Abstract
Introduction
Distal hereditary motor neuropathy (dHMN) causes distal-predominant weakness without prominent sensory loss. Myosin heavy chain disorders most commonly result in distal myopathy and cardiomyopathy with or without hearing loss, but a complex phenotype with dHMN, myopathy, hoarseness, and hearing loss was reported in a Korean family with a c.2822G>T mutation in MYH14.
Objective
To report phenotypic features in a North American family with the c.2822G>T in MYH14.
Methods
Clinical and molecular characterization was performed in a large, 6-generation, Caucasian family with MYH14 dHMN.
Results
A total of 11 affected and 7 unaffected individuals were evaluated and showed varying age of onset and severity of weakness. Genotypic concordance was confirmed with molecular analysis. Electrophysiological studies demonstrated distal motor axonal degeneration without myopathy in all affected subjects tested.
Conclusions
Mutation of MYH14 can result in a range of neuromuscular phenotypes that includes a dHMN and hearing loss phenotype with variable age of onset.
Keywords: Myosin, distal hereditary motor neuropathy, hearing loss, autosomal dominant, myopathy
Introduction
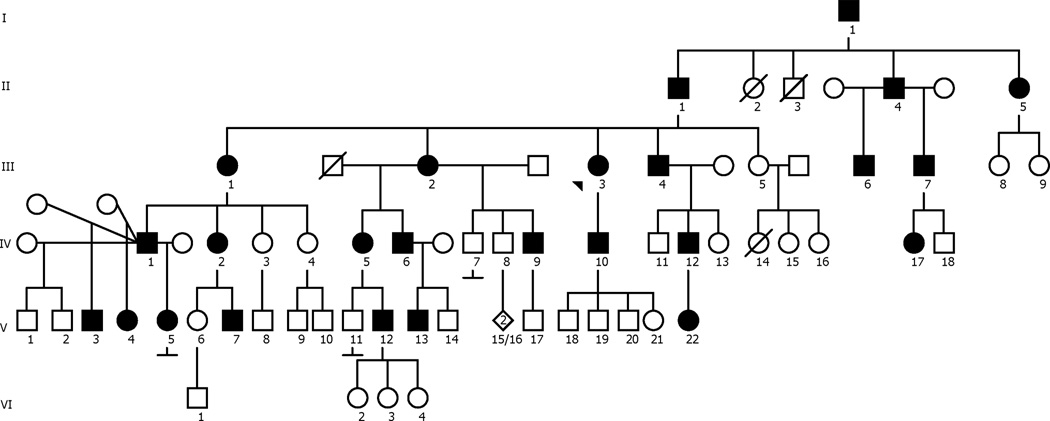
Distal weakness and muscle atrophy with complete or marked sensory sparing is caused by several genetic neuromuscular conditions including the distal hereditary motor neuropathies (dHMN) and distal myopathies. Myosin heavy chain disorders can result in distal myopathies and cardiomyopathies with (for example, MYH7, MYH14, MYH15 and MYH9) or without (for example, MYH3 and MYH8)1 hearing loss. Most of the myosin heavy chains associated with distal myopathies belong to the conventional or sarcomeric muscle myosin heavy chain or myosin complexes (including myosin light chains) (for example, MYL2 and MYL3)2. While conventional myosins contribute to the actin-myosin complex/actin-myosin crossbridge cycle in the sarcomere, the ‘unconventional’ (or non-muscle) myosins are important for many other cellular functions such as neuronal transport, cell division and ciliary motility, including the stereocilia of the ear3,4. Several mutations in MYH14 have been reported that result in autosomal dominant hearing loss5,6. Recently, a Korean family with a complex phenotype of dHMN, myopathy, hoarseness, and hearing loss associated with a dominant mutation in MYH14 (c.2822G>T) was described7. In this report, we describe the phenotype associated with the c.2822G>T mutation in the MYH14 gene in a 6-generation, 71-member Caucasian family (See pedigree, Figure 1).
Figure 1.
Pedigree of the affected family. Filled symbols indicate affected family members. Arrowhead indicates proband.
Methods
Standard Protocol Approvals, Registrations, and Patient Consents
The study was approved by the Institutional Review Board at The Ohio State University Wexner Medical Center, and written informed consent was obtained from all participants.
Subjects
The proband (Figure 1, III-3) presented with a 29 year history of progressive distal muscle weakness and atrophy associated with hearing loss. The family history was positive for a similar pattern of weakness in multiple family members. Eighteen clinically unaffected and affected family members (including the proband) were enrolled for clinical characterization and DNA linkage analysis. Clinical examinations were performed by a single examiner in 7 clinically affected and 5 clinically unaffected individuals prior to molecular analysis. Clinical, electrophysiological, and audiometric data were analyzed for phenotypic characterization. Genomic DNA was prepared from blood samples as previously described8.
Linkage analysis
DNA was hybridized to Human Mapping 500K SNP Set (Includes 250K Nsp Array and 250K Sty Array) Affymetrix™ GeneChips® to determine genotype at the loci. Genome wide genotyping calls were extracted using BRLMM algorithm9. The SNP call rate of each sample was >82% with an overall average call rate of 97%. Mendelian errors were identified (<1%) with the PEDSTAT program10. Parametric linkage analysis with autosomal dominant inheritance was performed using MERLIN assuming a penetrance of 0.9 and disease allele frequency of 0.0000111.
Whole exome sequencing
Exome sequencing was performed on 3 affected individuals (III-4, IV-1, and IV-2) and 1 unaffected individual (VI-1). DNA was prepared using the SOLiD exon capture kit, and SOLiD sequencing was performed (Thermo Fisher). Reads were aligned to the human hg19 genome using BWA12. The Genome Analysis Tool Kit pipeline was used for calling variants13. This includes steps for removal of duplicate reads, recalibration of base quality scores, and local realignment of reads with insertions or deletions. Finally, pVAAST was used to determine which variants segregated with the affected individuals but not unaffected controls14. ANNOVAR was used to annotate the candidate variants15. At the time of these efforts, the study by Choi et al, reported the c.2822G>T MYH147. We then searched for mutations of the MYH14 gene in the exome sequences of affected individuals in our pedigree.
Variant validation
We developed a PCR assay followed by restriction digest to identify the c.2822G>T mutation in MYH14. This was used to identify the mutation in 11 affected individuals and the absence of the mutation in 7 unaffected individuals. PCR primers FP: 5`CAAAGCAAGTTACAAGGCAGTAG, RP: 5`GTTTGCATTTGACGGCTGCAC were used to amplify 50ng of genomic patient DNA. The MYC14 mutation creates an SmlI site. PCR products were digested with SmlI (Catalogue Number: R0597S, NEB) for 2 hours at 37°C and run on a 2% agarose gel.
Results
The proband (III-3) initially presented at age 34 with progressive distal weakness and atrophy that began at age 5. Symptoms of progressive hearing loss began at age 20. She had no other pertinent medical history. Examination demonstrated bilateral hearing loss and distal muscle atrophy, greater in the legs than the arms. Strength testing (Medical Research Council grading) revealed shoulder abductors (5-/5, right and left respectively), elbow flexors (4/5), elbow extensors (4+/5), wrist extensors (4-/5), and hand intrinsic muscles (3/5). Neck flexors and extensors were 5/5. Lower extremity strength testing revealed hip flexors (4-/5), hip abductors (3-/5), knee extensors (5-/5), knee flexors (4/5), ankle dorsiflexors, evertors and invertors (0/5), and plantar flexors (1/5). Sensory examination and reflexes were normal. Electrodiagnostic testing (Table) demonstrated an absent fibular compound muscle action potential (CMAP), normal median motor and sural sensory nerve action potentials (SNAPs), and length-dependent active denervation (positive sharp waves and fibrillation potentials) associated with large-amplitude, long-duration motor unit action potentials with reduced recruitment patterns. Proximal limb or paraspinal muscle denervation and short-duration, small-amplitude motor unit action potentials were not noted. Audiometry confirmed sensorineural hearing loss that was preferentially reduced in the 1000 to 8000 Hz range.
Table.
Sensory and motor nerve conduction studies in affected family members.
| ID | Age (years) |
Median Motor | Peroneal Motor | Sural Sensory | ||
|---|---|---|---|---|---|---|
| Amp (mV) (≥4 mV) |
CV (m/s) (≥50 m/s) |
Amp (mV) (≥2 mV) |
CV (m/s) (≥40 m/s) |
Amp (µV) (≥10µV) |
||
| III-3 | 40 | 7.9 | 58 | NR | -- | 17 |
| III-7 | 30 | 14 | 56 | NR | -- | 16 |
| IV-5 | 47 | 7.2 | 54 | NR | -- | 31 |
| IV-10 | 29 | ND | ND | 0.1 | 47 | 14 |
ID – Pedigree identification, Amp - amplitude, CV - conduction velocity, mV - millivolts, µV - microvolts, ND - not done, NR - no response
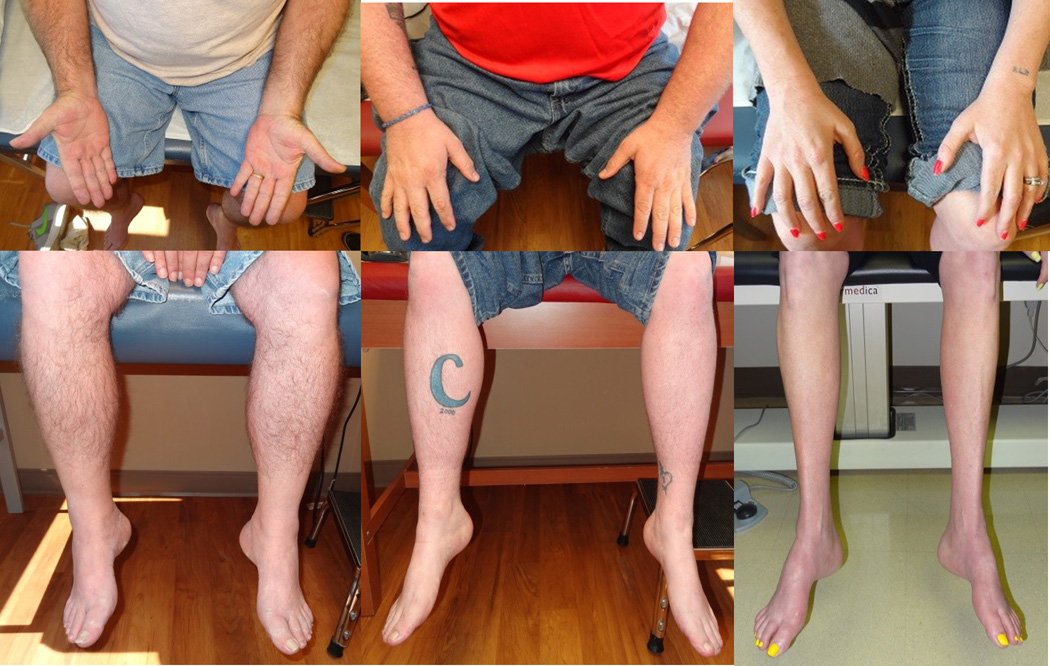
The proband had a family history that included 25 family members spanning 6 generations who were affected with symptoms of distal muscle atrophy and weakness (Figure 2) and sensorineural hearing loss. Clinical examination of 7 affected individuals by a single examiner showed symmetric motor deficits that were most prominent in the distal upper and lower limbs (Supplemental Table S1, available online). Distal muscles of the upper and lower limbs were equally affected in 5 of 7 individuals. Proximal weakness was present in 2 of the older individuals. Sensory examinations were normal. Muscle stretch reflexes were generally reduced in proportion to weakness. Short hands and fingers were noted in affected individuals (Figure 2), but mildly short fingers were also noted in an unaffected individual (VI-1) who lacked the MYH14 mutation. By history, and on examination as assessed by finger rub, all affected individuals had hearing loss. In addition to the proband, hearing loss was confirmed with formal audiometric testing in 2 other affected individuals. At age 7, IV-10 showed moderate to severe loss at 4–8 kHz frequencies, and IV-11 showed hearing loss at frequencies between 0.5–4 kHz frequencies at age 4.There was no history of cardiac abnormalities in any of the affected individuals enrolled in the study. There was no history of pulmonary abnormalities or ventilatory dysfunction in any affected family members except for the proband (III-3), who developed obstructive sleep apnea and chronic obstructive pulmonary disease later in life, which were felt to be related to obesity and smoking, respectively.
Figure 2.
Features of muscle atrophy in distal upper and lower limbs in 3 affected family members.
Variability of onset was noted in affected individuals (See Supplemental Table S1). Onset of symptoms was in late adolescence/early adulthood in 3 affected individuals and was pre-pubertal in 2 others. In 1 affected individual (III-1), late onset hearing loss (at age 67) was reported without complaints of weakness, but detailed examination demonstrated mild weakness of finger abduction. All other examined individuals had both motor and hearing symptoms, and the reported age of onset for motor weakness (mean: 14.0 ± 8.8 years, range 4 – 23, inter-quartile range 17.5) and hearing loss (mean: 13.7 ± 8.5 years, range 4 – 23, inter-quartile range 17.5) were strongly correlated such that individuals with earlier onset of weakness also experienced earlier onset of hearing loss (Pearson correlation coefficient r=0.996, P<0.001).
Electrodiagnostic studies were available in 4 affected individuals (Table). Fibular CMAPs were absent from the extensor digitorum brevis muscle in 3; 1 had a very low fibular CMAP amplitude from this muscle, with normal conduction velocity. Sural SNAPs were normal in all 4. Motor and sensory nerve conduction velocities were within reference ranges, excluding 2 patients with superimposed ulnar neuropathy at the elbow and 1 patient with bilateral median neuropathies at the wrist (not shown in Table). Needle electromyography demonstrated findings consistent with distal, chronic motor axonal loss in the upper and lower limbs in a length-dependent fashion. Fibrillation potentials, rare fasciculation potentials, and large-amplitude, long-duration motor unit action potentials with reduced recruitment were noted in distal muscles. Proximal muscles showed normal findings.
Parametric linkage analysis of the family members was performed which mapped the locus of interest to 19q13, specifically to a series of SNPs between SNP A-2141378-SNPA-2026352 which corresponds to the sequence position 45188471–53100377 on chromosome 19 using build 19 of the human genome. This position is 71.26–92.47 using the Decode sex averaged recombination map16. A series of SNPs in this region showed an LOD score of 4, and the HLOD for these markers was also 4. No other regions showed significant LOD scores. We next performed exome analysis on individuals IV-2 (affected), III-4 (affected), IV-1 (affected), using pVAAST to determine which variants segregated with the affected individuals. This identified a mutation in the MYH14 gene, a G-to-T change at position 2822, corresponding to a change in amino acid 941 (Arg941Leu). We extracted a list of all genes in the linkage region, and used Ingenuity Pathway Analysis to determine if any of these genes had a neurological phenotype17. Among the genes in the region were APOE, RTN2, FKRP, MED25, MYH14, and CEACAM16, all of which have been associated with a motor phenotype or hearing loss. However, only MYH14 had a variant present in all 3 affected individuals on exome sequencing.
The c.2822G>T change in the MYH14 gene was 100% concordant with the dHMN and hearing loss phenotype (present in 11 of 11 affected family members) and absent in 7 of 7 in unaffected family members (Supplementary figure S1).
Discussion
The pedigree presented here expands the dHMN and sensorineural hearing loss phenotype associated with MYH14 (c.2822G>T) and is similar to the phenotype described previously by Choi and colleagues7. Nevertheless, the range of severity and extent of tissue involvement appears to differ. Whereas the Korean pedigree demonstrated a more complex phenotype of peripheral neuropathy, hearing loss, and myopathy, the phenotype in this Caucasian pedigree had a pure dHMN with sensorineural hearing loss without overt findings of myopathy7. No myopathic features were noted on electrodiagnostic testing in our study. It is possible that subtle electromyographic changes of myopathic involvement could have been obscured by changes of chronic denervation, and our study did not include muscle biopsies.
Onset of muscle weakness in this family was more variable [14.0 ± 8.8 years (range: 4–23)] compared with the previously reported family [10.6 ± 2.4 years (range: 5–14 years)]7. Detailed examinations of affected individuals demonstrated distal limb weakness, but interestingly, in some patients the distal upper limbs were similarly involved compared to the lower limbs. This pattern of weakness argues against a classic length-dependent distribution. Yet, in the 4 individuals who underwent electrophysiological assessment, the findings supported length-dependent axonal loss. A similar distribution was noted in 2 of the patients previously reported by Choi et al7. Additionally, 1 affected individual in our pedigree reported only late-onset hearing loss without weakness, although mild intrinsic hand weakness was noted on examination. Short hands have been noted in patients who carry mutations in the MYH3 gene18–20. Short hands and fingers were noted in affected individuals (Figure 2), but this was also noted in an unaffected individual who lacked the MYH14 mutation. Thus, it is unclear whether this is an associated phenotypic characteristic.
The differences between the 2 pedigrees may be due to differences in genetic background, genetic modifiers, expression levels of the pathogenic protein in target tissues, or a combination of these factors. Individuals with this mutation may have long periods of subclinical disease given the wide range of age of symptom onset, thus diagnostic vigilance for the MYH14 mutation must be afforded to individuals with mild, chronic symptoms of hearing loss and/or muscle weakness and atrophy regardless of age of onset. When hearing loss and distal skeletal muscle weakness occur together, one should consider a MYH14 gene disorder in the differential diagnosis.
The functional consequence of the MYH14 (c.2822G>T, Arg941Leu) mutation is not known. MYH14 is a member of the ‘unconventional’ non-muscle myosin II family of molecular motors. The human MYH14 was cloned by RT-PCR strategies from sciatic nerve RNA21, and 2 splice variants were identified by Golomb and colleagues in 200422. MYH14 is expressed in skeletal muscle, brain, heart, colon, kidney, liver, small intestine, lung, and peripheral nerve22. MYH14 encodes a 1995-amino acid protein containing an N-terminal myosin ‘motor’ domain, the myosin head domain, 2 IQ domains, and a C-terminal myosin tail. The myosin tail domain is important for attachment to various intracellular vesicular cargoes. Five mutations in the MYH14 gene (Ser7Ter, Ser120Leu, Gly376Cys, Arg726Ser, and Leu976Phe) were found to be associated with an isolated phenotype of progressive hearing loss21,6. The arginine to leucine transition (Arg941Leu) seen in our pedigree would likely disrupt the secondary structure of the C-terminal domain, which is the cargo-binding domain, and this could alter the interaction with client/adaptor/cargo attachment proteins. The particular cargo proteins affected could determine the most affected tissues. Further biochemical studies to understand the functional consequence of this mutation are warranted and could identify potential therapeutic targets.
In conclusion, we have described the phenotypic characteristics of a large family with a dominant mutation in the MYH14 gene. The unique features of this familial condition are motor neuropathy and hearing loss without overt myopathy. Affected family members displayed a wide range of disease onset and severity. When hearing loss and distal skeletal muscle weakness occur together, one should consider a MYH14 gene disorder in the differential diagnosis.
Supplementary Material
Acknowledgments
W.D.A. was supported by grant funding from NIH-NICHD (5K12HD001097-17). S.J.K. was supported by NINDS (K08NS067282).
Abbreviations
- CMAP
compound muscle action potential
- dHMN
distal hereditary motor neuropathies
- SNAP
sensory nerve action potential
References
- 1.Tajsharghi H, Oldfors A. Myosinopathies: pathology and mechanisms. Acta neuropathologica. 2013;125(1):3–18. doi: 10.1007/s00401-012-1024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poetter K, Jiang H, Hassanzadeh S, Master SR, Chang A, Dalakas MC, Rayment I, Sellers JR, Fananapazir L, Epstein ND. Mutations in either the essential or regulatory light chains of myosin are associated with a rare myopathy in human heart and skeletal muscle. Nature genetics. 1996;13(1):63–69. doi: 10.1038/ng0596-63. [DOI] [PubMed] [Google Scholar]
- 3.Cheney RE, Mooseker MS. Unconventional myosins. Current Opinion in Cell Biology. 1992;4(1):27–35. doi: 10.1016/0955-0674(92)90055-h. [DOI] [PubMed] [Google Scholar]
- 4.O'Connell CB, Tyska MJ, Mooseker MS. Myosin at work: Motor adaptations for a variety of cellular functions. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2007;1773(5):615–630. doi: 10.1016/j.bbamcr.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 5.Donaudy F, Snoeckx R, Pfister M, Zenner HP, Blin N, Di Stazio M, Ferrara A, Lanzara C, Ficarella R, Declau F, Pusch CM, Nurnberg P, Melchionda S, Zelante L, Ballana E, Estivill X, Van Camp G, Gasparini P, Savoia A. Nonmuscle myosin heavy-chain gene MYH14 is expressed in cochlea and mutated in patients affected by autosomal dominant hearing impairment (DFNA4) American journal of human genetics. 2004;74(4):770–776. doi: 10.1086/383285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang T, Pfister M, Blin N, Zenner HP, Pusch CM, Smith RJ. Genetic heterogeneity of deafness phenotypes linked to DFNA4. American journal of medical genetics Part A. 2005;139(1):9–12. doi: 10.1002/ajmg.a.30989. [DOI] [PubMed] [Google Scholar]
- 7.Choi BO, Kang SH, Hyun YS, Kanwal S, Park SW, Koo H, Kim SB, Choi YC, Yoo JH, Kim JW, Park KD, Choi KG, Kim SJ, Zuchner S, Chung KW. A complex phenotype of peripheral neuropathy, myopathy, hoarseness, and hearing loss is linked to an autosomal dominant mutation in MYH14. Human mutation. 2011;32(6):669–677. doi: 10.1002/humu.21488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolb SJ, Snyder PJ, Poi EJ, Renard EA, Bartlett A, Gu S, Sutton S, Arnold WD, Freimer ML, Lawson VH, Kissel JT, Prior TW. Mutant small heat shock protein B3 causes motor neuropathy: utility of a candidate gene approach. Neurology. 2010;74(6):502–506. doi: 10.1212/WNL.0b013e3181cef84a. [DOI] [PubMed] [Google Scholar]
- 9.Affymetrix. BRLMM: an improved genotype calling method for the GeneChip® Human Mapping 500K array set. Revision version 1.0 ed. 2006 [Google Scholar]
- 10.Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21(16):3445–3447. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- 11.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nature genetics. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 12.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu H, Roach JC, Coon H, Guthery SL, Voelkerding KV, Margraf RL, Durtschi JD, Tavtigian SV, Shankaracharya, Wu W, Scheet P, Wang S, Xing J, Glusman G, Hubley R, Li H, Garg V, Moore B, Hood L, Galas DJ, Srivastava D, Reese MG, Jorde LB, Yandell M, Huff CD. A unified test of linkage analysis and rare-variant association for analysis of pedigree sequence data. Nature biotechnology. 2014;32(7):663–669. doi: 10.1038/nbt.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong A, Thorleifsson G, Gudbjartsson DF, Masson G, Sigurdsson A, Jonasdottir A, Walters GB, Jonasdottir A, Gylfason A, Kristinsson KT, Gudjonsson SA, Frigge ML, Helgason A, Thorsteinsdottir U, Stefansson K. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467(7319):1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- 17.Krämer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2013 doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall JG, Reed SD, Greene G. The distal arthrogryposes: delineation of new entities--review and nosologic discussion. American journal of medical genetics. 1982;11(2):185–239. doi: 10.1002/ajmg.1320110208. [DOI] [PubMed] [Google Scholar]
- 19.Kimber E, Tajsharghi H, Kroksmark AK, Oldfors A, Tulinius M. A mutation in the fast skeletal muscle troponin I gene causes myopathy and distal arthrogryposis. Neurology. 2006;67(4):597–601. doi: 10.1212/01.wnl.0000230168.05328.f4. [DOI] [PubMed] [Google Scholar]
- 20.Krakowiak PA, Bohnsack JF, Carey JC, Bamshad M. Clinical analysis of a variant of Freeman-Sheldon syndrome (DA2B) American journal of medical genetics. 1998;76(1):93–98. doi: 10.1002/(sici)1096-8628(19980226)76:1<93::aid-ajmg17>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 21.Leal A, Endele S, Stengel C, Huehne K, Loetterle J, Barrantes R, Winterpacht A, Rautenstrauss B. A novel myosin heavy chain gene in human chromosome 19q13.3. Gene. 2003;312:165–171. doi: 10.1016/s0378-1119(03)00613-9. [DOI] [PubMed] [Google Scholar]
- 22.Golomb E, Ma X, Jana SS, Preston YA, Kawamoto S, Shoham NG, Goldin E, Conti MA, Sellers JR, Adelstein RS. Identification and characterization of nonmuscle myosin II-C, a new member of the myosin II family. The Journal of biological chemistry. 2004;279(4):2800–2808. doi: 10.1074/jbc.M309981200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.