Abstract
White-rot fungi (WRF) and their ligninolytic enzymes (laccases and peroxidases) are considered promising biotechnological tools to remove lignin related Persistent Organic Pollutants from industrial wastewaters and contaminated ecosystems. A high diversity of the genus Ganoderma has been reported in Cuba; in spite of this, the diversity of ligninolytic enzymes and their genes remained unexplored. In this study, 13 native WRF strains were isolated from decayed wood in urban ecosystems in Havana (Cuba). All strains were identified as Ganoderma sp. using a multiplex polymerase chain reaction (PCR)-method based on ITS sequences. All Ganoderma sp. strains produced laccase enzymes at higher levels than non-specific peroxidases. Native-PAGE of extracellular enzymatic extracts revealed a high diversity of laccase isozymes patterns between the strains, suggesting the presence of different amino acid sequences in the laccase enzymes produced by these Ganoderma strains. We determined the diversity of genes encoding laccases and peroxidases using a PCR and cloning approach with basidiomycete-specific primers. Between two and five laccase genes were detected in each strain. In contrast, only one gene encoding manganese peroxidase or versatile peroxidase was detected in each strain. The translated laccases and peroxidases amino acid sequences have not been described before. Extracellular crude enzymatic extracts produced by the Ganoderma UH strains, were able to degrade model chromophoric compounds such as anthraquinone and azo dyes. These findings hold promises for the development of a practical application for the treatment of textile industry wastewaters and also for bioremediation of polluted ecosystems by well-adapted native WRF strains.
Keywords: white-rot fungi, Ganoderma, laccase, manganese peroxidase, versatile peroxidases, bioremediation, chromophoric compounds, Persistent Organic Compounds
Introduction
Environmental pollution with hazardous industrial wastes containing recalcitrant xenobiotics has become a major ecological issue. In contrast to naturally occurring organic compounds that are readily degraded upon their introduction into the environment, some synthetic substances are very resistant to biodegradation by indigenous microorganisms (Asgher et al., 2008). The development of cost effective and at the same time efficient methods for their removal from industrial wastewaters and also from water ecosystems and soils is very important. White-rot fungi (WRF) have the potential to be used as powerful biotechnological tools that can contribute to solve this problem. WRF possess an enzymatic system for lignin degradation, which, due to its broad substrate specificity, has been reported as responsible for the transformation and mineralization of lignin related compounds considered Persistent Organic Pollutants (POPs) such as industrial dyes, chlorophenols, polychlorinated biphenyls, polycyclic aromatic hydrocarbons (PAHs), pesticides and munition wastes (Rodríguez-Couto, 2009; Lanfermann et al., 2015). The main extracellular ligninolytic enzymes are laccases (EC 1.10. 3.2), manganese peroxidase (MnPs, EC 1.11.1.13), lignin peroxidases (LiPs, EC:1.11.1) and versatile peroxidases (VPLs, EC 1.11.1.16) (Janusz et al., 2013; Rivera-Hoyos et al., 2015). Fungal laccases use the redox ability of copper ions to catalyze the oxidation of a broad range of aromatic substrates concomitantly with the reduction of molecular oxygen to water (Giardina et al., 2010). LiPs are able to oxidize high redox-potential aromatic compounds and non-phenolic lignin model dimers while MnPs oxidizes Mn2+ to Mn3+ (Martínez, 2002). VPLs combine the substrate specificity characteristics of both LiPs and MnPs enzymes (Martínez, 2002; Pérez-Boada et al., 2005). A rapid and practical approach for verifying the presence of this non-specific enzymatic system involved in the degradation of xenobiotic compounds is to determine the capacity of the WRF strains to decolorize model chromophoric compounds (Maganhotto et al., 2005). Model dyes such as Remazol Brilliant Blue R (RBBR) (anthraquinone dye) and Reactive Black 5 (azo dye) have been conveniently used to screen large numbers of fungi with ligninolytic activity and degradative capacity (Levin et al., 2010; Jamal et al., 2011; Zeng et al., 2012). These model dyes have a chemical structure similar to different POPs, therefore these chromophoric compounds are used in biodegradation studies of these xenobiotic compounds.
A majority of the earlier studies focused on the lignin-degrading enzymes of Phanerochaete chrysosporium and Trametes versicolor. Nowadays there exists a growing interest in finding better lignin-degrading systems to use them in biotechnological applications. The genus Ganoderma was extensively investigated because some of its species possess medicinal properties (Park et al., 2012; Kües et al., 2015). However, the potentialities of its ligninolytic machinery have attracted little attention. Nevertheless, some authors reported interesting decolorization properties by some Ganoderma sp. strains (Murugesan et al., 2007; Zhuo et al., 2011; Manavalan et al., 2013). Furthermore, it has been described that the majority of the investigated Ganoderma strains are able to produce laccase enzymes at higher levels compared with peroxidases (D’Souza et al., 1999; Murugesan et al., 2007; Mendonça et al., 2008; Zhuo et al., 2011). Therefore, the laccase enzymes from different Ganoderma strains have been purified and characterized (Ko et al., 2001; Teerapatsakul et al., 2007; Kumar et al., 2015). In addition, there exist a few reports related with the detection of genes coding laccases (D’Souza et al., 1996; Joo et al., 2007; Zhuo et al., 2011; Manzano et al., 2013; You et al., 2013) and peroxidases (D’Souza et al., 1999) from Ganoderma lucidum. Zhuo et al. (2011) and You et al. (2013) reported the molecular cloning of a laccase gene from G. lucidum and their heterologous expression.
Several authors studied the bioremediation capacity of WRF strains deposited in public collections (Jaouani et al., 2003). However, there have been less investigations attempting to exploit directly local biodiversity (Pointing et al., 2000; Sanchez et al., 2008). However, this approach appears to be potentially productive for identifying new, promising strains for biotechnological applications (Pointing et al., 2003).
In case of the genus Ganoderma the studies have been performed using mainly single strains from culture collections and with diverse ecological origins, but the genetic potential and ligninolytic machinery of several well-adapted autochthonous Ganoderma strains have not been explored.
A high biodiversity of the genus Ganoderma (Minter et al., 2001) and the description of different species such as G. australe, G. zonatum, G. opacum, G. colossus, G. lucidum, G. coffeatum, G. flaviporum (Pérez and Camino, 2000; Cabarroi et al., 2008, 2014) and G. weberianum (Manzano et al., 2013) have been reported for Cuba. Moreover, Almaguer et al. (2014) described the presence of airborne spores of the Ganoderma genus in the atmosphere of Havana as one of the predominant spores of basidiomycetes. In spite of this, the diversity of ligninolytic enzymes and their genes in Cuban native strains of Ganoderma genus remain unexplored; only Manzano et al. (2013) described the presence of five new laccase genes and several laccase isozymes present in the strain Ganoderma weberianum B-18. Therefore, the study of native strains of Ganoderma sp. may lead to an untapped genetic potential for ligninolytic enzymes that could be applied for degradation of POPs. The main objectives of this work are: (1) To analyze the diversity of ligninolytic enzymes and genes of Cuban native strains from the genus Ganoderma. (2) To evaluate the contribution of the ligninolytic enzymes to the degradation of model chromophoric compounds.
Materials and Methods
Isolation and Identification of WRF Strains Belonging to the Genus Ganoderma
The fruiting bodies of basidiomycetes belonging to the genus Ganoderma were taken from the base of the trees or decayed wood located in different urban areas (such as parks and main streets) in Havana, Cuba during the years 2013 and 2014. They were identified as Ganoderma based on their typical morphology. Pure fungal cultures were isolated from the context of fruiting bodies by using Malt Extract Agar (Merck, Germany) according to the methodology of Guglielmo et al. (2007) and Manzano et al. (2011). Genomic DNA was isolated using Wizard Genomic DNA Purification Kit (Promega, United States). Taxonomic confirmation of the strains was done by means of multiplex polymerase chain reaction (PCR)-based on the amplification of the internal transcriber spacer region of ribosomal DNA (primers ITS 1, ITS 4, Table 1) and taxon-specific primers (Gano 2R, Table 1) for Ganoderma species (Guglielmo et al., 2007). The PCR conditions were as reported Guglielmo et al. (2007). Purified PCR products were sequenced at Macrogen, The Netherlands.
Table 1.
Sequence of primers used in the present study.
| Primers | Nucleotide sequences (5′–3′) | Reference | Specificity |
|---|---|---|---|
| ITS 1 | TCCGTAGGTGAACCTGCGG | Guglielmo et al., 2007 | ITS-5.8S rRNA gene |
| ITS 4 | TCCTCCGCTTATTGATATGC | ||
| Gano 2R | TATAGAGTTTGTGATAAACGCA | ||
| Cu 1F | CAT(C)TGGCAT(C)GGNTTT(C)TTT(C)CA | Luis et al., 2004 | Laccase gene |
| Cu 2R | G G(A)CTGTGGTACCAGAANGT NCC | ||
| pmnp 1 | ACCTTCCACGACGCTATT | Sarkar et al., 1997 | Manganese peroxidase gene |
| pmnp 2 | GACATCGGAGCAATCGAT |
The primers were synthesized at Biolegio (Nijmegen, Netherlands).
Determination of Ligninolytic Enzymes and Isozymes Produced by Ganoderma Strains
Ganoderma strains were cultured in 100 mL Erlenmeyer flasks with 20 mL of SB-U medium (33.9 mL.L-1 of sugarcane molasses, 3 g.L-1 urea, 1 g.L-1 KH2PO4 and 0.5 g.L-1 MgSO4.7 H2O, pH 5.5, Manzano et al., 2013). The incubation of the strains in SB-medium was performed in agitated conditions at 100 rpm. For determination of the activities of ligninolytic enzymes and isozyme analysis, extracellular crude enzymatic extracts were obtained after removal of the mycelium by filtration (Sartorius 0, 22 μm, United Kingdom) every 24 h.
The laccase activity was determined spectrophotometrically by examining the oxidation of 2 mmol.L-12,6-dimethoxyphenol (DMP) to 2,2′,6,6′-dimethoxydiphenoquinone in 100 mM acetate buffer, pH 5.0 at 468 nm (𝜀468 nm: 49 600 mmol-1.L.cm-1) (Teerapatsakul et al., 2007). The activity of non-specific peroxidase (NsP) was measured by the oxidation of 0.5 mM o-dianisidine in presence of 4 mM H2O2 in 50 mM of sodium acetate buffer, pH 5 at 445 nm (𝜀445 nm: 47 665 mmol-1.L.cm-1) (Claiborne and Fridovich, 1979). The enzymatic activities were defined as the amount of enzyme required to produce 1 μmol product.min-1 at 30°C and expressed as international units per liter of medium (U.L-1).
Isozyme analysis of extracellular crude enzyme extracts was performed by a native polyacrylamide gel electrophoresis (native-PAGE, 15%) (Murugesan et al., 2007). The staining of the gels was performed in 100 mM acetate buffer (pH 5.0) with 10 mM 2,6 dimethoxyphenol (Manzano et al., 2013). Dark orange bands indicated the presence of laccase activity. To detect peroxidase activity the gels were stained with 2, 6 dimethoxyphenol together with H2O2 (0.1 mmol.L-1) and MnSO4 (0.1 mmol.L-1).
Diversity of Laccase and Manganese Peroxidase Genes
The analysis of the diversity of laccase and MnPs genes of the Ganoderma strains was carried out by means of PCR amplification and sequence analysis. The primers were designed previously taking into account highly conserved regions in DNA sequences of these genes in basidiomycetes (Table 1). For the amplifications, 1 μL of the DNA extract was added to a 50 ml reaction mixture containing 5 μL of 10X High Fidelity PCR Buffer, 2 μL of 50 mM MgSO4, 1 μL of dNTP Mix (10 mM each) 1 μL of each primer (10 μM), and 0.2 μL of Platinum® TaqDNA Polymerase High Fidelity (5 U.μL-1). All the PCR components were obtained from Invitrogen Life Technologies, United States. Genomic DNA of strains Ganoderma weberianum B-18 and Phanerochaete chrysosporium MUCL 19343 were used as positive controls for the amplification of the laccase and mnp genes, respectively. PCRs were run on a Master cycler gradient system (Eppendorf, Hamburg, Germany) with an initial denaturation cycle (2 min at 94°C) followed by 35 cycles with denaturation (30 s at 94°C), annealing for laccase amplification (60 s at 50°C) and for mnp amplification (60 s at 53°C) and elongation (2 min at 68°C) and by a final elongation (10 min at 68°C). PCR reactions without DNA were used as negative controls.
The PCR products were separated by electrophoresis in 2% (w/v) agarose gels. PCR products purified from gel were cloned into TOP10 chemically competent Escherichia coli after their ligation to a pCR4-Topo Vector according to the manufacturer’s instructions for the TOPO TA Cloning Kit (Invitrogen Life Technologies, United States). The plasmid DNA, containing the PCR product, was extracted from Escherichia coli TOP10 by using the GeneJet Plasmid Miniprep Kit (Invitrogen Life Technologies, United States). The purified recombinant plasmids were sequenced using the universal primers (M13for and M13rev) of Macrogen Europe (The Netherlands). Fifteen clones per cloning reaction were sequenced and analyzed (Kellner et al., 2007).
Evaluation of Decolorization Capacity of Ganodermasp. Strains
The strains were tested for their in vitro ability to decolorize model chromophoric compounds such as anthraquinone dyes [Remazol Brilliant Blue R (RBBR), Intraacid Blue 62 (AB-62)] and azo dyes [Reactive Black 5 (RB-5) and Intraacid Navy (INT-R)]. For this purpose we used the extracellular crude enzymatic extracts obtained in SB-medium at the day of the maximun ligninolytic enzyme production. The experiment was carried out in reaction tubes containing 100 mM acetate buffer pH 5, 100 μL of crude enzymatic extracts and 100 mg.L-1 of each dye individually. The reaction tubes were incubated in agitation conditions at 100 rpm at 30°C and complete darkness during 12 h (Camarero et al., 2005).
All experiments were performed in triplicate. An abiotic control without enzyme addition was included and a control with the heat inactivated enzyme (autoclaved at 121°C, 15 min) was also used. Decolorization percentages were determined by the difference in absorbance between sample filtrates and abiotic control at the corresponding maximum wavelength for each dye.
Sequence Analysis
Identical/similar nucleotide sequences were identified by BLAST (Basic Local Alignment Search Tool) search on sequences available online in the databases GenBank, NCBI, EMBL, DDBJ, and PDB. All the sequences were edited and analyzed using CLC Main Work Bench 7.5.1 and Vector NTI 7 software.
To determine the intron positions of the amplified laccase and MnP genes, the obtained sequences were aligned with known cDNA coding laccase or MnP genes from basidiomycetes (e.g., T. versicolor; Accession No. L78077). Multiple alignment was performed by means of Clustal W (Thompson et al., 1994) using the following parameters: gap opening penalty = 10, gap extension penalty = 0.2, and Gonnet protein weight matrix. The introns were discarded following the intron splice junctions corresponding to the general eukaryotic rule –5′-GTA/G…..T/CAG-3′ (Kupfer et al., 2004; Luis et al., 2004; Hildén et al., 2005). The protein sequences translated with Vector NTI 7 software were used for phylogenetic analysis.
The phylogenetic analyses were performed by the Maximum Likelihood Analysis and Bayesian Analysis with software Mega 7.0 (Kumar et al., 2016) and BEAST (Bayesian Evolutionary Analysis Sampling Trees (BEAST) v 1.8.4, Drummond et al., 2012) respectively. The Tracer software v1.6.0 (Rambaut et al., 2014) was used to analyze graphically and quantitatively the empirical distributions of continuous parameters obtained after the analysis with BEAST. The FigTree application v 1.4.3 (Rambaut, 2014) was used for displaying the molecular phylogenies.
Statistical Analysis
Results are presented as the average of five replicates. Normality and variance homoscedasticity were investigated prior to carrying out the statistical analyses by means of the Kolmogorov-Smirnov test and the Bartlett test, respectively. Where data met these criteria, an analysis of variance of simple classification (ANOVA) and a parametric Tukey’s test were used. If these preliminary criteria were not fulfilled, the Kruskal–Wallis and SNK tests were used. All data were processed with the statistical package Statistic 7.0.
Results
Isolation of Indigenous White-Rot Fungi Belonging to the Genus Ganoderma
Thirteen native Ganoderma fruiting bodies were collected from decayed wood in Havana (Cuba) based on morphological characteristics of this genus such as the presence or absence of laccate or shiny appearance of the upper surface of the fruiting bodies (Zakaria et al., 2009).
The fungal isolates that were collected were named consecutively as UH-A until UH-M and were deposited in the Collection of Microbial Cultures of Faculty of Biology, University of Havana (CCMFB) with consecutive numbers from CCMFB-H714 until CCMFB-H726. The multiplex PCR showed that all the isolated strains were belonging to the genus Ganoderma (Figure 1).
FIGURE 1.
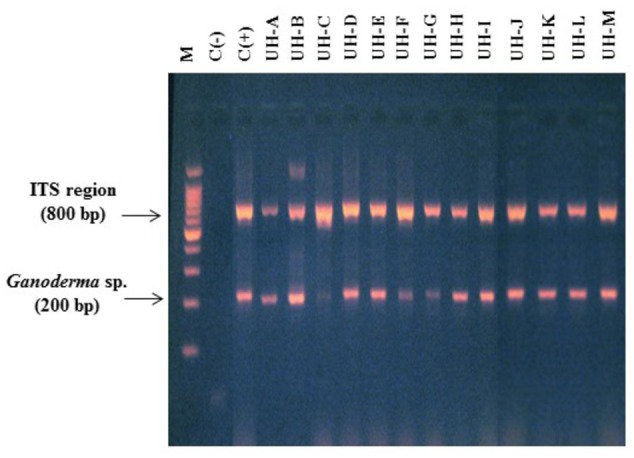
Multiplex polymerase chain reaction (PCR) (primers ITS 1, ITS 4, Gano 2R) for the identification of WRF strains. Lane M. Molecular marker 100 bp Promega. Lanes 1 and 2. Negative and positive controls. Lanes 3–15: strains UH-A until UH-M.
The sequencing of the ITS regions and the analysis of DNA sequences through the BLAST-N program with nucleotide sequences previously published in NCBI GenBank showed that all the ITS sequences obtained from the WRF strains were 99% identical to ITS sequences of different strains of Ganoderma (accession numbers in Gen Bank: JQ514104.1, GQ249880.1, JQ514105.1, HM192933.1, AF255133.1, KF963258.1, JN637827.1, KC884264.1). Table 2 shows the accession numbers of ITS sequences from Ganoderma strains.
Table 2.
Accession numbers of DNA sequences obtained in this work.
| Ganoderma sp. strains | Accession numbers EMBL |
|||
|---|---|---|---|---|
| ITS | Laccase | Manganese peroxidase | Versatile peroxidase | |
| Ganoderma weberianum B-18 CCMFB-H601 | JN637827 | lac I HE585213 | – | LT726744 |
| lac IIHE585214 | ||||
| lac III HE585215 | ||||
| lac IV HE585216 | ||||
| lac V HE585217 | ||||
| UH-A (CCMFB-H714) | LT726719 | lac I LN999878 | LT726718 | – |
| lac II LN999879 | ||||
| lac III LN999880 | ||||
| lac IV LN999881 | ||||
| UH-B (CCMFB-H715) | LT726720 | lac I LN999882 | – | LT726737 |
| lac II LN999883 | ||||
| lac III LN999884 | ||||
| lac IV LN999885 | ||||
| UH-C (CCMFB-H716) | LT726721 | lac I LN999886 | LT726732 | – |
| lac II LN999887 | ||||
| lac III LN999888 | ||||
| lac IV LN999889 | ||||
| UH-D (CCMFB-H717) | LT726722 | lac I LN999890 | – | LT726736 |
| lac II LN999891 | ||||
| UH-E (CCMFB-H718) | LT726723 | lac I LN999892 | – | LT726738 |
| lac IILN999893 | ||||
| UH-F (CCMFB-H719) | LT726724 | lac ILN999894 | – | LT726743 |
| lac IILN999895 | ||||
| lac III LN999896 | ||||
| UH-G (CCMFB-H720) | LT726725 | lac ILN999897 | – | LT726739 |
| lac IILN999898 | ||||
| lac IIILN999899 | ||||
| UH-H (CCMFB-H721) | LT726726 | lac ILN999900 | LT726733 | – |
| lac II LN999901 | ||||
| UH-I (CCMFB-H722) | LT726727 | lac ILN999902 | LT726734 | – |
| lac II LN999903 | ||||
| lac III LN999904 | ||||
| UH-J (CCMFB-H723) | LT726728 | lac I LT796160 | LT726735 | |
| lac II LT796161 | ||||
| lac III LT796157 | ||||
| UH-K (CCMFB-H724) | LT726729 | lac I LN999905 | – | LT726742 |
| lac II LN999906 | ||||
| lac III LN999907 | ||||
| UH-L (CCMFB-H725) | LT726730 | lac ILN999908 | – | LT726740 |
| UH-M (CCMFB-H726) | LT726731 | lac I LT796162 | LT726741 | |
| lac II LT796158 | ||||
| lac III LT 796163 | ||||
| lac IV LT 796159 | ||||
Determination of Ligninolytic Enzymes and Diversity of Laccase Isozymes
In order to determine the ligninolytic enzymes produced by the Ganoderma strains and also to define the day of the maximum production of these enzymes, a time course analysis of laccase and non-specific peroxidases (NsP) production was performed in SB-U medium with sugarcane molasses as carbon source and ligninolytic enzyme inducer (Manzano et al., 2013).
All strains were able to produce NsP and laccase enzymes (Figure 2). The activities of both enzymes were identified from the 2nd day of incubation. For most of the Ganoderma strains the highest productions of both ligninolytic enzymes were reached at the 4th day of incubation. The strains UH-E, B-18, UH-D, UH-M, UH-B, and UH-L showed the highest values for both enzymes. In all strains the levels of laccase activities (42.2–424.3 U.L-1) were one order of magnitude higher than those of the NsP (1.5–9.1 U.L-1).
FIGURE 2.
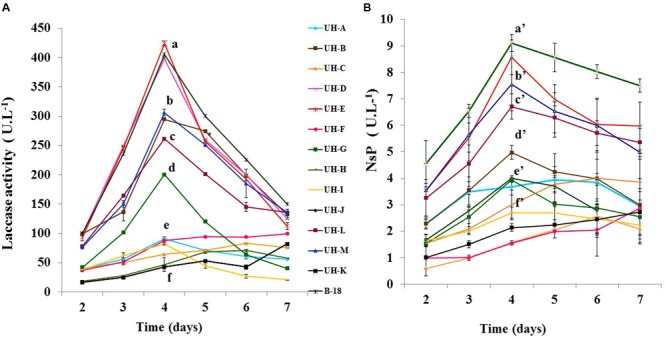
Time course analysis of (A) laccase (B) Non-specific peroxidases (NsP) production by Ganoderma sp. strains in SB-U medium. The cultures were kept at 30°C during 7 days. The results show the means ± SD of five independent cultures. Different letters indicate significant differences (p < 0.05) according to Tukey HSD test.
The analysis of extracellular crude enzymatic extracts of Ganoderma strains by native-PAGE and staining with an enzyme specific substrate allowed the detection of several laccase isozymes, with diverse isozymatic patterns that differ in numbers, mobility and intensity of the electrophoretic bands (Figure 3).
FIGURE 3.
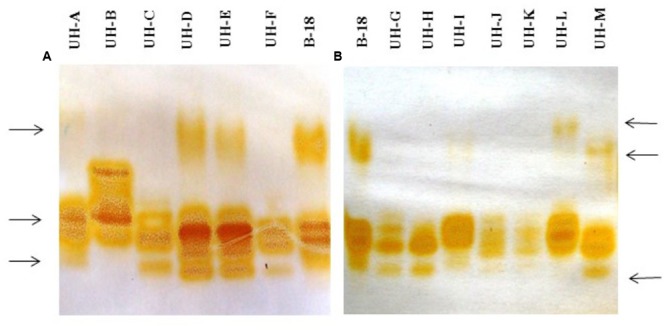
Native polyacrylamide gel electrophoresis analysis of extracellular crude enzymatic extracts of Ganoderma strains. (A) Ganoderma strains UH-A until UH-F. (B) Ganoderma strains UH-G until UH-M. Staining with 2,6 dimetoxyphenol.
Not all the strains produced the same laccase isozymes. The main differences were the following: there was a band with less electrophoretic mobility in the strains B-18, UH-A, UH-D, UH-E, UH-L, and UH-M; however, there were some differences in the electrophoretic mobility of this isoform between the strains UH-M and UH-L. There was a high-intensity band that it was only present in the strain UH-B. Finally, the strains B-18, UH-D, UH-E, UH-F, UH-G, UH-H, and UH-M produced the same band with a lower electrophoretic mobility.
Although NsP activity was detected in extracellular enzyme extracts, it was not possible to detect other orange zones when gels were stained with the 2,6 DMP assayed together with H2O2 and MnSO4, which are favorable conditions for the catalytic action of ligninolytic peroxidases.
Diversity of Genes Encoding Ligninolytic Enzymes of Ganoderma Strains
Amplification of Laccase Gene
Taking into account that all the Ganoderma strains produced several laccase isozymes and to obtain a better knowledge of the laccase system of Ganoderma strains, we investigated if the laccase isozymes detected in these strains are coded by one or more laccase genes and also examined the novelty of their sequences. PCR products obtained with the primers Cu1F/Cu2R showed DNA fragments of sizes ranging from 144 to 240 bp (Figure 4), which was consistent with the expected amplicon size (Luis et al., 2004). In some cases we obtained more than one band in the agarose gel electrophoresis.
FIGURE 4.
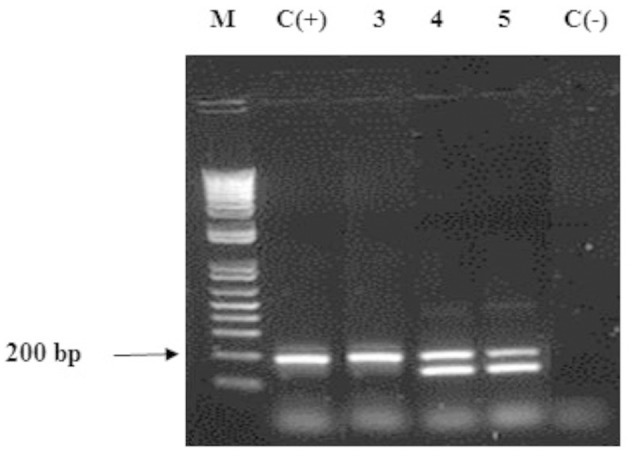
Agarose gel electrophoresis (2% w/v) of fragments of laccase genes amplified by PCR on Ganoderma strains. Lane M: DNA ladder mix (1kb, Invitrogen), lane 2: positive control Ganoderma weberianum B-18. Lanes 3–5: Ganoderma UH. strains (illustration of the 13 strains). Lane 6: negative control.
The analysis of DNA sequences through the BLAST-N program with nucleotide sequences published in NCBI GenBank allowed to distinguish different laccase genes in each Ganoderma UH strain (EMBL Nucleotide Sequence Database LN999879 - LN999908, Table 2). The laccase genes detected were 80–87% identical to laccase genes of Ganoderma weberianum (lac 1: HE585213.1, lac 2: HE585214.1, lac 4:HE585216.1), Ganoderma lucidum (KC507947.1, DQ914870.1, JN654464.1), Trametes sp. (AY846842.1), Flammulina velutipes (AY485826.1), Lenzites gibbosa (JF817353.1) and uncultured basidiomycetes (AJ420341.1, EU882652.1).
To analyze the diversity of the laccase genes detected, a phylogenetic tree (Figure 5) was constructed with the translated amino acid sequences of laccase gene fragments from Ganoderma strains and closely related laccase protein sequences retrieved from GenBank after the Blast P analysis. A representation of sequences belonging to the different laccase families (Sirim et al., 2011) was included in the analysis (Family B: ascomycete Fusarium oxysporum B1 uni A7LBK4, Family G: plant Arabidopsis thaliana G1 uni Q9FY79.1, Family C: insect Anopheles gambiae C1 uni Q8I8Y2 and Family K: the copper oxidase Streptomyces ghanaensis ATCC 14672 K. Phylogenetic trees obtained by Bayesian analysis and Maximum Likelihood correspond in terms of the general tree topology; here, we only present the Bayesian tree with the posterior probability in each node.
FIGURE 5.
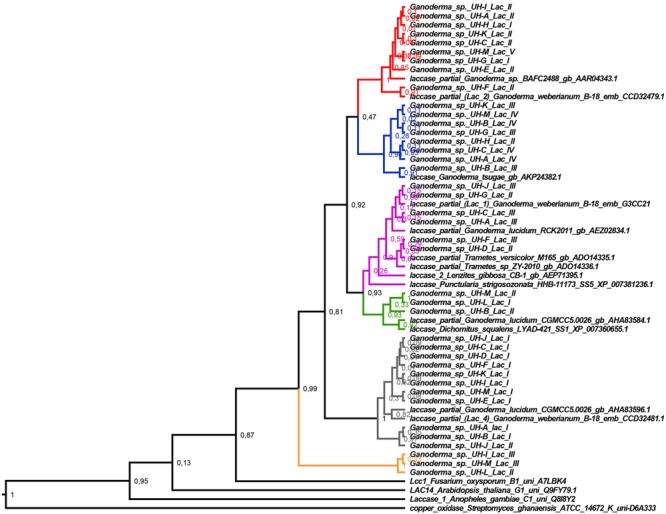
Bayesian tree of deduced amino acid sequences of laccase gene fragments from the Ganoderma strains and the closely related sequences retrieved from GenBank after the Blast P analysis. Posterior probability values are shown in each node. The red, blue, violet, green, gray, and orange branches indicate the different specific clades that are formed. The Bayesian tree was constructed with BEAST v 1.8.4 with the following parameters: substitution model: WAG model with Gamma distribution, tree prior: Yule process, Markov chain Monte Carlo: length of chain 20 000 000. The rest of parameters were set as default options.
Interestingly, the laccase genes detected in the Ganoderma strains did not group in a common clade. Six principal clades were formed where the laccase genes of different strains were grouped together with other laccase protein sequences belonging to the Family A of laccase (Basidiomycete Laccase) according to The Laccase Engineering Database1 (Sirim et al., 2011). The major clades that were formed are supported with high posterior probability values after the Bayesian analysis. This fact confirms the differences between these laccases.
A relatively high diversity of laccase genes was found among the Ganoderma strains. All strains have at least two highly distinctive laccase gene sequences. This supports the detection of several laccase isozymes in all Ganoderma strains. The laccase partial sequences grouped in each clade have identical amino acid sequences, with the exception of the sequences UH-K Lac II, UH-F Lac II, UH-B Lac II. This fact was confirmed by multiple alignment of these sequences and pairwise distances analysis.
Figure 6 illustrates the multiple alignment of the laccase deduced amino acid sequences of Ganoderma strains with 11 of the laccases presented in the phylogenetic tree. The alignment showed that all the laccase proteins detected in UH strains contained the conserved histidine residues involved in copper-binding regions I and II (T2 and T3-sites ligands) of the active site (Giardina et al., 2010) as well as the L1 region with the patterns [H-W-H-G-x(9)–D-G-x(5)–Q-C-P-I] and a section of the region L2 (GTFWYHS).
FIGURE 6.
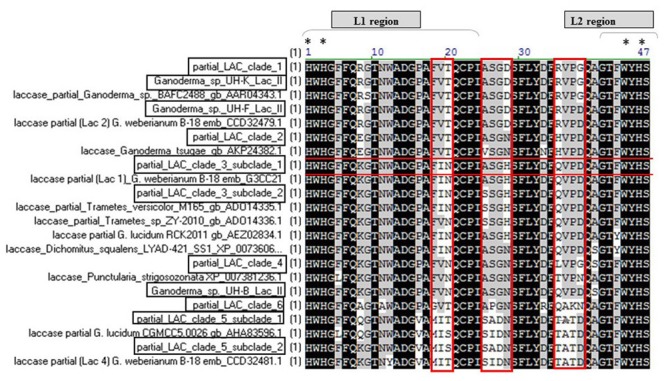
Extracted fragment from the multiple alignment the deduced amino acid sequences of laccase fragments from Ganoderma strains with the laccases presented in the phylogenetic tree. The position residues conserved in all sequences are highlighted in black. The positions conserved in 80 and 60% of the sequences, respectively, are highlighted in dark and light gray. The L1 region and a section of L2 region present in the studied fragments as well as the histidine residues involved in copper-binding (T2 and T3-sites ligands marked with asterisks) of the active site are indicated. LAC clade 1 (UH-I lac II, UH-A lac II, UH-H lac I, UH-K lac II, UH-C lac II, UH-M lac V, UH-G lac I, UH-E lac II), LAC clade 2 (UH-M lac IV, UH-B lac IV, UH-G lac III, UH-H lac II, UH-C lac IV, UH-A lac IV, UH-B lac III), LAC clade 3 subclade 1 (UH-C lac III, A lac III, G lac II), LAC clade 3 subclade 2 (UH-F lac III, UH-D Lac II), LAC clade 4 (UH-M lac II, UH-L lac I), LAC clade 5 subclade 1 (UH-J lac I, UH-C lac I, UH-D lac I, UH-F lac I, UH-K lac I, UH-I lac I, UH-M lac I, UH-E lac I), LAC clade 5 subclade 2 (UH-A lac I, UH-B lac I, UH-J lac II) LAC clade 6 (UH-I lac III, UH-M lac III, UH-L lac II).
In the L1 and L2 regions, considered as signature regions of laccase enzymes (Figure 6) the copper ligand residues HWHG and WYHS of the L1 and L2, respectively, are highly conserved in all the sequences, however, not all the amino acids of the L1 region are preserved in all laccase proteins from the Ganoderma strains. This is especially evident if we consider the residues FVT, ASGD and RVPG (marked in red) which are not conserved in all the laccase sequences of Ganoderma UH strains.
With the exception of partial laccases that are grouped in clade 3 subclade 2 (sequences UH-F lac III, UH-D Lac II, marked in red in Figure 6), which is 100% identical to a partial lac 2 sequence of Ganoderma weberianum B-18, the rest of the partial laccase protein sequences from the studied Ganoderma strains showed differences in their amino acid sequences with respect to laccases previously described in GenBank. The most divergent protein and with more novelty in its sequence are the partial laccase proteins grouped in clade 6 (sequences UH-I lac III, UH-M lac III, UH-L lac II).
Amplification of Gene Encoding Manganese Peroxidase
Polymerase chain reaction using primers pmnp1 and pmnp2 amplified the expected 1.0 kb fragment from the genomic DNA of all Ganoderma UH strains (Figure 7). The analysis using the BLAST-N program and multiple alignment of the cloned sequences revealed that in contrast to laccases, only one gene encoding peroxidases was present in all Ganoderma sp. strains (Table 2). In order to analyze the phylogenetic relationship with previously studied fungal peroxidases, a Bayesian tree (Figure 8) was constructed with the translated amino acid sequences of genes encoding peroxidases detected in the UH strains, the sequences retrieved from GenBank after the Blast P analysis and sequences that belong to class II peroxidases.
FIGURE 7.
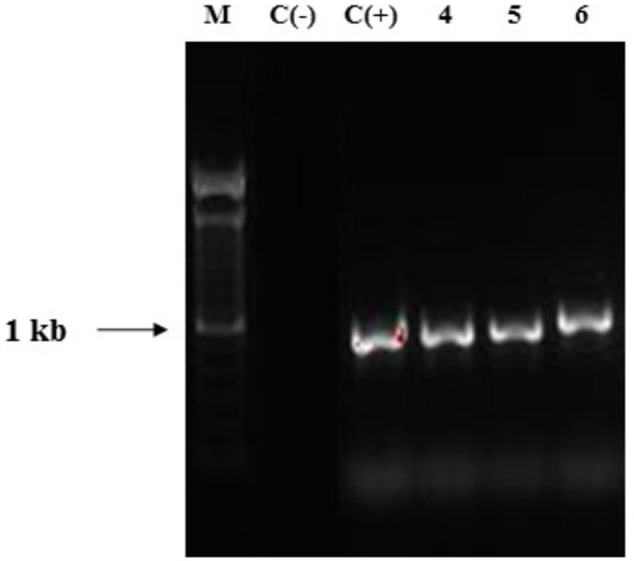
Agarose gel electrophoresis of PCR products obtained with primers pmnp1/ pmnp2. Lane 1: DNA ladder mix (1kb, Invitrogen), Lane 2: negative control. Lane 3: positive control Phanerochaete chrysosporium MUCL 19343 Lanes 4–6 Ganoderma strains (representative for the 13 strains).
FIGURE 8.
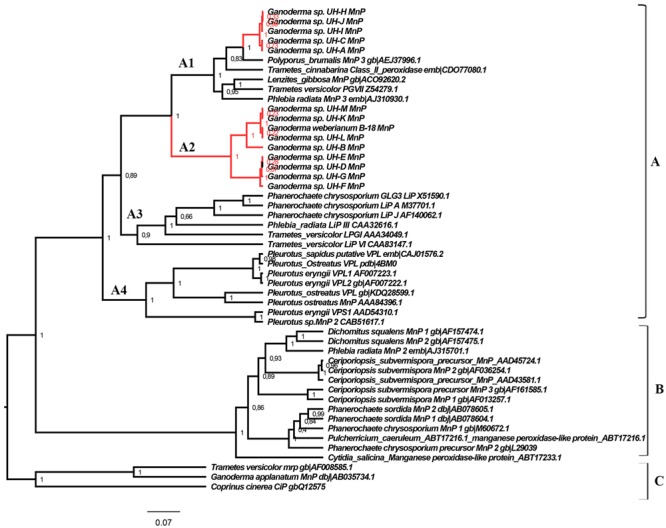
Phylogeny of the class II fungal secretory heme peroxidases. The main phylogenetic subgroups A, B, and C and translated peroxidases sequences of Ganoderma strains are illustrated. Posterior probability values are shown in each node. The Bayesian tree was constructed with BEAST v 1.8.4 with the following parameters: substitution model: LG model with Gamma distribution, tree prior: Yule process, Markov chain Monte Carlo: length of chain 20 000 000. The rest of parameters were set as default options.
The translated peroxidases enzymes detected in the Ganoderma strains were clustered in group A of class II peroxidases (Hildén et al., 2005), but in two different clades: clade A1 and clade A2. In group A we can also find sequences of LiPs (clade A3), VPL (clade A4) and the short-type hybrid MnPs variants (clades A1, A4) more related to LiPs and VPLs than to the classical long MnPs (group B). The sequences of group C are structurally related, but are functionally different from the ligninolytic enzymes forming the majority of class II fungal peroxidases.
Interestingly, the sequences of peroxidases detected in Ganoderma UH strains were related to short-type hybrid MnPs variants, but were also associated to LiPs sequences of Phanerochaete chrysosporium, T. versicolor, and Phlebia radiata (Figure 8). To determine the similarities that peroxidases identified in this work share with the LiPs and VPLs studied previously we performed a multiple alignment analysis (Figure 9) with sequences of these peroxidases grouped in different clades of the phylogenetic tree obtained. In the alignment we searched for the presence or absence of the active site residues for manganese binding characteristic of manganese dependent enzymes (MnPs) and the critical residues for the three different long-range electron transfer (LRET) pathways proposed for the substrate oxidation that LiPs and VPLs can perform (Pérez-Boada et al., 2005; Morgenstern et al., 2008).
FIGURE 9.
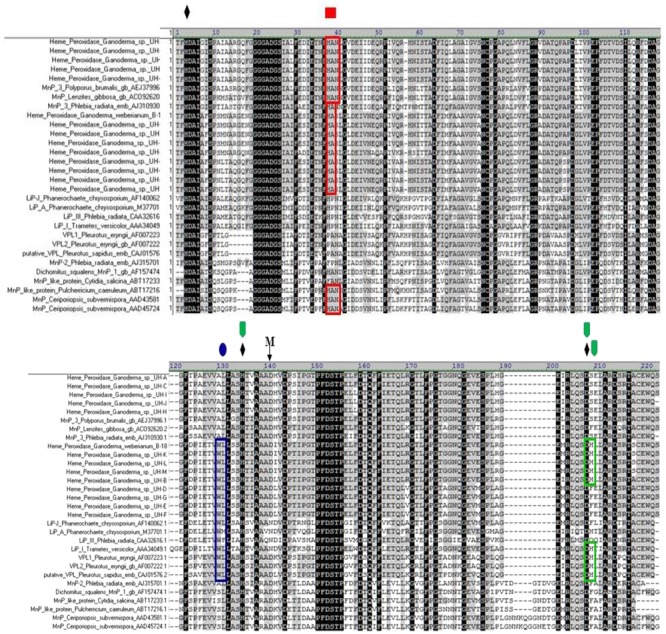
Multiple amino-acid sequence alignment of the deduced amino acid sequence of MnP from Ganoderma strains in comparison to other deduced fungal manganese peroxidases. Functional residues described in the text are marked with symbols:distal and proximal histidines, M- manganese binding residues,  and aminoacids marked in green;
and aminoacids marked in green;  and aminoacids marked in blue ;
and aminoacids marked in blue ;  and aminoacids marked in red are residues proposed to be active in the LRET I, LRET II and LRET III pathway respectively
and aminoacids marked in red are residues proposed to be active in the LRET I, LRET II and LRET III pathway respectively
In all the predicted amino acid sequences of Ganoderma UH strains, the catalytically indispensable amino acids within the heme environment were found. The two histidines (heme peroxidase proximal H132 and distal H3) and the aspartate D 206 were identified in the alignment as ♦. These residues are conserved in all class II peroxidases.
In all partial sequences of peroxidases from UH strains it was possible to identify one residue involved in Mn-binding, the D 138 (M in the alignment). The other two residues involved in Mn-binding were not amplified with the primers used. The amino acid (D138) is conserved in all the translated amino acid sequence obtained from UH strains as well in the sequence VPL1 and VPL2 of Pleurotus eryngii. This residue is not present in that position in the sequences of LiPs analyzed.
Residues required for the LRET I pathway are present only in the peroxidases sequences of the strains UH-L, UH-K, UH-M, B-18, and UH-B as well in all the sequences of LiPs of T. versicolor (AA34049.1) and in VPLs of Pleurotus eryngii (AF007223.1, AF007222.1) and Pleurotus ostreatus (CAJ015762) described previously. The critical tryptophan residue required for the LRET II pathway is present in all sequences of Ganoderma strains grouped in clade A2 as well as in all sequences of the LiPs and VPLs analyzed. However, it is missing from the sequences of clade A1 and from sequences of MnPs grouped in clade A1 and in group B. The residues hypothetically involved in the LRET III pathway are present in all the sequences analyzed in the alignment, however, there are some differences between the sequences from clade A1 and A3.
Taking into account that the peroxidases grouped in clade A2 from strains UH-L, UH-M, UH-K, UH-B, UH-D, UH-E, UH-G, UH-F, B-18 contain the typical manganese binding residues characteristics of MnPs and also the residues involved in the LRET II described experimentally in LiPs and VLPs, we can consider these sequences as putative versatile peroxidases (VLPs) at functional level because these enzymes combine the catalytic properties of LiPs and MnPs due to the presence of Mn-binding residues and oxidation sites such as LRET described previously.
The peroxidases sequences of Ganoderma sp. strains grouped in clade A1: strains UH-H, UH-I, UH-J, UH-C, UH-A have in their sequence the LRET III, however, this oxidation pathway has not been demonstrated experimentally in VPLs or LiPs. Therefore, the peroxidases of these strains can be classified as Manganese Peroxidases enzymes taking into account that in their sequence the Mn binding site is present.
Decolorization Capacity of Ganoderma sp. Strains
The extracellular crude enzymatic extracts, obtained at the day of maximum ligninolytic enzyme production by the Ganoderma strains, showed able to decolorize the model anthraquinone and azo dyes that were examined (Figure 10).
FIGURE 10.
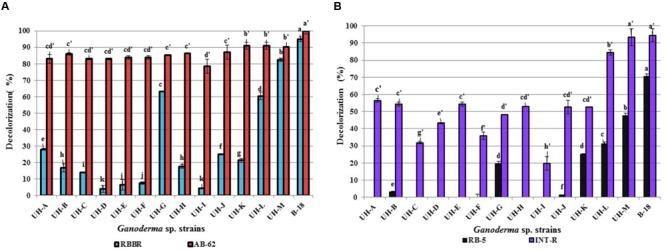
Decolorization percentages of 100 mg.L-1of (A) anthraquinone dyes (B) azo dyes, reached by extracellular crude enzymatic extracts produced by the Ganoderma strains. The results show the means ± SD of five independent cultures. Different letters indicate significant differences (p < 0.05) according to Tukey HSD test.
All Ganoderma strains proved capable to degrade differentially the four examined model dyes. The anthraquinone dye AB-62 was degraded to a large extent by all strains (78–100%) followed by the azo dye INT-R (19–94%). The anthraquinone dye RBBR was also degraded by all the strains, but with lower decolorization percentages. Only four strains (UH-G, UH-L, UH-M, and B-18) accomplished decolorization percentages higher than 60% for this dye. The azo dye RB-5 showed to be the most recalcitrant to degradation; only the strains UH-M and B-18 obtained 47 and 70% of decolorization, respectively, the other five strains (UH-B, UH-G, UH-J, UH-K, and UH-L) presented low decolorization values (between 1 and 31%).
Discussion
In the present study, we isolated 13 basidiomycetes belonging to the genus Ganoderma from urban ecosystems in Havana, Cuba. The native strains exhibited differences between the activities of ligninolytic enzymes, the isozyme profiles and the genes encoding for laccase, MnPs and VPLs enzymes. Extracellular crude enzymatic extracts produced by the Ganoderma strains showed able to degrade the model chromophoric compounds such as anthraquinone and azo dyes, making them candidates for further exploration in function of treatment of wastewater from the textile industry and/or for bioremediation of matrices polluted with POPs.
The strains were isolated in Havana because this city is one of the most contaminated urban areas in Cuba due to the presence of industrial activities, for example the Refinery of Crude Oil. Moreover, the air of the city is polluted with the exhaust of old cars and busses. For all these reasons, the WRF that grow in such kind of ecosystems are exposed to a whole range of chemical compounds (Lorenzo and Lázaro, 2003). These fungi should be adapted also to conditions of high temperatures and low humidity in comparison with natural ecosystems (Sanchez et al., 2008).
After identification of the WRF strains as Ganoderma sp., their degradative capacity was characterized taking into account their capability to produce high activity levels of ligninolytic enzymes, the diversity of the genes coding for these ligninolytic enzymes and the decolorization efficiency of these enzymes against model chromophoric compounds.
Some WRF produce all three ligninolytic enzymes while others produce only one or two of them (Wesenberg et al., 2003). The SB-U medium with sugarcane molasses as carbon source and inducer of ligninolytic enzymes (Manzano et al., 2013) allowed to grow the Ganoderma strains and also the production of ligninolytic enzymes. Sugarcane molasses is a byproduct of sugar production, it contains numerous compounds like sugars, amino acids, proteins, vitamins, minerals allowing fungal growth but also contain phenolic acids and aromatic compounds derived from lignin that stimulate production of ligninolytic enzymes (Otero et al., 2000). Due to the composition of the SB-U medium and the inducing effects of their components, the maximum ligninolytic production was reached after 4 days of incubation by all Ganoderma strains (Figure 2). From the point of view of an eventual industrial application, the use of the SB-U medium offers obvious economic advantages since it allows to reach in a shorter time the maximum ligninolytic production by WRF strains and its components such as urea, KH2PO4, MgSO4.7 H2O have low cost and the sugarcane molasses are available in all sugar producing countries.
All Ganoderma strains produced laccase enzymes at higher levels than non-specific peroxidases (Figure 2); this suggests that the Ganoderma strains secreted laccases as dominant ligninolytic enzymes in the conditions used for cultivation. Our results agree with earlier findings reporting high levels of laccase production in other Ganoderma strains (Teerapatsakul et al., 2007; Deswal et al., 2012; Fang et al., 2015). The laccase activities produced by Ganoderma strains B-18, UH-E, UH-D, UH-M, UH-B, UH-L, and UH-G in SB-U medium were similar and even higher in comparison to laccase activities produced in different culture conditions by other WRF basidiomycetes, including Ganoderma strains. For example Novotny et al. (2001), Kokol et al. (2007), and Šnajdr and Baldrian (2007) reported laccase values between 0.31 and 29.4 U.L-1 for promising WRF strains such as Irpex lacteus, Ischnoderma resinosum, T. versicolor, and Pleurotus ostreatus. Laccase activities of 27, 80, and 120 U.L-1 were produced by Ganoderma sp. (Maganhotto et al., 2005), Ganoderma australe (Mendonça et al., 2008), and Ganoderma sp. En3 (Zhuo et al., 2011). All these WRF strains were able to decolorize synthetic dyes.
Often, more than one isoform of ligninolytic enzymes is expressed by different WRF depending on the fungal species and the environmental conditions (Majeau et al., 2010; Yuan et al., 2016). The Ganoderma strains produced different types and numbers of laccase isozymes in the SB-U medium (Figure 3). The strains UH-B, UH-D, UH-E, UH-L, and UH-M presented higher numbers of electrophoretic bands in comparison with the other strains (UH-A, UH-C, UH-F, UH-G, UH-H, UH-J, and UH-K). Moreover, they showed a similar isoenzyme pattern to the Ganoderma weberianum B-18 strain (used as a positive control in this experiment). The presence of these laccase isoforms in the cathodic region of the electrophoresis gel could contribute to the higher levels of laccase enzyme detected in the strains UH-B, UH-D, UH-E, UH-L, and UH-M. Moreover, the presence of these laccase isoforms with lower electrophoretic mobility could reflect an adaptive value for the degradation of a wide range of complex substrates. The occurrence of a higher number of laccase isozymes in these Ganoderma strains indeed could be advantageous for the degradation of a broad spectrum of xenobiotic pollutants. Several authors (Teerapatsakul et al., 2007; Kumar et al., 2015) demonstrated that single Ganoderma strains produced different extracellular isoforms of laccases using diverse culture conditions and ligninolytic inducers. However, our study is the first concerning diversity of laccase isozymes in several strains of the genus Ganoderma isolated from urban ecosystems.
The diversity of laccase isozymes in our Ganoderma UH strains was associated with the presence of various laccase genes. Between two and five laccase genes were amplified from the genome of each strain (Figure 5) and at least one translated sequence of a laccase enzyme with novelty in its sequence was detected in each strain, especially in the region where the residues responsible to maintain a local three-dimensional fold characterizing the active site are located. For example, the residues of L1 region marked in red in the alignment analysis (Figure 6) are related with the formation of the one of water channels of laccase enzymes. These channels are very important for the catalytic activity of laccases because they allow the access of the molecular oxygen (final electron acceptor) to the active site of the enzyme and also allow the expel of the water molecules produced (Giardina et al., 2010). The fact that we found differences in these important residues may indicate that the laccase isozymes detected in the Ganoderma strains might have novel biochemical properties.
The existence of multiple genes encoding peroxidases has also been described in different ligninolytic fungi (Hofrichter et al., 2010; Järvinen et al., 2012; Janusz et al., 2013). However, in the Ganoderma strains only one peroxidase encoding gene was detected in each strain. The phylogenetic analysis (Figure 8) and multiple alignment examination of translated proteins (Figure 9) revealed differences between the peroxidase encoding genes of the different strains. The peroxidases detected in strains UH-H, UH-I, UH-J, UH-C, and UH-A were classified as manganese peroxidases, however, the translated peroxidases of the strains UH-L, UH-M, UH-K, UH-B, UH-D, UH-E, UH-G, UH-F, and B-18 were classified as putative versatile peroxidase taking into account the presence of residues involved in Mn-binding and residues related with the long-range electron transfer (LRET) pathways described for LiPs and VPLs (Pérez-Boada et al., 2005; Morgenstern et al., 2008).
The molecular structure of ligninolytic peroxidases comprises a heme cofactor situated at an internal cavity (the “heme pocket”) connected to the protein surface through two small access channels. The narrow heme channel prevents the direct contact between the large lignin polymer and the heme. This implies the participation of low molecular mass compounds as peroxidase redox mediators, like the fungal metabolite 3,4-dimethoxybenzyl alcohol (veratryl alcohol). However, even such small compounds appear to be too big to approach the heme through the above-mentioned channel(s). Therefore, LRET pathways from the exterior of the enzyme to the heme cofactor represent a reasonable alternative to explain oxidation of redox mediators, aromatic substrates, and even polymeric lignin performed by LiPs and VPLs enzymes (Pérez-Boada et al., 2005; Morgenstern et al., 2008). Different studies applying crystallographic models (Pérez-Boada et al., 2005; Sundaramoorthy et al., 2005) and site-directed mutagenesis (Gelpke et al., 2002; Pérez-Boada et al., 2005) demonstrated experimentally the occurrence of the LRET I and II pathways in VPLs from Pleurotus eryngii (Pérez-Boada et al., 2005; Ruiz-Dueñas et al., 2009) and in LiPs from Phanerochaete chrysosporium (Ruiz-Dueñas et al., 2001). However, the LRET III is absent from the crystal structure of the VPLs of Pleurotus eryngii and has not been confirmed experimentally in LiPs (Morgenstern et al., 2008). None of the above pathways exists in the crystal structure of MnP of P. chrysosporium. However, the peroxidase sequences of the present study that were classified as manganese peroxidases belonging to the strains UH-H, UH-I, UH-J, UH-C, and UH-A, the residues hypothetically involved in LRET III are present. Our results correspond with the analysis performed by Morgenstern et al. (2008) who found after Bayesian analysis and multiple alignment analysis of several MnPs retrieved from GenBank that some “classical” MnPs peroxidases (belonging to group B in the phylogeny of our work) contain the residues hypothetically involved in LRET III, for example the MnP from Ceriporiopsis subvermispora (AAD43581, AAD45724), Cytidia salicina (ABT17233), and Pulcherricium caeruleum (ABT17233). Morgenstern et al. (2008) postulated that if all of the predicted LRET pathways are functional (which still has to be demonstrated experimentally for the vast majority of sequences), then true MnPs may be less widespread than previously thought.
The deduced peroxidase proteins of strains UH-L, UH-M, UH-K, UH-B, UH-D, UH-E, UH-G, UH-F, and B-18 were classified as putative versatile peroxidases due to the presence of the residues related to Mn-binding and also the critical tryptophan residue related to LRET II pathway. These results are similar to the findings of Morgenstern et al. (2008) with a sequence of Ganoderma applanatum (AB035734.1) annotated as MnP in GenBank and two MnPs sequences of T. versicolor (AAB63460, CAG39281) that possess both manganese binding residues and residues for LRET II and, in fact, might be VPL. These results suggest that VPL might be much more widespread than presently assumed.
The sequences of the peroxidase encoding genes detected in this study have not been described before. These enzymes could be candidates for functional studies in order to determine more precisely the residues involved in the complex catalytic sites of these peroxidases. To the best of our knowledge, the diversity of genes coding ligninolytic enzymes such as laccases, MnPs, and VPLs of different indigenous strains of the genus Ganoderma isolated from Cuban urban ecosystems has not been characterized before.
The presence of different genes encoding laccase isozymes in Ganoderma strains may have an adaptive value for these WRF that grow on complex substrates such as hard wood, and also in changing environments (Luis et al., 2004; Shleev et al., 2007; Majeau et al., 2010) such as the urban ecosystems from where they were isolated. The presence of these laccase isozymes together with the peroxidases described in this study could provide to these strains multiple advantages to survive in their natural habitats, but might also be valuable for the degradation of a variety of recalcitrant environmental pollutants such as POPs.
Indeed, all crude enzymatic extracts produced by the Ganoderma strains could degrade, with diverse percentages, the model chromophoric compounds with chemical structure similar to different POPs compounds after 12 h of incubation (Figure 10). However, the dyes RBBR and RB-5 were the most recalcitrant to degradation. The observed differences in decolorization abilities of these WRF suggest differences in the catalytic properties of the ligninolytic enzymes produced. In addition, the differences in chemical structure between these dyes can affect their biodegradability (Levin et al., 2010). Generally, azo dyes are more recalcitrant to biodegradation due to the presence of sulfonate groups and azo bonds (Rodríguez-Couto, 2007). However, we found that the azo dye INT-R was easily degraded by Ganoderma strains in comparison to the anthraquinone dye RBBR. This is an interesting result, since it has been reported that azo dyes were recalcitrant to the decolorization or could only be decolorized to a limited extent (Nyanhongo et al., 2002; Levin et al., 2010).
Taking into account that Ganoderma strains B-18, UH-L, and UH-M reached the highest percentages of decolorization against the four assayed dyes (between 90 and 100% for AB-62; between 60 and 96% for RBBR, 85–95% for INT-R and 32–70% for RB-5), these strains can be considered as promising candidates for development of an efficient biological system for the treatment of textile industry wastewaters. The values of decolorization achieved by these strains are comparable or superior to those reported by other authors using promising WRF strains for dye degradation. Phanerochaete chrysosporium, Bjerkandera sp. and Irpex lacteus, for instance, were reported to decolorize the dye RBBR with values of 83, 65, and 90%, respectively (Máximo and Costa-Ferreira, 2004). Correspondingly, Zeng et al. (2011) reported that the extracellular crude extract produced by Trametes trogii decolorized the dye RBBR with 85%, but only in presence of the redox mediator 1-hydroxybenzotriazole.
Several studies investigating the decolorization of the diazo-dye RB-5demonstrated that this dye is extremely recalcitrant to degradation due to its complex chemical structure and high redox potential (Camarero et al., 2005; Murugesan et al., 2007). Laccases produced by Pycnoporus cinnabarinus and Trametes villosa showed able to degrade RB-5 with 70 and 80% of decolorization only in presence of the redox mediators syringaldehyde and acetosyringone (Camarero et al., 2005). In contrast, the extracellular crude enzymatic extracts produced by Ganoderma strains B-18, UH-M, and UH-L in our study realized efficient decolorization without the addition of a redox mediator. This is very important taking into account that the laccase-mediator system has yet to be applied on the industrial scale, the cost of mediators and the lack of studies that guarantee the absence of toxic effects of these compounds or their derivatives (Malarczyk et al., 2009).
The fact that all Ganoderma strains isolated in this work could degrade the model dyes under investigation might indicate that the ligninolytic machinery of these strains also possesses the capacity to degrade other xenobiotic compounds with similar chemical structures. These findings could open new perspectives for the development of practical applications for degradation of xenobiotic compounds by well-adapted native WRF strains.
Conclusion
Thirteen native WRF strains were isolated from trees in urban ecosystems in Havana (Cuba). Multiplex PCR with taxa-specific primers and sequence analysis provided strong evidence for classification of all strains as Ganoderma sp. All strains produced laccase as the main ligninolytic enzyme in the crude enzymatic extract although non-specific peroxidase activity was also detected. A PCR and cloning approach using specific primers for basidiomycetes allowed determining the diversity of genes encoding laccase and peroxidase enzymes. The Ganoderma UH strains possess at least one laccase gene with novelty in their sequence, especially in the region where the amino acid residues are involved in maintaining a local three-dimensional fold characterizing the active site of the enzyme. The diversity of the laccase genes detected in the Ganoderma strains indicates the occurrence of several laccase isozymes. In contrast, in each strain, only one gene encoding peroxidases such as MnPs or VPLs was detected. Most of the translated amino acid sequences obtained in the present study have not been described before. All Ganoderma UH strains possess the potential to degrade POPs. The present findings contribute to a better understanding of the properties of a WRF genus such as Ganoderma which have not been studied in details by other authors. These findings could also constitute a good incentive for better protection of fungal biodiversity.
Author Contributions
Design of the work: GT-F, AMML, MR-L, MIS-L, JC, GG, and JV. Conducting experiments: GT-F, AMML, LLLA, FR, and ST. Interpretation of data: GT-F, AMML, LLLA, FR, MR-L, MIS-L, GG, JC, ST, and JV. Drafting the work: GT-F, AMML, FR, JC, and JV. Final approval: GT-F, AMML, MR-L, GG, and JV.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to the financial support of the following grants: the BOF-BILA grant from Hasselt University for GT-F, the UHasselt Methusalem project 08M03VGRJ and to the International Foundation for Sciences (IFS, Sweden) (grant F/4442-1 and F/4442-2).
Funding. This work was supported by a BOF-BILA grant from Hasselt University for GT-F, by the UHasselt Methusalem project 08M03VGRJ and by International Foundation for Sciences (IFS, Sweden) (grant F/4442-2).
References
- Almaguer M., Rojas-Flores T., Rodríguez-Rajo J. Aira María-Jesús. (2014). Airborne basidiospores of Coprinus and Ganoderma in a Caribbean region. Aerobiologia 30197–204. 10.1007/s10453-013-9318-y [DOI] [Google Scholar]
- Asgher M., Bhatti H., Ashraf M., Legge R. (2008). Recent developments in biodegradation of industrial pollutants by white rot fungi and their enzyme system. Biodegradation 19 771–783. 10.1007/s10532-008-9185-3 [DOI] [PubMed] [Google Scholar]
- Cabarroi M., Maldonado S., Castillo L. (2008). Hongos del Jardín Botánico nacional de Cuba. I. basidiomycota. Rev. Jardín Bot. Nac. 29161–169. [Google Scholar]
- Cabarroi M., Maldonado S., Recio G., Camino M., Blanco N., Herrera S., et al. (2014). Catálogo de hongos y Mixomycetes del Jardín Botánico Nacional de Cuba. Washington, DC: Editorial Universitaria. [Google Scholar]
- Camarero S., Ibarra D., Martínez M. J., Martínez A. T. (2005). Lignin-derived compounds as efficient laccase mediators for decolorization of different types of recalcitrant dyes. Appl. Environ. Microbiol 71 1775–84. 10.1128/AEM.71.4.1775-1784.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claiborne A., Fridovich I. (1979). Purification of the o-dianisidine peroxidase from Escherichia coli. Physicochemical characterization and analysis of its dual catalatic and peroxidatic activities. J. Biol. Chem. 254 4245–4252. [PubMed] [Google Scholar]
- Deswal D., Sharma A., Gupta R., Kuhad R. C. (2012). Application of lignocellulolytic enzymes produced under solid state cultivation conditions. Bioresour. Technol. 115 249–54. 10.1016/j.biortech.2011.10.023 [DOI] [PubMed] [Google Scholar]
- Drummond A., Suchard M., Xie D., Rambaut A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29 1969–1973. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza T., Merritt C., Reddy A. (1999). Lignin-modifying enzymes of the white rot basidiomycete Ganoderma lucidum. Appl. Environ. Microbiol. 65 5307–5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza T. M., Boominathan K., Reddy C. A. (1996). Isolation of laccase gene-specific sequences from white rot and brown rot fungi by PCR. Appl. Environ. Microbiol. 62 3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z., Liu X., Chen L., Shen Y., Zhang X., Fang W., et al. (2015). Molecular cloning of a laccase gene from Ganoderma lucidum and heterologous expression in Pichia pastoris. J. Basic Microbiol. 53 S134–S141. [DOI] [PubMed] [Google Scholar]
- Gelpke M. D. S., Lee J., Gold M. H. (2002). Lignin peroxidase oxidation of veratryl alcohol: effects of the mutants H82A, Q222A, W171A, and F267L. Biochemistry 41 3498–3506. 10.1021/bi011930d [DOI] [PubMed] [Google Scholar]
- Giardina P., Faraco V., Pezzella C., Pisutelli A., Vanhulle S., Sannnia G. (2010). Laccases: a never-ending story. Cell Molec. Life Sci. 67 369–385. 10.1007/s00018-009-0169-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmo F., Bergemann S. E., Gonthier P., Nicolotti G., Garbelotto M. (2007). A multiplex PCR-based method for the detection and early identification of wood rotting fungi in standing trees. J. Appl. Microbiol. 103 1490–1507. 10.1111/j.1365-2672.2007.03378.x [DOI] [PubMed] [Google Scholar]
- Hildén K., Martinez A., Hatakka A., Lundell T. (2005). The two manganese peroxidases Pr-MnP2 and Pr-MnP3 of Phlebia radiata, a lignin-degrading basidiomycete, are phylogenetically and structurally divergent. Fungal Genet. Biol. 42 403–419. 10.1016/j.fgb.2005.01.008 [DOI] [PubMed] [Google Scholar]
- Hofrichter M., Ullrich R., Pecyna M., Liers C., Lundell T. (2010). New and classic families of secreted fungal heme peroxidases. Appl. Microbiol. Biotechnol. 87 871–897. 10.1007/s00253-010-2633-0 [DOI] [PubMed] [Google Scholar]
- Jamal F., Qidwai T., Prabhash K., Pandey R., Singh S. (2011). Azo and anthraquinone dye decolorization in relation to its molecular structure using soluble Trichosanthes dioica peroxidase supplemented with redox mediator. Catal. Commun. 12 1218–1223. 10.1016/j.catcom.2011.04.012 [DOI] [Google Scholar]
- Janusz G., Kucharzykb K., Pawlika A., Staszczaka M., Paszczynskic A. (2013). Fungal laccase, manganese peroxidase and lignin peroxidase: gene expression and regulation. Enzyme Microb. Technol. 52 1–12. 10.1016/j.enzmictec.2012.10.003 [DOI] [PubMed] [Google Scholar]
- Jaouani A., Sayadi S., Vanthournhout M., Penninckx M. (2003). Potent fungi for decolourisation of olive oil mill wastewater. Enzyme Microb. Technol. 33 802–809. 10.1016/S0141-0229(03)00210-2 [DOI] [Google Scholar]
- Järvinen J., Taskila S., Isomäki R., Ojamo H. (2012). Screening of white-rot fungi manganese peroxidases: a comparison between the specific activities of the enzyme from different native producers. AMB Express 2:62 10.1186/2191-0855-2-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo S., Ryu W., Park J., Yoo Y., Lee D., Hwang K.et al. (2007). Molecular cloning and expression of a laccase from Ganoderma lucidum, and its antioxidative properties. Mol. Cells 25 112–118. [PubMed] [Google Scholar]
- Kellner H., Luis P., Francois B. (2007). Diversity of laccase-like multicopper oxidase genes in Morchellaceae: identification of genes potentially involved in extracellular activities related to plant litter decay. FEMS Microbiol. Ecol. 61 153–163. 10.1111/j.1574-6941.2007.00322.x [DOI] [PubMed] [Google Scholar]
- Ko E.-M., Leem Y.-E., Choi H. T. (2001). Purification and characterization of laccase isozymes from the white-rot basidiomycete Ganoderma lucidum. Appl. Microbiol. Biotechnol. 57 98–102. 10.1007/s002530100727 [DOI] [PubMed] [Google Scholar]
- Kokol V., Doliska A., Eichlerova I., Baldrian P., Nerud F. (2007). Decolorization of textile dyes by whole cultures of Ischnoderma resinosum and by purified laccase and Mn-peroxidase. Enzyme Microb. Technol. 40 1673–1677. 10.1016/j.enzmictec.2006.08.015 [DOI] [Google Scholar]
- Kües U., Nelson D., Liu C., Yu G., Zhang J., Li J., et al. (2015). Genome analysis of medicinal Ganoderma spp. with plant-pathogenic and saprotrophic life-styles. Phytochemistry 114 18–37. 10.1016/j.phytochem.2014.11.019 [DOI] [PubMed] [Google Scholar]
- Kumar A., Kant K., Kumar P., Ramchiary N. (2015). Laccase isozymes from Ganoderma lucidum MDU-7: isolation, characterization, catalytic properties and differential role during oxidative stress. J. Mol. Catal. Enzymat. 113 68–75. 10.1016/j.molcatb.2015.01.010 [DOI] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfer D. M., Drabenstot S. D., Buchanan K. L., Lai H., Zhu H., Dyer D. W., et al. (2004). Introns and splicing elements of five diverse fungi. Eukaryot. Cell 3 1088–1100. 10.1128/EC.3.5.1088-1100.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfermann I., Linke D., Nimtz M., Berger R. (2015). Manganese peroxidases from Ganoderma applanatum degrade β-carotene under alkaline conditions. Appl. Biochem. Biotechnol. 175 3800–3812. 10.1007/s12010-015-1548-8 [DOI] [PubMed] [Google Scholar]
- Levin L., Melignani E., Ramos A. (2010). Effect of nitrogen sources and vitamins on ligninolytic enzyme production by some white-rot fungi. Dye decolorization by selected culture filtrates. Bioresour. Technol. 101 4554–4563. 10.1016/j.biortech.2010.01.102 [DOI] [PubMed] [Google Scholar]
- Lorenzo A., Lázaro E. (2003). “La educación ambiental comunitaria a través de la radio y su contribución al desarrollo de una cultura general integral,” in Proceedings of the IV Congreso Iberoamericano de Educación Ambiental Habana. [Google Scholar]
- Luis P., Walther G., Kellner H., Martin F., Buscot F. O. (2004). Diversity of laccase genes from basidiomycetes in a forest soil. Soil Biol. Biochem. 36 1025–1036. 10.1016/j.soilbio.2004.02.017 [DOI] [Google Scholar]
- Maganhotto C. M., Soares de Melo I., Oliveira P. R. (2005). Ligninolytic enzyme production by Ganoderma spp. Enzyme Microb. Technol. 37 324–329. 10.1016/j.enzmictec.2004.12.007 [DOI] [Google Scholar]
- Majeau J. A., Brar S. K., Tyagi R. (2010). Laccases for removal of recalcitrant and emerging pollutants. Bioresour. Technol. 101 2331–2350. 10.1016/j.biortech.2009.10.087 [DOI] [PubMed] [Google Scholar]
- Malarczyk E., Kochmanska-Rdest J., Jarosz-Wilkolazka A. (2009). Influence of very low doses of mediators on fungal laccase activity – nonlinearity beyond imagination. Nonlinear Biomed. Phys. 3:10 10.1186/1753-4631-3-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavalan T., Manavalan A., Kalaichelvan P., Thangavelua P., Heesed K. (2013). Characterization of optimized production, purification and application of laccase from Ganoderma lucidum. Biochem. Eng. J. 70 106–114. 10.1016/j.bej.2012.10.007 [DOI] [Google Scholar]
- Manzano A. M., Ramos-Leal M., Domínguez O., Sánchez-López M. I., Chinea R., Batista A., et al. (2011). Isolation and selection of white-rot fungi for decolorization of industrial dyes. Rev. CENIC Cienc. Biol. 42 17–23. [Google Scholar]
- Manzano A. M., Torres G., González A., Banguela A., Ramos-González P. L., Valiente P. A., et al. (2013). Role of laccase isozymes in textile dye decolorization and diversity of laccase genes from Ganoderma weberianum B-18. J. Appl. Sci. Environ. Sanit. 8 237–242. [Google Scholar]
- Martínez A. T. (2002). Molecular biology and structure-function of lignin-degrading heme peroxidases. Enzyme Microb. Technol. 30 425–444. 10.1016/S0141-0229(01)00521-X [DOI] [Google Scholar]
- Máximo C., Costa-Ferreira M. (2004). Decolorization of reactive textile dyes by Irpex lacteus and lignin modifying enzymes. Proc. Biochem. 39 1475–1479. 10.1016/S0032-9592(03)00293-0 [DOI] [Google Scholar]
- Mendonça T., Jara J., González V., Elissetche J. P., Freer J. (2008). Evaluation of the white-rot fungi Ganoderma australe and Ceriporiopsis subvermispora in biotechnological applications. J. Ind. Microbiol. Biotechnol. 35 1323–1330. 10.1007/s10295-008-0414-x [DOI] [PubMed] [Google Scholar]
- Minter D. W., Rodríguez M., Mena J. (2001). Fungi of the Caribbean. An Annotated Checklist. London: PDMS Publishing. [Google Scholar]
- Morgenstern I., Klopman S., Hibbett D. (2008). Molecular evolution and diversity of lignin degrading heme peroxidases in the agaricomycetes. J. Mol. Evol. 66 243–57. 10.1007/s00239-008-9079-3 [DOI] [PubMed] [Google Scholar]
- Murugesan K., In-Hyun N., Kim Y., Chang Y. (2007). Decolorization of reactive dyes by a thermostable laccase produced by Ganoderma lucidum in solid state culture. Enzyme Microb. Tech. 4 1662–1672. 10.1016/j.enzmictec.2006.08.028 [DOI] [Google Scholar]
- Novotny C., Rawal B., Bhatt M., Patel M., Sasek V., Molitoris H. (2001). Capacity of Irpex lacteus and Pleurotus ostreatus for decolorization of chemically different dyes. J. Biotechnol. 89 113–122. 10.1016/S0168-1656(01)00321-2 [DOI] [PubMed] [Google Scholar]
- Nyanhongo G. S., Gomes J., Gübitz G. M., Zvauya R., Read J., Steiner W. (2002). Decolorization of textile dyes by laccases from a newly isolated strain of Trametes modesta. Water Res. 36 1449–1456. 10.1016/S0043-1354(01)00365-7 [DOI] [PubMed] [Google Scholar]
- Otero M. A., Reyes A., Sáenz T., Blanco G. (2000). Manual de los Derivados de la Caña de Azúcar 3th Edn. Habana: MINAZ. [Google Scholar]
- Park Y., Kwon O., Son E., Yoon D., Han W., Jae-Young N., et al. (2012). Genetic diversity analysis of Ganoderma species and development of a specific marker for identification of medicinal mushroom Ganoderma lucidum. Afr. J. Microbiol. 6 5417–5425. [Google Scholar]
- Pérez J. M., Camino V. (2000). Riqueza micológica en un sitio natural del Jardín Botánico Nacional. Rev. Jardín Bot. Nac. 21 133–137. [Google Scholar]
- Pérez-Boada M., Ruiz-Dueñas F., Pogni R., Basosi R., Choinowski T., María Jesús Martínez M., et al. (2005). Versatile peroxidase oxidation of high redox potential aromatic compounds: site-directed mutagenesis, spectroscopic and crystallographic investigation of three long-range electron transfer pathways. J. Mol. Biol. 354 385–402. 10.1016/j.jmb.2005.09.047 [DOI] [PubMed] [Google Scholar]
- Pointing S. B., Bucher V. V. C., Vrijmoed L. L. P. (2000). Dye decolorization bysubtropical basidiomycetous fungi and the effect of metals on dye degradation. World J. Microbiol. Biotechnol. 16 199–205. 10.1023/A:1008910113322 [DOI] [Google Scholar]
- Pointing S. B., Parungao M. M., Hyde K. D. (2003). Production of wood-decay enzymes, mass loss and lignin solubilization in wood by tropical Xylariaceae. Mycol. Res. 107 231–235. 10.1017/S0953756203007329 [DOI] [PubMed] [Google Scholar]
- Rambaut A. (2014). Tree Figure Drawing Tool Version 1.4.2. Available at: http://tree.bio.ed.ac.uk/ [Google Scholar]
- Rambaut A., Suchard M. A., Xie D., Drummond A. J. (2014). Tracer v1.6. Available at: http://beast.bio.ed.ac.uk/Tracer [Google Scholar]
- Rivera-Hoyos C., Morales-Álvarez E., Poveda-Cuevas D., Reyes-Guzmán E., Poutou-Piñales A., Reyes-Montaño E., et al. (2015). Computational analysis and low-scale constitutive expression of laccases synthetic genes gllcc1 from Ganoderma lucidum and poxa 1b from Pleurotus ostreatus in Pichia pastoris. PLoS ONE 10:e0116524 10.1371/journal.pone.0116524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Couto S. (2007). Decolouration of industrial azo dyes by crude laccase from Trametes hirsuta. J. Hazard. Mater. 148 768–770. 10.1016/j.jhazmat.2007.06.123 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Couto S. (2009). Dye removal by immobilised fungi. Biotechnol. Adv. 27 227–235. 10.1016/j.biotechadv.2008.12.001 [DOI] [PubMed] [Google Scholar]
- Ruiz-Dueñas F., Morales M., García E., Miki Y., Martıńez M., Martínez A. (2009). Substrate oxidation sites in versatile peroxidase and other basidiomycete peroxidases. J. Exp. Bot. 60 441–452. 10.1093/jxb/ern261 [DOI] [PubMed] [Google Scholar]
- Ruiz-Dueñas S., Camarero M., Perez-Boada M., Martinez J., Martinez A. (2001). A new versatile peroxidase from Pleurotus. Biochem. Soc. Trans. 29(Pt 2) 116–22. 10.1042/bst0290116 [DOI] [PubMed] [Google Scholar]
- Sanchez M. I., Vanhulle S., Mertens V., Guerra G., Herrera F., Decock C., et al. (2008). Autochthonous white rot fungi from the tropical forest: potential of Cuban strains for dyes and textile industrial effluents decolourisation. Afr. J. Biotechnol. 7 1983–1990. 10.5897/AJB08.042 [DOI] [Google Scholar]
- Sarkar S., Martínez A., Martínez M. J. (1997). Biochemical and molecular characterization of a manganese peroxidase isoenzyme from Pleurotus ostreatus. Biochim. Biophys. Acta 1339 23–30. 10.1016/S0167-4838(96)00201-4 [DOI] [PubMed] [Google Scholar]
- Shleev S., Nikitina O., Christenson A., Reimann C. T., Yaropolov A. I., Ruzgas T., et al. (2007). Characterization of two new multiforms of Trametes pubescens laccase. Bioorg. Chem. 35 35–49. 10.1016/j.bioorg.2006.08.001 [DOI] [PubMed] [Google Scholar]
- Sirim D., Wagner F., Wang L., Schmid R. D., Pleiss J. (2011). The laccase engineering database: a classification and analysis system for laccases and related multicopper oxidases. Database 2011:bar006 10.1093/database/bar006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šnajdr J., Baldrian P. (2007). Temperature affects the production, activity and stabilityof ligninolytic enzymes in Pleurotus ostreatus and Trametes versicolor. Folia Microbiol. 52 498–502. 10.1007/BF02932110 [DOI] [PubMed] [Google Scholar]
- Sundaramoorthy M., Youngs H. L., Gold M. H., Poulos T. L. (2005). High resolution crystal structure of manganese peroxidase: substrate and inhibitor complexes. Biochemistry 44 6463–6470. 10.1021/bi047318e [DOI] [PubMed] [Google Scholar]
- Teerapatsakul C., Abe N., Bucke C., Chitradon L. (2007). Novel laccases of Ganoderma sp. KU-Alk4, regulated by different glucose concentration in alkaline media. World J. Microbiol. Biotechnol. 23 1559–1567. 10.1007/s11274-007-9401-z [DOI] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acids Res. 22 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesenberg D., Kyriakides I., Agathos S. (2003). White-rot fungi and their enzymes for the treatment of industrial dye effluents. Biotechnol. Adv. 22 161–187. 10.1016/j.biotechadv.2003.08.011 [DOI] [PubMed] [Google Scholar]
- You L., Liu Z. M., Lin J. F., Guo L. Q., Huang X. L., Yang H. X. (2013). Molecular cloning of a laccase gene from Ganoderma lucidum and heterologous expression in Pichia pastoris. J. Basic Microbiol. 53 S134–S141. [DOI] [PubMed] [Google Scholar]
- Yuan X., Tian G., Zhao Y., Zhao L., Wang H., Bun T. (2016). Biochemical characteristics of three laccase isoforms from the basidiomycete Pleurotus nebrodensis. Molecules 21:E203 10.3390/molecules21020203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakaria L., Ali N. S., Salleh B., Zakaria M. (2009). Molecular analysis of Ganoderma species from different hosts in Peninsula Malaysia. J. Biol. Sci. 9 12–20. 10.3923/jbs.2009.12.20 [DOI] [Google Scholar]
- Zeng X., Cai Y., Liao X., Luo S., Zhang D. (2012). Anthraquinone dye assisted the decolorization of azo dyes by a novel Trametes trogii laccase. Proc. Biochem. 47 160–163. 10.1016/j.procbio.2011.10.019 [DOI] [Google Scholar]
- Zeng X., Cai Y., Liao X., Zeng X., Li W., Zhang D. (2011). Decolorization of synthetic dyes by crude laccase from a newly isolated Trametes trogii strain cultivated on solid agro-industrial residue. J. Hazard. Mater. 187 517–525. 10.1016/j.jhazmat.2011.01.068 [DOI] [PubMed] [Google Scholar]
- Zhuo R., Ma L., Fan F., Gong Y., Wan X., Jiang M., et al. (2011). Decolorization of different dyes by a newly isolated white-rot fungi strain Ganoderma sp.En3 and cloning and functional analysis of its laccase gene. J. Hazard. Mater. 192 855–873. 10.1016/j.jhazmat.2011.05.106 [DOI] [PubMed] [Google Scholar]


