Abstract
The role of heparin as an anticoagulant is well defined; however, its role in tumorigenesis and tumor progression is not clear yet. Some studies have shown that anticoagulant treatment in cancer patients improve overall survival, however, recent clinical trials have not shown a survival benefit in cancer patients receiving heparin treatment. In our previous studies we have shown the inhibitory effects of heparin on Hepatocyte Growth Factor (HGF)-induced invasion and migration in hepatocellular carcinoma (HCC) cells. In this study, we showed the differential effects of heparin on the behaviors of HCC cells based on the presence or absence of HGF. In the absence of HGF, heparin activated HGF/c-Met signaling and promoted motility and invasion in HCC cells. Heparin treatment led to c-Met receptor dimerization and activated c-Met signaling in an HGF independent manner. Heparin-induced c-Met activation increased migration and invasion through ERK1/2, early growth response factor 1 (EGR1) and Matrix Metalloproteinases (MMP) axis. Interestingly, heparin modestly decreased the proliferation of HCC cells by inhibiting activatory phosphorylation of Akt. The inhibition of c-Met signaling reversed heparin-induced increase in motility and invasion and, proliferation inhibition. Our study provides a new perspective into the role of heparin on c-Met signaling in HCC.
Electronic supplementary material
The online version of this article (doi:10.1007/s12079-016-0368-0) contains supplementary material, which is available to authorized users.
Keywords: c-Met, Heparin, Hepatocellular carcinoma, Invasion, Proliferation
Introduction
The c-Met proto-oncogene, also called HGF receptor, was first identified as a fusion gene (tpr-met) in a chemically transformed human osteosarcoma cell line (Cooper et al. 1984). HGF/c-Met signaling activates several biological responses including cell proliferation, survival, migration, and angiogenesis in various types of cells including hepatocytes (Gherardi et al. 2012; Spina et al. 2015). It has been shown that the conditional inactivation of c-Met in mouse hepatocytes causes deficient liver regeneration (Grant et al. 1993). In addition to its role in liver development and regeneration, abnormalities in HGF/c-Met signaling were reported to be linked to unfavorable clinical-pathological status, including high proliferation index, low degree differentiation, vascular invasion and metastasis in several cancer types including HCC (Spina et al. 2015; Bozkaya et al. 2012; Kaposi-Novak et al. 2006; Korhan et al. 2014). Thus, the HGF/c-Met signaling pathway has recently gained considerable attention as a target for targeted cancer therapies (You et al. 2011; Eder et al. 2009; Furlan et al. 2014; Garber 2014; Peters and Adjei 2012; Sakai et al. 2015). The HGF-dependent autocrine loop has also been reported in the acquired sorafenib resistance in hepatocellular carcinoma (Firtina Karagonlar et al. 2016).
HGF binding to c-Met results in receptor dimerization and phosphorylation of Y1234 and Y1235 located within the catalytic domain of c-Met. Then the tyrosines within the multifunctional docking site (MDS) become phosphorylated and recruit signaling effectors, such as the adaptor proteins growth factor receptor-bound protein 2 (Grb2) and the effector molecules such as phosphatidylinositol 3-kinase (PI3K). Furthermore, association of activated c-Met with a multi-adaptor protein Grb2-associated binding protein 1 (Gab1) leads to its phosphorylation, forming binding sites for more downstream adaptors, which causes activation of the mitogen-activated protein kinase (MAPK) and protein kinase B (PKB)/Akt signaling (Gherardi et al. 2012; Spina et al. 2015; Kaposi-Novak et al. 2006). In addition, ligand-independent kinase activity for c-Met is the most frequent cause of the constitutive activation of c-Met in human tumors, occurring by several genetic and epigenetic mechanisms (Gherardi et al. 2012; Korhan et al. 2014; Cappuzzo et al. 2009). Besides the role of Heparan Sulfate Proteoglycans (HSPGs) and Dermatan Sulfates (DSs) on the activation of c-Met has been determined (Lyon et al. 2002).
Heparin is a highly sulfated and negatively charged glycosaminoglycan (GAG) and has been used as an anti-coagulant agent over the last 60 years (Lever and Page 2002). Since cancer increases the risk of thromboembolic events in patients, heparin treatment has been used for preventing mortality, pulmonary embolism and deep venous thrombosis in patients with cancer. Initial preclinical and case control studies showed that anticoagulant treatment in cancer patients improve overall survival. However, recent clinical trials did not show a survival benefit in cancer patients receiving heparin treatment (Sanford and Lazo-Langner 2014; Spek et al. 2015).
Much of the data that exists on the controversial role of heparin in tumor progression is context dependent. It has been shown that heparin increases tumor growth and metastasis in colon cancer, however, it reduces metastasis in fibrosarcomas, lung, prostate and mammary carcinomas (Takeuchi et al. 2013; Lim et al. 2015; Zhong et al. 2015). These data support that heparin may affect survival in cancer patients, with mechanisms that are different from its anti-coagulant effect, but are linked to the ability of influencing tumor biology. The underlying mechanisms by which heparin modulates tumor progression need to be elucidated.
It is known that, apart from the anti-coagulant action, the highly negative charge leads heparin to interact with a large number of proteins including growth factors, growth factor receptors and proteases, and thus enables it to have a variety of biological activities. In this way, heparin influences differentiation, proliferation, migration and invasion in various cell types via a receptor or co-receptor role for different ligands (Lim et al. 2015; Zhong et al. 2015).
In our previous studies we have shown that heparin represses HGF-induced c-Met signaling activation as well as HGF/c-Met mediated cell invasion and cell migration in HCC cell lines (Ozen et al. 2012). Our recent data supported the differential effects of heparin on the behaviors of HCC cells based on the presence or absence of HGF. In our present study, we showed that heparin increased the expression and activation of c-Met by inducing receptor dimerization in HCC cells in HGF independent manner. To understand the biological significance of heparin mediated c-Met activation, we examined the effect of heparin on proliferation, motility and invasion of HCC cells. Furthermore, we demonstrated the effects of c-Met inhibition on heparin mediated biological responses. Consequently, our study supports the role of heparin on the ligand-independent activation of c-Met signaling and c-Met-induced proliferation, invasion and migration of HCC cells. It also provides new insights into the context dependent roles of heparin on tumor progression in HCC and may provide an explanation on the differential effects of heparin treatment in cancer patients.
Materials and methods
Reagents
All cell culture supplies were obtained from Biological Industries (Israel). Heparin was from Applichem (A3004). Anti-EGR1 (15F7) cs 4351, anti-phosho-Met (Y-1234/1235) cs 3129, p21/Cip1/waf1 (DCS60) cs 2946 and anti-phosho-42/44 MAPK (Thr202/Tyr204) (19G2) cs 4377, PTEN (138G6) cs 9559 antibodies were from Cell Signaling Technology (Danvers, MA, USA). Anti-Met (C-28) sc- 161, anti-ERK1 (C-16) sc-93, anti-p27 (F-8) sc-1641, anti-CycE (HE12) sc-247, anti-CycA (BF683) sc-239, anti-CycD1 (M-20) sc-718, anti-CDK2 (D-12) sc-6248, anti-CDK4 (H-22) sc-601 and anti-CDK8 (C-4) sc-7344 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA USA). Anti- p85 (06–496), anti-Gab1 (06–579) antibodies were from Upstate (Germany). HRP-anti-mouse/rabbit secondary antibodies were from Pierce (IL, USA). ECL detection system was from Pierce (IL, USA). c-Met inhibitor SU11274 was from Calbiochem (Darmstadt, Germany). Crosslinking agent, Sulfo-EGS (21566) was form Pierce (IL, USA) (Rockford, USA). Sulphorhodamine B (SRB) (86183) was from Sigma-Aldrich (St. Louis, MO, USA).
Cell culture
Human HCC cell lines; HuH-7, SNU-449 and SK-HEP-1, were cultivated as described previously (Bozkaya et al. 2012). Authentication of cell lines was done by DNA profiling at the University of Colorado Cancer Center (UCCC) DNA Sequencing & Analysis Shared Resource (CO, USA) using Applied Biosystem’s Identifier kit (PN 4322288). Cells were treated with different concentrations (1, 10, 100 μg/ml) of heparin at indicated time points after 16 h serum starvation in DMEM with 2% FBS. For the inhibition of c-Met, SU11274 (0, 2500 nM) was used. In preliminary studies, the effects of different heparins (Applichem A3004, Biochrom L651, Calbiochem 3,750,973) were tested and similar results were observed with all types of heparins.
In vitro motility and invasion assays
In vitro motility and invasion assays were performed as described (Cokakli et al. 2009). Briefly, 70% confluent cells were starved overnight with Dulbecco’s Modified Eagle Medium (DMEM) containing 2% Fetal Bovine Serum (FBS) and cell lines were treated with/without heparin at the indicated concentrations. Invasion inserts were washed, fixed and stained with Diff Quick kit (Siemens Healthcare Diagnostics) after 24 h of incubation. Cells that had traversed through the membrane were counted for each chamber using a bright-field inverted microscope. Measurements were performed for at least three technical and two biological replicates. Fold differences were calculated by dividing experimental results by control. In addition invasion and motility of cells were also monitored using a real-time cell electronic sensing RT-CES system (xCELLigence- Roche Aplied Science) (Ozen et al. 2012). For real time motility analysis, the lower chambers filled with DMEM containing 2% FBS, to which heparin was added at the indicated concentrations. Measurements of cell index were taken every 30 min. For invasion studies, the membrane on the bottom of the top chamber of CIM plates were coated with 0.25 mg/ml of Matrigel (BD Biosciences) in serum free media and incubated at 37 °C for 2 h. Then 104 cells/per chamber were added to the upper chamber in DMEM with/without 2% FBS, which contained heparin at the indicated concentrations. Subsequent steps were performed in the same manner as described for cell migration assays.
Luciferase reporter assay
The pMET 0.2-driven firefly luciferase reporter plasmid was used for (Liu et al. 1998; Zhou et al. 2011) reporter gene analysis, SK-HEP-1 cells were seeded at 25,000 cells/per well in a 12 well plate for 24 h before transfection. Cells were then transfected with the pMET 0.2 plasmid construct by using Fugene HD Transfection Reagents Roche (Mannheim, Germany) according to manusfacturer’s instructions. For normalization, pRL-TK Renila luciferase reporter was co-transfected. After transfection, cells were starved overnight then treated with heparin (1, 10, 100 μg/ml Calbiochem) for 2 h. Cells were harvested in luciferase lysis buffer and the luciferase activity was analyzed in each cell lysate using Dual-Glo Luciferase Assay system (Promega, Madison, WI, USA) with a luminometer (Turner Designs 20/20n Sunnyvale, CA). Relative luciferase unit (RLU) represents firefly luciferase normalized against Renilia luciferase activity.
Crosslinking analysis
After overnight serum starvation, cells were treated with 0, 10 μg/ml of heparin for 2 h. The media was removed and replaced with 1X Phosphate Buffered Saline (PBS). Cells were then treated with the crosslinking agent, SULFO EGS at 4 °C for 2 h, according to the manufacturer’s instructions. 500 μg of crosslinked lysate was immunoprecipitated with c-Met antibody, analyzed on 8% Tris-glycine gels, transferred to polyvinylidene fluoride (PVDF) membranes, immunoblotted with anti-Met antibody. Proteins were detected with ECL-Plus.
Co-immunoprecipitation
After 2 h of heparin treatment, treated and control SK-HEP-1 cells media were removed and washed with ice-cold 1X PBS including 1 mM NaF and 1 mM Na3VO4. Cells were scraped on ice with 1X PBS supplemented with protease and phosphatase inhibitor cocktails (Roche, Germany) and centrifuged. The pellets were resuspended with ice cold modified RIPA buffer (50 mM Tris-CI pH 7.4, 150 mM NaCI, 1 mM EDTA pH 8.0, 1% NP-40, 1X protease inhibitor cocktail, 1 mM NaF, 1 mM Na3VO4) and lysed for 30 min on ice, with occasional mixing. To remove the cell debris, samples were centrifuged for 20 min at 4 °C at 18000 g, supernatants were transferred into new tubes. For preclearance, lysates were rotated with prewashed protein G sepharose beads (GE Healthcare) for 20 min and centrifuged. Protein concentration was determined by Bicinchoninic Acid (BCA) assay and immunoprecipitation was initiated with 1 mg protein lysate. After overnight rotation at 4 °C with anti-c-Met antibody (Sc-161, Santa Cruz), prewashed protein G sepharose beads were incubated with antibody-protein complex for 3 h at +4 °C on a rotating platform. The beads were then washed with ice-cold modified RIPA buffer including protease and phosphatase inhibitors three times. Finally bead-protein conjugates were collected, resuspended in 2X Sodium Dodecyl Sulphate (SDS) sample buffer, boiled at 95 °C for 10 min and samples were centrifuged for 5 min before loading for Western Blot analysis.
Immunoblotting
Total cell lysates were prepared from SK-HEP-1 cell line using modified RIPA buffer. Protein concentrations were determined by BCA assay according to the manufacturer’s instructions (Pierce, IL, USA). Equal volumes of lysates were loaded onto 10% SDS polyacrylamide gels for electrophoretic analysis. The proteins in the gel were transferred to PVDF membranes (Pierce), which were first blocked with Tris buffered saline/ 0.1% Tween-20 containing 5% nonfat dry milk (NFDM) for 1 h at room temperature. Membranes then were blotted with primary antibodies against phospho-Met, EGR1, Calnexin, phospho-ERK1/2, ERK1/2, phospho-Akt, Akt and PTEN in TBST containing 3% NFDM, c-Met in 1X PBS with 0.1% Tween-20 containing 3% bovine serum albumin for overnight at +4 °C. Proteins were detected by HRP-conjugated anti-rabbit and anti-mouse secondary antibodies, with visualization by the ECL detection system. The specific bands were recorded on X-ray film. Equal loading and transfer were confirmed by probing with calnexin. Band intensities were quantified as pixels using ImageJ software.
Gelatin-zymography
Gelatin zymography assay was performed as mentioned previously (Ozen et al. 2012). The activities of MMP-2, 9 in conditioned medium were measured by gelatin-zymography. Briefly, cells were grown in serum-free medium then treated with heparin (0, 10 μg/ml) for 24 h in the absence and presence of SU11274 and cultured supernatant was directly used for detection of secreted MMPs. Cultured cell media were prepared with SDS sample buffer without boiling or reduction, and then equal amounts of medium for each condition were loaded on 10% polyacrylamide gels containing 0.1% SDS and 1 mg/ml gelatin. After electrophoresis the gels were washed with 2.5% Triton X-100 and then incubated in 50 mM Tris-CI (pH 7.0) solution containing 10 mM CaCl2 and 150 mM NaCI at 37 °C overnight. Then, the gels were stained with 0.25% Coomassie Brilliant Blue R-250 (Bio-RAD) solution and destained. Gelatinolytic activity was visualized as a transparent band against a blue background. Relative densities of MMP-2 and MMP-9 bands were measured by scanning the photographic negatives and quantified as pixels by using ImageJ software.
Sulphorhodamine B assay
2000 cells/well were inoculated into 96-well plates in 100 μL of media and incubated in 37 °C incubator containing 5% CO2 and 95% air. After a 24 h incubation cells were treated with heparin 0, 10 μg/ml in the absence and presence of SU11274 and incubated in 37 °C incubator containing 5% CO2 and 95% air for 72 h. Following the termination of the incubation period after heparin treatment, the cells were fixed with 100 μL 10% ice-cold trichloroacetic acid (TCA) and incubated in the dark at 4 °C for 1 h. Then the TCA was washed away with dH2O five times and the plates were left to air dry. For the final step, the plates were stained with 100 μL of 0.4% Sulforhodamine B (SRB) solution in 1% acetic acid solution. Following staining, the plates were incubated in dark for 10 min at room temperature. The unbound dye was washed away using 1% acetic acid and the plates were left to air dry. To measure the absorbance, the bound stain was then solubilized using 200 μL of 10 mM Tris-Base. The OD values were obtained at 515 nm.
Time-dependent cellular response profiles by cell electronic sensing system (xCELLigence)
Real-time cell growth monitoring was analyzed with the Real-Time Cell Analyzer, using the xCELLigence System (Roche, Germany) as described previously (Ozen et al. 2012). Briefly 103 cells/well HuH-7, SK-HEP-1, SNU449 were plated in 16-well E-Plate and placed onto the multi-plate Real-Time Cell Analyzer located inside a tissue culture incubator. Electrical impedance data of first 4 h were collected for cellular adhesion analysis while following 96-h data was collected with 15 min periods and used for proliferation analysis.
Flow cytometry
Cells were incubated with 0, 10 μg/ml heparin after overnight serum starvation for 24 h. Cells were washed with 1X PBS, and trypsinized. Then cells were washed by centrifugation at 200×g, 5 min, 4 °C and resuspended as 1 × 106/ml cells in 1 ml PBS, vortexed gently, slowly adding the cell suspension dropwise to 3 ml of 100% ethanol in a 15 ml polypropylene centrifuge tube. Cells were washed with cold PBS and centrifuged 200 x g, 10 min, at 4 °C. Resuspended cells were stained in Propidium iodide (PI)/RNase staining solution and incubated at 37 °C for 15 min. The cell cycle profile was analysed by the Modfit software (BD FACSCanto II).
Quantitative real time-PCR (qPCR)
RNA isolation was performed after cells were treated with 10 μg/ml of heparin for 2 h. 1 μg total RNA was reverse transcribed into cDNA with RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA) by using thermal cycler. cDNA samples were diluted by adding equal volume of ultrapure water and used for qPCR analysis with Power Sybr Green PCR Master Mix (Life Thecnologies, USA) in StepOnePlus Real Time PCR System (Applied Biosystems, USA). Ribosomal Protein L41 (RPL41) expression was used as internal control. Primers used in qPCR reactions are listed in Supplementary Table 1.
Statistical analysis
Statistical analysis was performed using the GraphPad Prism 6. Statistical methods included Analysis of variance (ANOVA), Student’s t-test, p < 0.05 was considered statistically significant.
Results
Heparin activated c-Met signaling in a time dependent manner in HCC cell lines
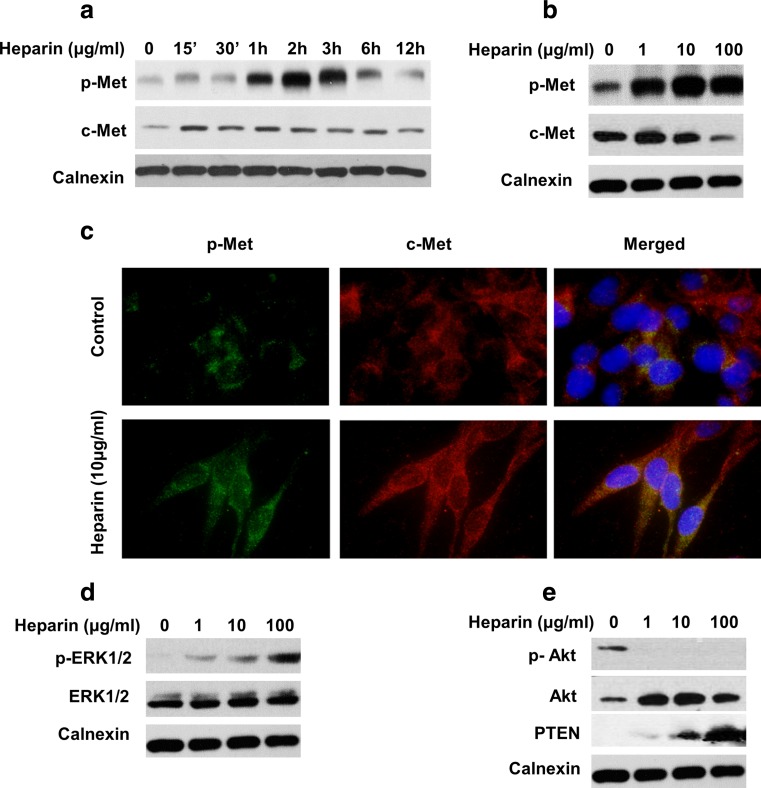
To determine the effects of heparin on c-Met signaling, we examined expression and activatory phosphorylation of c-Met (Y-1234/1235) in the presence of heparin in a time and dose dependent manner. Western blot analysis showed that heparin treatment significantly increased c-Met activation with respect to untreated controls (Fig. 1a). In addition, 1 μg/ml and 10 μg/ml heparin treatment increased c-Met expression levels, whereas 100 μg/ml heparin caused a moderate decrease in c-Met expression in SK-HEP-1 cells (Fig. 1b). Similar data was observed with SNU-449 and HuH-7 HCC cell lines (Supp Fig. 1A).
Fig. 1.
Heparin regulated expression, activation and downstream signaling of c-Met. a and b The effect of heparin on expression and activation of c-Met was determined by western blot, following serum starvation, SK-HEP-1 cells were treated with heparin in a time (0, 15′, 30′, 1 h, 2 h, 3 h, 6 h, 12 h) and dose (0, 1, 10, 100 μg/ml) dependent manner. Calnexin was used as a loading control. c Immunofluorescence was performed to examine expression and localization of p-Met (Y-1234/1235 and c-Met after 10 μg/ml heparin treatment for 2 h. d and e p-ERK1/2 (Thr202/Tyr204), p-Akt Ser473, total ERK1/2, total Akt and PTEN protein levels were determined by western blot after heparin treatment (0, 1, 10, 100 μg/ml) for 2 h. Calnexin levels were used as loading control
We then examined the effects of heparin on c-Met localization. Since c-Met activation was observed to be highest at 10 μg/ml heparin, a 10 μg/ml concentration was used for immunofluorescence analysis. As showed in Fig. 1c, c-Met activatory phosphorylation, c-Met protein levels and perinuclear localization were all increased after heparin treatment. We further investigated the effects of heparin on downstream effectors of c-Met, namely ERK1/2 and Akt. ERK1/2 (T202/Y204) phosphorylation was increased after heparin treatment in a dose dependent manner, whereas heparin abolished S473 phosphorylation of Akt (Fig. 1d, e). Moreover, heparin treatment caused an increased expression of PTEN, which is a well-known inhibitor of PI3K/Akt (Fig. 1e). These data support the idea that heparin upregulated c-Met signaling through increasing c-Met expression and activation, as well as activation of its downstream effector molecules, ERK 1/2.
Heparin treatment caused an increase in receptor dimerization and transcriptional activation of c-Met
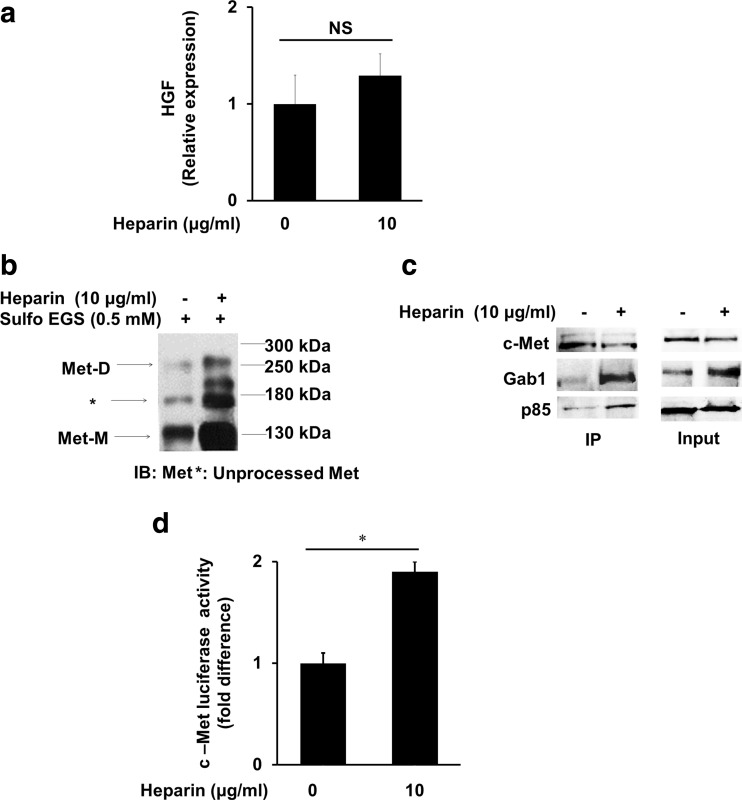
To determine whether heparin regulates c-Met activation in an autocrine manner in HCC cell lines, we examined HGF expression by RT-PCR. We found that heparin did not change HGF mRNA level (Fig. 2a). Basal secretion and protein expression levels of HGF were measured by ELISA and western blotting in HCC cell lines and none of these cells were found to express or secrete HGF protein (Data not shown).
Fig. 2.
Dimerization and transcriptional activation of c-Met was upregulated by heparin. a Cells were treated with 10 μg/ml heparin for 2 h and HGF expression investigated by RT-PCR. b Western blot analysis of c-Met receptor dimerization from the lysates of serum starved SK-HEP-1 cells incubated with 10 μg/ml heparin for 2 h followed by treatment of Sulfo-EGS for 1 h at +4 °C. c When treated with 10 μg/ml heparin for 2 h, interaction of c-Met with its binding partners Gab1 and p85 was determined by co-immunoprecipitation assay and western blot analysis. d c-Met transcriptional activity was determined by luciferase reporter assay. After serum starvation SK-HEP-1 cells were left untreated or treated with 10 μg/ml heparin for 2 h. Data are presented as the mean ± SD from 3 independent experiments. *p < 0.05
To investigate further the mechanisms of c-Met activation by heparin, we tested the effects of heparin on c-Met receptor dimerization by using Sulfo-EGS, a membrane impermeable crosslinker. It has been reported that the molecular weight of c-Met monomer is about 130 kDa, unprocessed Met is about 180 kDa and dimeric form of c-Met is about 250 kDa (Kong-Beltran et al. 2004). As shown in Fig. 2b, treatment of SK-HEP-1 cells with heparin, induced dimerization of c-Met receptor tyrosine kinase. Furthermore, monomeric and unprocessed forms of c-Met were increased by heparin treatment. In addition, heparin treatment increased binding of p85 unit of PI3K and Gab1 to c-Met (Fig. 2c). We then tested the effect of heparin on transcriptional activity of c-Met by luciferase reporter assay using pMET 0.2 construct, which contains EGR1 binding site (Liu et al. 1998). We found that transcriptional activation of c-Met was doubled when SK-HEP-1 cells were treated with heparin (Fig. 2d).
Heparin stimulated c-Met mediated invasion and migration of HCC cells through EGR1-MMP axis
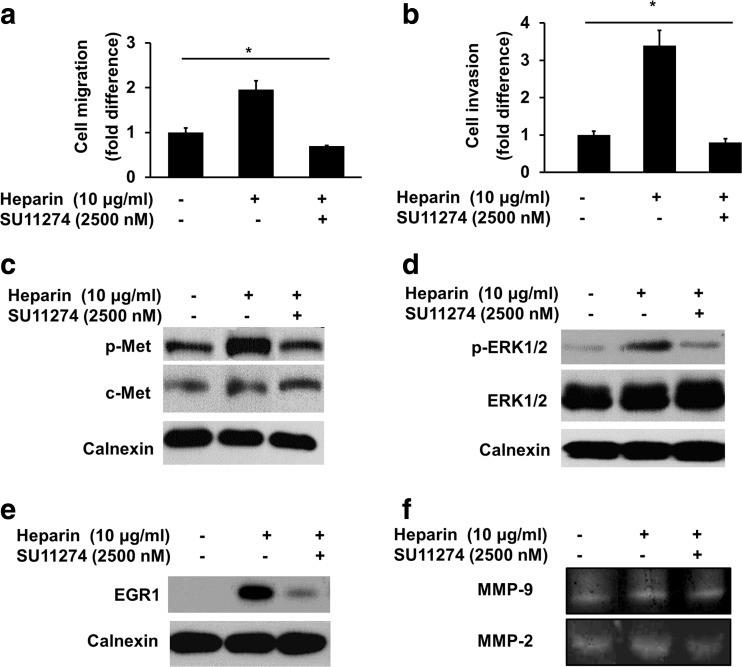
Since it is known that HGF induces c-Met activation, leading to invasion and migration in HCC cells (Bozkaya et al. 2012; Ozen et al. 2012) we tested the effect of heparin-mediated c-Met activation on invasion and migration of HCC cell lines. Following overnight serum starvation, cell invasion and migration assays were performed in the absence or presence of heparin and/or c-Met inhibitor SU11274, using the modified Boyden chamber method. Similar to HGF actions, heparin significantly increased the migration and invasion of SK-HEP-1 cells, whereas c-Met inhibitor SU11274 significantly decreased heparin mediated migration and invasion (p < 0.05) (Fig. 3a, b). Similar effects of heparin on cell migration and invasion were obtained using SNU-449 and HuH-7 HCC cell lines (Supp Fig. 1B, C).
Fig. 3.
Heparin increased migration, invasion of SK-HEP-1 cells and ERK1/2-EGR1-MMPs activation through c-Met activation. a and b The effect of heparin mediated migration and invasion of SK-HEP-1 cells were investigated when cells were treated with 10 μg/ml heparin in the presence and absence of SU11274. Migrated and invaded cells were determined by staining with eosin red and crystal violet. Data are presented as the mean ± SD from 10 independent fields of 3 independent experiments. *p < 0.05. c and d The effect of SU11274 on heparin induced c-Met (Y-1234/1235) and ERK1/2 (Thr202/Tyr204) phosphorylation was determined after western blot analysis of lysates from 10 μg/ml heparin treated or non-treated SK-HEP-1 cells incubated in the presence and absence of SU11274. e EGR1 protein level was determined by western blotting when cells were left untreated or treated with 10 μg/ml heparin for 2 h, with or without SU11274. Calnexin levels were used as loading control. f MMP-2, 9 activities were determined by gelatin zymography. After serum starvation, SK-HEP-1 cells were treated with 10 μg/ml heparin, in the presence and absence of SU11274, for 24 h and their growth medium were collected for performing the assay
Previous studies showed that HGF-induced c-Met activation increases cell invasion via EGR1/MMP-2/9 axis in HCC cells (Ozen et al. 2012). We therefore analyzed the expression levels of EGR1, in the absence or presence of heparin. We found that heparin-induced c-Met activation increased EGR1 expression in parallel to the activation observed with c-Met and ERK1/2 compared to untreated controls, while SU11274 abolished these responses (Fig. 3c, d and e). In order to analyze MMP-2 and MMP- 9 activation, SK-HEP-1 cells were cultured in the absence or presence of heparin, and conditioned media were used to determine MMP activation by the zymography assay. We found that heparin increased activation of both MMP-2 and -9, whereas SU11274 decreased the activation (Fig. 3f). Our data thus suggested that c-Met mediated EGR1 overexpression is important for heparin induced motility and invasion of HCC cells.
Since c-Met signaling pathway upregulates EMT through modulating EMT-markers expression, we investigated EMT marker (E-cadherin, Slug, Snail, Zeb1, Zeb2, AFP, Albumin, N-cadherin, Fibronectin) expression by RT-PCR after cells were treated with heparin at indicated time and observed no significant change in the EMT markers’ expression (data not shown).
Heparin decreased proliferation of HCC cells by inhibiting activatory phosphorylation of Akt
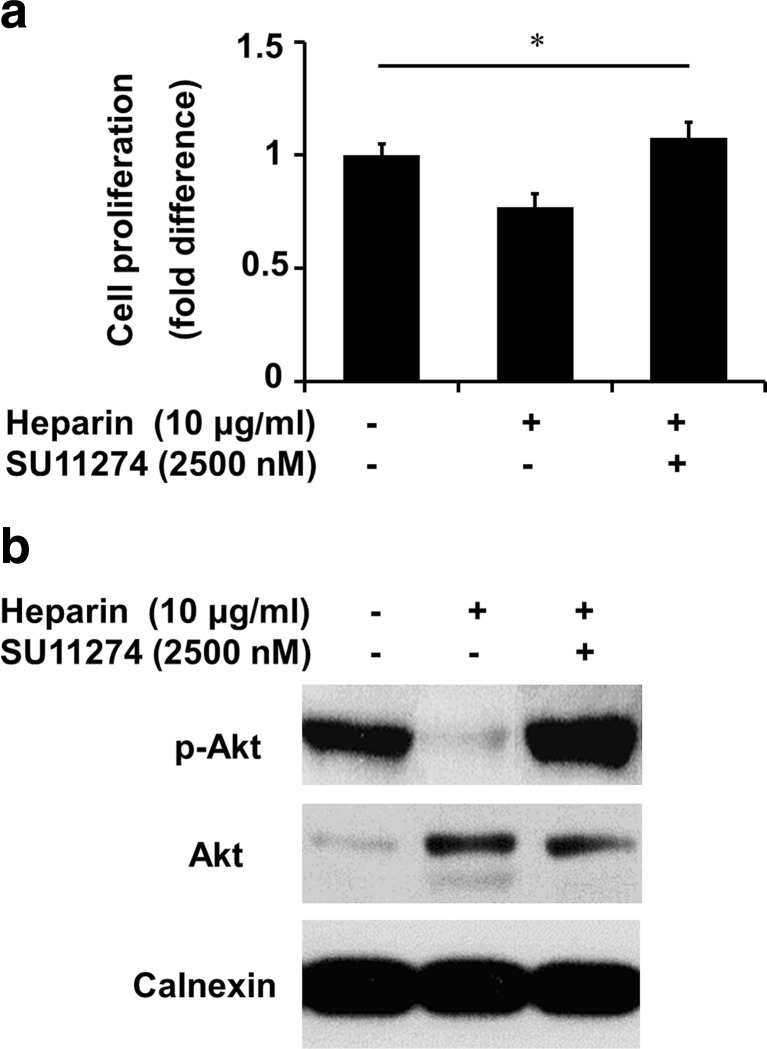
In addition to the role of heparin on invasion and migration of HCC cell lines, we examined the effect of heparin on cell proliferation by using a SRB assay. In contrast to our observed increase in cellular motility and invasion by heparin treatment, proliferation of SK-HEP-1 cells was moderately inhibited (Fig. 4a). Similar data was obtained using SNU-449 and HuH-7 cells (Supp. Fig. 1D). The effect of 10 ng/ml HGF treatment on proliferation also showed similar results, there is no significant change in proliferation of HCC cells SK-HEP-1, HuH-7, Hep3B and Mahlavu upon HGF treatment (Supp. Fig. 2).
Fig. 4.
Heparin modestly inhibited cell proliferation and Akt phosphorylation. a Cell proliferation was determined by Sulforhodamine B (SRB) assay, in response to treatment of SK-HEP-1 cells with, 10 μg/ml heparin for 48 h. The modest inhibition on proliferation was reversed with c-Met inhibitor SU11274 treatment. Results represent three or more independent experiments performed in triplicates. (Bars indicate standard error of the mean (SEM), asterisks indicate statistically significant differences between the indicated groups) *p < 0.05 b Western blot was performed for phosphorylated Akt, total Akt after treatment with 10 μg/ml heparin for 2 h in the presence or absence of c-Met inhibitor SU11274. Calnexin was used as loading control
To clarify the mechanisms underlying the decrease of cell proliferation by heparin in HCC cells, we next examined the effects of heparin on expression and activation of Akt. Our results showed that S473 phosphorylation of Akt disappeared when cells were treated with heparin. When we used c-Met inhibitor, we observed that along with inhibition of c-Met activation, heparin-mediated inhibition of cell proliferation and p-Akt activation were antagonized (Fig. 4b).
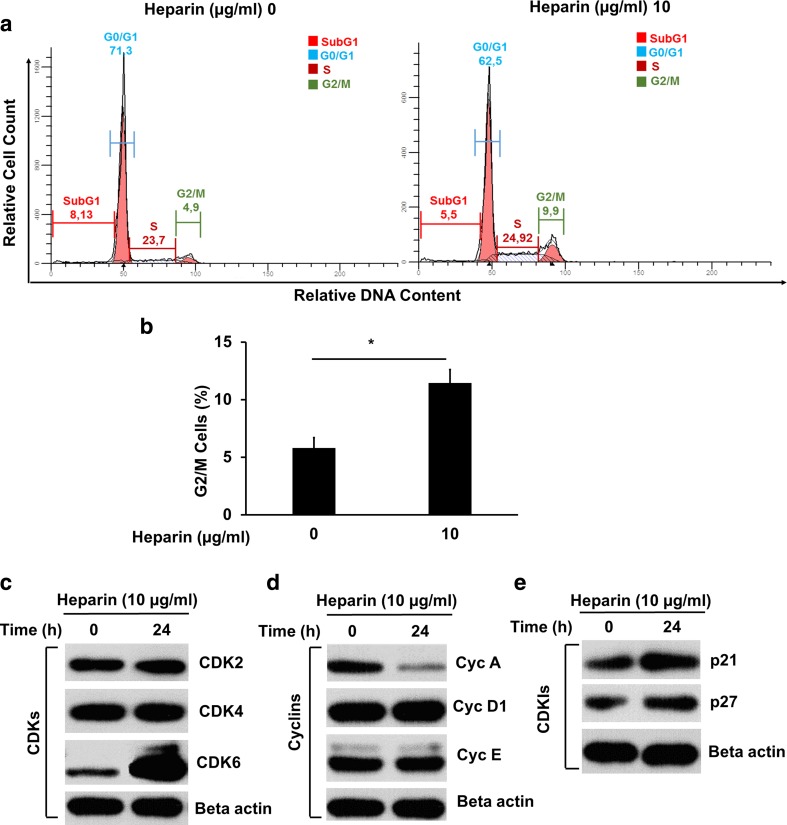
Heparin regulated cell cycle progression as well as expression of cell cycle molecules in SK-HEP-1 cells
To investigate the effects of heparin on cell cycle progression, cell-cycle analysis was performed by flow cytometer on cells stained with propidium iodide. Heparin caused a two-fold increase in the percentage of G2/M phase cells, in comparison to the heparin untreated cells. However, heparin did not significantly affect the cell number in G1 and S phases (Fig. 5a, b). To examine the molecular mechanisms of G2/M arrest following heparin treatment, we analyzed expression of cell-cycle-regulatory proteins. Our western blot analysis showed that protein levels of CDK6 were increased, whereas levels of Cyclin A were decreased in heparin treated SK-HEP-1 cells, compared with control cells (Fig. 5c and d). We also found that heparin activated the expression of cyclin dependent kinase inhibitors, p21 and p27 (Fig. 5e).
Fig. 5.
Heparin regulated G2/M cycle arrest and cell cycle molecules’ expression. a and b The cells were grown in the absence or presence of 10 μg/ml heparin for 24 h and then stained with propidium iodide and analysed by flow cytometry. The cell cycle was analyzed by Modfit software. Results represent three or more independent experiments performed in triplicates. (Bars indicate standard error of the mean (SEM), asterisks indicate statistically significant differences between the indicated groups) *p < 0.05. c, d and e The effects of heparin on CDKs’ (CDK2, CDK4, CDK6), Cyclins’ (Cyc D1, Cyc E, Cyc A) and CDKIs’ (p21, p27) expressions were determined by western blotting following cells were treated with 10 μg/ml of heparin for 24 h
Discussion
Much progress has been made in our understanding of the role of c-Met signaling in invasion, migration, metastasis and drug resistance in cancer. Preclinical studies have confirmed the prognostic significance of c-Met expression and activation in HCC and highlight potential inhibitors for c-Met signaling pathway, targeting ligand-receptor interaction, c-Met-adaptor protein interaction and c-Met kinase activity (Gao et al. 2011; Giordano and Columbano 2014). Recent data for c-Met kinase inhibitors, from preclinical trials and phase II studies, have shown that inhibition of c-Met signaling is a promising therapeutic strategy in HCC (Lee et al. 2015; LIovet and Bruix 2008). Tivantinib is a selective c-Met inhibitor which has been shown in randomized controlled phase II trial to benefit especially patients with MET-high tumors (Rimassa et al. 2014).
It has been reported that ligand independent activation of c-Met is a common event in several cancers, including HCC. In the study reported here, we found that heparin as an activator of c-Met signaling pathway. Although the role of heparin on c-Met signaling has been reported in several publications, many of them mostly focus on the effect of heparin under HGF treated conditions. Rubin et al. reported that heparin treatment enhances HGF-induced proliferation and migration in Heparan Sulphate (HS) deficient murine interleukin 3-dependent hematopoietic 32D/c-Met cells (Rubin et al. 2001). Similarly, it has been suggested that heparin binds to the NK1 kringle domain of HGF, as well as Sema domain of c-Met, inducing stabilization of HGF-c-Met complex (Gherardi et al. 2003; Holmes et al. 2007). In our previous study, we examined the effects of heparin on HGF/c-Met signaling and showed that when HCC cells were treated with heparin under HGF treatment, HGF-induced c-Met activation; in addition, HGF-induced invasion and migration were abolished. Furthermore, we showed that heparin inhibits HGF-mediated responses by decreasing HGF/c-Met interaction (Ozen et al. 2012). It was shown that heparin binds to HGF through its NK1 domain with high affinity (Gherardi et al. 2003; Holmes et al. 2007). In this way HGF binds to heparin rather than c-Met. In this study we found that in the absence of HGF, heparin increased expression and activation of c-Met in a time and dose dependent manner. As we reported previously, HGF treatment increases c-Met phosphorylation up to 10-fold in 5 min in SK-HEP-1 cells (Ozen et al. 2012). In this study, we observed that heparin induces c-Met activation to the same level in 2 h. Gherardi et al., have shown that HGF and heparin bind to the same site on c-Met, between the amino acids 25–519, they also reported that binding affinity of heparin to c-Met is lower than the binding affinity to HGF (Gherardi et al. 2003).
Our data supports the differential effects of heparin on c-Met signaling based on the presence or absence of HGF. In the absence of HGF, heparin interacted with c-Met and activated it through receptor dimerization. In addition we also tested whether heparin caused c-Met activation via HGF expression. We found that heparin treatment did not change HGF expression.
The Sema domain of c-Met, which carries structural similarity with other Semaphorins and Plexin family members, has been shown to have a critical role in c-Met receptor activation through inducing receptor dimerization following ligand binding (Gherardi et al. 2003; Rubin et al. 2001). It has been reported that heparin binding to c-Met Sema domain induces c-Met receptor dimerization (Gherardi et al. 2003). To test the hypothesis that heparin enhances c-Met activation through receptor dimerization, we examined c-Met receptor dimerization under heparin treated conditions. We observed that heparin did enhance c-Met activation via increasing c-Met dimerization. We thus showed that a novel mechanism of c-Met activation in the presence of heparin was via receptor dimerization. Our results are consistent with those of Murray et al., who suggested that heparin is a ligand for ALK and activates it via receptor dimerization in neuroblastoma cells (Murray et al. 2015).
To our knowledge, this is the first study to report that heparin enhanced c-Met activation through receptor dimerization in HCC cell lines. It is known that following c-Met activation, several downstream signaling pathways, including ERK1/2 and PI3K-Akt signaling, are activated. When we investigated the activation status of ERK1/2 and Akt following heparin treatment, we found that heparin increased ERK1/2 activation, but suppressed Akt activation. Kermorgant et al. suggested that endocytic trafficking of c-Met to the perinuclear area enhances cell migration through ERK1/2 activation and its translocation to focal adhesion (Kermorgant et al. 2004). We therefore examined the effect of heparin on c-Met perinuclear localization and observed that in accordance with ERK1/2 activation, heparin enhanced c-Met perinuclear localization of c-Met in heparin treated HCC cells.
To investigate further the effect of heparin-mediated c-Met activation on HCC we examined the biological significance of heparin. Strikingly, heparin increased cell invasion and migration through induction of the EGR1-MMPs axis, similar to the pattern observed by HGF/c-Met signaling. We suggested that c-Met activated EGR1 as a key molecule in the regulation of migration and invasion of HCC cell lines (Ozen et al. 2012). EGR1 is a c-Met target gene, and the level of EGR1 expression by heparin is context dependent.
We previously showed that in the presence of HGF heparin decreased binding affinity of c-Met to HGF, while in this study we observed that in the absence of HGF, heparin induced c-Met activation via receptor dimerization. The results presented in this study support the hypothesis that the effects of heparin on c-Met signaling in HCC cells are context dependent and the response of HCC cells to heparin is affected by the presence or absence of HGF. Whether to test the effect of heparin is HCC specific, we have tested the effect of heparin in different cancer cell lines such as cervix cancer cell line HeLa, bladder cancer cell line T24, breast cancer cell line MCF7, Burkitt’s lymphoma cell line Raji and colon cancer cell line SW-620. We investigated c-Met protein level and activation upon 10 μg/ml heparin treatment for 2 h by western blotting (Supp Fig. 3). Interestingly there is moderate c-Met activation in response to heparin in HeLa, T24, MCF7 and Raji cells. In contrary in SW-620 cells, although basal c-Met activatory phosphorylation is high, heparin treatment resulted in inhibition of c-Met activation. In the literature, we could not see basal HGF secretion in the indicated cancer cell lines except SW-620 (Jia et al. 2006). The presence of secreted HGF results in basal c-Met activity in SW-620 and correlating with our previous findings that shows inhibitory effects of heparin on HGF-induced c-Met activation (Ozen et al. 2012), HGF induced c-Met activation is inhibited. In conclusion, the effect of heparin on c-Met expression and activation is cell type and context dependent and, the presence of HGF in tumor microenvironment mediates heparin induced responses.
In this study, we also found that heparin treatment decreased proliferation and induced G2/M cell cycle arrest of HCC cells through upregulation of PTEN expression and downregulation of Akt activation. In another study we showed that Akt activation is important for proliferation of HCC cell lines. When we repressed Akt activation using LY294002 or dominant negative Akt, cell proliferation is decreased in HCC cell lines (Kunter et al. 2014). In addition, the inhibition of AKT signaling does not affect activation of MAPK signaling in HCC cell lines. An intriguing study by Ganepola et al. demonstrated that expression of cell cycle molecules and cell proliferation markers are down regulated in metastatic colon tumors when compared to primary counter parts (Ganepola et al. 2010). Thus, it seems that heparin might regulate metastatic ability of HCC cells by inducing migration and invasion while repressing proliferation.
In summary in this study, we demonstrated that in the absence of HGF, heparin activated the c-Met signaling pathway, increasing invasion and migration, while also suppressing proliferation (Fig. 6). Our results provide a novel perspective for the role of heparin in hepatocarcinogenesis through regulation of the HGF/c-Met signaling pathway. Regarding the fact that heparin is frequently used in the prevention of cancer-associated thrombosis, the use of heparin in patients with HCC, overexpressing c-Met might worsen the course of disease by inducing metastasis.
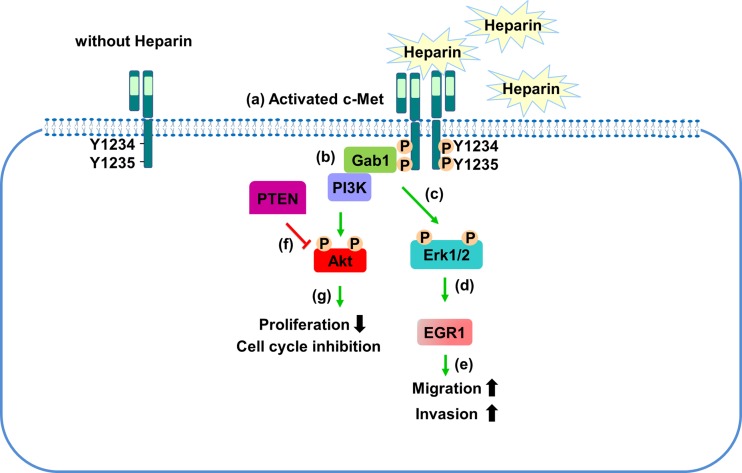
Fig. 6.
The mechanisms of heparin mediated c-Met signaling upregulation in HCC. Heparin activated c-Met receptor through receptor dimerization (a), recruited Gab1 and PI3K to the receptor (b) heparin-induced c-Met activation led to ERK1/2 activation (c), and EGR1 up-regulation (d), EGR1 activation caused induction in migration, invasion (e), heparin induced PTEN expression caused downregulation of Akt activation (f), Akt inhibition caused proliferation repression and cell cycle arrest (g)
Thus, our data provide support for the use of c-Met as a biomarker for stratification of patients with HCC who might benefit or the opposite, from treatment with heparin and support the hypothesis that the effects of heparin on behavior of HCC cells are context dependent and the response of HCC cells to heparin is affected by the presence or absence of HGF.
Electronic supplementary material
(DOCX 13 kb)
(GIF 250 kb)
(GIF 69 kb)
(GIF 158 kb)
Acknowledgments
We thank Prof. Brian Carr for critically reading the manuscript and improving the English. This work was supported by The Scientific and Technological Research Council of Turkey (Project # 110S349).
Abbreviations
- ANOVA
Analysis of variance
- BCA
Bicinchoninic acid
- DSs
Dermatan sulfates
- DMEM
Dulbecco’s modified eagle medium
- EGR1
Early growth response factor 1
- ERK1/2
Extracellular signal-regulated kinases
- FBS
Fetal bovine serum
- GAG
Glycosaminoglycan
- GPC3
Glypican 3
- Gab1
Grb2 associated binder 1
- Grb2
Growth factor receptor-bound protein 2
- HSPGs
Heparan sulfate proteoglycans
- HCC
Hepatocellular carcinoma
- HGF
Hepatocyte growth factor
- MMPs
Matrix metalloproteinases
- MAPK
Mitogen-activated protein kinase
- NFDM
Nonfat dry milk
- PBS
Phosphate buffer saline
- PI
Propidium iodide
- PI3K
Phosphoinositide-3 kinase
- PVDF
Polyvinylidene fluoride
- SDS
Sodium dodecyl sulphate
- SEM
Standard error of the mean
- SRB
Sulforhodamin b
- TCA
Trichloroacetic acid
- TBST
Tris buffered saline/Tween-20
Compliance with ethical standards
Conflict of interest
The authors have no conflicts of interest to declare
Footnotes
Evin Iscan and Aysim Gunes contributed equally to this work.
Electronic supplementary material
The online version of this article (doi:10.1007/s12079-016-0368-0) contains supplementary material, which is available to authorized users.
References
- Bozkaya G, Korhan P, Cokakli M, Erdal E, Sagol O, Karademir S, Korch C, Atabey N. Cooperative interaction of MUC1 with the HGF/c-Met pathway during hepatocarcinogenesis. Mol Cancer. 2012;11:64. doi: 10.1186/1476-4598-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappuzzo F, Marchetti A, Skokan M, Rossi E, Gajapathy S, Felicioni L, Del Grammastro M, Sciarrotta MG, Buttitta F, Incarbone M, Toschi L, Finocchiaro G, Destro A, Terracciano L, Roncalli M, Alloisio M, Santoro A, Varella-Garcia M. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol. 2009;27:1667–1674. doi: 10.1200/JCO.2008.19.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokakli M, Erdal E, Nart D, Yilmaz F, Sagol O, Kilic M, Karademir S, Atabey N. Differential expression of Caveolin-1 in hepatocellular carcinoma: correlation with differentiation state, motility and invasion. BMC Cancer. 2009;2407:9–65. doi: 10.1186/1471-2407-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper CS, Park M, Blair DG, Tainsky MA, Huebner K, Croce CM, Vande Woude G. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature. 1984;311:29–33. doi: 10.1038/311029a0. [DOI] [PubMed] [Google Scholar]
- Eder JP, Vande Woude GF, Boerner SA, LoRusso PM. Novel therapeutic inhibitors of the c-Met signaling pathway in cancer. Clin Cancer Res. 2009;15:2207–2214. doi: 10.1158/1078-0432.CCR-08-1306. [DOI] [PubMed] [Google Scholar]
- Firtina Karagonlar Z, Koc D, Iscan E, Erdal E, Atabey N. Elevated HGF expression as an autocrine c-Met activation mechanism in acquired resistance to sorafenib in HCC cells. Cancer Sci. 2016 doi: 10.1111/cas.12891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlan A, Kherrouche Z, Montagne R, Copin MC, Tulasne D. Thirty years of research on met receptor to move a biomarker from bench to bedside. Cancer Res. 2014;74:6737–6744. doi: 10.1158/0008-5472.CAN-14-1932. [DOI] [PubMed] [Google Scholar]
- Ganepola GA, Mazziotta RM, Weeresinghe D, Corner GA, Parish CJ, Chang DH, Tebbutt NC, Murone C, Ahmed N, Augenlicht LH, Mariadason JM. Gene expression profiling of primary and metastatic colon cancers identifies a reduced proliferative rate in metastatic tumors. Clin Exp Metastasis. 2010;1:1–9. doi: 10.1007/s10585-009-9295-2. [DOI] [PubMed] [Google Scholar]
- Gao JJ, Inagaki Y, Xue X, Qu XJ, Tang W. C-met: a potential therapeutic target for hepatocellular carcinoma. Drug Discov Ther. 2011;1:2–11. doi: 10.5582/ddt.2011.v5.1.2. [DOI] [PubMed] [Google Scholar]
- Garber K. MET inhibitors start on road to recovery. Nat Rev Drug Discov. 2014;13:563–565. doi: 10.1038/nrd4406. [DOI] [PubMed] [Google Scholar]
- Gherardi E, Youles ME, Miguel RN, Blundell TL, Iamele L, Goug J, Bandyopadhyay A, Hartmann G, Butler PJG. Functional map and domain structure of MET, the product of the c-Met protooncogene and receptor for hepatocyte growth factor/scatter factor. PNAS. 2003;100:12039–12044. doi: 10.1073/pnas.2034936100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. Targeting MET in cancer: rationale and progress. Nat Rev Cancer. 2012;12:89–103. doi: 10.1038/nrc3205. [DOI] [PubMed] [Google Scholar]
- Giordano S, Columbano A. Met as a therapeutic target in HCC: facts and hopes. J Hepatol. 2014;60:442–452. doi: 10.1016/j.jhep.2013.09.009. [DOI] [PubMed] [Google Scholar]
- Grant DS, Kleinman HK, Goldbergt ID, Bhargava MM, Nickoloff BJ, Kinsella JL, Polverini P, Rosen EM. Scatter factor induces blood vessel formation in vivo. PNAS. 1993;90:1937–1941. doi: 10.1073/pnas.90.5.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes O, Pillozzi S, Deakin JA, Carafoli F, Kemp L, Butler PJG, Lyon M, Gherardi E. Insights into the structure / function of hepatocyte growth factor / scatter factor from studies with individual domains. J Mol Biol. 2007;367:395–408. doi: 10.1016/j.jmb.2006.12.061. [DOI] [PubMed] [Google Scholar]
- Jia Y, Zhang L, Li Y, Wang Y, Guo W, Cao L, Li Z. Effects on proliferation and migration of the human colon carcinoma cell line SW620 by silencing of hepatocyte growth factor expression. Clin Oncol Cancer Res. 2006;7:277–283. doi: 10.1007/s11805-010-0531-y. [DOI] [Google Scholar]
- Kaposi-Novak P, Lee JS, Gomez-Quiroz L, Coulouarn C, Factor VM, Thorgeirsson SS. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest. 2006;116:1582–1595. doi: 10.1172/JCI27236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermorgant S, Zicha D, Parker PJ. PKC controls HGF-dependent c-Met traffic, signaling and cell migration. EMBO J. 2004;23:3721–3734. doi: 10.1038/sj.emboj.7600396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong-Beltran M, Stamos J, Wickramasinghe D. The Sema domain of met is necessary for receptor dimerization and activation. Cancer Cell. 2004;6:75–84. doi: 10.1016/j.ccr.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Korhan P, Erdal E, Atabey N. MiR-181a-5p is downregulated in hepatocellular carcinoma and suppresses motility, invasion and branching-morphogenesis by directly targeting c-met. Biochem Biophys Res Commun. 2014;4:1304–1312. doi: 10.1016/j.bbrc.2014.06.142. [DOI] [PubMed] [Google Scholar]
- Kunter I, Erdal E, Nart D, Yılmaz F, Karademir S, Sagol O, Atabey N. Active form of AKT controls cell proliferation and response to apoptosis in hepatocellular carcinoma. Oncol Rep. 2014;31:573–580. doi: 10.3892/or.2013.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJX, Chan JC, Choo SP. Clinical development of c-Met inhibition in hepatocellular carcinoma. Diseases. 2015;3:306–324. doi: 10.3390/diseases3040306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever R, Page CP. Novel drug development opportunities for heparin. Nat Rev Drug Discov. 2002;1:140–148. doi: 10.1038/nrd724. [DOI] [PubMed] [Google Scholar]
- Lim HC, Multhaupt HAB, Couchman JR. Cell surface heparan sulfate proteoglycans control adhesion and invasion of breast carcinoma cells. Mol Cancer. 2015;27:1. doi: 10.1186/s12943-014-0279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology. 2008;48:1312–1327. doi: 10.1002/hep.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Rangnekar VM, Adamson E, Mercola D. Suppression of growth and transformation and induction of apoptosis by EGR-1. Cancer Gene Ther. 1998;5:3–28. [PubMed] [Google Scholar]
- Lyon M, Deakin JA, Gallagher JT. The mode of action of heparan and dermatan sulfates in the regulation of hepatocyte growth factor/scatter factor. J Biol Chem. 2002;277:1040–1046. doi: 10.1074/jbc.M107506200. [DOI] [PubMed] [Google Scholar]
- Murray PB, Lax I, Reshetnyak A, Ligon GF, Lillquist JS, Natoli EJ, Jr, Shi X, Folta-Stogniew E, Gunel M, Alvarado D, Schlessinger J. Heparin is an activating ligand of the orphan receptor tyrosine kinase ALK. Sci Signal. 2015;360:rab6. doi: 10.1126/scisignal.2005916. [DOI] [PubMed] [Google Scholar]
- Ozen E, Gozukizil A, Erdal E, Uren A, Bottaro DP, Atabey N. Heparin inhibits hepatocyte growth factor induced motility and invasion of hepatocellular carcinoma cells through early growth response protein 1. PLoS One. 2012;7:e42717. doi: 10.1371/journal.pone.0042717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters S, Adjei AA. MET: a promising anticancer therapeutic target. Nat Rev Clin Oncol. 2012;9:314–326. doi: 10.1038/nrclinonc.2012.71. [DOI] [PubMed] [Google Scholar]
- Rimassa L, Porta C, Borbath I, Daniele B, Finn RS, Raoul JL, Schwartz LH, He AR, Trojan J, Peck-Radosavljevic M, Abbadessa G, Goldberg T, Santoro A, Bruix J. Tivantinib in MET-high hepatocellular carcinoma patients and the ongoing phase III clinical trial. Hepatic Oncol. 2014;2:181–188. doi: 10.2217/hep.14.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin JS, Day RM, Breckenridge D, Atabey N, Taylor WG, Stahl SJ, Wingfield PT, Kaufman JD, Schwall R, Bottaro DP. Dissociation of heparan sulfate and receptor binding domains of hepatocyte growth factor reveals that heparan sulfate-c-met interaction facilitates signaling. J Biol Chem. 2001;276:32977–32983. doi: 10.1074/jbc.M105486200. [DOI] [PubMed] [Google Scholar]
- Sakai K, Aoki S, Matsumoto K. Hepatocyte growth factor and met in drug discovery. J Biol Chem. 2015;157:271–284. doi: 10.1093/jb/mvv027. [DOI] [PubMed] [Google Scholar]
- Sanford D, Lazo-Langner A. The effect of low molecular weight heparin on survival in cancer patients: an updated systematic review and meta-analysis of randomized trials: reply. J Thromb Haemost. 2014;12:1574–1575. doi: 10.1111/jth.12666. [DOI] [PubMed] [Google Scholar]
- Spek CA, Versteeg HH, Borensztajn KS. Anticoagulant therapy of cancer patients: will patient selection increase overall survival? Thromb Haemost. 2015;114:530–536. doi: 10.1160/TH15-02-0124. [DOI] [PubMed] [Google Scholar]
- Spina A, De Pasquale V, Cerulo G, Cocchiaro P. HGF/c-MET axis in tumor microenvironment and metastasis formation. Biomed. 2015;3:71–88. doi: 10.3390/biomedicines3010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi A, Yamamoto Y, Munesue S, Harashima A, Watanabe T, Yonekura H, Yamamoto H, Tsuchiya H. Low molecular weight heparin suppresses receptor for advanced glycation end products-mediated expression of malignant phenotype in human fibrosarcoma cells. Cancer Sci. 2013;104:740–749. doi: 10.1111/cas.12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You H, Ding W, Dang H, Jiang Y, Rountree CB. c-Met represents a potential therapeutic target for personalized treatment in hepatocellular carcinoma. Hepatology. 2011;54:879–889. doi: 10.1002/hep.24450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong GX, Gong Y, Yu CJ, Wu SF, Ma QP, Wang Y, Ren J, Zhang XC, Yang WH, Zhu W. Significantly inhibitory effects of low molecular weight heparin (Fraxiparine) on the motility of lung cancer cells and its related mechanism. Tumour Biol. 2015;36:4689–4697. doi: 10.1007/s13277-015-3117-8. [DOI] [PubMed] [Google Scholar]
- Zhou AX, Toylu A, Nallapalli RK, Nilsson G, Atabey N, Heldin CH, Borén J, Bergo MO, Akyürek LM. Filamin a mediates HGF/c-Met signaling in tumor cell migration. Int J Cancer. 2011;128:839–846. doi: 10.1002/ijc.25417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 13 kb)
(GIF 250 kb)
(GIF 69 kb)
(GIF 158 kb)