Abstract
While Transforming growth factor-βs (Tgf-βs) have been known to play an important role in liver fibrosis through an activation of Hepatic Stellate Cells (HSC), their fibrotic role on hepatocytes in liver damage has not been addressed thoroughly. To shed more light on the hepatocyte-specific role of Tgf-β signaling during liver fibrosis, we generated transgenic mice expressing constitutively active Tgf-β type I receptor Alk5 under the control of albumin promoter. Uninjured mice with increased Tgf-β/Alk5 signaling in hepatocytes (caAlk5/Alb-Cre mice) did not show characteristics related to hepatocyte death, fibrosis and inflammation. When subjected to thioacetamide (TAA) treatment, caAlk5/Alb-Cre mice exhibited more severe liver injury, when compared to control littermates. After TAA administration for 12 weeks, an increase in pathological changes was evident in caAlk5/Alb-Cre livers, with higher number of infiltrating cells in the portal and periportal area. Immunohistochemistry for F4/80, myeloperoxidase and CD3 showed that there was an increased accumulation of macrophages, neutrophils and T-lymphocytes, respectively, in caAlk5/Alb-Cre livers. Coincidently, we observed an exacerbated liver damage as seen by increases in serum aminotransferase level and number of apoptotic hepatocytes in caAlk5/Alb-Cre mice. Sirius staining of collagen demonstrated that the fibrotic response was worsened in caAlk5/Alb-Cre mice. The enhanced fibrosis in mutant livers was associated with marked production of α-SMA-positive myofibroblast. Hepatic expression of genes indicative of HSC activation was greater in caAlk5/Alb-Cre mice. In conclusion, our data indicated that elevation of Tgf-β signaling via Alk5 in hepatocytes is not sufficient to induce liver pathology but plays an important role in amplifying TAA-induced liver damage.
Keywords: Pathology, Toxicology
1. Introduction
Hepatic injury contributes to the pathogenesis of several liver diseases (Mengshol et al., 2007; Natori et al., 2001). Following an injury, Hepatic cells that include Hepatocytes, Hepatic Stellate Cells (HSCs), Kupffer cells, and endothelial cells communicate with each other to trigger the healing response which leads to the tissue repair. This cell-to-cell interaction is critical for promoting liver regeneration and maintaining organ function (Diehl, 2012; Forbes and Newsome, 2016; Friedman et al., 2013). The communication among cells can be achieved by various types of tropic factor, hormone, extracellular matrix (ECM) protein, proteolytic enzyme and extracellular nucleic acid (de Oliveira et al., 2017; Seifert et al., 2015; Tian et al., 2016; Zheng et al., 2016). Better understanding of how these molecules govern cell behaviors will lead to a new strategy to improve organ repair and reduce the risk of developing liver disease in individual exposed to hepatotoxin.
Transforming growth factor-βs (Tgf-βs) are widely distributed, multifunctional cytokines that are released from compromised epithelial cells and stimulated immune cells during organ injury (Dooley and ten Dijke, 2012; Liu et al., 2006; Meng et al., 2016). Tgf-β ligands exert their effects via a family of transmembrane serine/threonine kinase receptors, Tgf-β type II receptor TβRII and Tgf-β type I receptor TβRI (Massague, 2012; Shi and Massague, 2003; Weiss and Attisano, 2013). Upon ligand binding, the TβRII activates the kinase catalytic domain of the TβRI (Alk5 or Alk1), leading to phosphorylation of Smad proteins. While the Alk1 receptor phosphorylates Smad1, Smad5 and Smad8, the Alk5 receptor causes phosphorylation of Smad2 and Smad3 (Massague and Chen, 2000; ten Dijke and Hill, 2004). The phosphorylated Smad proteins subsequently form a complex with Smad4 and translocate to nucleus to regulate transcription of specific genes (Macias et al., 2015).
In liver, Tgf-βs are known as a key HSC activator that promotes fibrosis after liver injury (Dooley and ten Dijke, 2012; Fabregat et al., 2016). Accumulating evidence from studies of genetically engineered mouse models revealed that Tgf-βs also act on hepatocytes to amplify hepatic fibrogenesis. Dooley and coworkers have reported that blunting Tgf-β signaling in hepatocytes with antagonistic Smad7 attenuated carbon tetrachloride (CCl4)-induced fibrosis (Dooley et al., 2008). In line with this observation, Yang et al. have shown that Tgf-β signaling in hepatocytes was essential for the development of spontaneous liver fibrosis in Tak1 knockout mice (Yang et al., 2013). However, this idea is currently challenged by results from recent work employing mutant mice lacking TβRII in hepatocytes. Mu et al. demonstrated that deletion of TβRII in hepatocytes did not affect the development of liver fibrosis in several mouse models of liver injury (Mu et al., 2016). In this study, we investigated the role of Tgf-β signaling via Alk5 in hepatocytes during thioacetamide(TAA)-induced injury in mice.
2. Materials and methods
2.1. Experimental mice
Alb-Cre mice were purchased from Jackson laboratories (Bar Harbor, ME). Mice expressing the constitutively active (ca) Alk5 transgene (caAlk5 mice) have been previously described (Bartholin et al., 2008). These mouse lines were crossed to generate mice with following genotypes: CaAlk5+/Alb-Cre+, CaAlk5+/Alb-Cre−, CaAlk5−/Alb-Cre+ and CaAlk5−/Alb-Cre− on C57Bl/6 background. CaAlk5+/Alb-Cre+ female mice were used as mutants in our experiments, while CaAlk5+/Alb-Cre−, CaAlk5−/Alb-Cre+ and CaAlk5−/Alb-Cre− were considered as control mice. Genotype of animals was identified by PCR analysis (VioTwinPack PCR kit, Viogene Biotech) on DNA extracted from mouse tail using DirectPCR lysis reagent (Viagen). The following primers were used for PCR reaction: caAlk5, 5′-AGAAAGAACAATCAAGGGTCC-3’ and TTGTGAACAGAAGTTAAGGC-3′; Cre, 5′-AGGTGTAGAGAAGGCACTTAGC-3′ and 5′-CTAATCGCCATCTTCCAG CAGG-3′. To confirm the Cre-mediated removal of stop sequence from caAlk5 gene, PCR analysis of genomic DNA extracted from mouse liver were performed using the following primers (Vincent et al., 2010): 5′-GACAGCAACGGAATTCGATATC-3′ and 5′-CTCTAGAGCCTCTGCTAACC-3′. The caAlk5 mRNA expression was demonstrated by quantitative RT-PCR performed on total RNA from livers of control and caAlk5/Alb-Cre mice. RT-PCR primer set for caAlk5 was previously described (Bartholin et al., 2008).
2.2. Induction liver injury
The protocol for thioacetamide-induced liver damage was adapted from those that previously described (Hayashi and Sakai, 2011). In brief, adult (8–12 weeks) mice were injected intraperitoneally with TAA (Sigma) at 100 μg/g body weight, three times a week for one week followed by one-week treatment at 150 μg/g body weight. Starting on the third week, mice received three TAA injections (200 μg/g body weight) per week for 4 weeks or 10 weeks. A the end of the treatment, liver tissues from each lobe were fixed in Bouin solution (Bio-Optica) or snap frozen in liquid nitrogen and stored at −80 °C until analysis. All animal studies were conducted with the approval of Animal Ethics Committee on Use and Care of animals at Faculty of Science, Mahidol University, Thailand (Protocol number 257).
2.3. Histochemistry and fibrosis quantification
Liver sections were cut to 6 μm and stained with hematoxylin-eosin (Bio-Optica). To access collagen deposition, sections were stained with Sirius Red (saturated picric acid containing 0.1% direct red and 0.1% Fast green FCF, Sigma). Five Sirius Red-stained slides per mouse were generated, with six 10x pictures taken randomly per slide for a total of 30 images per mouse. The images were analyzed for Sirius Red-positive area using ImageJ program. Five mice per experimental group were used for collagen quantification.
2.4. Immunoperoxidase and immunofluorescence staining
For immunohistochemistry, paraffin-embedded, bouin-fixed sections were deparaffinized, rehydrated and boiled in citrate buffer for 15 min. After protein blocking step, sections with antibodies for p-smad2/3 (SC-11769R, Santa Cruz Biotech.), myeloperoxidase (PA5-16672, Thermo Fisher Sci.), CD3 (A0452, Dako) and cleaved caspase-3 (#9664, Cell signaling Tech.) were applied to sections for 2 h. Slides were rinsed in PBS and incubated in either biotinylated donkey anti-goat IgG (#705-065-147, Jackson Immunoresearch Labs.) or biotinylated donkey anti-rabbit IgG (#711-065-152, Jackson Immunoresearch Labs.). For F4/80 and α–SMA immunostaining, Frozen-sections were stained with anti-α–SMA (M085, Dako) and F4/80 antibody (MCA497G, AbD Serotec) followed by incubation with Alexa fluor 594 goat anti-mouse IgG2a (invitrogen) or biotinylated donkey anti-rat IgG (#712-065-153, Jackson Immunoresearch Labs). Immunoreactive cells were scored in ten high-power fields (20x) per mouse. At least five mice from control and mutant groups were analyzed in each immunostaining.
2.5. RT-PCR analysis
RNA was prepared from snap-frozen tissue using RNeasy mini kit (Qiagen). QuantiNova RT kit (Qiagen) and Omniscript RT kit (Qiagen) was used to synthesize cDNA from 1 μg of RNA. For RT-PCR reactions, cDNA samples were amplified with Hotstar Taq DNA polymerase (Qiagen). Quantitative RT-PCRs were carried out with Applied Biosystems real-time PCR 7500 using iTaq Universal SYBR green supermix (Bio-Rad Lab). Data were represented as the relative expression of gene after normalizing to β-actin. All primer pairs have been previously described (Bartholin et al., 2008; Basciani et al., 2004; Bopp et al., 2013; Puche et al., 2013; Stewart et al., 2014; Xiang et al., 2012; Yang et al., 2013).
2.6. Statistical analysis
All experiments were performed with five animals per experimental group and representative data were presented as means ± SD. Comparisons were performed by ANOVA test and differences were considered significant if P < 0.05.
3. Results
3.1. Alb-Cre-mediated expression of caAlk5 enhanced Tgf-β signaling in hepatocytes
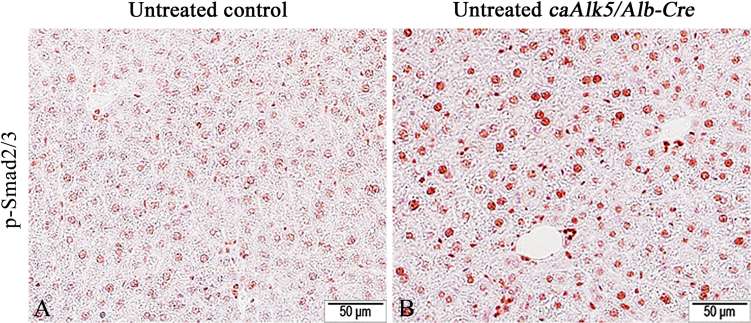
To study the role of Tgf-β signaling to hepatocytes during liver injury, we generated caAlk5/Alb-Cre mice expressing constitutively active (ca) Alk5 in hepatocytes by crossing Alb-Cre mice with caAlk5 mice. We verified that the Alb-Cre transgene expression was able to induce Cre-mediated removal of stop sequence upstream of caAlk5 coding sequence in liver parenchyma by using PCR analysis with pCAG/pTβRI primers (Vincent et al., 2010) (Fig. 1A). The DNA of control mice displayed a 1.93-kb-PCR product amplified from inactive caAlk5 transgene, while DNA from the mutant mice showed a single band of 350-bp-PCR product representing a product of recombined caAlk5 transgene (Fig. 1B). Quantitative RT-PCR experiments revealed that caAlk5 mRNA was highly expressed in the liver caAlk5/Alb-Cre mice (Fig. 1C). To demonstrate that expression of caAlk5 could lead to the increased Tgf-β signaling in mutant livers, we performed immunohistochemistry using p-Smad2/3 antibody. The results showed that a higher number of p-Smad2/3 positive hepatocytes were found in caAlk5/Alb-Cre livers compared to those in control (Fig. 2A and B). H&E staining demonstrated comparable hepatic architecture between control and caAlk5/Alb-Cre mice (Fig. 3A and C). No sign of liver damage and fibrosis was observed in caAlk5/Alb-Cre mice and control littermates (Fig. 3B and D). These findings indicated that expression of caAlk5 in hepatocytes is not sufficient to induced liver damage in mice.
Fig. 1.
Analysis of caAlk5 transgenes in liver of control and caAlk5/Alb-Cre mice. (A) PCR strategy to assess the DNA recombination of caAlk5 transgene. (B) Genomic DNA extracted from control and caAlk5/Alb-Cre livers were used for PCR analysis with pCAG and pTβRI primers. (C) Detection of caAlk5 mRNA by quantitative RT-PCR performed on total RNA from livers of control and caAlk5/Alb-Cre mice.
Fig. 2.
Representative images of immunohistochemical staining for phospho-Smad2 and 3 (pSmad2/3). While pSmad2/3 immunoreactivity was barely detected in control liver (A), nucleus of hepatocytes were clearly stained with pSmad2/3 antibody in caAlk5/Alb-Cre livers (B). Immunochemistry with NovaRed substrate, counterstained with hematoxylin.
Fig. 3.
Histochemical analysis of caAlk5/Alb-Cre mice. Hematoxylin and Eosin-stained liver sections obtained from control (A) and caAlk5/Alb-Cre mice (B). Sirius Red/Fast green-stained liver sections obtained from control (C) and caAlk5/Alb-Cre mice (D).
3.2. Exposure to TAA led to increased pathological changes in caAlk5/Alb-Cre livers
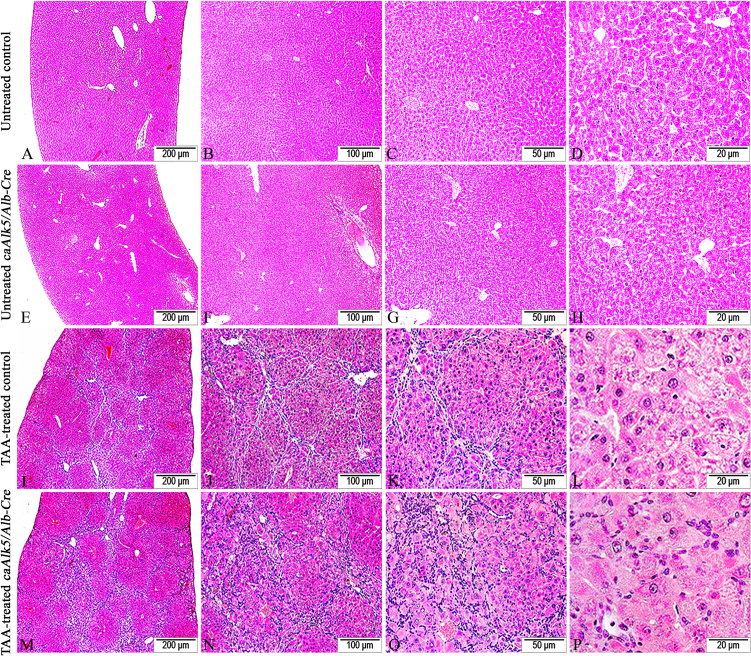
Control and caAlk5/Alb-Cre mice were administrated intraperitoneally with TAA for 12 weeks to examine pathological response following expression of caAlk5 in hepatocytes. While TAA-treated control mice showed the presence of hematoxylin-positive cells (likely inflammatory cells) in portal and periportal area (Fig. 4I-L), caAlk5/Alb-Cre mutants displayed a notable increase in number of these cells in their livers (Fig. 4M-P). In addition, severe swelling of hepatocytes was noticeable in livers of caAlk5/Alb-Cre mice (Fig. 4P), whereas less swollen hepatocytes were noted in liver of control mice (Fig. 4L). These data suggested that hepatocyte degeneration was more pronounced in mutants than those in control mice after TAA treatment.
Fig. 4.
Histological changes of TAA-induced liver injury in control and caAlk5/Alb-Cre mice. Following TAA exposure for 12 weeks, control liver developed fibrotic lesions that connected among centrilobular- centrilobular, centrilobular-portal and portal-portal area (I-L).The fibrotic lesions were populated by a number of small hematoxylin-positive stromal cells. While fibrous bridgings were also seen in liver of TAA-treated caAlk5/Alb-Cre mice, hematoxylin-positive cells were more marked along fibrotic lesions (M-P).
3.3. caAlk5/Alb-Cre mice showed increased hepatic immune cell infiltration following TAA exposure
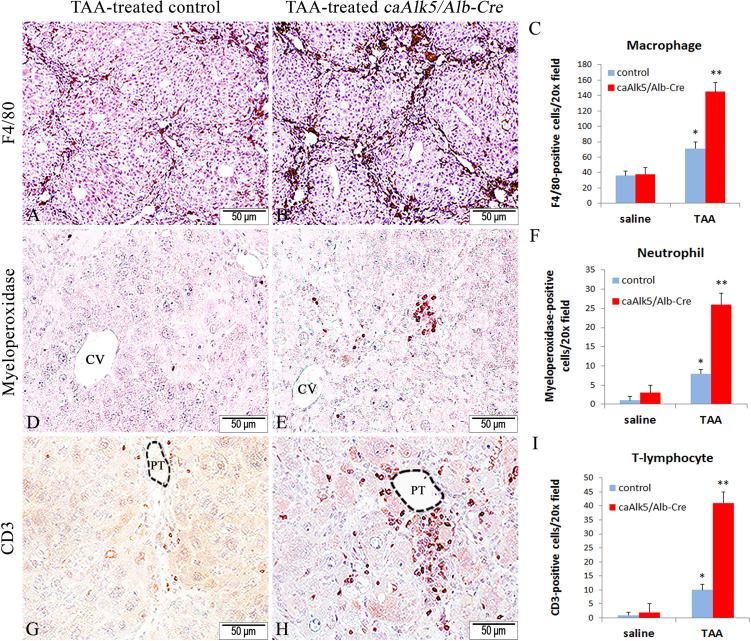
Because TAA treatment lead to an increased number of cells that exhibited inflammatory cell appearance (Fig. 4), we next performed immunostaining to determine whether which type of inflammatory cells are increased in mutant liver after TAA exposure. F4/80 immunohistochemistry revealed an increased number of Kupffer cells/macrophages in liver of caAlk5/Alb-Cre mice treated with TAA (Fig. 5A, B and C). Similarly, immunochemistry for neutrophil marker myeloperoxidase showed that more neutrophils were accumulated in caAlk5/Alb-Cre liver (Fig. 5D, E and F). In addition, we assessed T-lymphocyte infiltration into liver by anti-CD3 immunostaining. While some CD3 positivity can be detected in control livers, caAlk5/Alb-Cre mutants displayed an increased amount of CD3-positive cells surrounding hepatic blood vessel (Fig. 5G, H and I). Our result suggested that enhanced Tgf-β signaling in hepatocytes promoted recruitment of macrophages, neutrophils and T cell into liver after TAA treatment.
Fig. 5.
Immunohistochemistry for macrophages: F4/80 (A and B), for neutrophils: myeoperoxidase (D and E) and for T lymphocytes: CD3 (G and H) in livers of control (A, D, G) and caAlk5/Alb-Cre mice (B, E, H) treated with TAA for 12 weeks. Quantification of F4/80-(C), myeloperoxidase-(F), and CD3-(I) positive cells in livers from saline-treated and TAA-treated mice. Data represent means ± SD, n = 5 mice per experimental group, ten 20x fields per mouse. There is a significant difference after TAA treatment when compared with genotype-specific saline control (*P < 0.05). There is a significant difference when compared with genotype-specific saline control and between genotypes after TAA treatment (**P < 0.05). Immunochemistry with NovaRed substrate, counterstained with hematoxylin. CV denotes the central vein. PT denotes the portal tract.
3.4. Expression of caAlk5 was associated with reduced liver function after TAA exposure
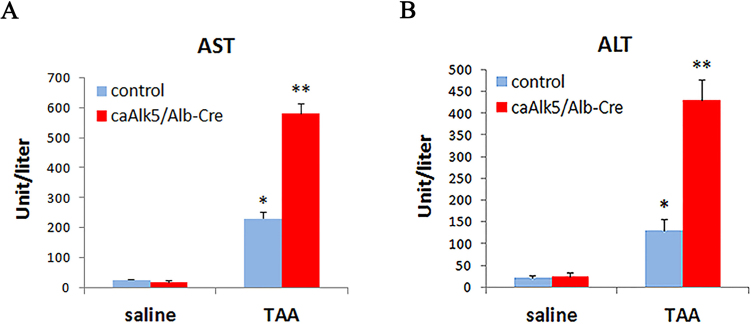
To address the level of tissue injury induced by TAA treatment in control and caAlk5/Alb-Cre mice, serum aminotransferase was quantified. Our results showed that serum Aspartate transaminase (AST) and Alanine transaminase (ALT) were higher in TAA-treated caAlk5/Alb-Cre mice than those in TAA-treated control (Fig. 6A-B). Since AST/ALT is also expressed in cells of other organs, such as, heart, skeletal muscle and kidney, damage to these organs can also lead to increased level of serum AST and ALT. To validate whether TAA-induced liver injury is increased in caAlk5/Alb-Cre mice, we performed immunohistochemistry for cleaved-caspase-3 to detect apoptotic cells in liver of TAA-treated mice. The data showed that a higher number of apoptotic hepatocytes were found in livers of caAlk5/Alb-Cre mice compared to those of control (Fig. 7). These findings suggested that caAlk5/Alb-Cre mice exhibited more severity of hepatic damage than control mice after TAA treatment.
Fig. 6.
Serum level of aspartate transaminase (AST) and alanine transaminase (ALT) in control and caAlk5/Alb-Cre mice treated with saline or TAA for 12 weeks. AST (A) and ALT (B) level was higher in caAlk5/Alb-Cre mice compared to those of control mice. Data are means ± SD of n = 5 per experimental group. There is significant difference compared with saline control group (*P < 0.05). There is a significant difference when compared with genotype-specific saline control and between genotypes after TAA treatment (**P < 0.05).
Fig. 7.
Immunohistochemistry for cleaved caspase 3 (CC3) in liver sections from TAA-treated control and caAlk5/Alb-Cre mice. While CC3 positivity was infrequently seen in liver parenchyma of control mice (A and B), increased number of CC3-positive cells were observed in centrilobular area of caAlk5/Alb-Cre liver (C-D). (E) Quantification of CC#-positive hepatocytes at 12 weeks of saline or TAA treatment. There is a significant difference after TAA treatment when compared with genotype-specific saline control (*P < 0.05). There is a significant difference when compared with genotype-specific saline control and between genotypes after TAA treatment (**P < 0.05). CV denotes the central vein.
3.5. caAlk5/Alb-Cre mutants displayed severe liver fibrosis induced by TAA treatment
To determine whether expression of caAlk5 in hepatocytes could affect TAA-induced fibrogenesis, Sirius red/fast green staining was used to assess the degree of liver fibrosis. After TAA exposure for 6 weeks, sparse collagen fibers were detected in livers of control mice (Fig. 8A-B). The higher quantity of collagen was found in caAlk5/Alb-Cre mice compared with control mice (Fig. 8C, D). TAA treatment for 12 weeks resulted in widened portal area forming fibrous septa throughout the liver parenchyma of control mice (Fig. 8E and F). TAA-treated caAlk5/Alb-Cre mice demonstrated increased portal fibrosis, bridging fibrosis and increased scar thickness than in TAA-treated control mice (Fig. 8G and H). Striking collagen deposition was observed in the periportal area and fibrous septa of caAlk5/Alb-Cre mice (Fig. 8H), whereas less Sirius Red-stained collagen fibers were detected in control livers (Fig. 8F). Morphometric analysis of Sirius red-stained sections showed an increase in collagen deposition in caAlk5/Alb-Cre livers after 6-week and 12-week TAA exposure (Fig. 8I).
Fig. 8.
Hepatic fibrosis after TAA exposure. After 6-week (A-D) and 12-week (E-H) treatment, mice were sacrificed and their livers were analyzed with Sirius red staining. Images are representative of n = 5 mice per experimental group and 10 fields per mouse. (I) Morphometric analysis of Sirius red density from mice exposed to saline or TAA. Data are means ± SD of n = 5 per experimental group. *P < 0.05, significantly different compared with saline control for the same genotype. There is a significant difference after TAA treatment when compared with genotype-specific saline control and between genotypes at the same time point (**P < 0.05).
3.6. Increased production of myofibroblast caAlk5/Alb-Cre mutants
Collagen and other extracellular matrix proteins in fibrotic livers are mainly produced from myofibroblast. To determine an amount of myofibroblast in livers of TAA-treated mice, we performed anti-α-SMA Immunohistochemistry using α-SMA antibody (Fig. 9). The result showed that a notable increase of myofibroblasts was observed in liver of TAA-treated caAlk5/Alb-Cre mice (Fig. 9E). This suggested an involvement of Tgf-β signaling in hepatocytes during myofibroblast production. It is possible that enhanced Tgf-β signaling could stimulate the release of fibrogenic factors from hepatocytes leading to myofibroblast differentiation via HSC activation. To test this idea, quantitative RT-PCR analysis was performed to measure mRNA expression of well-known fibrogenic factors. As shown in Fig. 10, caAlk5/Alb-Cre displayed an increased expression of these factors such as PDGF-A, PDGF-B or Tgf-β in their livers compared to those in control livers (Fig. 10A-C). Of interest, a hepatocyte-derived CTGF had been implicated in Tgf-β-mediated liver fibrosis; its mRNA expression in mutant liver was also increased (Fig. 10D).
Fig. 9.
Immunohistochemistry for myofibroblast: α-smooth muscle actin (α-SMA) in liver sections from TAA-treated control and caAlk5/Alb-Cre mice. While α-SMA-positive cells were seen in the interface of portal area and liver parenchyma (A and B), more immunoreactivity for α-SMA were found in the fibrous septa of caAlk5/Alb-Cre livers (C and D). (E) Quantification of α-SMA-positive cells in liver sections from TAA-treated control and caAlk5/Alb-Cre mice. Data are means ± SD of n = 5 per experimental group. *P < 0.05, significantly different compared with saline control for the same genotype. There is a significant difference when compared with genotype-specific saline control and between genotypes after TAA treatment (**P < 0.05). CV denotes the central vein.
Fig. 10.
Hepatic expression of fibrogenic factors after chronic TAA exposure. Relative mRNA expression of PDGF-A (A), PDGF-B (B), Tgf-β (C) and CTGF (D) was determined quantitative RT-PCR. Expression level was normalized to β-actin. Five mice from each experimental group were used for the analysis. There is a significant change in gene expression when compared with genotype-specific saline control (*P < 0.05). There is a significant difference when compared with genotype-specific saline control and between genotypes after TAA treatment (**P < 0.05).
4. Discussion
The relationship between Tgf-β signaling and hepatic cell damage has been previously described in many situations (Dooley and ten Dijke, 2012; Liu et al., 2006). Tgf-β was found to induce cell death by stimulating expression of proteins in apoptotic machinery (Jang et al., 2002; Perlman et al., 2001; Ramesh et al., 2008). Overexpression of Tgf-β ligand was shown to enhance liver injury in mouse model of alcoholic liver disease (Preisegger et al., 1999). Inhibition of Tgf-β signaling by over expression of antagonistic Smad7 protected mice against CCl4–induced liver injury (Dooley et al., 2008), whereas abrogation of Smad7 exacerbated CCl4-dependent liver injury (Hamzavi et al., 2008). These findings suggested that Tgf-β signaling promotes hepatic cell injury following experimentally induced liver damage. In this study, we characterized caAlk5/Alb-Cre mice to determine whether elevation of Tgf-β/Alk5 signaling in hepatocytes could lead to spontaneous liver damage. We did not find any histological and biochemical feature of hepatic cell death in the transgenic mice. This observation indicated that activation of Alk5-mediated Tgf-β signaling was not sufficient to cause hepatocellular damage in mice.
Although expression of caAlk5 in hepatocytes did not seem to be enough to cause liver injury, it made liver more susceptible to a toxicological insult. Following TAA treatment, caAlk5/Alb-Cre mice showed a higher level of serum liver enzyme when compared to control littermates. In accordance with this data, a number of apoptotic cells were higher in the liver of transgenic mice. Our data would support the notion that, while augmentation of Tgf-β signaling in hepatocytes does not affect the integrity of cells, hepatocytes with increased Tgf-β/Alk5 signaling are more prone to chemical-induced cell damage. This reflects cooperation between Tgf-β/Alk5 signaling and other signaling pathways in amplifying liver damage caused by toxic stimuli. Further studies are necessary to identify a liver-damaging factor that may act synergistically with Tgf-β to promote liver injury.
In the past years, some efforts have been made to identify the role of hepatocyte-dependent Tgf-β signaling in the fibrotic liver response (Dooley et al., 2008; Mu et al., 2016; Yang et al., 2013). Yang et al. reported that ablation of TβRII in hepatocytes decreased spontaneous liver fibrosis developed in Tak knockout mice (Yang et al., 2013). This suggested that TβRII-dependent signaling in hepatocytes was essential for the progression of hepatic fibrosis. A recent study by Mu and coworkers, however demonstrated that the deletion of TβRII in hepatocytes did not affect the development of CCl4-induced fibrosis (Mu et al., 2016), indicating that epithelial Tgf-β signaling did not contribute to liver fibrogenesis. Our results showed that expression of caAlk5 in hepatocytes led to increased myofibroblasts production and enhanced interstitial collagen accumulation in liver of caAlk5/Alb-Cre mice treated with TAA. Concomitantly, there was increased mRNA expression of potent fibrogenic factors such as PDGF-A, PDGF-B and Tgf-β in mutant liver compared to those in control. Of interest, connective tissue growth factor (CTGF) that was known to be regulated by Tgf-β signaling (Gressner et al., 2007) was also increased in caAlk5/Alb-Cre livers. Since previous report indicating that Alk5-dependent Tgf-β signaling mediated the up-regulation of CTGF expression in hepatocytes (Weng et al., 2007), we speculate that increased expression of CTGF in caAlk5/Alb-Cre liver might be, at least, partially responsible for the enhancement of fibrogenesis. While the definite mechanism of how Tgf-β signaling in hepatocytes amplifies fibrosis needs further investigation, results from our study support the view of hepatocyte as an important player in Alk5-mediated fibrogenic response following liver injury.
Declarations
Author contribution statement
Wanthita Kongphat: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data.
Arnon Pudgerd: Performed the experiments.
Somyoth Sridurongrit: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This study was supported by TRF grant number TRG5780070 (From Thailand Research Fund in collaboration with Mahidol University), and by funds for new principal investigators from Mahidol University. This research is also partially supported by Central Instrument Facility (CIF), Faculty of Science, Mahidol University.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We would like to thank Dr. Laurent Bartholin (INSERM, University of Lyon, France) for caAlk5 mice.
References
- Bartholin L. Generation of mice with conditionally activated transforming growth factor beta signaling through the TbetaRI/ALK5 receptor. Genesis. 2008;46:724–731. doi: 10.1002/dvg.20425. [DOI] [PubMed] [Google Scholar]
- Basciani S. Expression of platelet-derived growth factor (PDGF) in the epididymis and analysis of the epididymal development in PDGF-A PDGF-B, and PDGF receptor beta deficient mice. Biol. Reprod. 2004;70:168–177. doi: 10.1095/biolreprod.103.019232. [DOI] [PubMed] [Google Scholar]
- Bopp A. Rac1 modulates acute and subacute genotoxin-induced hepatic stress responses, fibrosis and liver aging. Cell Death Dis. 2013;4 doi: 10.1038/cddis.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira F.L. Galectin-3, histone deacetylases, and Hedgehog signaling: Possible convergent targets in schistosomiasis-induced liver fibrosis. PLoS Negl. Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl A.M. Neighborhood watch orchestrates liver regeneration. Nat. Med. 2012;18:497–499. doi: 10.1038/nm.2719. [DOI] [PubMed] [Google Scholar]
- Dooley S. Hepatocyte-specific Smad7 expression attenuates TGF-beta-mediated fibrogenesis and protects against liver damage. Gastroenterology. 2008;135:642–659. doi: 10.1053/j.gastro.2008.04.038. [DOI] [PubMed] [Google Scholar]
- Dooley S., ten Dijke P. TGF-beta in progression of liver disease. Cell Tissue Res. 2012;347:245–256. doi: 10.1007/s00441-011-1246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat I. TGF-beta signalling and liver disease. FEBS J. 2016;283:2219–2232. doi: 10.1111/febs.13665. [DOI] [PubMed] [Google Scholar]
- Forbes S.J., Newsome P.N. Liver regeneration − mechanisms and models to clinical application. Nat. Rev. Gastroenterol. Hepatol. 2016;13:473–485. doi: 10.1038/nrgastro.2016.97. [DOI] [PubMed] [Google Scholar]
- Friedman S.L. Therapy for fibrotic diseases: nearing the starting line. Sci. Transl. Med. 2013;5:167sr1. doi: 10.1126/scitranslmed.3004700. [DOI] [PubMed] [Google Scholar]
- Gressner O.A. Differential effects of TGF-beta on connective tissue growth factor (CTGF/CCN2) expression in hepatic stellate cells and hepatocytes. J. Hepatol. 2007;47:699–710. doi: 10.1016/j.jhep.2007.05.015. [DOI] [PubMed] [Google Scholar]
- Hamzavi J. Disruption of the Smad7 gene enhances CCI4-dependent liver damage and fibrogenesis in mice. J. Cell Mol. Med. 2008;12:2130–2144. doi: 10.1111/j.1582-4934.2008.00262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H., Sakai T. Animal models for the study of liver fibrosis: new insights from knockout mouse models. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;300:G729–738. doi: 10.1152/ajpgi.00013.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C.W. TGF-beta induces apoptosis through Smad-mediated expression of DAP-kinase. Nat. Cell Biol. 2002;4:51–58. doi: 10.1038/ncb731. [DOI] [PubMed] [Google Scholar]
- Liu X. Therapeutic strategies against TGF-beta signaling pathway in hepatic fibrosis. Liver Int. 2006;26:8–22. doi: 10.1111/j.1478-3231.2005.01192.x. [DOI] [PubMed] [Google Scholar]
- Macias M.J. Structural determinants of Smad function in TGF-beta signaling. Trends Biochem. Sci. 2015;40:296–308. doi: 10.1016/j.tibs.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. TGF beta signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J., Chen Y.G. Controlling TGF-beta signaling. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- Meng X.M. TGF-beta: the master regulator of fibrosis. Nat. Rev. Nephrol. 2016;12:325–338. doi: 10.1038/nrneph.2016.48. [DOI] [PubMed] [Google Scholar]
- Mengshol J.A. Mechanisms of Disease: HCV-induced liver injury. Nat. Clin. Pract. Gastroenterol. Hepatol. 2007;4:622–634. doi: 10.1038/ncpgasthep0961. [DOI] [PubMed] [Google Scholar]
- Mu X. Epithelial Transforming Growth Factor-beta Signaling Does Not Contribute to Liver Fibrosis but Protects Mice From Cholangiocarcinoma. Gastroenterology. 2016;150:720–733. doi: 10.1053/j.gastro.2015.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natori S. Hepatocyte apoptosis is a pathologic feature of human alcoholic hepatitis. J. Hepatol. 2001;34:248–253. doi: 10.1016/s0168-8278(00)00089-1. [DOI] [PubMed] [Google Scholar]
- Perlman R. TGF-beta-induced apoptosis is mediated by the adapter protein Daxx that facilitates JNK activation. Nat. Cell Biol. 2001;3:708–714. doi: 10.1038/35087019. [DOI] [PubMed] [Google Scholar]
- Preisegger K.H. Atypical ductular proliferation and its inhibition by transforming growth factor beta1 in the 3,5-diethoxycarbonyl-1,4-dihydrocollidine mouse model for chronic alcoholic liver disease. Lab Invest. 1999;79:103–109. [PubMed] [Google Scholar]
- Puche J.E. A novel murine model to deplete hepatic stellate cells uncovers their role in amplifying liver damage in mice. Hepatology. 2013;57:339–350. doi: 10.1002/hep.26053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh S. TGF beta-mediated BIM expression and apoptosis are regulated through SMAD3-dependent expression of the MAPK phosphatase MKP2. EMBO Rep. 2008;9:990–997. doi: 10.1038/embor.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert L. Dectin-1 Regulates Hepatic Fibrosis and Hepatocarcinogenesis by Suppressing TLR4 Signaling Pathways. Cell Rep. 2015;13:1909–1921. doi: 10.1016/j.celrep.2015.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Stewart R.K. A novel mouse model of depletion of stellate cells clarifies their role in ischemia/reperfusion- and endotoxin-induced acute liver injury. J. Hepatol. 2014;60:298–305. doi: 10.1016/j.jhep.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dijke P., Hill C.S. New insights into TGF-beta-Smad signalling. Trends Biochem. Sci. 2004;29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Tian X.F. Activation of the miR-34a/SIRT1/p53 Signaling Pathway Contributes to the Progress of Liver Fibrosis via Inducing Apoptosis in Hepatocytes but Not in HSCs. PLoS One. 2016;11 doi: 10.1371/journal.pone.0158657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent D.F. A rapid strategy to detect the recombined allele in LSL-TbetaRICA transgenic mice. Genesis. 2010;48:559–562. doi: 10.1002/dvg.20653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A., Attisano L. The TGFbeta superfamily signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 2013;2:47–63. doi: 10.1002/wdev.86. [DOI] [PubMed] [Google Scholar]
- Weng H.L. Profibrogenic transforming growth factor-beta/activin receptor-like kinase 5 signaling via connective tissue growth factor expression in hepatocytes. Hepatology. 2007;46:1257–1270. doi: 10.1002/hep.21806. [DOI] [PubMed] [Google Scholar]
- Xiang X. Grhl2 determines the epithelial phenotype of breast cancers and promotes tumor progression. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L. Transforming growth factor-beta signaling in hepatocytes promotes hepatic fibrosis and carcinogenesis in mice with hepatocyte-specific deletion of TAK1. Gastroenterology. 2013;144:1042–1054. doi: 10.1053/j.gastro.2013.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S. Inhibition of Notch Signaling Attenuates Schistosomiasis Hepatic Fibrosis via Blocking Macrophage M2 Polarization. PLoS One. 2016;11 doi: 10.1371/journal.pone.0166808. [DOI] [PMC free article] [PubMed] [Google Scholar]