Abstract
Introduction
The growing public threat of Alzheimer's disease (AD) has raised the urgency to quantify the degree of cognitive decline during the conversion process of mild cognitive impairment (MCI) to AD and its underlying genetic pathway. The aim of this article was to test genetic common variants associated with accelerated cognitive decline after the conversion of MCI to AD.
Methods
In 583 subjects with MCI enrolled in the Alzheimer's Disease Neuroimaging Initiative (ADNI; ADNI-1, ADNI-Go, and ADNI-2), 245 MCI participants converted to AD at follow-up. We tested the interaction effects between individual single-nucleotide polymorphisms and AD diagnosis trajectory on the longitudinal Alzheimer's Disease Assessment Scale-Cognition scores.
Results
Our findings reveal six genes, including BDH1, ST6GAL1, RAB20, PDS5B, ADARB2, and SPSB1, which are directly or indirectly related to MCI conversion to AD.
Discussion
This genome-wide association study sheds light on a genetic mechanism of longitudinal cognitive changes during the transition period from MCI to AD.
Keywords: Alzheimer's disease, GWAS, Mild cognitive impairment, Cognitive decline, Longitudinal study
1. Introduction
Alzheimer's disease (AD) is a progressive, neurodegenerative disorder that imposes social, psychological, and financial burden on both patients and their caregivers. Hebert et al. predict that the total number of individuals with AD dementia will be 13.8 million by 2050 [1]. This prediction, coupled with a lack of disease-modifying treatments, is estimated to have cumulative costs of more than $20 trillion by 2050 for the care of AD patients [2]. To date, extensive efforts have been made to delineate a set of risk factors that affect the development of AD. In particular, genome-wide association studies (GWASs) have been used to identify genetic variants that may contribute to AD. Most GWASs of AD have focused on the detection of single-nucleotide polymorphisms (SNPs) that are associated with the susceptibility of developing AD [3], [4], [5], [6], [7], [8]. The early onset of AD is known to result from mutations in one of three genes: the amyloid precursor protein (APP), presenilin 1 (PSEN1), or presenilin 2 (PSEN2) [9], [10]. The inheritance of the ɛ4 allele of the apolipoprotein E (APOE) has a substantial impact on the late onset of sporadic AD [11], [12]. Recent GWASs have identified additional AD-related genes, including clusterin (CLU), complement receptor 1 (CR1), and phosphatidylinositol-binding clathrin assembly protein gene (PICALM) [13], [14], whereby those genes alter production of the amyloid β peptide (Aβ) [15], [16], [17].
In the present study, we aimed to detect genetic common variants associated with accelerated cognitive decline after the conversion of mild cognitive impairment (MCI) to AD. MCI is a clinical syndrome characterized by insidious onset and progression of cognitive impairments. It is often considered as a transitional stage between normal aging and AD, because approximately 50% of MCI patients develop AD in 5 years from diagnosis [18]. Critically, therapeutic interventions and disease-modifying drugs appear to be more effective during the MCI or early stage of AD than at the more severe stages of AD [19], [20], [21]. As such, it is an ongoing quest to delineate a set of risk factors that affect conversion from MCI to AD. Recent GWASs have focused on the following phenotypes related to MCI-AD progression: binary outcome indicating MCI-AD conversion, time to conversion from MCI to AD, and cognitive decline. For example, Hu et al. (2011) found novel loci to be associated with longitudinal cognitive changes of MCI patients [22] and those loci were also associated with time to conversion from MCI to AD. Here, we only focus on an MCI-AD conversion group to identify genetic common variants contributing to rapid cognitive decline after an MCI patient develops AD.
We analyzed the Alzheimer's Disease Neuroimaging Initiative (ADNI; ADNI-1, ADNI-2, and ADNI-GO) cohort data. Among them, there remained 245 participants who converted to AD at follow-up. Longitudinal Alzheimer's Disease Assessment Scale-Cognition (ADAS-Cog) scores were used to measure cognitive function including memory, language, praxis, and orientation domain scores indicated greater cognitive impairment. To identify SNPs associated with longitudinal cognitive changes, we tested interaction effects between individual SNPs and AD diagnosis trajectory (MCI = 0, AD = 1) on the longitudinal ADAS-Cog scores. To account for individual variability of (1) baseline ADAS-Cog score and (2) the effect size of AD diagnosis trajectory, random effects for intercept and slope of AD diagnosis trajectory were incorporated in our model.
2. Materials and methods
2.1. Alzheimer's Disease Neuroimaging Initiative
The study population was obtained from the ADNI database (www.loni.usc.edu/ADNI). The ADNI study has aimed to detect and monitor the early stage of AD by investigating serial magnetic resonance imaging, positron emission tomography, genetic, biochemical biomarkers, and neuropsychological and clinical assessment. The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California, San Francisco. The ADNI began in 2004 and recruited 400 subjects with MCI, 200 subjects with early AD, and 200 cognitively normal elderly from more than 50 sites across the United States and Canada. This multisite, longitudinal study was financially supported as $67 million by National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration, and 13 private pharmaceutical companies. This initial phase, called ADNI-1, was extended with ADNI-GO in 2009. ADNI-GO investigated the existing ADNI-1 cohort and included 200 participants diagnosed as having early MCI. In 2011, ADNI-2 began to study participants from the ADNI-1/ADNI-GO and added 150 elderly control subjects, 100 early MCI participants, 150 late MCI participants, and 150 MCI patients. For up-to-date information, see www.adni-info.org.
2.2. Data description
In the sample of this study, we considered White participants who had developed AD from MCI at the baseline. During 120 months of follow-up, 245 MCI patients progressed to AD before study completion and the remaining 338 MCI patients did not convert to AD before study end. We considered only 245 patients who developed AD by their follow-up. The data included basic demographic and clinical information at the baseline: age, education length, and gender. Their ADAS-Cog scores were recorded about every 9 months in average with the mean follow-up duration of 48 months. Genotyping for the ADNI-1, ADNI-GO, and ADNI-2 data was performed using the Human 610-Quad BeadChip, Illumina Human Omni Express BeadChip, Illumina Omni 2.5 M (whole-genome sequencing [WGS] Platform), respectively. It was completed on all ADNI participants using the genotyping protocol whose details are described in [23] and http://adni.loni.usc.edu/methods/genetic-data-methods.
2.3. Quality control and genotype imputation
We performed quality control (QC) steps on the raw genotype data to ensure that only high-quality data were included in the final analysis. QC procedures included (1) call rate check per subject and per SNP marker, (2) gender check, (3) sibling pair identification, (4) the Hardy-Weinberg equilibrium test, (5) marker removal by the minor allele frequency, and (6) population stratification. We also calculated an inbreeding coefficient (F) that represents the expected percentage of homozygosity. There were no subjects with excessive heterozygosity (|F| > 0.15) [53]. The second line preprocessing steps included removal of SNPs with (1) more than 5% missing values, (2) minor allele frequency smaller than 5%, and (3) Hardy-Weinberg equilibrium P value < 10−6. The previously mentioned procedures were carried out in PLINK version 1.9 [24] with visualization performed in R (http://www.r-project.org/) using the qqman package (http://cran.r-project.org/web/packages/qqman/).
Genotype imputation was conducted to estimate unobserved genotypes. MaCH software [25] was used with NCBI 1000 Genomes build 37 (UCSC hg19) as the reference panel. We used different imputation quality metric R2 for different minor allele frequency categories by following the recommendations given in Beecham et al. [26]. After the imputation step, 242 subjects and 8,092,642 SNPs remained in the present study. Strict QC procedures were followed: (1) removal of individuals with 10% missing genotypes, removal of SNPs with (2) more than 2% missing values, (3) minor allele frequency smaller than 5%, and (4) Hardy-Weinberg equilibrium P value < 10−3. After the further QC, 242 subjects and 5,908,215 SNPs remained. Their demographic information is summarized in Supplementary Table 1.
2.4. Statistical analysis
To identify the SNPs that were associated with ADAS-Cog score changes over time after AD conversion from MCI, we ran linear mixed-effects models with ADAS-Cog score trajectory measured across time as longitudinal responses. Fixed main effects included gender (1 = male; 0 = female), length of education, age at the baseline, follow-up time trajectory, AD diagnosis trajectory (0 = MCI; 1 = AD), and each SNP. We included a random intercept to account for individual variability of baseline ADAS-Cog scores. A random slope of the AD diagnosis trajectory was added to take into account its variability across subjects. We tested a (fixed) interaction effect between each SNP and the AD diagnosis trajectory. To test the interaction effect, we used an approximate F-test based on the Kenward-Roger approach [27]. The first five PC scores from all the 22 chromosomes were also included to address the population stratification issue [28]. The principal component analysis was conducted using PLINK package. Quantile-quantile (QQ) plots and Manhattan plots were produced using the qqman package in R. Regional plots (see Supplementary Figs. 2–6) were generated to graphically show the GWAS results within a given genomic region using Locus Zoom 1.1 [29]. All SNPs within 200–400 kb of identified SNPs by our GWAS were plotted with their −log10(P) value. Their degree of linkage disequilibrium (LD) with each SNP was color-coded.
We ran sensitivity analyses to examine if our results were robust to model specification. We compared the estimation and test results for the top meaningful SNPs (Table 1) by changing the included covariates. We considered two reduced models: all covariates (1) without the length of education and (2) without the length of education and gender. We also conducted a sensitivity analysis using the full model on a subsample consisting of subjects whose baseline ages were greater than 60.
Table 1.
Meaningful single-nucleotide polymorphisms (SNPs) associated with accelerated cognitive decline and the corresponding regression coefficients for the interaction effects. Minor allele frequency (MAF) is the frequency where the second most common allele occurs in a population
| SNP | Chr | Gene | MAF | Coefficient | SE | F value | P value |
|---|---|---|---|---|---|---|---|
| rs17090219 | 18 | 0.093 | 5.15 | 0.93 | 30.54 | 9.48 × 10−8 | |
| rs3936289 | 3 | ST6GAL1 | 0.256 | 3.34 | 0.64 | 26.83 | 4.92 × 10−7 |
| rs56378310 | 13 | RAB20 | 0.424 | −2.98 | 0.60 | 24.40 | 1.52 × 10−6 |
| rs192470679 | 13 | PDS5B | 0.300 | 2.89 | 0.59 | 24.36 | 1.55 × 10−6 |
| rs11121365 | 1 | SPSB1 | 0.062 | 5.76 | 1.18 | 24.00 | 1.92 × 10−6 |
| rs2484 | 3 | BDH1 | 0.095 | 4.53 | 0.94 | 23.03 | 2.83 × 10−6 |
| rs10903488 | 10 | ADARB2 | 0.083 | 4.58 | 0.96 | 22.80 | 3.24 × 10−6 |
NOTE. The SNPs mentioned in table have been reported to be related to Alzheimer's disease (AD) directly/indirectly. The regression coefficient, SE (standard error), and F-test statistic value are shown for the interaction effect of each of the SNPs and the AD diagnosis trajectory. A positive coefficient value implies that cognitive decline gets accelerated after AD conversion as the number of minor alleles of the corresponding SNP increases.
3. Result
3.1. Participant characteristics
To avoid the possible bias caused by population stratification, this study was restricted to White participants from the ADNI-1 and ADNI-2/ADNI-GO cohorts. There were 242 White MCI-AD converters after the QC steps. Supplementary Table 1 lists the characteristics of the study participants.
3.2. SNPs associated with the time to conversion
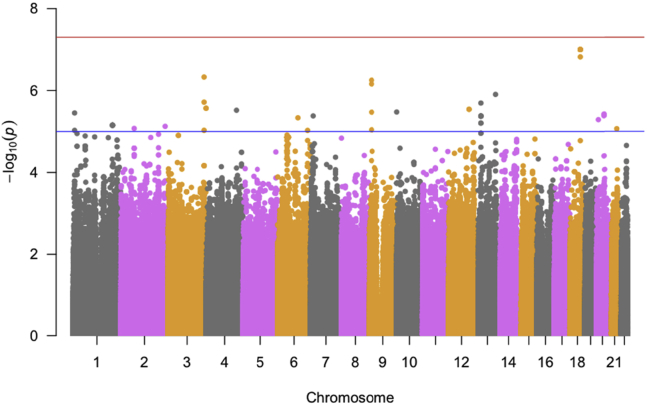
After running a set of linear mixed-effects models for each SNP, we examined if there existed population stratification. The genomic inflation factor λ is estimated as 1.000, which suggests that there is no evidence of population stratification. The QQ plot (Supplementary Fig. 1) shows no evidence of population stratification, as most of the observed P values do not deviate from the expected line (red line). The plot implies that identified SNPs by our longitudinal GWAS do not have a spurious association with the ADAS-Cog score trajectory. Also, as the Manhattan plot shows in Fig. 1, we observed that 56 SNPs had P values that were smaller than a suggestive significance level in GWAS (P < 1 × 10−5). Within the same gene, we only present one of the most significant genotyped SNPs in Supplementary Table 2. Supplementary Figs. 2–6 and Fig. 2 present the regional plots of the identified SNPs in Table 1. Sensitivity analyses (Supplementary Table 3) showed that estimated coefficient values and the corresponding P values in the primary analysis were similar to those in the reduced models and the full model on the subsample (age > 60).
Fig. 1.
The Manhattan plot shows the individual −log10(P values) against base pair positions of 5,908,215 SNPs. Horizontal red and blue lines indicate genome-wide (P < 5 × 10−8) and suggestive (P < 1 × 10−5) significance levels, respectively. Abbreviation: SNPs, single-nucleotide polymorphisms.
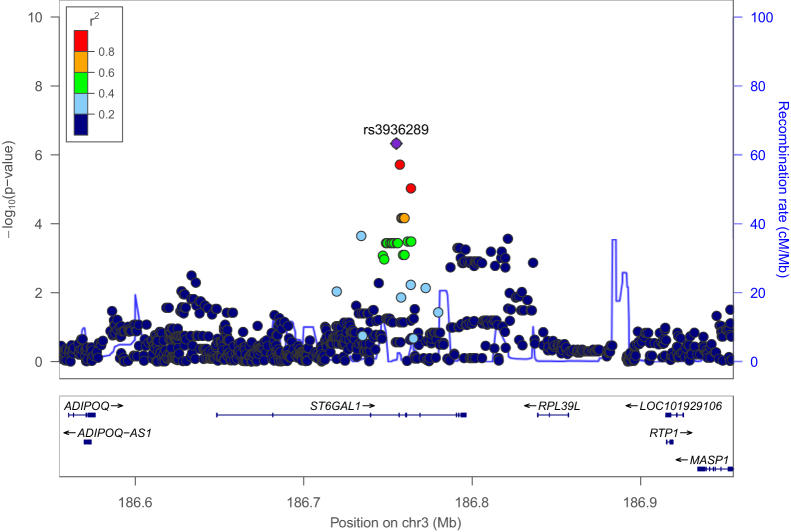
Fig. 2.
A regional plot for association with cognitive decline. The purple dot is the most significant SNP rs3936289 among SNPs in the region surrounding it within 200 kb. Dots are colored according to the range r2 to show their degree of LD with rs3936289. The blue line shows the estimated recombination rate. Abbreviations: LD, linkage disequilibrium; SNP, single-nucleotide polymorphism.
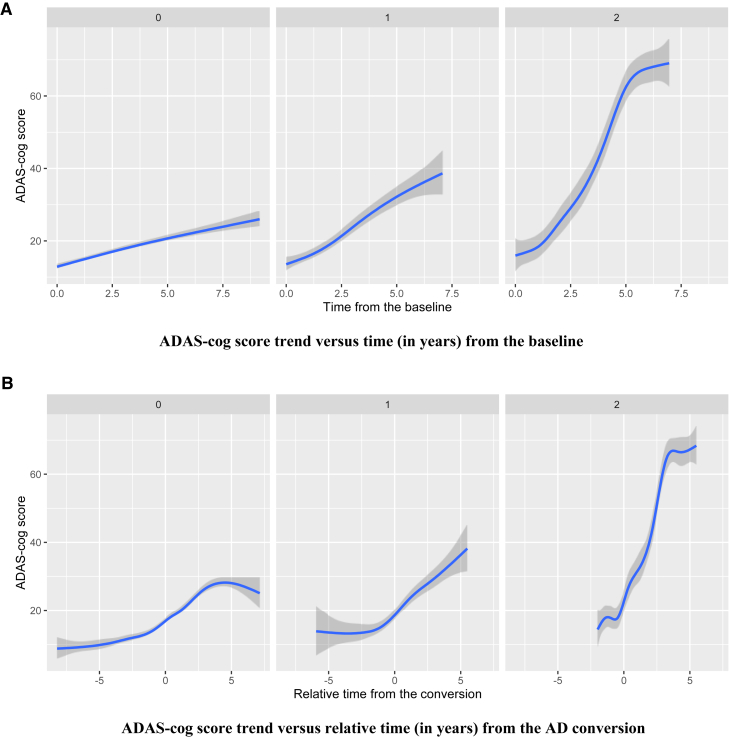
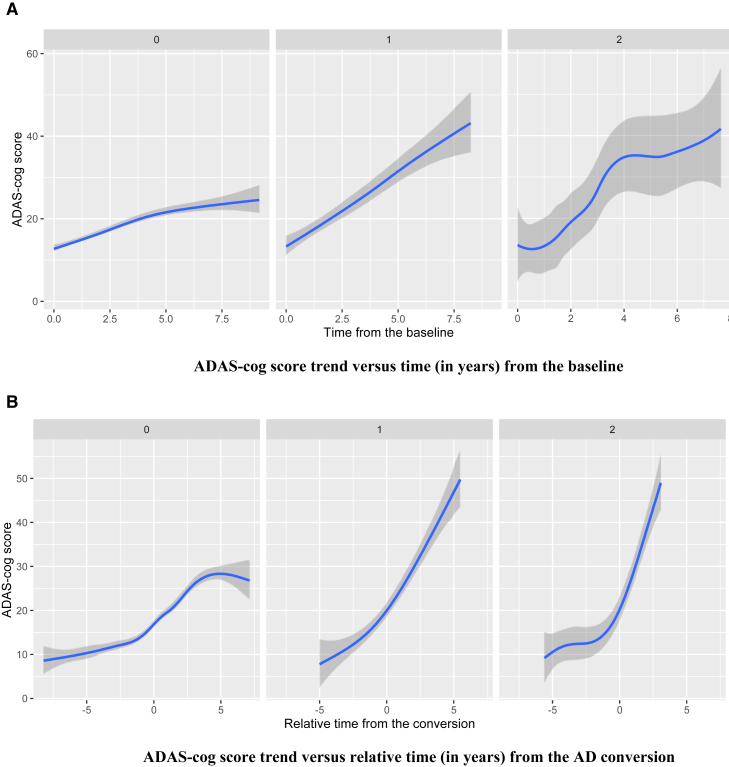
There were eight SNPs, including rs17090219 in chromosome 18, that had the most significant interaction effect with the presence of AD measured at every visit (P = 9.48 × 10−8). The corresponding estimated coefficient is 5.16 (Table 1), which suggests that the cognitive decline tends to be accelerated after AD conversion as the number of minor allele copies in rs17090219 increases. To visualize the mean ADAS-Cog score trend for different SNP groups (the number of copies of minor allele is 0, 1, or 2), we plotted fitted locally polynomial regression (loess) curves. Fig. 3 shows loess plots of three different groups: 0, 1, or 2 for rs17090219. Fig. 3A suggests that cognitive performance rapidly decreases as time goes by for subjects with two copies of the minor allele in rs17090219. Other subject groups had similar ADAS-Cog score trends to each other. Fig. 3B shows that cognitive dysfunction is accelerated after AD conversion for the two copies group. The main and interaction effects for rs17090219 suggest that the mean ADAS-Cog score increases by 6.64 for an additional copy of the minor allele, for any fixed change in the other covariates. This is a novel locus that has not been previously identified in other AD-related articles showing direct/indirect association with AD.
Fig. 3.
(A) The mean trend of ADAS-Cog score was plotted against time (in years) from the baseline for each group (the number of minor alleles of rs17090219 = 0, 1, or 2). (B) The mean trend of ADAS-Cog score was plotted against relative time (in years) from the AD conversion for each group. Abbreviation: ADAS-Cog, Alzheimer's Disease Assessment Scale-Cognition.
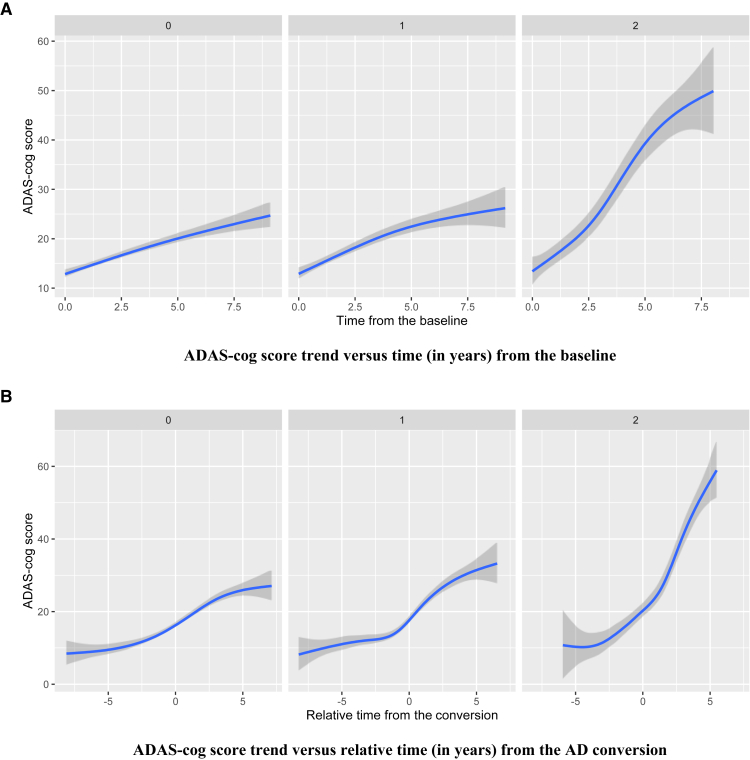
Our GWAS identified 11 SNPs located within the PDS5 cohesin associated factor B (PDS5B) gene. One of the significant SNPs within the PDS5B gene was rs192470679 (P = 1.55 × 10−6) with the corresponding estimated coefficient 2.89 (Table 1). Fig. 4 shows the corresponding loess plots of the three groups: 0, 1, or 2 for rs192470679. Fig. 4A shows that cognitive decline is accelerated as time goes by, especially for subjects who have two copies of the minor allele in rs192470679. Inspection of the bottom figure shows a clear difference among the mean ADAS-Cog score trajectories on the relative time to AD conversion. For subjects having zero or one copy of the minor allele, cognitive decline is increased until AD conversion and it tends to plateau after the conversion. ADAS-Cog scores keep rapidly increasing in their changes for subjects who have two copies of the minor allele. In particular, the additional copy of the minor allele of rs192470679 is associated with a 3.28 increase in the average ADAS-Cog score.
Fig. 4.
(A) The mean trend of ADAS-Cog score was plotted against time (in years) from the baseline for each group (the number of minor alleles of rs192470679 within PDS5B = 0, 1, or 2). (B) The mean trend of ADAS-Cog score was plotted against relative time (in years) from the AD conversion for each group. Abbreviations: AD, Alzheimer's disease; ADAS-Cog, Alzheimer's Disease Assessment Scale-Cognition.
Also, there was an SNP rs10903488 (P = 3.24 × 10−6) within a gene known as adenosine deaminase, RNA-specific, B2 (ADARB2). Its regression coefficient was estimated as 4.58 (Table 1), which implies a greater rate of cognitive decline as the number of minor alleles of rs10903488 increases. Fig. 5A shows that although cognitive deficit increases for all participants over time, the rate of deficit accrual is faster for participants with at least one copy of the minor allele. There is a clear difference in the ADAS-Cog score trend between one and two copies of the minor allele groups on the relative time to conversion domain. Cognitive decline tends to plateau after AD conversion in the zero copy group, whereas the one or two copies groups have accelerated progression of cognitive dysfunction. The average ADAS-Cog score increases by 5.61 as the number of minor alleles of rs10903488 increases by 1.
Fig. 5.
(A) The mean trend of ADAS-Cog score was plotted against time (in years) from the baseline for each group (the number of minor alleles of rs10903488 within ADARB2 = 0, 1, or 2). (B) The mean trend of ADAS-Cog score was plotted against relative time (in years) from the AD conversion for each group. Abbreviations: AD, Alzheimer's disease; ADAS-Cog, Alzheimer's Disease Assessment Scale-Cognition.
4. Discussion
In this GWAS of ADNI-1, ADNI-2, and ADNI-GO data, we identified significant SNPs associated with ADAS-Cog score changes over time after AD conversion from MCI at the 1 × 10−5 suggestive significance level. We found six genes including BDH1, ST6GAL1, RAB20, PDS5B, ADARB2, and SPSB1 that were directly or indirectly related to AD.
There are two genes, PDS5B and ADARB2, that have direct association with AD. The PDS5B gene has been found to be significantly associated with brain atrophy and episodic memory scores [30], [31] for cognitively normal, MCI, and AD subjects. ADARB2 is one of the genes that influences magnetic resonance imaging–derived temporal lobe volume measures [32], where temporal lobe volume significantly changes in aging and in AD [33]. Together these studies demonstrate that the two genes affect cognitive deficit of MCI patients through brain atrophy and brain volume changes when the MCI patients develop AD.
There are four genes, BDH1, ST6Gal1, RAB20, and SPSB1 that are associated with AD indirectly.
Our GWAS identified two SNPs located within the 3-hydroxybutyrate dehydrogenase, type 1 (BDH1) gene. Shi et al. [34] identified that the BDH1 gene was related to aging. Aging is an important factor in AD studies, because risks of age-related diseases, such as hypertension, diabetes mellitus, heart attack, stroke, can be decreased in offspring of long-lived parents [35]. The BDH1 gene has effects on both human aging and cognitive decline within the MCI patients.
Aβ, the main component of amyloid plaques, plays an important role in developing AD. Sequential cleavage of APP forms Aβ through β-secretase (BACE1, β-site APP-cleaving enzyme 1) and γ-secretase [36], [37], [38], [39], [40]. We found three SNPs within the gene ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 (ST6Gal1). It has been demonstrated that cleavage and secretion of ST6Gal1 is affected by BACE1 and overexpression of ST6Gal1 increases soluble APP secretion [40], [41]. It suggests that ST6Gal1 plays an important role in the APP pathway by affecting Aβ generation and being produced within the process of Aβ formation.
Wyss-Coray et al. [42] demonstrated that certain inflammatory mediators strongly drive AD by their mouse models. Genetic studies in mouse models suggest that multiple inflammatory mediators have great effects on AD-like pathogenesis. More reviews can be found in Cummings et al. [42]. One SNP, rs56378310, in a gene RAB20 (RAB20, member Ras oncogene family) was identified at the suggestive level (P < 1 × 10−5). Liang et al (2012) found that RAB20 and RAB32 (member Ras oncogene family) were strongly upregulated in the acute phase of inflammation in mice, which suggested that these RABs might participate in subsequent inflammatory responses in the brain [43].
Our GWAS identified two SNPs including rs11121365 and rs12069701 located in a gene called, SplA/ryanodine receptor domain and SOCS box containing 1 (SPSB1). SPSB1, SPSB2, and SPSB4 regulate ubiquitination and proteasomal degradation of inducible nitric oxide (NO) synthase in activated macrophages [44]. They, in result, negatively control NO production and limit cellular toxicity. Togo et al. (2004) suggest that NO pathways contribute to pathogenesis of neurodegeneration in AD and other neurodegenerative dementias by involving in microvasculopathy and neuroinflammation [45]. A large amount of NO is produced by the inducible NO synthase [46], which thus has been linked to AD, asthma, cancer, cerebral infarction, inflammatory bowel disease, arthritis, and endotoxin shock [47], [48]. It shows that SPSB1 contributes to the development of AD through NO pathways.
Other than the identified SNPs at the 1 × 10−5 significance level in a region of LD of rs3936289 (see Fig. 2), two SNPs including rs114805931 have P values as 1.25 × 10−5. Fig. 2 shows regional association for the regions around rs3936289 within ST6GAL1. Their P values are plotted based on the −log 10 scales, and the color represents the range in r2 to show their degree of LD with rs3936289. They are located within the gene CACNA1D whose disruption caused by copy number variations may result in a dysregulation in Ca2+ homeostasis and affect the development of AD [49]. Also, Kim and Rhim (2011) found that CACNA1D was upregulated by Aβ25–35, which suggested that CaV1.3 had an impact on the pathogenesis of AD [50].
In 2011, a GWAS in South Asians identified that SNPs at ST6GAL1 might increase risk to develop type II diabetes by being involved in post-translational modification of cell-surface components by glycosylation, glycosylation through addition of sialic acid residues is reported to influence both insulin action and cell surface trafficking [51]. Whitmer et al. (2009) found that among older patients with type II diabetes, a history of severe hypoglycemic episodes was associated with a greater risk of dementia [52]. It suggests that ST6GAL1 may be related to the pathogenesis of AD through type II diabetes.
After identifying the relevant genes to cognitive decline, we examined if the genes are associated with the risk of MCI by running logistic regression using the baseline diagnosis of ADNI-1, ADNI-GO, and ADNI-2 subjects. We included age, gender, length of education, and ADAS-Cog score at the baseline as covariates. Among the identified SNPs, 11 SNPs within the PDS5B gene were associated (P < .05) with the risk of MCI with/without considering the presence of APOE ɛ4 allele as an additional covariate. In particular, the P values of rs192470679 were .024 and .028, respectively, for the APOE ɛ4 allele excluded and included models. It suggests that the PDS5B gene is associated with cognitive dysfunction in both early and transition stages of MCI.
In summary, we detected six genes that putatively associated with accelerated cognitive decline of MCI subjects after they developed AD. The genes had been implicated in other AD or AD-related disease studies. This GWAS sheds light on what genetic factors have impact on the ADAS-Cog score changes within MCI-AD conversion subjects. It allows better understanding of the underlying mechanism and pathology of accelerated cognitive impairment of AD patients within their transition period from MCI, which further may help to develop future treatments targeting the identified genetic loci. Although the associations were significant at the 1 × 10−5 suggestive level, a bigger sample size could ensure more significant results. Also, a follow-up study is recommended as future works to examine if the identified genes are replicated in other samples.
Research in Context.
-
1.
Systematic review: We used PubMed and Google Scholar to review the literature related to genetic association analyses of Alzheimer's disease (AD). Also, we used them to search for articles showing any interesting relationship between our significant loci to AD and AD-related diseases.
-
2.
Interpretation: We identified new six genes that could be associated with accelerated cognitive decline of mild cognitive impairment (MCI) subjects after their conversion to AD. The genes had been implicated in other AD or AD-related disease studies. Our study suggests that the genetic factors have affected the ADAS-Cog score changes within MCI-AD conversion subjects. It helps to understand the underlying mechanism of accelerated cognitive impairment of AD patients within their transition period from MCI.
-
3.
Future directions: A follow-up study should be taken as future works to examine if the identified genes are replicated in other and bigger samples.
Acknowledgments
This work was supported in part by National Institutes of Health (NIH) grants MH086633, National Science Foundation (NSF) grants SES-1357666 and DMS-1407655, and a grant from Cancer Prevention Research Institute of Texas to H.Z. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH and NSF. Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at http://adni.loni.usc.edu/wpcontent/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Data collection and sharing for this project was funded by the ADNI (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer's Association; Alzheimer's Drug Discovery Foundation; BioClinica, Inc; Biogen Idec Inc; Bristol-Myers Squibb Company; Eisai Inc; Elan Pharmaceuticals, Inc; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC; Medpace, Inc; Merck & Co., Inc; Meso Scale Diagnostics, LLC; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc; Piramal Imaging; Servier; Synarc Inc; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuroimaging at the University of Southern California.
P.M.D. has received research grants and/or advisory fees from several government agencies, advocacy groups, and pharmaceutical/imaging companies. P.M.D. received a grant from ADNI to support data collection for this study and he owns stock in Sonexa, Maxwell, Adverse Events, and Clarimedix, whose products are not discussed here.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.dadm.2017.04.004.
Supplementary data
References
- 1.Hebert L.E., Weuve J., Evans D.A., Scherr P.A. Alzheimer disease in the United States (2010-2050) estimated using the 2010 census. Neurology. 2013;80:1778–1783. doi: 10.1212/WNL.0b013e31828726f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alzheimer's Association . Alzheimer's Association; Chicago, IL: 2010. Changing the trajectory of Alzheimer's disease: A National Imperative. [Google Scholar]
- 3.Seshadri S., Fitzpatrick A.L., Ikram M.A., DeStefano A.L., Gudnason V., Boada M. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beecham G.W., Martin E.R., Li Y.J., Slifer M.A., Gilbert J.R., Haines J.L. Genome-wide Association Study Implicates a Chromosome 12 Risk Locus for Late-Onset Alzheimer Disease. Am J Hum Genet. 2009;84:35–43. doi: 10.1016/j.ajhg.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naj A.C., Beecham G.W., Martin E.R., Gallins P.J., Powell E.H., Konidari I. Dementia Revealed: Novel Chromosome 6 Locus for Late-Onset Alzheimer Disease Provides Genetic Evidence for Folate-Pathway Abnormalities. PLoS Genet. 2010;6:e1001130. doi: 10.1371/journal.pgen.1001130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H., Wetten S., Li L., Jean P.L.S., Upmanyu R., Surh L. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008;65:45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- 7.Carrasquillo M.M., Zou F., Pankratz V.S., Wilcox S.L., Ma L., Walker L.P. Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer's disease. Nat Genet. 2009;41:192–198. doi: 10.1038/ng.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naj A.C., Jun G., Beecham G.W., Wang L.S., Vardarajan B.N., Buros J. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheuner D., Eckman C., Jensen M., Song X., Citron M., Suzuki N. Secreted amyloid β–protein similar to that in the senile plaques of Alzheimer's disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer's disease. Nat Med. 1996;2:864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 10.Goate A., Chartier-Harlin M.C., Mullan M., Brown J., Crawford F., Fidani L. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 11.Saunders A.M., Strittmatter W.J., Schmechel D., George-Hyslop P.S., Pericak-Vance M.A., Joo S.H. Association of apolipoprotein E allele ε4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 12.Corder E.H., Saunders A.M., Strittmatter W.J., Schmechel D.E., Gaskell P.C., Small G. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 13.Harold D., Abraham R., Hollingworth P., Sims R., Gerrish A., Hamshere M.L. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambert J.C., Heath S., Even G., Campion D., Sleegers K., Hiltunen M. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 15.Coon K.D., Myers A.J., Craig D.W., Webster J.A., Pearson J.V., Lince D.H. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer's disease. J Clin Psychiatry. 2007;68:613–618. doi: 10.4088/jcp.v68n0419. [DOI] [PubMed] [Google Scholar]
- 16.Walsh D.M., Selkoe D.J. Aβ oligomers-a decade of discovery. J Neurochem. 2007;101:1172–1184. doi: 10.1111/j.1471-4159.2006.04426.x. [DOI] [PubMed] [Google Scholar]
- 17.Xiao Q., Gil S.H., Yan P., Wang Y., Han S., Gonzales E. Role of phosphatidylinositol clathrin assembly lymphoid-myeloid leukemia (PICALM) in intracellular amyloid precursor protein (APP) processing and amyloid plaque pathogenesis. J Biol Chem. 2012;287:21279–21289. doi: 10.1074/jbc.M111.338376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petersen R., Smith G., Waring S., Ivnik R., Tangalos E., Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 19.Olazaran J., Muniz R., Reisberg B., Peña-Casanova J., Del Ser T., Cruz-Jentoft A.J. Benefits of cognitive-motor intervention in MCI and mild to moderate Alzheimer disease. Neurology. 2004;63:2348–2353. doi: 10.1212/01.wnl.0000147478.03911.28. [DOI] [PubMed] [Google Scholar]
- 20.Cummings J.L., Doody R., Clark C. Disease-modifying therapies for Alzheimer disease Challenges to early intervention. Neurology. 2007;69:1622–1634. doi: 10.1212/01.wnl.0000295996.54210.69. [DOI] [PubMed] [Google Scholar]
- 21.Buschert V.C., Friese U., Teipel S.J., Schneider P., Merensky W., Rujescu D. Effects of a newly developed cognitive intervention in amnestic mild cognitive impairment and mild Alzheimer's disease: a pilot study. J Alzheimers Dis. 2011;25:679–694. doi: 10.3233/JAD-2011-100999. [DOI] [PubMed] [Google Scholar]
- 22.Hu X., Pickering E.H., Hall S.K., Naik S., Liu Y.C., Soares H. Genome-wide association study identifies multiple novel loci associated with disease progression in subjects with mild cognitive impairment. Transl Psychiatry. 2011;1:e54. doi: 10.1038/tp.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saykin A.J., Shen L., Foroud T.M., Potkin S.G., Swaminathan S., Kim S. Alzheimer's Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimers Dement. 2010;6:265–273. doi: 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu E.Y., Buyske S., Aragaki A.K., Peters U., Boerwinkle E., Carlson C. Genotype Imputation of MetabochipSNPs Using a Study-SpecificReference Panel of∼ 4,000 Haplotypes in African Americans From the Women's Health Initiative. Genet Epidemiol. 2012;36:107–117. doi: 10.1002/gepi.21603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kenward M.G., Roger J.H. Small sample inference for fixed effects from restricted maximum likelihood. Biometrics. 1997;53:983–997. [PubMed] [Google Scholar]
- 28.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 29.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furney S.J., Simmons A., Breen G., Pedroso I., Lunnon K., Proitsi P. Genome-wide association with MRI atrophy measures as a quantitative trait locus for Alzheimer’s disease. Mol Psychiatry. 2011;16:1130–1138. doi: 10.1038/mp.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan J., Kim S., Nho K., Chen R., Risacher S.L., Moore J.H. Hippocampal transcriptome-guided genetic analysis of correlated episodic memory phenotypes in Alzheimer's disease. Front Genet. 2015;6 doi: 10.3389/fgene.2015.00117. http://doi.org/10.3389/fgene.2015.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohannim O., Hibar D.P., Stein J.L., Jahanshad N., Hua X., Rajagopalan P. Discovery and Replication of Gene Influences on Brain Structure Using LASSO Regression. Front Neurosci. 2012;6 doi: 10.3389/fnins.2012.00115. http://doi.org/10.3389/fnins.2012.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith C.D., Malcein M., Meurer K., Schmitt F.A., Markesbery W.R., Pettigrew L.C. MRI temporal lobe volume measures and neuropsychologic function in Alzheimer's disease. J Neuroimaging. 1999;9:2–9. doi: 10.1111/jon1999912. [DOI] [PubMed] [Google Scholar]
- 34.Shi H., Belbin O., Medway C., Brown K., Kalsheker N., Carrasquillo M. Genetic variants influencing human aging from late-onset Alzheimer's disease (LOAD) genome-wide association studies (GWAS) Neurobiol Aging. 2012;33 doi: 10.1016/j.neurobiolaging.2012.02.014. 1849.e5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atzmon G., Schechter C., Greiner W., Davidson D., Rennert G., Barzilai N. Clinical phenotype of families with longevity. J Am Geriatr Soc. 2004;52:274–277. doi: 10.1111/j.1532-5415.2004.52068.x. [DOI] [PubMed] [Google Scholar]
- 36.Hussain I., Powell D., Howlett D.R., Tew D.G., Meek T.D., Chapman C. Identification of a novel aspartic protease (Asp 2) as β-secretase. Mol Cell Neurosci. 1999;14:419–427. doi: 10.1006/mcne.1999.0811. [DOI] [PubMed] [Google Scholar]
- 37.Sinha S., Anderson J.P., Barbour R., Basi G.S., Caccavello R., Davis D. Purification and cloning of amyloid precursor protein β-secretase from human brain. Nature. 1999;402:537–540. doi: 10.1038/990114. [DOI] [PubMed] [Google Scholar]
- 38.Vassar R., Bennett B.D., Babu-Khan S., Kahn S., Mendiaz E.A., Denis P. β-Secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 39.Cai H., Wang Y., McCarthy D., Wen H., Borchelt D.R., Price D.L. BACE1 is the major β-secretase for generation of Aβ peptides by neurons. Nat Neurosci. 2001;4:233–234. doi: 10.1038/85064. [DOI] [PubMed] [Google Scholar]
- 40.Kitazume S., Tachida Y., Oka R., Shirotani K., Saido T.C., Hashimoto Y. Alzheimer's β-secretase, β-site amyloid precursor protein-cleaving enzyme, is responsible for cleavage secretion of a Golgi-resident sialyltransferase. Proc Natl Acad Sci U S A. 2001;98:13554–13559. doi: 10.1073/pnas.241509198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakagawa K., Kitazume S., Oka R., Maruyama K., Saido T.C., Sato Y. Sialylation enhances the secretion of neurotoxic amyloid-β peptides. J Neurochem. 2006;96:924–933. doi: 10.1111/j.1471-4159.2005.03595.x. [DOI] [PubMed] [Google Scholar]
- 42.Wyss-Coray T. Inflammation in Alzheimer disease: driving force, bystander or beneficial response? Nat Med. 2006;12:1005–1015. doi: 10.1038/nm1484. [DOI] [PubMed] [Google Scholar]
- 43.Liang Y., Lin S., Zou L., Zhou H., Zhang J., Su B. Expression profiling of Rab GTPases reveals the involvement of Rab20 and Rab32 in acute brain inflammation in mice. Neurosci Lett. 2012;527:110–114. doi: 10.1016/j.neulet.2012.08.039. [DOI] [PubMed] [Google Scholar]
- 44.Nishiya T., Matsumoto K., Maekawa S., Kajita E., Horinouchi T., Fujimuro M. Regulation of inducible nitric-oxide synthase by the SPRY domain-and SOCS box-containing proteins. J Biol Chem. 2011;286:9009–9019. doi: 10.1074/jbc.M110.190678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Togo T., Katsuse O., Iseki E. Nitric oxide pathways in Alzheimer's disease and other neurodegenerative dementias. Neurol Res. 2004;26:563–566. doi: 10.1179/016164104225016236. [DOI] [PubMed] [Google Scholar]
- 46.Griffith O.W., Stuehr D.J. Nitric oxide synthases: properties and catalytic mechanism. Annu Rev Physiol. 1995;57:707–734. doi: 10.1146/annurev.ph.57.030195.003423. [DOI] [PubMed] [Google Scholar]
- 47.Guo F.H., Comhair S.A., Zheng S., Dweik R.A., Eissa N.T., Thomassen M.J. Molecular mechanisms of increased nitric oxide (NO) in asthma: evidence for transcriptional and post-translational regulation of NO synthesis. J Immunol. 2000;164:5970–5980. doi: 10.4049/jimmunol.164.11.5970. [DOI] [PubMed] [Google Scholar]
- 48.Nathan C. Inducible nitric oxide synthase: what difference does it make? J Clin Invest. 1997;100:2417. doi: 10.1172/JCI119782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villela D., Suemoto C., Pasqualucci C., Grinberg L.T., Rosenberg C. Do copy number changes in CACNA2D2, CACNA2D3 and CACNA1D constitute a predisposing risk factor for Alzheimer’s disease? Front Genet. 2016;7:107. doi: 10.3389/fgene.2016.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim S., Rhim H. Effects of amyloid-β peptides on voltage-gated L-type CaV1. 2 and CaV1. 3 Ca2+ channels. Mol Cells. 2011;32:289–294. doi: 10.1007/s10059-011-0075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kooner J.S., Saleheen D., Sim X., Sehmi J., Zhang W., Frossard P. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet. 2011;43:984–989. doi: 10.1038/ng.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitmer R.A., Karter A.J., Yaffe K., Quesenberry C.P., Selby J.V. Hypoglycemic episodes and risk of dementia in older patients with type 2 diabetes mellitus. JAMA. 2009;301:1565–1572. doi: 10.1001/jama.2009.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.