognostic relevance in renal cell carcinoma. J Urol. 2009;182:2920–2925. doi: 10.1016/j.juro.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 6.Avissar-Whiting M, Koestler DC, Houseman EA, Christensen BC, Kelsey KT, Marsit CJ. Polycomb group genes are targets of aberrant DNA methylation in renal cell carcinoma. Epigenetics. 2011;6:703–709. doi: 10.4161/epi.6.6.16158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 8.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 9.Kim E, Kim M, Woo D-H, Shin Y, Shin J, Chang N, et al. Phosphorylation of EZH2 Activates STAT3 Signaling via STAT3 Methylation and Promotes Tumorigenicity of Glioblastoma Stem-like Cells. Cancer Cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He A, Shen X, Ma Q, Cao J, von Gise A, Zhou P, et al. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes & development. 2012;26:37–42. doi: 10.1101/gad.173930.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JM, Lee JS, Kim H, Kim K, Park H, Kim JY, et al. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Molecular cell. 2012;48:572–586. doi: 10.1016/j.molcel.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez ME, Moore HM, Li X, Toy KA, Huang W, Sabel MS, et al. EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proc Natl Acad Sci U S A. 2014;111:3098–3103. doi: 10.1073/pnas.1308953111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Comet I, Riising EM, Leblanc B, Helin K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat Rev Cancer. doi: 10.1038/nrc.2016.83. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 15.Kim KH, Roberts CWM. Targeting EZH2 in cancer. Nat Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang X, Milyavsky M, Shats I, Erez N, Goldfinger N, Rotter V. Activated p53 suppresses the histone methyltransferase EZH2 gene. Oncogene. 2004;23:5759–69. doi: 10.1038/sj.onc.1207706. [DOI] [PubMed] [Google Scholar]
- 17.Fujii S, Tokita K, Wada N, Ito K, Yamauchi C, Ito Y, et al. MEK-ERK pathway regulates EZH2 overexpression in association with aggressive breast cancer subtypes. Oncogene. 2011;30:4118–28. doi: 10.1038/onc.2011.118. [DOI] [PubMed] [Google Scholar]
- 18.Tiwari N, Tiwari Vijay K, Waldmeier L, Balwierz Piotr J, Arnold P, Pachkov M, et al. Sox4 Is a Master Regulator of Epithelial-Mesenchymal Transition by Controlling Ezh2 Expression and Epigenetic Reprogramming. Cancer Cell. 2013;23:768–83. doi: 10.1016/j.ccr.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 19.Nolan KD, Franco OE, Hance MW, Hayward SW, Isaacs JS. Tumor secreted Hsp90 subverts Polycomb function to drive prostate tumor growth and invasion. J Biol Chem. 2015 doi: 10.1074/jbc.M115.637496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gandellini P, Folini M, Longoni N, Pennati M, Binda M, Colecchia M, et al. miR-205 Exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cepsilon. Cancer Res. 2009;69:2287–2295. doi: 10.1158/0008-5472.CAN-08-2894. [DOI] [PubMed] [Google Scholar]
- 22.Tzatsos A, Paskaleva P, Lymperi S, Contino G, Stoykova S, Chen Z, et al. Lysine-specific demethylase 2B (KDM2B)-let-7-enhancer of zester homolog 2 (EZH2) pathway regulates cell cycle progression and senescence in primary cells. J Biol Chem. 2011;286:33061–33069. doi: 10.1074/jbc.M111.257667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin L, Zheng Y, Tu Y, Wang Z, Liu H, Lu X, et al. MicroRNA-144 suppresses tumorigenesis and tumor progression of astrocytoma by targeting EZH2. Hum Pathol. 2015;7:971–980. doi: 10.1016/j.humpath.2015.01.023. [DOI] [PubMed] [Google Scholar]
- 24.Riising EM, Boggio R, Chiocca S, Helin K, Pasini D. The polycomb repressive complex 2 is a potential target of SUMO modifications. PloS one. 2008;3:e2704. doi: 10.1371/journal.pone.0002704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Y-L, Chou R-H, Shyu W-C, Hsieh S-C, Wu C-S, Chiang S-Y, et al. Smurf2-mediated degradation of EZH2 enhances neuron differentiation and improves functional recovery after ischaemic stroke. EMBO Mol Med. 2013;5:531–547. doi: 10.1002/emmm.201201783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahasrabuddhe AA, Chen X, Chung F, Velusamy T, Lim MS, Elenitoba-Johnson KS. Oncogenic Y641 mutations in EZH2 prevent Jak2/beta-TrCP-mediated degradation. Oncogene. 2014;4:445–454. doi: 10.1038/onc.2013.571. [DOI] [PubMed] [Google Scholar]
- 27.Chu CS, Lo PW, Yeh YH, Hsu PH, Peng SH, Teng YC, et al. O-GlcNAcylation regulates EZH2 protein stability and function. Proc Natl Acad Sci U S A. 2014;111:1355–1360. doi: 10.1073/pnas.1323226111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan J, Li B, Lin B, Lee PT, Chung TH, Tan J, et al. EZH2 phosphorylation by JAK3 mediates a switch to non-canonical function in natural killer/T-cell lymphoma. Blood. 2016;7:948–58. doi: 10.1182/blood-2016-01-690701. [DOI] [PubMed] [Google Scholar]
- 29.Knutson SK, Warholic NM, Wigle TJ, Klaus CR, Allain CJ, Raimondi A, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci U S A. 2013;110:7922–7927. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 31.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Visconte V, Makishima H, Jankowska A, Szpurka H, Traina F, Jerez A, et al. SF3B1, a splicing factor is frequently mutated in refractory anemia with ring sideroblasts. Leukemia. 2012;26:542–545. doi: 10.1038/leu.2011.232. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. The New England journal of medicine. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2012;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 36.Chen K, Chen S, Huang C, Cheng H, Zhou R. TCTP increases stability of hypoxia-inducible factor 1alpha by interaction with and degradation of the tumour suppressor VHL. Biol Cell. 2013;105:208–218. doi: 10.1111/boc.201200080. [DOI] [PubMed] [Google Scholar]
- 37.Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- 38.Chen K, Yu G, Gumireddy K, Li A, Yao W, Gao L, et al. ZBRK1, a novel tumor suppressor, activates VHL gene transcription through formation of a complex with VHL and p300 in renal cancer. Oncotarget. 2015;6:6959–6976. doi: 10.18632/oncotarget.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grzenda A, Lomberk G, Svingen P, Mathison A, Calvo E, Iovanna J, et al. Functional characterization of EZH2beta reveals the increased complexity of EZH2 isoforms involved in the regulation of mammalian gene expression. Epigenetics & chromatin. 2013;6:3. doi: 10.1186/1756-8935-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 42.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ketel CS, Andersen EF, Vargas ML, Suh J, Strome S, Simon JA. Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Mol Cell Biol. 2005;25:6857–6868. doi: 10.1128/MCB.25.16.6857-6868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nature genetics. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J, Manley JL. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov. 2013;3:1228–1237. doi: 10.1158/2159-8290.CD-13-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 47.Yu G, Yao W, Wang J, Ma X, Xiao W, Li H, et al. LncRNAs expression signatures of renal clear cell carcinoma revealed by microarray. PLoS One. 2012;7:e42377. doi: 10.1371/journal.pone.0042377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci. 2006;119:2635–2641. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- 49.Chang CJ, Hung MC. The role of EZH2 in tumour progression. Br J Cancer. 2012;106:243–247. doi: 10.1038/bjc.2011.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee SC, Phipson B, Hyland CD, Leong HS, Allan RS, Lun A, et al. Polycomb repressive complex 2 (PRC2) suppresses Emu-myc lymphoma. Blood. 2013;122:2654–2663. doi: 10.1182/blood-2013-02-484055. [DOI] [PubMed] [Google Scholar]
- 51.Chase A, Cross NC. Aberrations of EZH2 in cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:2613–2618. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- 52.Kemp CD, Rao M, Xi S, Inchauste S, Mani H, Fetsch P, et al. Polycomb repressor complex-2 is a novel target for mesothelioma therapy. Clin Cancer Res. 2012;18:77–90. doi: 10.1158/1078-0432.CCR-11-0962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee SR, Roh YG, Kim SK, Lee JS, Seol SY, Lee HH, et al. Activation of EZH2 and SUZ12 regulated by E2F1 predicts the disease progression and aggressive characteristics of bladder cancer. Clin Cancer Res. 2015;21:5391–5403. doi: 10.1158/1078-0432.CCR-14-2680. [DOI] [PubMed] [Google Scholar]
- 54.Wu K, Xie D, Zou Y, Zhang T, Pong R-C, Xiao G, et al. The Mechanism of DAB2IP in Chemoresistance of Prostate Cancer Cells. Clinical Cancer Research. 2013;19:4740–4749. doi: 10.1158/1078-0432.CCR-13-0954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smits M, van Rijn S, Hulleman E, Biesmans D, van Vuurden DG, Kool M, et al. EZH2-Regulated DAB2IP Is a Medulloblastoma Tumor Suppressor and a Positive Marker for Survival. Clinical Cancer Research. 2012;18:4048–4058. doi: 10.1158/1078-0432.CCR-12-0399. [DOI] [PubMed] [Google Scholar]
- 56.Fillmore CM, Xu C, Desai PT, Berry JM, Rowbotham SP, Lin YJ, et al. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature. 2015;520:239–242. doi: 10.1038/nature14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gibaja V, Shen F, Harari J, Korn J, Ruddy D, Saenz-Vash V, et al. Development of secondary mutations in wild-type and mutant EZH2 alleles cooperates to confer resistance to EZH2 inhibitors. Oncogene. 2015;35:558–66. doi: 10.1038/onc.2015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 59.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 60.Qi W, Chan H, Teng L, Li L, Chuai S, Zhang R, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci U S A. 2012;109:21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Konze KD, Ma A, Li F, Barsyte-Lovejoy D, Parton T, Macnevin CJ, et al. An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1. ACS chemical biology. 2013;8:1324–1334. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bradley WD, Arora S, Busby J, Balasubramanian S, Gehling VS, Nasveschuk CG, et al. EZH2 inhibitor efficacy in non-Hodgkin’s lymphoma does not require suppression of H3K27 monomethylation. Chem Biol. 2014;21:1463–1475. doi: 10.1016/j.chembiol.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 63.Knutson SK, Kawano S, Minoshima Y, Warholic NM, Huang KC, Xiao Y, et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol Cancer Ther. 2014;13:842–854. doi: 10.1158/1535-7163.MCT-13-0773. [DOI] [PubMed] [Google Scholar]
- 64.Verma SK, Tian X, LaFrance LV, Duquenne C, Suarez DP, Newlander KA, et al. Identification of Potent, Selective, Cell-Active Inhibitors of the Histone Lysine Methyltransferase EZH2. ACS Med Chem Lett. 2012;3:1091–1096. doi: 10.1021/ml3003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 66.Poirier JT, Gardner EE, Connis N, Moreira AL, de Stanchina E, Hann CL, et al. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene. 2015;34:5869–5878. doi: 10.1038/onc.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 68.Harbour JW, Roberson ED, Anbunathan H, Onken MD, Worley LA, Bowcock AM. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet. 2013;45:133–135. doi: 10.1038/ng.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Te Raa GD, Derks IA, Navrkalova V, Skowronska A, Moerland PD, van Laar J, et al. The impact of SF3B1 mutations in CLL on the DNA damage response. Leukemia. 2014;29:1133–1142. doi: 10.1038/leu.2014.318. [DOI] [PubMed] [Google Scholar]
- 70.Schilling B, Bielefeld N, Sucker A, Hillen U, Zimmer L, Schadendorf D, et al. Lack of SF3B1 R625 mutations in cutaneous melanoma. Diagn Pathol. 2013;8:87. doi: 10.1186/1746-1596-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Furney SJ, Pedersen M, Gentien D, Dumont AG, Rapinat A, Desjardins L, et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer discovery. 2013;3:1122–1129. doi: 10.1158/2159-8290.CD-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rossi D, Bruscaggin A, Spina V, Rasi S, Khiabanian H, Messina M, et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood. 2011;118:6904–6908. doi: 10.1182/blood-2011-08-373159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ogawa S. Splicing factor mutations in AML. Blood. 2014;123:3216–3217. doi: 10.1182/blood-2014-04-566752. [DOI] [PubMed] [Google Scholar]
- 74.Malcovati L, Karimi M, Papaemmanuil E, Ambaglio I, Jadersten M, Jansson M, et al. SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood. 2015;126:233–241. doi: 10.1182/blood-2015-03-633537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mupo A, Seiler M, Sathiaseelan V, Pance A, Yang Y, Agrawal AA, et al. Hemopoietic-specific Sf3b1-K700E knock-in mice display the splicing defect seen in human MDS but develop anemia without ring sideroblasts. Leukemia. doi: 10.1038/leu.2016.251. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dolatshad H, Pellagatti A, Liberante FG, Llorian M, Repapi E, Steeples V, et al. Cryptic splicing events in the iron transporter ABCB7 and other key target genes in SF3B1-mutant myelodysplastic syndromes. Leukemia. doi: 10.1038/leu.2016.149. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Inoue D, Bradley RK, Abdel-Wahab O. Spliceosomal gene mutations in myelodysplasia: molecular links to clonal abnormalities of hematopoiesis. Genes Dev. 2016;30:989–1001. doi: 10.1101/gad.278424.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Darman RB, Seiler M, Agrawal AA, Lim KH, Peng S, Aird D, et al. Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3′ Splice Site Selection through Use of a Different Branch Point. Cell Rep. 2015;13:1033–1045. doi: 10.1016/j.celrep.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 79.De Arras L, Alper S. Limiting of the innate immune response by SF3A-dependent control of MyD88 alternative mRNA splicing. PLoS Genet. 2013;9:e1003855. doi: 10.1371/journal.pgen.1003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Visconte V, Rogers HJ, Singh J, Barnard J, Bupathi M, Traina F, et al. SF3B1 haploinsufficiency leads to formation of ring sideroblasts in myelodysplastic syndromes. Blood. 2012;120:3173–3186. doi: 10.1182/blood-2012-05-430876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kajiwara T, Matsushita K, Itoga S, Tamura M, Tanaka N, Tomonaga T, et al. SAP155-mediated c-myc suppressor far-upstream element-binding protein-interacting repressor splicing variants are activated in colon cancer tissues. Cancer Sci. 2013;104:149–156. doi: 10.1111/cas.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, et al. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gokmen-Polar Y, Neelamraju Y, Goswami CP, Gu X, Nallamothu G, Janga SC, et al. Expression levels of SF3B3 correlate with prognosis and endocrine resistance in estrogen receptor-positive breast cancer. Mod Pathol. 2015;28:677–685. doi: 10.1038/modpathol.2014.146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.