Abstract
Heart diseases are the most significant cause of morbidity and mortality in the world. De novo generated cardiomyocytes (CMs) are a great cellular source for cell-based therapy and other potential applications. Direct cardiac reprogramming is the newest method to produce CMs, known as induced cardiomyocytes (iCMs). During a direct cardiac reprogramming, also known as transdifferentiation, non-cardiac differentiated adult cells are reprogrammed to cardiac identity by forced expression of cardiac-specific transcription factors (TFs) or microRNAs. To this end, many different combinations of TFs (±microRNAs) have been reported for direct reprogramming of mouse or human fibroblasts to iCMs, although their efficiencies remain very low. It seems that the investigated TFs and microRNAs are not sufficient for efficient direct cardiac reprogramming and other cardiac specific factors may be required for increasing iCM production efficiency, as well as the quality of iCMs. Here, we analyzed gene expression data of cardiac fibroblast (CFs), iCMs and adult cardiomyocytes (aCMs). The up-regulated and down-regulated genes in CMs (aCMs and iCMs) were determined as CM and CF specific genes, respectively. Among CM specific genes, we found 153 transcriptional activators including some cardiac and non-cardiac TFs that potentially activate the expression of CM specific genes. We also identified that 85 protein kinases such as protein kinase D1 (PKD1), protein kinase A (PRKA), calcium/calmodulin-dependent protein kinase (CAMK), protein kinase C (PRKC), and insulin like growth factor 1 receptor (IGF1R) that are strongly involved in establishing CM identity. CM gene regulatory network constructed using protein kinases, transcriptional activators and intermediate proteins predicted some new transcriptional activators such as myocyte enhancer factor 2A (MEF2A) and peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PPARGC1A), which may be required for qualitatively and quantitatively efficient direct cardiac reprogramming. Taken together, this study provides new insights into the complexity of cell fate conversion and better understanding of the roles of transcriptional activators, signaling pathways and protein kinases in increasing the efficiency of direct cardiac reprogramming and maturity of iCMs.
Keywords: Direct cardiac reprogramming, Induced cardiomyocyte, Gene regulatory network, Transcriptional activators, Transcription factors, And protein kinase
Introduction
The mammalian heart lacks an adequate capacity to generate new cardiomyocyte (CMs) and restore its normal function after damage. Accordingly, cardiovascular diseases (CVDs) are the most significant cause of morbidity and mortality worldwide, representing 30% of all global deaths (17.3 million deaths annually) (Talkhabi et al. 2016). On the other hand, CVDs constitute a huge cost for national health systems. In 2010, the estimated global cost of CVDs was 863 billion dollars (Talkhabi et al. 2016). To this end, several medical interventions including drug therapy, organ transplantation and cell therapy have been developed to treat patients with cardiac diseases. It seems that cell therapy has more advantages in restoring the cardiac normal functions after damages. In the last years, different types of human cell, including bone marrow-derived mononuclear cells, mesenchymal stem cells (MSCs), C-kit + cardiac progenitor cells, early endothelial progenitor cells (EPCs), cardiospheres-derived cells (CDCs) and induced pluripotent stem cells (iPSCs) have been studied clinically and experimentally to enhance the cardiac regeneration (Jakob and Landmesser 2013). These cells have yielded mixed results with respect to effects on cardiac function. Therefore, it is necessary to develop a new approach to produce CMs required in cardiac cell-based therapy and other potential applications.
iPSCs technology opened a new avenue for generating different cell types from differentiated somatic cells, only by overexpressing cell type specific transcription factors (TFs) or microRNAs. Today this approach is called transdifferentiation, direct cell-fate conversion, or direct reprogramming, in which the identity of one type of somatic cells is changed to other adult cell types without intermediate reversion to a pluripotent state (Talkhabi et al. 2015). In 2010, for the first time Ieda et al. directly reprogrammed mouse cardiac fibroblasts (CFs) and tail tip fibroblasts (TTFs) into beating CMs, known as induced CMs (iCMs), by overexpressing only three CM-specific TFs: GATA binding protein 4 (GATA4), myocyte enhancer factor 2C (MEF2C), and T-Box 5 (TBX5) (Ieda et al. 2010). It seems that GATA4, MEF2C, and TBX5 (GMT) to be the “master regulators” for direct cardiac reprogramming. As the first study, their efficiency was low (~0.5% of the αMHC-EGFP+/cTnT+ cells were capable of beating), which caused they and many other groups worked on increasing the efficiency of iCM generation in vitro and in vivo. Very soon after the first study, several combinations were reported, indicating that other combinations such as GMT plus HAND2 (Song et al. 2012), myocardin, MEF2C and TBX5 (Protze et al. 2012), MESP1 and ETS2 (Islas et al. 2012), GATA4, HAND2, myocardin, TBX5, miR-1 and miR-133 (Nam et al. 2013), could reprogram mouse or human fibroblasts into iCMs. Their efficiencies were around 20% as measured using a single CM marker such as αMHC, but the functional properties such as spontaneous contraction were observed rarely (<1% of total cell population). Surprisingly, unlike iPSCs technology in which a invariable combination of four mammalian TFs - OCT4, SOX2, KLF4 and c-Myc- is able to generate iPSCs from different types of somatic cells in different species including mouse, humans, birds, fish, and fly (Rosselló et al. 2013), there is no common combination of cardiac-specific factors for direct cardiac reprogramming of somatic cells derived from different species. The preliminary findings have served to intensify interest in understanding the molecular basis of direct cardiac reprogramming and the potential uses of iCMs such as in cell-based therapy. However, before such potential applications can be routinely and safely developed, numerous crucial issues must be addressed. In particular, although different combinations have been tested and reported for direct cardiac reprogramming in mammals, scientists still are investigating different factors (TFs, signaling pathways, microRNAs) (Sadahiro et al. 2015) and the most efficient combination for direct cardiac reprogramming of fibroblasts have yet to be fully determined. Importantly, we showed that fibroblasts are derived from different origins, even in the same organ such as heart, which might affect the efficiency of direct cardiac reprogramming (Ali et al. 2014). Recently, we found some mechanisms affecting the efficiency of direct cardiac reprogramming of mouse fibroblast derived from different origins and different organs (unpublished data). During the direct cardiac reprogramming, the identity of starting cells (e.g. CFs) is converted to the target cells (e.g. CMs). Although both cell types have virtually identical genome, each one has a unique transcriptome reflecting the expression of a subset of genes that control morphological, developmental, physiological and pathological behaviors of each cell. Therefore, transcriptome analysis within a direct reprogramming process allows us to access the gene regulatory network at a whole-genome scale to identify genes and pathways that underlie this process.
Here, we used previously published gene expression data (Qian et al. 2012) and analyzed the transcriptome of CFs, iCMs and aCMs to find new regulators including transcriptional activators and protein kinases boosting direct reprogramming of mouse CFs to iCMs. The up-regulated genes in CMs (iCMs and aCMs) than CFs were determined as cardiac specific genes, while fibroblast specific genes were determined based on downregulation in CMs (aCMs and iCMs) than CFs. Estrogen related receptor beta (ESRRB), GATA4, peroxisome proliferator activated receptor gamma (PPARG), TATA-box binding protein associated factor 7 like (TAF7L), nuclear receptor subfamily 0 group B member 1 (NR0B1), E1A binding protein p300 (EP300), Nk2 homeobox 5 (NKX2–5), ASH2 like histone lysine methyltransferase complex subunit (ASH2L), E2F transcription factor 1 (E2F1) and B cell leukemia/lymphoma 3 (BCL3) were found to be the most important transcriptional activators that potentially could activate the expression of cardiac specific genes during direct cardiac reprogramming. We also found that many proteins, called as intermediate proteins, directly interacted with the important transcriptional activators. Importantly; we found a catalog of protein kinases might influence the expression of downstream cardiac specific genes through phosphorylation the intermediate proteins interacting with the important transcriptional activators. CM gene regulatory network constructed using important transcriptional activators, important kinases and intermediate proteins predicted that myocyte enhancer factor 2A (MEF2A), peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PPARGC1A), EP300, activating transcription factor 3 (ATF3) and melanogenesis associated transcription factor (MITF) could be potential candidates to join GMT to increase the efficiency and maturity of iCMs generated through direct reprogramming. Moreover, we identified that the activation of protein kinase D1 (PKD1), protein kinase A (PRKA), calcium/calmodulin-dependent protein kinase (CAMK), protein kinase C (PRKC), and insulin like growth factor 1 receptor (IGF1R) might be very important for enhancement of direct cardiac reprogramming and increasing the quality of iCMs.
Materials and methods
Re-analysis of microarray data
A literature review for high-throughput gene expression studies was performed to identify gene expression differences in mouse CFs and iCMs. A study reported by Srivastava’s group was selected for a precise computational analysis (Qian et al. 2012). The microarray data was obtained from NCBI’s Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) with GEO accession number GSE49192, composed of overall 13 different mouse and human cell types. In the current study, only mouse related cell types were analyzed including wild type CFs, iCMs and wild type aCMs. Data was analyzed by GEO2R (www.ncbi.nlm.nih.gov/geo/geo2r) tools to identify significantly upregulated and downregulated genes in CMs (iCMs plus aCMs) against CFs. Differentially expressed genes were selected with univariate tests according to the p-value (p < 0.01) and log-fold change (FC ≥ 1).
Functional analysis of differentially expressed genes
The functions of differentially expressed genes (DEGs) were analyzed by the database for annotation, visualization and integrated discovery (DAVID) (http://david.abcc.ncifcrf.gov) and BioCarta (https://cgap.nci.nih.gov/Pathways/BioCarta_Pathways). In addition, DEGs were analyzed using BioGPS database (http://biogps.org/#goto=welcome), a well-known gene-expression tissue atlas, to check out the specificity of their expression to fibroblasts or cardiac linage cells.
Construction and analysis of CM gene regulatory network
n order to identify regulators of DEGs involved during the direct cardiac reprogramming of mouse CFs into iCMs, we used a web based interactive application called ChIP Enrichment Analysis (ChEA) (Essaghir et al. 2010; Lachmann et al. 2010). The ChEA database contains results from high throughput data regarding TFs binding sites provides the opportunity to analyze a list of mammalian DEGs at the same time. This database contains 458,471 interactions, manually extracted from different ChIP-based experimental procedure data including ChIP-chip, ChIP-seq, ChIP-PET, and DamID-seq, describing the binding of more than 200 TFs to their target genes. The TFs obtained from the ChEA database were selected based on the p-value and their expression (p-value < 0.05, fold change ≥1). To find the intermediate proteins, we analyzed the differentially expressed TFs using Gene2Networks a powerful software system integrates the content of ten mammalian interaction network datasets to find relationships between genes and proteins from the list of interest, as well as to predict additional genes or proteins that may play key roles in common pathways or protein complexes (Berger et al. 2007). We also analyzed protein kinases using KEA database that is a web-based tool with an underlying database providing ability to link lists of mammalian proteins/genes with the kinases that phosphorylate them (Lachmann and Ma’ayan 2009). In order to construct the CM and CF gene regulatory networks representing the selected transcriptional activators, protein kinases and intermediate proteins, we used yEd (version 3.10.2) graph editor software.
Result
Cardiac specific genes are involved in different cardiac processes
In the current study, we analyzed the microarray data of three different cell types including CFs (two replicates), aCMs (three replicates) and iCMs (three replicates). In the original study, the CFs was directly reprogrammed to iCMs using GMT, which are known as the main regulators in mouse cardiac reprogramming. The gene expression data of iCMs and aCMs was pooled together and then was compared with the gene expression data of CFs (as the starting cells). We found that 226 genes were significantly upregulated in CMs (iCMs and aCMs) than CFs. Here, these genes were determined as cardiac specific genes. Whereas, the expression of 200 genes was significantly decreased in CMs than CFs, which was determined as the fibroblast specific genes. Myosin light chain 2 (Myl2) and annexin A1 (Anxa1) were identified as the most upregulated and downregulated genes with 6.3 and 5.6 log-fold changes in CMs, respectively. CM specific genes function in different cardiac related processes including cardiac development and disease, secretion, extracellular related processes, signal transduction, and glycosylation.
CM gene regulatory network relies on cardiac specific transcriptional activators, protein kinases and intermediate proteins
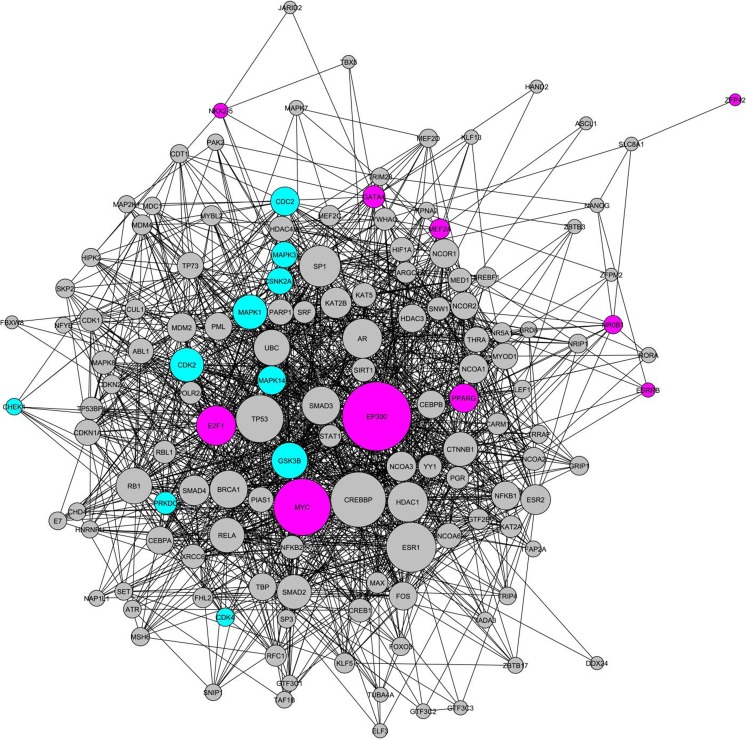
In order to construct the CM gene regulatory network establishing cardiac identity in CFs, we analyzed transcriptional activators might govern the expression of DEGs. The promotor and upstream regions of DEGs were analyzed using ChEA to find the probable transcriptional activators might work on these regions. We found that 153 transcriptional activators could bind to the promotor and upstream regions of CMs specific genes (genes upregulated in CM). While, 224 transcriptional activators were identified to work on the promotor and upstream regions of CF specific genes (downregulated genes in CMs). Among the transcriptional activators that were related to the upregulated and downregulated genes in CMs, 10 transcriptional activators were selected as the most important transcriptional activators, which had activating interactions with a high number of DEGs (Table 1). ESRRB, GATA4, PPARG, TAF7L, NR0B1, EP300, NKX2–5, ASH2L, E2F1, and BCL3 were the most effective transcriptional activators that could bind the promotor or upstream region of CM specific genes. Our analysis indicated that the first three transcriptional activators were the most important transcriptional activators in activating the expression of upregulated genes. For example, ESRRB could interact with 73 CM specific genes (Table 1). While retinoic acid receptor gamma (RARG), NF-E2-related factor 2 (NRF2), nuclear factor, erythroid 2 like 2 (NFE2L2), E2F1, sry-box 17 (SOX17), transcription factor 3 (TCF3), clock circadian regulator (CLOCK), yes associated protein 1 (YAP1), sry-box 2 (SOX2) and peroxisome proliferator activated receptor gamma (PPARG) had the high number of interactions with downregulated genes in CMs (Table 1), suggesting that these transcriptional activators as the main transcriptional activators could be involved in stablishing or retaining CF identity. Next, 10 most important transcriptional activators, which presumably interacted with the upregulated and downregulated genes, were analyzed using Genes2Newtork to find intermediate proteins that could have protein-protein interactions with 10 important transcriptional activators. we found that 117 and 284 intermediate proteins for CM and CF specific genes, respectively. Moreover, analysis of these intermediate proteins using KEA (Kinase Expression Analysis) resulted in the identification of some protein kinases. Accordingly, our analysis showed that 85 protein kinases interacted with 117 intermediate proteins to upregulate the expression of CM specific genes. Among these protein kinases, protein kinase CAMP-activated catalytic subunit alpha (PRKACA), protein kinase CAMP-activated catalytic subunit gamma (PRKACG) and calcium/calmodulin dependent protein kinase II alpha (CAMK2A) had highest score, indicating they could be very vital in establishing or maintaining CM identity (Table 2). Whereas 92 protein kinases were identified to work with 284 intermediate proteins to upregulate the expression of CF specific genes (downregulated genes in CMs). It seems that mitogen-activated protein kinase 1 (MAPK1), cyclin dependent kinase 2 (CDK2) and glycogen synthase kinase 3 beta (GSK3B) are the most important protein kinases, as determined by combining different scores (Table 2). In order to clearly show how the analyzed factors interact with each other, the CM and CF gene regulatory networks were constructed using CM and CF specific transcriptional activators, intermediate proteins, and protein kinases, respectively. Accordingly, 10 most important transcriptional activators (Table 1), 117 intermediate proteins and 10 most important protein kinases (Table 2) were used for construction CM gene regulatory network (Fig. 1). These three different groups of proteins totally create 1432 protein-protein interactions in the constructed gene regulatory network. It was identified that EP300 had the highest number of interactions with other components of the network (20 direct interactions and 11 indirect interactions), suggesting that EP300 is highly involved in establishing and/or maintaining CM identity.
Table 1.
Important potential transcriptional activators of upregulated and downregulated genes in CMs
| Term | Overlap | P-value | Adjusted P-value | Combined score | |
|---|---|---|---|---|---|
| TFS activating the expression of CM specific genes | ESRRB | 73 | 3.24E-54 | 2.01E-51 | 238.0692 |
| GATA4 | 46 | 5.51E-19 | 6.86E-17 | 75.45215 | |
| PPARG | 16 | 5.11E-13 | 1.91E-11 | 75.39176 | |
| TAF7L | 26 | 3.1E-13 | 1.29E-11 | 70.3822 | |
| NR0B1 | 41 | 5.25E-18 | 3.92E-16 | 68.08592 | |
| EP300 | 25 | 1.9E-11 | 4.44E-10 | 62.14964 | |
| NKX2–5 | 34 | 3.75E-14 | 1.75E-12 | 60.82659 | |
| ASH2L | 61 | 8.45E-21 | 1.58E-18 | 42.91397 | |
| E2F1 | 64 | 4.67E-18 | 3.92E-16 | 41.71403 | |
| BCL3 | 24 | 3.94E-12 | 1.22E-10 | 41.42514 | |
| TFs activating the expression of CF specific genes | RARG | 21 | 2.68E-16 | -2.45541 | 88.04412 |
| NRF2 | 30 | 1.50E-15 | -1.70414 | 58.16592 | |
| NFE2L2 | 30 | 1.50E-15 | -1.70414 | 58.16592 | |
| E2F1 | 64 | 3.31E-19 | -1.18513 | 50.43121 | |
| SOX17 | 37 | 1.15E-13 | -1.57483 | 46.92527 | |
| TCF3 | 40 | 5.29E-14 | -1.52041 | 46.47912 | |
| CLOCK | 16 | 9.67E-11 | -1.7818 | 41.08756 | |
| YAP1 | 42 | 1.04E-14 | -1.26895 | 40.86082 | |
| SOX2 | 42 | 2.45E-13 | -1.38276 | 40.14992 | |
| PPARG | 54 | 8.78E-16 | -1.06914 | 37.066 |
Table 2.
Important protein kinases phosphorylating the CM and CF specific intermediate proteins
| Term | Overlap | P-value | Z-score | Combined score | |
|---|---|---|---|---|---|
| Protein kinases interacting with CM specific intermediate proteins | PRKACA | 14 | 0.000379 | −2.17102 | 7.458062 |
| PRKACG | 8 | 0.002131 | −1.92501 | 5.631611 | |
| CAMK2A | 6 | 0.001314 | −1.91366 | 5.598411 | |
| PRKD1 | 4 | 0.002524 | −1.58338 | 4.632169 | |
| RPS6KA3 | 9 | 0.028299 | −1.86183 | 2.412492 | |
| PRKCE | 3 | 0.025067 | −1.43824 | 1.863615 | |
| IGF1R | 4 | 0.028394 | −1.39841 | 1.812008 | |
| PRKCA | 10 | 0.060151 | −1.26071 | 1.547892 | |
| RPS6KA1 | 4 | 0.057566 | −0.97664 | 1.199122 | |
| PRKACB | 4 | 0.0609 | −0.94454 | 1.159706 | |
| Protein kinases interacting with CF specific intermediate proteins | MAPK1 | 32 | 7.57E-08 | -2.42103 | 39.69706 |
| CDC2 | 44 | 4.92E-07 | -2.20551 | 32.03513 | |
| CDK2 | 42 | 7.59E-07 | -2.14405 | 30.21185 | |
| GSK3B | 48 | 1.41E-06 | -2.13994 | 28.83151 | |
| HIPK2 | 12 | 4.32E-07 | -1.7411 | 25.51699 | |
| CDK7 | 11 | 3.10E-07 | -1.6357 | 24.51265 | |
| PRKDC | 26 | 5.00E-06 | -1.97809 | 24.14623 | |
| MAPK14 | 36 | 4.36E-05 | -1.90853 | 19.1636 | |
| CSNK2A1 | 24 | 2.54E-05 | -1.80883 | 19.137 | |
| ATM | 17 | 7.91E-05 | -1.49518 | 14.12089 |
Fig. 1.
CM gene regulatory network. The network is consisted of 10 important transcriptional activators of CM specific genes (pink), 117 intermediate proteins (gray) and 10 important protein kinases (bluegreen). 1432 protein-protein interactions exist between the components, in which the highest number of interactions belongs to EP300. MAPKs are the most abundant kinases involved in the network
Moreover, CF gene regulatory network was constructed using 10 most important transcriptional activators (Table 1), 10 most important protein kinases (Table 2) and 284 intermediate proteins. CF gene regulatory network consisted of 2534 interactions (Fig. 2). E2F1 had the highest number of interactions with other components (30 direct connections and 19 indirect connections), suggesting that E2F1 has an important role in CFs.
Fig. 2.
CF gene regulatory network. The network is consisted of 10 important transcriptional activators of CF specific genes(pink), 284 intermediate proteins (gray) and 10 important protein kinases (bluegreen) creating 2534 protein-protein interactions
New transcriptional activators might contribute to direct cardiac reprogramming
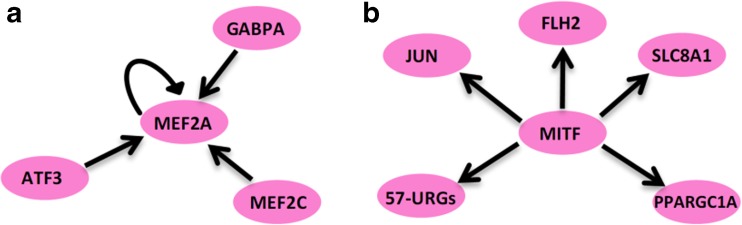
In this study, we analyzed the gene expression data of iCMs that had been directly reprogrammed by overexpression of master regulators of direct cardiac reprogramming (GATA4, MEF2C and TBX5). To find the potential transcriptional activators with ability to induce/activate the expression of CM genes in CFs, we reanalyzed the CM gene regulatory network. Our analysis showed that ATF3, GA binding protein transcription factor alpha subunit (GABPA), Jun proto-oncogene (JUN), MEF2A, lysine acetyltransferase 2B (KAT2B), MEF2C, and MAX interactor 1 (MXI1) could actively be involved in the CM gene regulatory network. Interestingly, ATF3, GABPA, JUN, MEF2A, and MITF were in the list of genes that were upregulated in CMs. Moreover, the all components of CM gene regulatory network including TFs, intermediated proteins and protein kinases were compared with the genes upregulated in CMs, and it was identified that some components including MEF2A, TATA-box binding protein associated factor, RNA polymerase I, B (TAF1B), KAT2B, MEF2C, PPARGC1A, tubulin- alpha 4A (TUBA4A) and FHL2 existed in both groups (Table 3). This finding suggests that these transcriptional activators, particularly ATF3, JUN, MEF2A, KAT2B, could have important roles in CM gene regulatory network. These factors activate the expression of themselves or other cardiac specific factors through different types of interactions (Fig. 3). For example, the expression of MEF2A is induced by GABPA, ATF3, MEF2C and MEF2A itself, and it can induce the expression of its target genes such as MAPK1, EP300 and MYOD1. Taken together, these data suggest that MEF2A and other CM indicated transcriptional activators could be strongly involved in direct cardiac reprogramming of CFs to iCMs.
Table 3.
Candidate transcriptional activators for enhancing direct cardiac reprogramming and increasing the maturity of iCMs
| Proteins | UP | G-TF | CM-Net | Net-TF | Score |
|---|---|---|---|---|---|
| MEF2A | ✓ | ✓ | ✓ | ✓ | 4 |
| JUN | ✓ | ✓ | ✓ | 3 | |
| KAT2B | ✓ | ✓ | ✓ | 3 | |
| ATF3 | ✓ | ✓ | ✓ | 3 | |
| GABPA | ✓ | ✓ | ✓ | 3 | |
| MEF2C | ✓ | ✓ | ✓ | 3 | |
| GATA4 | ✓ | ✓ | ✓ | 3 | |
| PPARGC1A | ✓ | ✓ | 2 | ||
| TUBA4A | ✓ | ✓ | 2 | ||
| FHL2 | ✓ | ✓ | 2 | ||
| SLC8A1 | ✓ | ✓ | 2 | ||
| MXI1 | ✓ | ✓ | 2 | ||
| MITF | ✓ | ✓ | 2 | ||
| TAF1B | ✓ | ✓ | 2 | ||
| TBX5 | ✓ | ✓ | 2 |
UP: genes upregulated in CMs,
G-TF: transcriptional activators of the genes upregulated in CMs,
CM-Net: CM gene regulatory network
Net-TF: TFs activating the CM gene regulatory network
✓ indicates the presence of factors in following items:
Fig. 3.
Transcriptional regulatory motifs and CM gene expression relationships. a Multi-input motif and autoregulation. MEF2A is activated by three CM specific transcriptional activators (MEF2C, GABPA and ATF3) and by itself. b Single input motif. MITF as CM important transcriptional activators induces the expression of many other CM specific transcriptional activators
Discussion
To this end, different combinations of TFs and miRNAs have been used for direct cardiac reprogramming of mouse fibroblast. It has been identified that many factors work as barrier or booster for direct cardiac reprogramming, indicating that direct cardiac reprogramming is a very complicated process, which might be affected by many internal and external factors. Today, GMT is used as the main combination for direct cardiac reprogramming in most mouse studies. It has been demonstrated that fibroblast growth factor 2 (FGF2), fibroblast growth factor 10 (FGF10) and vascular endothelial growth factor (VEGF) (Yamakawa et al. 2015), serum free media (Christoforou et al. 2013; Yamakawa et al. 2015), inhibition of TGFβ and WNT signaling (Mohamed et al. 2016), valproic acid and JAK inhibitor (Jayawardena et al. 2012; Christoforou et al. 2013), miR-133 (Muraoka et al. 2014), thymosin β4 (Qian et al. 2012), and hypoxia (Wang et al. 2016b) work as boosters to increase the effect of GMT on direct cardiac reprogramming, but the efficiency of de novo produced beating iCMs is approximately less than 1%, which is still very far from cardiac cell-based therapy and other potential applications. It seems that GMT alone is not sufficient for efficient generation of mature functional iCMs. Here we compared the transcriptome of CMs (aCMs and iCMs generated through GMT overexpression) with CFs to find new players (particularly TFs and protein kinases) that might join GMT to increase the efficiency of direct cardiac reprogramming and the maturity/quality of iCMs. Since iCMs resemble immature and neonatal CMs, we pooled the transcriptome data of iCMs with aCMs to avoid losing any important factors required for establishing the identity of mature and normal CMs. We considered the upregulated genes in CMs as CM specific genes, which are involved in establishment and maintenance of CM identity. While, the downregulated genes in CM were determined as CF specific genes that were postulated to be involved in establishing CF identity. Firstly, we analyzed the CM and CF specific genes to find the potential transcriptional activators with ability to bind the upstream sites of CM and CF specific genes, respectively. We found that 153 transcriptional activators that potentially induce the expression of CM specific genes. We listed 10 most important transcriptional activators including some well-known cardiac TFs (such as NKX2.5) and little-known cardiac TFs (such as ESRRB). Surprisingly, ESRRB was found to transcriptionally activate the highest number of CM specific genes. ESRRB is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal (Martello et al. 2012). It has been also reported that ESRRB is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells (Festuccia et al. 2012). Recently, it has been shown that, miR combo (miRNA 1, 133, 208, and 499) significantly increased Nanog expression during direct cardiac reprogramming (Wang et al. 2016a). On the other hand, the expression of cardiac gene (HAND2, TBX5, and MESP1) were significantly reduced following Nanog knockdown (Wang et al. 2016b). More recently, Kamp’s group reported that Nanog expression is elevated during direct reprogramming of fibroblast to induced cardiac progenitor cells (iCPCs), where it might inhibit BMP signaling to prevents further cardiac differentiation of iCPCs (Lalit et al. 2016). These data suggest that ESRRB might function in direct cardiac reprogramming by substituting for Nanog function. Importantly, our initial list also contains some epigenetic related factors such as EP300, which functions as histone acetyltransferase and regulates transcription via chromatin remodeling. Mice that carry a global loss-of-function mutation in the gene Ep300 die between E12.5 and E15.5, possibly because of the development of severe heart defects (Paige et al. 2015). Moreover, it has been shown that EP300 directly acetylates TBX5 (Lewandowski et al. 2014) and GATA4 (Yanazume et al. 2003) to enhance its transcriptional activity, suggesting an essential role for this factor during cardiac reprogramming induced by GMT. It has been also reported that EP300 is involved in some pathways such as Hippo signaling pathway to control CM physiology, suggesting a role for this pathway during cardiac reprogramming (Zhou et al. 2015b). In consistence with our findings, recently many studies have discovered that manipulation of epigenetic regulators including enhancer of zeste homolog 2 (Ezh2; a key subunit of the polycomb repressive complex 2 which induces di- and trimethylation of Lys27 on histone H3) (Hirai et al. 2013), Bmi1 (major component of polycomb repressive complex 1 which binds to histone H3K27me3 and monoubiquitinates H2A on K119 that leads to chromatin compaction and gene silencing) (Zhou et al. 2016), and Mll1 H3K4 methyltransferase (Liu et al. 2016) are actively involved in direct cardiac reprogramming.
In addition to transcriptional activators that directly promote the expression of cardiac specific genes, protein kinases are indirectly involved in many cardiac related processes such as CM survival, function and maturation. Here we analyzed 117 intermediate proteins interacting with 10 most transcriptional activators to find the important protein kinases might be involved in the direct cardiac reprogramming. Our analysis showed that 85 protein kinases directly affect 10 important transcriptional activators through phosphorylating intermediate proteins. Protein kinase D1 (PKD1), protein kinase A (PRKA), calcium/calmodulin-dependent protein kinase (CAMK), protein kinase C (PRKC), IGF1R (with tyrosine kinase activity) and MAPK-activated protein kinase 1b (also known as RPS6KA3) were found to strongly interact with CM specific intermediate proteins. Our results are consistent with the previous findings that protein kinases could work as boosters of direct cardiac reprogramming. For example, Olson’s group recently identified that overexpression of constitutively activated AKT serine/threonine kinase 1 (Akt1) highly enhanced the efficiency of GATA4, HAND2, MEF2C and TBX5 (GHMT)-mediated direct cardiac reprogramming from various types of mouse fibroblasts including TTFs, CFs and mouse embryonic fibroblasts (MEFs) (Zhou et al. 2015a). They showed that IGF1R and phosphoinositide 3-kinase (PI3K) are the upstream and mechanistic target of rapamycin complex 1 (mTORC1) and forkhead box O3a (FOXO3A) are the downstream mediators of Akt1 in boosting direct cardiac reprogramming from mouse fibroblast. Moreover, Ieda’s group showed that FGF2, FGF10, VEGF (FFV), increased the efficiency of cardiac reprogramming through activation of p38MAPK and PI3K/AKT (Yamakawa et al. 2015). These studies support our findings regarding the importance of kinases in cardiac reprogramming and it will be very interesting to manipulate PKD1, PRKA, CAMK, PRKC, IGF1R and RPS6KA3 during direct cardiac reprogramming induced by GMT or other combination of cardiac TFs and/or miRNAs.
Importantly, the gene regulatory network constructed using 10 most important transcriptional activators, 10 protein kinases, and intermediate proteins gave us more interesting information regarding transcriptional activators that might be involved in enhancing GMT-mediated direct cardiac reprogramming. We found that transcriptional activator EP300 has 20 direct and 11 indirect connections in CM gene regulatory network, indicating that this factor strongly is involved in establishing CM identity. More importantly, the analysis of CM gene regulatory network showed that MEF2A, JUN, ATF3, GABPA and MITF are the most important factors could be candidate for joining GMT to efficiently reprogram mouse CFs to iCMs. The reasons for candidating these factors are; 1) these 5 factors are significantly upregulated in CMs compared with CFs, 2) they have the ability to transcriptionally activate the CM specific genes and themselves as well, indicating the existence of autoregulatory mechanism in CMs, 3) they are strongly involved in CM gene regulatory network constructed by 10 important transcriptional activators, 10 important kinases and their intermediate proteins, 4) they potentially and theoretically can activate almost the entire CM gene regulatory network. Interestingly, different types of transcriptional motifs are determined between these factors (Fig. 3). For example, single input motif in which MITF can activate other transcriptional activators such as SLC8A1, FHL2, JUN and PPARGC1A that is the main TF connecting the CM gene regulatory network to PPAR signaling (Xu et al. 2009). Multi-input motif and autoregulation in which MEF2A is activated by MEF2C, ATF3, GABPA, and MEF2A itself (Fig. 3).
In addition to increase the efficiency of direct cardiac reprogramming, the candidate transcriptional activators also seem to be involved in enhancing the maturity of iCM. For example, mice deficient in MEF2A, which is the predominantly expressed form of MEF2 in postnatal cardiac muscle, develop a dilated cardiomyopathy after birth with a high probability for sudden death; surviving animals show mitochondrial deficiency and reduced mitochondrial content (Goffart et al. 2004). It seems that MEF2A is involved in upregulation of mitochondrial function in postnatal development and the maintenance of mitochondrial content in the heart. On the other hand, several studies have been reported that MEF2A is one of the main cardiac TFs involved in gene regulatory network of CM cell line HL-1 and ESC-derived CM (Xu et al. 2009; Schlesinger et al. 2011). PPARGC1A was another candidate transcriptional activator that is directly related to mitochondrial biogenesis, CM metabolism and maturation. It enhances the transcriptional activation of CM specific genes by recruiting histone acetyltransferases coactivators like CBP/P300, SRC-1 (a tyrosine protein kinase), or KAT2B (Puigserver et al. 1999). PPARGC1A and other PPARs (PPARA, PPARD, and PPARG) control myocardial metabolism by transcriptionally regulating genes encoding enzymes involved in fatty acid and glucose utilization (Rowe et al. 2010). A functional interlink between PPAR signaling and the CM transcription factor network via PPARGC1A has been discovered in ESC-derived CM (Xu et al. 2009). Interestingly, MEF2A directly interacts with PPARGC1A to maintain the CM identity and increase its maturity (Xu et al. 2009). Moreover, it has been shown that overexpression of PPARGC1A stimulates mitochondrial biogenesis in cultured cells (Wu et al. 1999) and in skeletal muscle of transgenic mice (Lin et al. 2002). Therefore, the novel transcriptional activators such as MEF2A and PPARGC1A might enhance both direct cardiac reprogramming and maturation of iCM through activation of CM genes involved in different cardiac process such as mitochondrial oxidative metabolism and maturation.
Moreover, we constructed a gene regulatory network for CF, which consisted of different kinase, transcriptional activators and intermediate proteins. Targeting the important components of CF gene regulatory network could be an alternative strategy to increase the efficiency of cardiac direct reprogramming. Therefore, activation CM gene regulatory network and destruction of CFs gene regulatory network could effectively increase the efficiency and the quality of iCM, which are required for potential applications of iCMs.
It is a well-known fact that for normal CM development, pluripotent epiblast cells undergo a series of sequential differentiation steps including mesoderm, cardiogenic mesoderm, common cardiovascular progenitors, cardiac progenitors, immature CMs and mature CMs (Später et al. 2014). This complex trajectory of CM development is orchestrated by an evolutionarily conserved regulatory network of transcription factors, miRNAs and signaling pathways that control specification, maturation and maintenance of each of the multiple highly specialized myocardial lineages (ventricular, atrial and conduction system cells) (Talkhabi et al. 2016). There are some differences between CM generation via development/differentiation and direct reprogramming. For example, all or most of the sequential steps of CM development are omitted in direct cardiac reprogramming. In GMT-mediated direct cardiac reprogramming, starting cells (e.g. CFs and TTFs) are directly converted to iCMs without first becoming intermediate pluripotent stem cells or early cardiac stem/progenitor cells, reducing the probability of existence of sequential cardiogenic steps in direct reprogramming. It was shown that during iCMs generation from TTFs and CFs, MESP1 (a marker for cardiac mesoderm) and ISL1 (a marker for early CPCs) did not express, demonstrating that the iCMs did not pass the cardiac mesoderm and CPC stage, but rather they were directly reprogrammed into iCMs (Ieda et al. 2010). In order to include the CM development sequential steps in direct cardiac reprogramming process, fibroblasts must be directly reprogrammed to earlier stages of cardiac development such as cardiac mesoderm or CPCs. To achieve this goal, the overexpression of early stage specific TFs/miRNAs, as well as manipulation of some signaling pathways and epigenetic state might be required. Recently, Kamp’s group showed that GATA4, TBX5, MESP1, NKX2.5 and BAF60C directly reprogram mouse fibroblasts to expandable iCPCs, which can pass some of sequential steps of CM development, which might increase the quality and maturity of produced iCMs (Lalit et al. 2016). It is important to note that the pluripotent stem cell-derived CMs pass all cardiac development steps, but they resemble to neonatal CM and are still immature based on gene expression profile and functionality such as beating rate (Fonoudi et al. 2015). Expandability of iCPCs makes these cells as a candidate for many applications. Accordingly, these cells can be expanded for several passages, preparing a large number of cells that can be differentiated into CMs, smooth muscle, and endothelial cells that are required for cardiac cell-based therapy and other potential applications. The accessibility to a large number of iCMs and other heart cells is a great progress, but the quality and maturity of iCMs are also very important. Here, we analyzed the iCMs and aCMs that resulted in finding some novel transcriptional activators such as MEF2A that is a late stage related TF, rather than an early cardiogenic TF (Ewen et al. 2011). Therefore, it seems that addition of the novel transcriptional activators found in the present study to GMT directly reprograms fibroblasts to iCMs, without first entering to early cardiac progenitor stage. In order to directly reprogram the fibroblasts into expandable iCPCs, it seems that one or more early stage specific TFs (e.g. MESP1/2, ISL1 or MIXL1) and epigenetic regulating factors (such as BAF60C) must be added to our provided novel transcriptional activators. Based on different analysis we performed in the present study, the addition of novel transcriptional activators and activation protein kinases seems to increase the efficiency and maturity of iCMs. Currently, the efficiency of iCM generation by different combinations of cardiac TFs is low (beating iCMs is less than 2%) and the maturity of iCMs is similar, but not identical, to neonatal CM. Unfortunately, the efficiency of iCM generation is routinely measured based on expression of a single cardiac factor such as cTnT or αMHC. While, the final efficiency must be measured via calculating the percentage of functional iCMs, which will be very low in comparison to cTnT+ cells or αMHC+ cells. The maturity of iCM derived from GMT-mediated direct cardiac reprogramming is not sufficient or suitable for cardiac cell-based therapy, as well as for other applications such as drug screening or toxicity assays. Interestingly, most of the novel transcriptional activators found in the present study such as MEF2A and PPARGC1A are related to CM maturation, metabolism and mitochondrial function. Scientists working in different fields (e.g. neural reprogramming, pluripotent stem cell reprogramming and diffentiation) are eagerly studying the roles of metabolism and mitochondria in cell fate specification and conversion (Prigione et al. 2014; Ghosh et al. 2015; Mlody and Prigione 2016). It seems that the addition of the novel transcriptional activators such as MEF2A and PPARGC1A to GMT and activation of protein kinases such as PKD1, PRKA, CAMK, PRKC and IGF1R significantly increase the maturity of iCMs that can be more similar to adult mature CMs. Moreover, the novel transcriptional activators found in the present study also might increase the maturity of pluripotent stem cell-derived CMs. it was recently shown that GMT are sufficient to activate the expression of cardiac specific genes and promote mouse embryonic stem cell-derived CM differentiation (Bai et al. 2015). Interestingly, the overexpression of cardiac specific TFs in pluripotent stems cells direct the cells to cardiac lineages and do not skip any stages of cardiac development. For example, transient expression of GATA4, TBX5, NKX2.5, and BAF60c (GTNB) for 5–7 days is necessary to activate the cardiogenesis through progenitor intermediates in a manner consistent with normal heart development (Dixon et al. 2011). It seems that during direct cardiac reprogramming (from fibroblasts) or forward cardiac programming (from pluripotent stem cells), joining the novel transcriptional activators to GMT and activation of novel protein kinases might increase the efficiency of CM generation, as well as increase the maturity of produced CMs. It would be very interesting to study the novel transcriptional activators and protein kinases in 1) direct reprogramming of human fibroblasts to iCMs; 2) in vivo cardiac reprogramming in mouse model and 3) direct cardiac reprogramming in 3D culture system. Direct cardiac reprogramming is currently far from cardiac cell-based therapy. Improving the efficiency and maturity is not sufficient, and for producing a large number of CMs lost after a myocardial infarction, 2D culture systems must move to 3D culture systems. Therefore, finding novel transcriptional activators and protein kinases increasing the efficiency and maturity of iCMs in 2D culture systems could pave the way for producing a large number of iCMs required in cell-based therapy.
Acknowledgements
This work was completed with financial supported by Systems Biology of Next Generation Company (SBNGC) [grant no. SBNGC13950231], Qom, Iran.
Compliance with ethical standards
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Footnotes
Mahmood Talkhabi and Ali Salari contributed equally to this work.
Contributor Information
Mahmood Talkhabi, Phone: +98 21 29902720, Email: m_talkhabi@sbu.ac.ir.
Ali Salari, Phone: +98 25 37740903, Email: asalari1365@gmail.com.
References
- Ali SR, Ranjbarvaziri S, Talkhabi M, Zhao P, Subat A, Hojjat A, Kamran P, Müller AM, Volz KS, Tang Z. Developmental heterogeneity of cardiac fibroblasts does not predict pathological proliferation and ActivationNovelty and significance. Circ Res. 2014;115(7):625–635. doi: 10.1161/CIRCRESAHA.115.303794. [DOI] [PubMed] [Google Scholar]
- Bai F, Lim CH, Jia J, Santostefano K, Simmons C, Kasahara H, Wu W, Terada N, Jin S. Directed differentiation of embryonic stem cells into cardiomyocytes by bacterial injection of defined transcription factors. Sci Rep. 2015;5:15014. doi: 10.1038/srep15014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SI, Posner JM, Ma’ayan A. Genes2Networks: connecting lists of gene symbols using mammalian protein interactions databases. BMC Bioinf. 2007;8(1):1. doi: 10.1186/1471-2105-8-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforou N, Chellappan M, Adler AF, Kirkton RD, Wu T, Addis RC, Bursac N, Leong KW. Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming. PLoS One. 2013;8(5):e63577. doi: 10.1371/journal.pone.0063577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JE, Dick E, Rajamohan D, Shakesheff KM, Denning C. Directed differentiation of human embryonic stem cells to interrogate the cardiac gene regulatory network. Mol Ther. 2011;19(9):1695–1703. doi: 10.1038/mt.2011.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essaghir A, Toffalini F, Knoops L, Kallin A, van Helden J, Demoulin J-B. Transcription factor regulation can be accurately predicted from the presence of target gene signatures in microarray gene expression data. Nucleic Acids Res. 2010;38(11):e120–e120. doi: 10.1093/nar/gkq149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen EP, Snyder CM, Wilson M, Desjardins D, Naya FJ. The Mef2A transcription factor coordinately regulates a costamere gene program in cardiac muscle. J Biol Chem. 2011;286(34):29644–29653. doi: 10.1074/jbc.M111.268094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festuccia N, Osorno R, Halbritter F, Karwacki-Neisius V, Navarro P, Colby D, Wong F, Yates A, Tomlinson SR, Chambers I. Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell Stem Cell. 2012;11(4):477–490. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonoudi H, Ansari H, Abbasalizadeh S, Larijani MR, Kiani S, Hashemizadeh S, Zarchi AS, Bosman A, Blue GM, Pahlavan S. A universal and robust integrated platform for the scalable production of human cardiomyocytes from pluripotent stem cells. Stem Cells Transl Med. 2015;4(12):1482–1494. doi: 10.5966/sctm.2014-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh JC, Siegelin MD, Vaira V, Faversani A, Tavecchio M, Chae YC, Lisanti S, Rampini P, Giroda M, Caino MC. Adaptive mitochondrial reprogramming and resistance to PI3K therapy. J Natl Cancer Inst. 2015;107(3):dju502. doi: 10.1093/jnci/dju502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffart S, von Kleist-Retzow J-C, Wiesner RJ. Regulation of mitochondrial proliferation in the heart: power-plant failure contributes to cardiac failure in hypertrophy. Cardiovasc Res. 2004;64(2):198–207. doi: 10.1016/j.cardiores.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Hirai H, Katoku-Kikyo N, Keirstead SA, Kikyo N. Accelerated direct reprogramming of fibroblasts into cardiomyocyte-like cells with the MyoD transactivation domain. Cardiovasc Res. 2013 doi: 10.1093/cvr/cvt167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Fu J-D, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142(3):375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islas JF, Liu Y, Weng K-C, Robertson MJ, Zhang S, Prejusa A, Harger J, Tikhomirova D, Chopra M, Iyer D. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc Natl Acad Sci. 2012;109(32):13016–13021. doi: 10.1073/pnas.1120299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob P, Landmesser U. Current status of cell-based therapy for heart failure. Curr Heart Fail Rep. 2013;10(2):165–176. doi: 10.1007/s11897-013-0134-z. [DOI] [PubMed] [Google Scholar]
- Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, Zhang Z, Rosenberg P, Mirotsou M, Dzau VJ. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to CardiomyocytesNovelty and significance. Circ Res. 2012;110(11):1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann A, Ma'ayan A. KEA: kinase enrichment analysis. Bioinformatics. 2009;25(5):684–686. doi: 10.1093/bioinformatics/btp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma'ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26(19):2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalit PA, Salick MR, Nelson DO, Squirrell JM, Shafer CM, Patel NG, Saeed I, Schmuck EG, Markandeya YS, Wong R. Lineage reprogramming of fibroblasts into proliferative induced cardiac progenitor cells by defined factors. Cell Stem Cell. 2016;18(3):354–367. doi: 10.1016/j.stem.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski SL, Janardhan HP, Smee KM, Bachman M, Sun Z, Lazar MA, Trivedi CM. Histone deacetylase 3 modulates Tbx5 activity to regulate early cardiogenesis. Hum Mol Genet. 2014;23(14):3801–3809. doi: 10.1093/hmg/ddu093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang C-Y, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature. 2002;418(6899):797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Liu L, Lei I, Karatas H, Li Y, Wang L, Gnatovskiy L, Dou Y, Wang S, Qian L, Wang Z. Targeting Mll1 H3K4 methyltransferase activity to guide cardiac lineage specific reprogramming of fibroblasts. Cell Discovery. 2016;2:16036. doi: 10.1038/celldisc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Sugimoto T, Diamanti E, Joshi A, Hannah R, Ohtsuka S, Göttgens B, Niwa H, Smith A. Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell. 2012;11(4):491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlody B, Prigione A. A glycolytic solution for pluripotent stem cells. Cell Stem Cell. 2016;19(4):419–420. doi: 10.1016/j.stem.2016.09.005. [DOI] [PubMed] [Google Scholar]
- Mohamed, T M, Stone NR, Berry EC, Radzinsky E, Huang Y, Pratt K, Ang Y-S, Yu P, Wang H Tang S (2016). Chemical enhancement of in vitro and in vivo direct cardiac reprogramming. Circulation: Circulationaha. 116.024692 [DOI] [PMC free article] [PubMed]
- Muraoka N, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Isomi M, Nakashima H, Akiyama M, Wada R, Inagawa K. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014;33(14):1565–1581. doi: 10.15252/embj.201387605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y-J, Song K, Luo X, Daniel E, Lambeth K, West K, Hill JA, DiMaio JM, Baker LA, Bassel-Duby R. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci. 2013;110(14):5588–5593. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige SL, Plonowska K, Xu A, Wu SM. Molecular regulation of cardiomyocyte differentiation. Circ Res. 2015;116(2):341–353. doi: 10.1161/CIRCRESAHA.116.302752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigione A, Rohwer N, Hoffmann S, Mlody B, Drews K, Bukowiecki R, Blümlein K, Wanker EE, Ralser M, Cramer T. HIF1α modulates cell fate reprogramming through early glycolytic shift and upregulation of PDK1–3 and PKM2. Stem Cells. 2014;32(2):364–376. doi: 10.1002/stem.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protze S, Khattak S, Poulet C, Lindemann D, Tanaka EM, Ravens U. A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J Mol Cell Cardiol. 2012;53(3):323–332. doi: 10.1016/j.yjmcc.2012.04.010. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O'Malley B, Spiegelman BM. Activation of PPARγ coactivator-1 through transcription factor docking. Science. 1999;286(5443):1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu J-d, Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485(7400):593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosselló RA, Chen C-C, Dai R, Howard JT, Hochgeschwender U, Jarvis ED. Mammalian genes induce partially reprogrammed pluripotent stem cells in non-mammalian vertebrate and invertebrate species. elife. 2013;2:e00036. doi: 10.7554/eLife.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe GC, Jiang A, Arany Z. PGC-1 coactivators in cardiac development and disease. Circ Res. 2010;107(7):825–838. doi: 10.1161/CIRCRESAHA.110.223818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadahiro T, Yamanaka S, Ieda M. Direct cardiac reprogramming progress and challenges in basic Biology and clinical applications. Circ Res. 2015;116(8):1378–1391. doi: 10.1161/CIRCRESAHA.116.305374. [DOI] [PubMed] [Google Scholar]
- Schlesinger J, Schueler M, Grunert M, Fischer JJ, Zhang Q, Krueger T, Lange M, Tönjes M, Dunkel I, Sperling SR. The cardiac transcription network modulated by Gata4, Mef2a, Nkx2. 5, Srf, histone modifications, and microRNAs. PLoS Genet. 2011;7(2):e1001313. doi: 10.1371/journal.pgen.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Nam Y-J, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485(7400):599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Später D, Hansson EM, Zangi L, Chien KR. How to make a cardiomyocyte. Development. 2014;141(23):4418–4431. doi: 10.1242/dev.091538. [DOI] [PubMed] [Google Scholar]
- Talkhabi M, Aghdami N, Baharvand H. Human cardiomyocyte generation from pluripotent stem cells: a state-of-art. Life Sci. 2016;145:98–113. doi: 10.1016/j.lfs.2015.12.023. [DOI] [PubMed] [Google Scholar]
- Talkhabi M, Pahlavan S, Aghdami N, Baharvand H. Ascorbic acid promotes the direct conversion of mouse fibroblasts into beating cardiomyocytes. Biochem Biophys Res Commun. 2015;463(4):699–705. doi: 10.1016/j.bbrc.2015.05.127. [DOI] [PubMed] [Google Scholar]
- Wang X, Hodgkinson CP, Lu K, Payne AJ, Pratt RE, Dzau VJ (2016a) Selenium augments microRNA directed reprogramming of fibroblasts to cardiomyocytes via Nanog. Sci Rep 6. doi:10.1038/srep2301 [DOI] [PMC free article] [PubMed]
- Wang Y, Shi S, Liu H, Meng L. Hypoxia enhances direct reprogramming of mouse fibroblasts to cardiomyocyte-like cells. Cellular Reprogramming (Formerly" Cloning and Stem Cells") 2016;18(1):1–7. doi: 10.1089/cell.2015.0051. [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98(1):115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Xu XQ, Soo SY, Sun W, Zweigerdt R. Global expression profile of highly enriched cardiomyocytes derived from human embryonic stem cells. Stem Cells. 2009;27(9):2163–2174. doi: 10.1002/stem.166. [DOI] [PubMed] [Google Scholar]
- Yamakawa H, Muraoka N, Miyamoto K, Sadahiro T, Isomi M, Haginiwa S, Kojima H, Umei T, Akiyama M, Kuishi Y. Fibroblast growth factors and vascular endothelial growth factor promote cardiac reprogramming under defined conditions. Stem Cell Rep. 2015;5(6):1128–1142. doi: 10.1016/j.stemcr.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanazume T, Hasegawa K, Morimoto T, Kawamura T, Wada H, Matsumori A, Kawase Y, Hirai M, Kita T. Cardiac p300 is involved in myocyte growth with decompensated heart failure. Mol Cell Biol. 2003;23(10):3593–3606. doi: 10.1128/MCB.23.10.3593-3606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Dickson ME, Kim MS, Bassel-Duby R, Olson EN. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc Natl Acad Sci. 2015;112(38):11864–11869. doi: 10.1073/pnas.1516237112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Li L, Zhao B, Guan K-L. The hippo pathway in heart development, regeneration, and diseases. Circ Res. 2015;116(8):1431–1447. doi: 10.1161/CIRCRESAHA.116.303311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wang L, Vaseghi HR, Liu Z, Lu R, Alimohamadi S, Yin C, Fu J-D, Wang GG, Liu J. Bmi1 is a key epigenetic barrier to direct cardiac reprogramming. Cell Stem Cell. 2016;18(3):382–395. doi: 10.1016/j.stem.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]