Abstract
Little is known about hematopoietic stem cell (HSC) development from mesoderm. To gain more information on the intraembryonic HSC site of origin, we purified multipotent hematopoietic progenitors from the aorta–gonads–mesonephros (AGM) of mice. This population, expressing c-Kit, AA4.1, CD31, and CD41, but not Flk1, and mainly negative for CD45, proved capable of long-term reconstitution in sublethally irradiated Rag2γc-/- recipients. We assigned the expression of GATA-2, GATA-3, and lmo2 to AGM-HSC, whereas erythromyeloid progenitors express only GATA-2. This unique combination of surface markers and transcription factors could be allocated in the AGM to the intraaortic clusters and the subaortic patches underlying aortic endothelial cells. Taken together, those data indicate that embryonic HSCs (i) differ from their fetal liver and adult counterpart by the low expression of CD45, (ii) do not colocalize with aortic endothelial cells as previously thought, and (iii) are localized, at 10.5 days postcoïtum, in the splanchnic mesoderm underlying aortic endothelial cells, within GATA-3+CD31+ cell clusters.
Keywords: aorta–gonads–mesonephros, yolk sac
During mouse development, the first hematopoietic cells, consisting of precursors with erythroid and/or myeloid potential (1), appear in the yolk sac (YS) blood islands, from 7 days postcoïtum (dpc). Before the establishment of circulation (8 dpc), the first progenitors endowed with long-term reconstitution (LTR) activity are generated in the intraembryonic splanchnopleura and are absent from the YS (2, 3). After circulation is established, multipotent precursors with LTR activity (4, 5) can be found in the blood (6) and in the YS (7). After 10 dpc the paraaortic splanchnopleura, which now comprises the aorta, gonads, and mesonephros, is called AGM. Hematopoietic precursors, present in the AGM after 10.5 dpc, have LTR capacity when transferred in adult normal recipient mice (4).
It was previously suggested that the floor of the aorta and/or the underlying splanchnic mesoderm is the site of origin of intraembryonic hematopoietic stem cells (HSCs) (8–12). Clusters of basophilic cells located within the floor of the aorta were detected from 10 to 10.5 dpc, further characterized as hematopoietic cells, and taken as a sign of ongoing hemogenesis. These hematopoietic intraaortic clusters (HIACs) have been described in all vertebrate embryos, including humans (for review, see ref. 13). Other structures potentially involved in intraembryonic HSC generation are the subaortic patches (SAPs), located below the aortic floor, which express the transcription factor GATA-3 (14) and the AA4.1 antigen (15). These SAPs are also present in human embryos (16, 17). In contrast to HIACs, which are detected only at the peak of intraembryonic HSC production (8), the SAPs are present in the intraembryonic hemogenic site for the duration of HSC generation. An involvement of both SAPs and HIACs in HSC generation is strengthened by the fact that both structures disappear (11, 14) concomitantly with the cessation of AGM-HSC production at 12 dpc (8).
The understanding of the process leading to intraembryonic HSC generation is currently a matter of debate that can be clarified by precisely locating the structure where this generation occurs. In the avian and murine models as well as in human embryos, AGM-hematopoietic activity has been shown to derive from cells harboring markers shared by endothelial and hematopoietic lineages, which also lack the panhematopoietic marker CD45 (18–20). It has thus been proposed that the endothelium of the aortic floor displays “hemogenic” activity and is able to give rise to HSCs.
Here we correlated surface phenotype, in vitro differentiation potential, LTR activity, and gene expression of 10.5-dpc AGM populations. We show that CD45+ cells contain only macrophage precursors. CD45-/loc-Kit+AA4.1+ cells from either AGM or YS are differentially enriched for two distinct types of progenitors. Erythromyeloid precursors are found at a frequency of 1:2 in YS. Multipotent precursors are present at frequencies higher than 1:3 in AGM, representing more than 80% of total multipotent cells in this site. LTR activity in alymphoid recipients was strictly ascribed to CD45-/loc-Kit+AA4.1+ AGM cells that can therefore be considered as HSCs.
HSCs purified according to these criteria express a number of cell surface markers (CD31, c-Kit, and AA4.1) and transcription factors (lmo2 and GATA-2) common to cells of both hematopoietic and endothelial lineages. However, they may be distinguished from endothelial cells (ECs) by the lack of Flk1 and the expression of CD41 (21–23) and GATA-3. The phenotype and gene expression profile of purified HSCs were compared with those obtained by in situ hybridization and immunostaining of the AGM (ref. 14 and present work). Coexpression of GATA-2, GATA-3, Lmo2, CD41, and CD31 was observed exclusively in two distinct anatomical structures, the HIAC and the SAP (14). We propose that HSCs are generated in the SAP before they form the aortic clusters and before they enter blood circulation.
Materials and Methods
Animals and Dissections. Two C57BL/6 congenic lines (H2b) bearing the Ly5.1 and Ly5.2 alleles of the CD45 marker and their F1 progeny were used in this study. Rag2γc-/- B10BR mice (H2k) were used as recipients for LTR assays. Embryonic development was estimated by considering the day of vaginal plug observation as 0.5 dpc. Embryos between somite stages (S) 30S and 40S (10.5 dpc) were staged by somite counting. Dissections of 10.5-dpc YS and AGM were done as described (8).
Flow Cytometry Analysis and Cell Sorting. Flow cytometric analyses were performed in a LSR with the cellquest software (Becton Dickinson). The following antibodies (Pharmingen) were used: CD45.1 (A20), c-Kit (2B8), AA4.1 (493), CD41 (MWReg30), CD31 (MEC13.3), Flk1 (Avas12α1), Gr-1 (RB6-8C5), Mac-1 (M1/70), B220 (RA3-6B2), CD19 (1D3), Ter119, H2-Kb (AF6-88.5), NK1.1 (PK136), TcRβ (H57), and TcRγδ (GL3). To test acetylated low-density lipoprotein (AcLDL), uptake cells conjugated with Alexa Fluor 488 (Molecular Probes) were incubated for 2 h at 37°C with 10 μM AcLDL. Cell sorting was performed with a MoFlo cell sorter (Cytomation, Ft. Collins, CO). The single-cell dispenser robot (Cyclone) was used for deposition of single cells on culture plates.
Differentiation Potential Assay. Sorted YS and AGM cells were cultured for 14 days on irradiated S17 stromal cells (kindly provided by K. Dorshkind, University of California, Riverside) (2, 3) or OP9 stromal cells (from S.-I. Nishikawa and T. Nakano, Riken Center for Developmental Biology, Kobe, Japan), supplemented with Flt3-ligand, a kind gift from R. Rottapel (Ontario Cancer Institute, Toronto). All cultures were done in 96-well plates in medium supplemented with cytokines, as described (2, 3).
Reconstitution Experiments. Sorted 10.5-dpc AGM cells were injected into Rag2γc-/- mice. For LTR analysis, recipients were killed at 6 months, and the bone marrow, spleen, and intestines were analyzed by cytometry for the presence of H2b T, B, natural killer, and myeloid cells. Animals that had donor-derived myeloid and B cell precursors in the bone marrow were considered long-term reconstituted.
Gene Expression Analysis. Cells were lysed in TRIzol (GIBCO/BRL), and total RNA was extracted according to the manufacturer's protocol. Oligo(dT)-primed cDNA was prepared from 6,000–25,000 cells by using avian myeloblastosis virus reverse transcriptase (GIBCO/BRL) in a 20-μl reaction volume. cDNA from 15-dpc Ter119-depleted fetal liver cells was used as a standard for the transcript quantification. cDNA from S17 cells was used as a negative control.
Quantitative RT-PCRs were performed on the GeneAmp 5700 Sequence Detection System (Applied Biosystems), in a 25-μl total volume, using 1 μl of cDNA, 1 μl of each primer (10 μM), and 12.5 μl of Sybr Green PCR Master Mix 2× (Applied Biosystems). Each sample was tested in triplicate. For each independent experiment, hypoxanthine phosphoribosyltransferase (HPRT) expression was scored in each population. Finally, the signals detected in each population for each transcript were normalized to HPRT and shown as percentages of the expression found in control populations (considered as 100%). Primers: HPRT-for, 5′-GACTGAAAGACTTGCTCGAG-3′; HPRT-rev, 5′-CCAGCAAGCTTGCAACCTTAACCA-3′; L-plastin-for, 5′-ACATCAGCTGCAATGAGC-3′; L-plastin-rev, 5′-TATCCAGTTGACGAAGGC-3′; CD45-for, 5′-AACACCTACACCCAGTGATG-3′; and CD45-rev, 5′-TTGGCTGCTGAATGTCTGAG-3′. Other primers are described elsewhere: GATA-2 (24), GATA-3 (25), Lmo2 (26), and EpoR (27).
In Situ Hybridization and Immunostaining. Digoxygenin-labeled (Boehringer Mannheim) riboprobes were obtained from PCR fragments. In situ hybridization on cryostat sections was performed as described in ref. 14. For multiple labeling, sections (from 10 embryos) were incubated overnight with anti-CD41-FITC, anti-CD31-PE, and/or anti-CD45 [clone 30-F11 coupled with either CyChrome or phycoerythrin (PE)], and mounted with VECTASHIELD Hardset (Vector Laboratories). Image stacks were collected with a LSM 510 laser scanning confocal microscope (Zeiss) using a ×20/numerical aperture 0.75 apochromat plan objective. Images were acquired by using BP505–530, BP560–615, and LP650 filters. Z-projection of slices was in real time by using LSM image examiner software (Zeiss). Images were processed by using photoshop 6.0 software (Adobe Systems, San Jose, CA).
Results
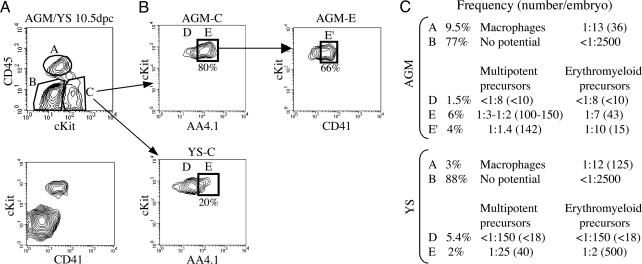
Phenotypic Characterization of Hematopoietic Cells in 10.5-dpc AGM. We analyzed the surface expression of the following marker combinations to ascribe a phenotype to hematopoietic precursors in the AGM: AA4.1 (28), shown to be expressed in paraaortic splanchnopleura/AGM (7, 15); CD45, panhematopoietic antigen; c-Kit (29, 30); Flk1, present in ECs and their precursors; CD31, expressed by ECs and hematopoietic cells; and CD41, expressed by early hematopoietic precursors but not by ECs (21–23). We fractionated 10.5-dpc AGM and YS cells, at the time of maximum multipotent cell generation (8), according to the expression of CD45, c-Kit, AA4.1, and CD41 (Fig. 1A). Sorted cells were tested in vitro in a single-cell assay for the potential to differentiate into erythroid (Ter119+), myeloid (CD11b+), and lymphoid (B220+) cells. Two types of precursors were identified: erythromyeloid precursors that differentiate only into erythroid and myeloid cells and multipotent precursors that also generate lymphoid cells. The frequency of each precursor in the different fractions is shown in Fig. 1C.
Fig. 1.
Distribution of multipotent and erythromyeloid precursors in 10.5-dpc AGM and YS. (A) AGM and YS cells at 10.5 dpc were fractionated according to the expression of CD45, c-Kit, and CD41. We could identify three fractions, A (CD45+), B (c-Kit-), and C (c-Kit+CD45-/lo). All c-Kit+ cells also express CD41. (B)In AGM and YS, fraction C was subdivided into D (c-Kit+AA4.1-) and E (c-Kit+AA4.1+), shown on the plots. AGM-E cells were further fractionated with CD41; E′ represents 66% of AGM fraction E. (C) Studies in single-cell cultures and limiting dilution analysis provided the frequency of multipotent and bipotent (erythromyeloid) precursors in each fraction. Absolute numbers (frequency × number of cells) presented here are calculated according to the total number of cells in AGM and YS, estimated to be 5,000 and 50,000, respectively.
CD45+ cells (fraction A) exclusively contain macrophage precursors, at a frequency of 1:13. The progeny of these cells (Mac-1+F4/80+) shows characteristic macrophage morphology after May–Grünwald Giemsa staining (data not shown). Fraction B, CD45-c-Kit-, is devoid of hematopoietic activity. Lymphomyeloid potential is restricted to CD45-/loc-Kit+ cells (fraction C). CD45 expression was considered negative to low, because 15–20% of these cells within this population expressed low levels of the protein. Cells from fraction C were subdivided into fractions D and E, AA4.1- and AA4.1+, respectively (Fig. 1B).
Within AGM fraction E (CD45-/loc-Kit+AA4.1+), multipotent precursors were detected at a frequency of 1:3–1:2, corresponding to 100–150 multipotent precursors per embryo, a result that correlates with our previous quantification of multipotent precursors in unfractionated 10.5-dpc AGM (8). The frequency of multipotent cells was further enriched to 1:1.4 (72% of seeded wells) when CD41+ cells were isolated within the fraction E (Fig. 1). Although all c-Kit+ cells also express CD41, this enrichment was obtained with the CD41-brightest cells (fraction E′), corresponding to 66% of fraction E.
In contrast to AGM, YS fraction E was enriched for erythromyeloid precursors at a frequency of 1:2 sorted cells, corresponding to 500 erythromyeloid precursors per YS (10 times higher than in the AGM), likely originated in situ. We conclude that AGM fraction E is a population highly enriched for hematopoietic precursors, multipotent, at the clonal level.
We ascertained that AGM fraction E also contained HSCs by an in vivo LTR assay into sublethally irradiated Rag2γc-/- mice (Table 1, which is published as supporting information on the PNAS web site). The CD45-/loc-Kit+AA4.1+ 10.5-dpc AGM fraction E, characterized as containing most multipotent precursors, appears to be the only fraction in AGM providing multilineage LTR. These cells can be considered HSCs.
The phenotype of AGM-HSCs determined above, the low levels of expression of CD45, and their capacity to take up acetylated low-density lipoprotein (Fig. 6, which is published as supporting information on the PNAS web site) makes them indistinguishable from ECs, macrophages, and other AGM cell types. We therefore analyzed the expression of two additional markers in this population. All c-Kit+ cells also express CD31 (Fig. 7, which is published as supporting information on the PNAS web site). Both Flk1 and CD41 stain subpopulations of c-Kit+ cells (Figs. 2A and 7). Hematopoietic differentiation analysis of both sorted populations indicated that CD41 stains all multipotent cells in the AGM, whereas no hematopoietic progeny was obtained from the Flk1+ cells (>200 cells analyzed; data not shown). Also consistent with the hematopoietic nature of AGM fraction E is the expression of L-plastin (31) and low levels of CD45 mRNA (Fig. 2C).
Fig. 2.
Distribution of endothelial and hematopoietic markers in cells from YS and AGM. (A) Flk1 (Center) and CD41 (Right) expression within the two gated c-Kit+AA4.1+ populations (Left), after exclusion of CD45+ cells. (B) CD45-/loc-Kit+AA4.1+ and CD45-c-Kit- cells from AGM and YS were sorted and analyzed for expression of Lmo2, GATA-3, and GATA-2. The transcript expression is represented, after normalization to hypoxanthine phosphoribosyltransferase, by the percentage of its expression in the standard population (15-dpc fetal liver depleted of erythrocytes). Levels of expression were divided into four groups according to the percentage of expression relative to the standard. Means and standard deviations on triplicates from two individual experiments are shown. No background shading, <25%; yellow shading, 25–100%; red shading, >250%. (C) CD45 and L-plastin transcript distribution within YS and AGM fractions E and B.
We show that, at this stage of development, HSCs express c-Kit, AA4.1, CD41, and CD31, whereas expression of Flk1 is undetectable. Flk1+CD31+ cells that express variable levels of c-Kit and AA4.1 do not generate a hematopoietic progeny and according to this expression pattern likely correspond to ECs.
At 10.5 dpc, Fraction E Cells Are Localized Within Intraaortic Clusters and SAPs. We next analyzed by RT-PCR the expression, within fraction E, of genes shared by hematopoietic cells and ECs [GATA-2 (32) and lmo2 (14)] and genes not expressed by ECs [GATA-3 (33)]. Fig. 2B shows that whereas GATA-2, GATA-3, and lmo2 are highly expressed in AGM fraction E, only GATA-2 is expressed at a notable level in the YS. Considering that HSCs are mostly represented in AGM fraction E and that erythromyeloid precursors are predominant in the YS, we conclude that erythromyeloid precursors are likely to express only high levels of GATA-2, whereas HSCs express GATA-2, GATA-3, and lmo2.
To locate in the AGM the various subpopulations characterized by flow cytometry, in vitro and in vivo potential assays, and gene expression analysis, we performed in situ hybridization (GATA-2 and GATA-3) and multiple immunostainings (CD41, CD31, and CD45) on 10- to 10.5-dpc (30–40S) AGM sections to discriminate between the various fractions.
Fraction A cells, corresponding to macrophages, were identified by their CD45+CD31+CD41- phenotype (Fig. 1). CD45+ cells were found surrounding the aorta (Figs. 3A and 4C).
Fig. 3.
Localization of AGM hematopoietic subsets in 10- to 10.5-dpc (30–35S) embryos. Confocal expression analyses (A) allow one to discriminate between macrophages (CD45+CD31+CD41-, arrowheads) and HSCs (CD41+CD31+CD45-/lo, arrows). HSCs are located within the intraaortic clusters (stars) and within the SAPs (asterisks). (B) CD31 is expressed by ECs and by GATA-3+ SAPs (asterisk). (Bars, 50 μm.)
Fig. 4.
Localization of AGM-HSCs and their environment in 10.5-dpc (35–40S) embryos. From 10.5 dpc (35–40S), the subendothelial CD31+GATA-3+ layer evolves into more compact SAPs (asterisks), which express GATA-2 mRNA (A) in addition to GATA-3 (B). Confocal analyses (C and D) show that HSCs are located within the SAPs. CD45+ macrophages (arrowheads) do not express CD41. XZ projection of the image stack (D) that comprises the top section in C shows that CD31+CD41+ HSCs are present only in the ventral part of the AGM (below the straight line joining the cardinal veins (CV)). Their distribution forms a continuum from the SAPs (asterisks) to the aortic floor. Note that CD31+CD41+ HSCs constitute only a minority of the CD31+GATA-3+ SAP cells. Stainings in A–C were performed on alternate sections of the same embryo. (Bar, 50 μm.)
CD41+CD31+CD45-/lo cells, which correspond to HSCs, were present only in the ventral side of the AGM, either in SAPs below the aortic floor (Figs. 3 and 4; see also Fig. 8, which is published as supporting information on the PNAS web site) or within the HIAC (Figs. 3 and 8). This labeling pattern colocalizes with GATA-3 expression (Fig. 3). At later stages, when HSCs are more numerous (8), SAPs are better individualized (Fig. 4). GATA-3 is expressed only in SAPs, where it colocalizes with GATA-2, also expressed by endothelial cells (Fig. 4 A and B). CD41+CD31+CD45- cells appeared as individual cells within the SAP (Fig. 4C). Lateral examination (Fig. 4D) of the confocal sections stack shows that CD31+CD41+ HSCs, present only in the ventral part of the AGM, are distributed along a continuum from the SAP to the aortic floor. Moreover, 3D confocal analysis of AGM explants (n = 6) established that the CD31+ cells (some of which coexpress CD41) forming the SAP are restricted to the ventral region of the aorta and are distinct from the vascular network (Fig. 9, which is published as supporting information on the PNAS web site).
The various approaches undertaken here converge to allocate intraembryonic HSCs to the SAP and some cells of the HIAC. They do not colocalize with the ventral aortic ECs.
Discussion
In this study, we define a population of CD45-/loc-Kit+AA4.1+ cells, obtained from prehepatic sites (YS and AGM) in 10.5-dpc embryos, which also express CD31 and CD41 but not Flk1. This CD45-/loc-Kit+AA4.1+CD41+ AGM fraction contains multipotent cells at a frequency of 1:1.4, which accounts for the bulk of multipotent precursors at this stage (8). It is noteworthy that, because of a 10–20% error in the single-cell seeding procedure, the 70% positive multipotent clones obtained correspond to an actual enrichment close to 1:1. The YS counterpart is mainly enriched in erythromyeloid precursors (with a frequency of 1:2). Sorted CD45+ AGM and YS cells are committed macrophage precursors, likely equivalent to “primitive macrophages,” described in vertebrate embryos (34–36).
Consistent with the in vitro data, LTR was exclusively obtained when we injected as few as 250 AGM CD45-/loc-Kit+AA4.1+ cells in sublethally irradiated alymphoid recipients. No donor-derived hematopoietic cells were obtained when CD45+ or c-Kit- AGM cells were used for LTR, illustrating the absence of HSCs in these two subsets. Within the fraction E, discrete differences in CD45 expression could be observed. Whether LTR activity is an exclusive property of the CD45lo subpopulation was not addressed here. Most AGM cells will express detectable levels of CD45 by 11.5 dpc, and 10.5-dpc fraction E might acquire CD45 expression shortly after transfer in the irradiated recipients.
The expression of CD41 in immature hematopoietic precursors, independent, of its expression within the megakaryocytic lineage has been documented (21, 37–39). More recently, CD41 was shown to constitute the earliest hematopoietic-specific marker during the determination of mesodermal cells toward this lineage, because it does not label ECs (21–23) and is expressed by hematopoietic precursors before CD45 (23), a result in total agreement with the present phenotyping of AGM-HSCs.
HSCs have been previously shown to express different surface markers depending on the hematopoietic organ where they reside. In contrast to bone marrow HSCs, fetal liver HSCs express Mac-1, AA4.1, CD41, and CD31 (40). We here show that AGM-HSCs express markers similar to the ones found in fetal liver (AA4.1, CD31, c-Kit, and CD41) but differ by the lack/low expression of CD45. No cells with hematopoietic precursor activity were found in the CD41-, CD31-, or c-Kit- fractions. Because of the strict hematopoietic representation of CD41, we conclude that the combination of CD41 with any of the above-mentioned markers allows discrimination between hematopoietic and any other cell type.
We here show that purified CD45-/loc-Kit+AA4.1+CD31+ multipotent precursors that also contain the majority of CD41+ cells in this location are the only hematopoietic AGM subsets expressing high levels of GATA-3 and GATA-2. The expression patterns of these transcription factors and cell surface markers completely correlate to locate AGM-HSCs within both the HIAC and the SAP (Figs. 3, 4, 5A, 8, and 9). We recently described the existence of SAPs in the ventral region of the aorta (14). These structures, identified by the expression of GATA-3 and AA4.1, do not contain CD45+ cells. After 10 dpc, HIACs appear in the aortic floor. Cells expressing GATA-3 and lmo2 are detected at the base of these clusters (14), which also express AA4.1 (15), CD31 (41), and CD41 (42) and contain a few CD45+ cells (14).
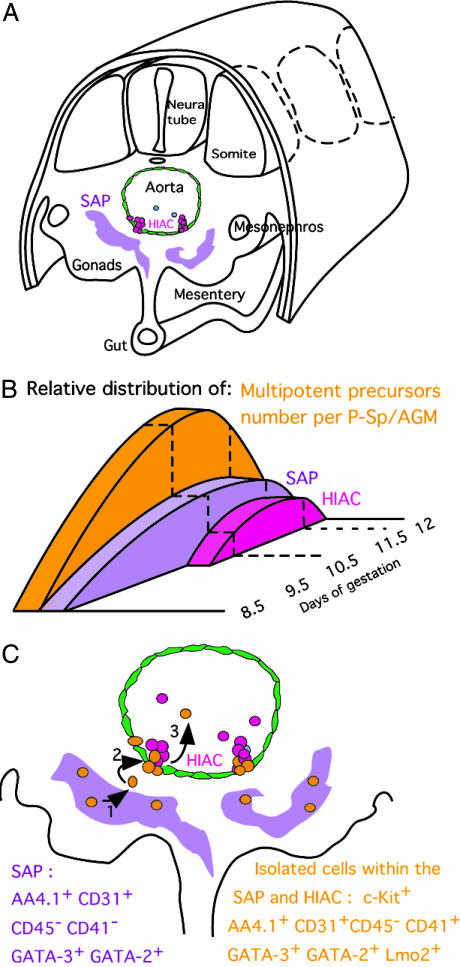
Fig. 5.
Toward a model for emergence and migration of embryonic HSCs. (A) Localization of hemogenic-related structures, SAPs (purple), and HIAC (pink) within a 10.5-dpc embryo. (B) Relative distribution of multipotent precursors (orange), SAPs (purple), and HIACs (pink) in 8.5- to 12-dpc embryos. Whereas GATA-3+ SAPs appear below the aortic floor as soon as the first multipotent precursor is generated, HIACs are present only at the AGM stage, when the number of precursors generated reaches a peak. (C) Model for emergence and migration of embryonic hematopoietic precursors: HSCs (orange) generated in the SAP (1) migrate toward the aortic floor (2). HSC translocation, which contributes to HIAC development (3), is followed by the release of HIACs into the circulation.
Several lines of evidence point to the SAP, rather than the HIAC or aortic floor, as the primary site of origin of intraembryonic HSCs in the embryo:
The GATA-3+ SAP become detectable concomitantly with the production of the first intraembryonic precursors (8.5–9 dpc) (7), whereas HIACs appear only at the AGM stage (10 dpc on) (11), when the number of precursors generated has significantly increased (8) (Fig. 5B).
Lateral examination of confocal sections (Fig. 4D) shows that HSCs form a continuum between the SAP and the HIAC, suggesting that these cells migrate from one site to the other. The most likely direction of migration appears to be from SAP to HIAC and finally into the aortic blood flow, because it contains a significant number of multipotent precursors at that stage (6). This model nevertheless requires in vivo cell tracking experiments to be confirmed.
The combinatorial approach undertaken here allowed us to distinguish between ECs and hematopoietic cells, by GATA-3, CD41, and Flk1 differential expression. The 3D examination of SAP anatomy also distinguishes SAP cells from ECs because the CD31+ cell clusters located below the aortic floor do not harbor a lumen and do not connect with the vascular network (Fig. 9).
Recent reports suggesting that the aortic floor in 11-dpc AGM (18, 20) could be the source of HSCs were based on the fact that CD31+CD45- cells, considered as ECs, display a hematopoietic activity. Through the use of additional markers and colocalization analysis, we here show that HSCs and aortic floor ECs are not overlapping populations.
We thus propose that HSCs are generated in the SAP, migrate toward the ventral region of the aorta, and protrude through the endothelium, contributing to the formation of the HIAC and leading to their release into circulation, from where they colonize the fetal liver (Fig. 5C).
In the course of our colocalization studies, we encountered only a limited number of such CD31+CD41+CD45-/lo cells in 10.5-dpc embryo sections, a result that concurs with the number of HSCs, previously estimated at ≈80–100 per embryo (8). It thus appears that the relative size of cells expressing GATA-3 and/or CD31 in the SAP greatly exceeds the number of HSCs present in the embryo. SAPs may constitute the environment suitable for the emergence of intraembryonic HSCs. Our results show that SAPs, which can be characterized by the expression of CD31 and AA4.1 and lack of CD45, contain HSCs that also express CD41. Further characterization of the SAP, now made possible by this observation, will be necessary to understand the relationship of SAP cells and native HSCs and to assess whether the SAP modulates HSC generation and self-renewal.
Supplementary Material
Acknowledgments
We thank J. P. DiSanto (Pasteur Institute, Paris) for providing B10BR Rag2γc-/- breeding pairs, A. Louise for cell sorting, A. Bandeira and O. Burlen-Defranoux for injecting and bleeding mice, and W. Vainchenker for critical reading of the manuscript. The work was supported by grants from the French Ministry of Research (Action Concertée Incitative Developmental Biology) to A.C. and I.G., from the Association pour la Recherche sur le Cancer (Grant 4441) and Institut Gustave Roussy (Contrat de Recherche Clinque Grant 73 308H) to I.G., and from the European Union Framework 6 Program EuroStem Cell and the Pasteur Institute through the Strategic Horizontal Program (Grand Programme Horizontal) on Stem Cells (to A.C.). J.Y.B. was supported by the French Ministry of Research and Education.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: dpc, days postcoïtum; AGM, aorta–gonads–mesonephros; YS, yolk sac; HSC, hematopoietic stem cell; EC, endothelial cell; HIAC, hematopoietic intraaortic cluster; SAP, subaortic patch; LTR, long-term reconstitution.
References
- 1.Palis, J., Chan, R. J., Koniski, A., Patel, R., Starr, M. & Yoder, M. C. (2001) Proc. Natl. Acad. Sci. USA 98, 4528-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cumano, A., Dieterlen-Lievre, F. & Godin, I. (1996) Cell 86, 907-916. [DOI] [PubMed] [Google Scholar]
- 3.Cumano, A., Ferraz, J. C., Klaine, M., Di Santo, J. P. & Godin, I. (2001) Immunity 15, 477-485. [DOI] [PubMed] [Google Scholar]
- 4.Muller, A. M., Medvinsky, A., Strouboulis, J., Grosveld, F. & Dzierzak, E. (1994) Immunity 1, 291-301. [DOI] [PubMed] [Google Scholar]
- 5.Yoder, M. C., Hiatt, K., Dutt, P., Mukherjee, P., Bodine, D. M. & Orlic, D. (1997) Immunity 7, 335-344. [DOI] [PubMed] [Google Scholar]
- 6.Delassus, S. & Cumano, A. (1996) Immunity 4, 97-106. [DOI] [PubMed] [Google Scholar]
- 7.Godin, I., Dieterlen-Lievre, F. & Cumano, A. (1995) Proc. Natl. Acad. Sci. USA 92, 773-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godin, I., Garcia-Porrero, J. A., Dieterlen-Lievre, F. & Cumano, A. (1999) J. Exp. Med. 190, 43-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Bruijn, M. F., Speck, N. A., Peeters, M. C. & Dzierzak, E. (2000) EMBO J. 19, 2465-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.North, T. E., de Bruijn, M. F., Stacy, T., Talebian, L., Lind, E., Robin, C., Binder, M., Dzierzak, E. & Speck, N. A. (2002) Immunity 16, 661-672. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Porrero, J. A., Godin, I. E. & Dieterlen-Lievre, F. (1995) Anat. Embryol. 192, 425-435. [DOI] [PubMed] [Google Scholar]
- 12.Wood, H. B., May, G., Healy, L., Enver, T. & Morriss-Kay, G. M. (1997) Blood 90, 2300-2311. [PubMed] [Google Scholar]
- 13.Godin, I. & Cumano, A. (2002) Nat. Rev. Immunol. 2, 593-604. [DOI] [PubMed] [Google Scholar]
- 14.Manaia, A., Lemarchandel, V., Klaine, M., Max-Audit, I., Romeo, P., Dieterlen-Lievre, F. & Godin, I. (2000) Development (Cambridge, U.K.) 127, 643-653. [DOI] [PubMed] [Google Scholar]
- 15.Petrenko, O., Beavis, A., Klaine, M., Kittappa, R., Godin, I. & Lemischka, I. R. (1999) Immunity 10, 691-700. [DOI] [PubMed] [Google Scholar]
- 16.Marshall, C. J., Kinnon, C. & Thrasher, A. J. (2000) Blood 96, 1591-1593. [PubMed] [Google Scholar]
- 17.Marshall, C. J., Moore, R. L., Thorogood, P., Brickell, P. M., Kinnon, C. & Thrasher, A. J. (1999) Dev. Dyn. 215, 139-147. [DOI] [PubMed] [Google Scholar]
- 18.de Bruijn, M. F., Ma, X., Robin, C., Ottersbach, K., Sanchez, M. J. & Dzierzak, E. (2002) Immunity 16, 673-683. [DOI] [PubMed] [Google Scholar]
- 19.Jaffredo, T., Gautier, R., Eichmann, A. & Dieterlen-Lievre, F. (1998) Development (Cambridge, U.K.) 125, 4575-4583. [DOI] [PubMed] [Google Scholar]
- 20.Oberlin, E., Tavian, M., Blazsek, I. & Peault, B. (2002) Development (Cambridge, U.K.) 129, 4147-4157. [DOI] [PubMed] [Google Scholar]
- 21.Ferkowicz, M. J., Starr, M., Xie, X., Li, W., Johnson, S. A., Shelley, W. C., Morrison, P. R. & Yoder, M. C. (2003) Development (Cambridge, U.K.) 130, 4393-4403. [DOI] [PubMed] [Google Scholar]
- 22.Li, W., Johnson, S. A., Shelley, W. C., Ferkowicz, M., Morrison, P., Li, Y. & Yoder, M. C. (2003) Blood 102, 4345-4353. [DOI] [PubMed] [Google Scholar]
- 23.Mikkola, H. K., Fujiwara, Y., Schlaeger, T. M., Traver, D. & Orkin, S. H. (2003) Blood 101, 508-516. [DOI] [PubMed] [Google Scholar]
- 24.Chen, D. & Zhang, G. (2001) Exp. Hematol. 29, 971-980. [DOI] [PubMed] [Google Scholar]
- 25.Kawamoto, H., Ikawa, T., Ohmura, K., Fujimoto, S. & Katsura, Y. (2000) Immunity 12, 441-450. [DOI] [PubMed] [Google Scholar]
- 26.Lowsky, R., DeCoteau, J. F., Reitmair, A. H., Ichinohasama, R., Dong, W. F., Xu, Y., Mak, T. W., Kadin, M. E. & Minden, M. D. (1997) Blood 89, 2276-2282. [PubMed] [Google Scholar]
- 27.Pharr, P. N., Hankins, D., Hofbauer, A., Lodish, H. F. & Longmore, G. D. (1993) Proc. Natl. Acad. Sci. USA 90, 938-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jordan, C. T., McKearn, J. P. & Lemischka, I. R. (1990) Cell 61, 953-963. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez, M. J., Holmes, A., Miles, C. & Dzierzak, E. (1996) Immunity 5, 513-525. [DOI] [PubMed] [Google Scholar]
- 30.Spangrude, G. J., Heimfeld, S. & Weissman, I. L. (1988) Science 241, 58-62. [DOI] [PubMed] [Google Scholar]
- 31.Arpin, M., Friederich, E., Algrain, M., Vernel, F. & Louvard, D. (1994) J. Cell Biol. 127, 1995-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, M. E., Temizer, D. H., Clifford, J. A. & Quertermous, T. (1991) J. Biol. Chem. 266, 16188-16192. [PubMed] [Google Scholar]
- 33.George, K. M., Leonard, M. W., Roth, M. E., Lieuw, K. H., Kioussis, D., Grosveld, F. & Engel, J. D. (1994) Development (Cambridge, U.K.) 120, 2673-2686. [DOI] [PubMed] [Google Scholar]
- 34.Herbomel, P., Thisse, B. & Thisse, C. (1999) Development (Cambridge, U.K.) 126, 3735-3745. [DOI] [PubMed] [Google Scholar]
- 35.Naito, M., Umeda, S., Yamamoto, T., Moriyama, H., Umezu, H., Hasegawa, G., Usuda, H., Shultz, L. D. & Takahashi, K. (1996) J. Leukocyte Biol. 59, 133-138. [DOI] [PubMed] [Google Scholar]
- 36.Smith, S. J., Kotecha, S., Towers, N., Latinkic, B. V. & Mohun, T. J. (2002) Mech. Dev. 117, 173-186. [DOI] [PubMed] [Google Scholar]
- 37.Emambokus, N. R. & Frampton, J. (2003) Immunity 19, 33-45. [DOI] [PubMed] [Google Scholar]
- 38.Mitjavila-Garcia, M. T., Cailleret, M., Godin, I., Nogueira, M. M., Cohen-Solal, K., Schiavon, V., Lecluse, Y., Le Pesteur, F., Lagrue, A. H. & Vainchenker, W. (2002) Development (Cambridge, U.K.) 129, 2003-2013. [DOI] [PubMed] [Google Scholar]
- 39.Ody, C., Vaigot, P., Quere, P., Imhof, B. A. & Corbel, C. (1999) Blood 93, 2898-2906. [PubMed] [Google Scholar]
- 40.Yokota, T., Kouro, T., Hirose, J., Igarashi, H., Garrett, K. P., Gregory, S. C., Sakaguchi, N., Owen, J. J. & Kincade, P. W. (2003) Immunity 19, 365-375. [DOI] [PubMed] [Google Scholar]
- 41.Garcia-Porrero, J. A., Manaia, A., Jimeno, J., Lasky, L. L., Dieterlen-Lievre, F. & Godin, I. E. (1998) Dev. Comp. Immunol. 22, 303-319. [DOI] [PubMed] [Google Scholar]
- 42.Corbel, C. & Salaun, J. (2002) Dev. Biol. 243, 301-311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.