Abstract
Key message
Among the qhir11 and qhir12 sub-regions of a major QTL qhir1, only qhir11 has significant effect on maternal haploid induction, segregation distortion and kernel abortion.
Abstract
In vivo haploid induction in maize can be triggered in high frequencies by pollination with special genetic stocks called haploid inducers. Several genetic studies with segregating populations from non-inducer x inducer crosses identified a major QTL, qhir1, on chromosome 1.04 contributing to in vivo haploid induction. A recent Genome Wide Association Study using 51 inducers and 1482 non-inducers also identified two sub-regions within the qhir1 QTL region, named qhir11 and qhir12; qhir12 was proposed to be mandatory for haploid induction because the haplotype of qhir11 was also present in some non-inducers and putative candidate genes coding for DNA and amino acid binding proteins were identified in the qhir12 region. To characterize the effects of each sub-region of qhir1 on haploid induction rate, F2 recombinants segregating for one of the sub-regions and fixed for the other were identified in a cross between CML269 (non-inducer) and a tropicalized haploid inducer TAIL8. To quantify the haploid induction effects of qhir11 and qhir12, selfed progenies of recombinants between these sub-regions were genotyped. F3 plants homozygous for qhir11 and/or qhir12 were identified, and crossed to a ligueless tester to determine their haploid induction rates. The study revealed that only the qhir11 sub-region has a significant effect on haploid induction ability, besides causing significant segregation distortion and kernel abortion, traits that are strongly associated with maternal haploid induction. The results presented in this study can guide fine mapping efforts of qhir1 and in developing new inducers efficiently using marker assisted selection.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-017-2873-9) contains supplementary material, which is available to authorized users.
Introduction
Large-scale production and utilization of doubled haploid (DH) lines has become common practice in maize breeding programs during the last decade owing to the associated acceleration and cost reduction in development of inbred lines and deployment of hybrid varieties (Melchinger et al. 2013). In vivo maternal haploid induction (HI) is the backbone of DH line production in maize (Prigge et al. 2012b), which involves pollination of desired populations with special genetic stocks called haploid inducers that induce relatively high frequencies of haploid seeds in the progeny (Coe 1959; Chaikam 2012; Prigge and Melchinger 2012). The phenomenon of in vivo maternal HI is unique to maize and has not been reported in other plant species so far (Hu et al. 2016), although its physiological and molecular bases are still elusive. Elimination of inducer chromosomes after fertilization (Zhang et al. 2008; Li et al. 2009; Xu et al. 2013a; Qiu et al. 2014) and single fertilization followed by parthenogenesis (Sarkar and Coe 1966; Bylich and Chalyk 1996; Barret et al. 2008; Swapna and Sarkar 2012) were proposed to be involved in the production of seeds with haploid embryos and normal triploid endosperms.
To understand the genetic basis of HI, several studies have been conducted. HI was determined to be a quantitatively inherited trait, controlled by a small number of genes and improvable through selection (Lashermes and Beckert 1988). It was also suggested that additive and epistatic gene action affect the HI process (Prigge et al. 2011). In first QTL mapping studies on HI with segregating progeny of crosses between non-inducers and inducers, a major QTL on chromosome 1 was identified in bin 1.04 (Deimling et al. 1997; Barret et al. 2008). An extensive QTL mapping study with four bi-parental populations involving inducers CAUHOI and UH400 detected two major QTL, named qhir1 and qhir8, and several minor QTL (Prigge et al. 2012b). The major QTL qhir1 on chromosome 1.04 was the same as reported in the previous studies and explained 66% of the genotypic variance. Besides its effect on HI, qhir1 has also been associated with segregation distortion (SD) and has a strong selective disadvantage (Barret et al. 2008; Prigge et al. 2012b; Dong et al. 2013; Xu et al. 2013a). It was also noted that in vivo HI is associated with embryo and endosperm abortion (Prigge et al. 2012b; Xu et al. 2013a). Less pronounced than the effect of qhir1 was the effect of the second major QTL found by Prigge et al. (2012b), qhir8, which maps to chromosome 9 and explained only 20% of the genotypic variance. However, all these linkage mapping studies resulted in large support intervals for the detected QTL.
To delineate the map position and to identify closely linked markers more useful for marker-assisted selection in development of new inducers, qhir1 was fine-mapped to a 243 kb region (Dong et al. 2013) and qhir8 to a 789 kb region (Liu et al. 2015). Considering the confirmation of qhir1 in multiple studies, qhir1 may be considered mandatory for HI ability (Prigge et al. 2012b), while other loci like qhir8 may enhance the function of qhir1 to increase the HIR (Liu et al. 2015).
Recently, the large qhir1 support interval described by Prigge et al. (2012b) was dissected by Hu et al. (2016) into two closely linked regions, named qhir11 and qhir12, using a novel type of genome wide association study (GWAS) to detect selective sweeps and address the problem of perfect confounding between population structure and trait expression, as in the case of inducers (cases) and non-inducer (controls). Sub-region qhir11 harbored the 243 kb interval fine-mapped by Dong et al. (2013) and had one major haplotype present in the majority of the inducers and one minor haplotype present only in two inducers studied. The latter occurred also in several non-inducers whose HIR was similar to spontaneous occurrence of haploids. Hence, the minor haplotype of qhir11 was deemed to be neither diagnostic for differentiating inducers and non-inducers nor effective for conditioning HI ability in maize. However, no conclusions were drawn about the major haplotype of qhir11 based on this study. By comparison, qhir12 had a single haplotype allele found in all the 53 inducers and absent in all 1482 non-inducers included in the study and was proposed to harbor three candidate genes related to putative functions involved in HI. To further determine the effects of the qhir12 and qhir11 haplotypes, the authors suggested testing the effect of these haplotypes on HI in near-isogenic lines or selfed progenies of recombinants that segregate at one locus while the other is fixed.
The main objective of our study is to adopt this strategy and test the effects of qhir11 and qhir12 haplotypes on HIR using selfed progenies of recombinants in a huge F2 population derived by crossing a non-inducer with a tropically adapted haploid inducer. In addition, we examined which of the specific sub-regions of qhir1 is specifically associated with segregation distortion and kernel abortion, traits associated with maternal haploid induction.
Materials and methods
Notation of the genotypes
We denote henceforth the qhir11 and qhir12 sub-regions as A and B, respectively. We use the following notations for the various genotypes possible for each sub-region: AA = homozygous for the putative inducer allele at all markers assayed in the qhir11 sub-region; aa = homozygous for the putative non-inducer allele at all markers in the qhir11 sub-region; BB = homozygous for the putative inducer allele at all markers assayed in the qhir12 sub-region; bb = homozygous for the putative non-inducer allele at all markers in the qhir12 sub-region; Aa = heterozygous at all markers assayed in the qhir11 sub-region; and Bb = heterozygous for all markers assayed in the qhir12 sub-region.
Genetic material
One tropically adapted inducer, TAIL8, and one tropically adapted non-inducer, CML269, were used as parents in this study. TAIL8, harboring the A and B alleles in homozygous state has a mean HIR of 9.9% (Chaikam et al. 2016). CML269 has no HI ability and harbors the a and b alleles in homozygous state. The non-inducer (CML269) x inducer (TAIL8) cross was made in the winter season of 2011 at CIMMYT’s experimental station at Agua Fria, Mexico (20.26°N, 97.38°W) to generate the F1 generation. From the F1, 100 seeds were planted and selfed to generate the F2 generation in the summer season of 2011. A total of 7160 F2 seeds of good quality were genotyped as described below. Recombinants between the qhir11 and qhir12 sub-regions identified on the basis of the marker assays were grouped into four F2 genotype classes: AABb; aaBb, AaBB, Aabb, and used for further assays.
From each of the four F2 genotype classes of recombinants, 10 individual plants were randomly selected for selfing to obtain F2:3 families segregating for the heterozygous sub-region. Only F3 seeds homozygous for the segregating sub-region were planted in the field at Agua Fria in the winter season of 2016. Hybrid (PDH3 × PDH8), homozygous for liguleless gene lg2 (Prigge et al. 2012a; Chaikam et al. 2016; Melchinger et al. 2016), was used as a female tester to produce testcross seed for evaluating the HIR. The tester was stagger-planted four times at weekly intervals to synchronize flowering with the F3 plants. Each F3 plant that produced pollen was crossed on to 10–15 tester plants (based on pollen availability) and was also self-pollinated. Some F3 plants were found to be haploids based on their weak plant stature, narrow and erect leaves and sterile tassels (Prigge et al. 2011; Chaikam et al. 2016) and were therefore not pollinated. Some plants could not be used for testcrossing because of severe virus infection. Testcross seed was bulked from all the tester plants pollinated by the same F3 plant. A graphical representation of the scheme followed for developing the genetic material is shown in Fig. 1.
Fig. 1.
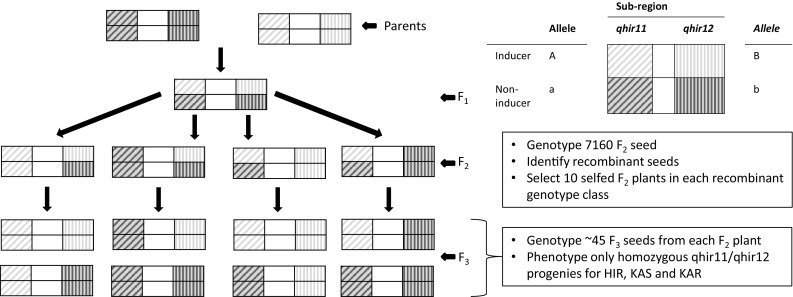
Strategy used for genetic delineation of qhir1 and analysis of the effects of qhir11 and qhir12 sub-regions on HIR in maize
Markers delineating the qhir11 and qhir12 sub-regions
According to Hu et al. (2016), the physical boundaries for qhir11 are between SNPs PZE-101,081,177 (physical co-ordinate: 1: 68,134,633) and SYN25793 (physical co-ordinate 1: 68,670,617). For qhir12, the borders are between SYN4966 (physical co-ordinate 1: 71,795,509) and PZA00714.1 (physical co-ordinate 1: 75,768,235). All the physical co-ordinates of the SNPs assayed are with reference to B73 AGP V2 (http://ensembl.gramene.org/Zea_mays). Sets of six markers covering the qhir11 sub-region and eight markers covering the qhir12 sub-region were used to genotype each sub-region (Supplementary table 1). Based on the selected SNPs, the haplotypes of TAIL8 and CML269 at each sub-region were compared with the large set of non-inducers and inducers reported by Hu et al. (2016) and verified. All markers used in this study were genotyped using KASP assays (LGC Genomics, UK) developed from the Illumina MaizeSNP50 BeadChip (Ganal et al. 2011), except for one SNP developed from HapMap V.2 (Suppl. Table 1).
Analysis of the F2 population
DNA was extracted from 7160 individual seeds of the F2 population of cross TAIL8 × CML269 following standard procedures (Gao et al. 2008) and genotyped with the above-mentioned SNPs. Among the polymorphic SNPs between the two parents available to CIMMYT for the qhir11 and qhir12 sub-regions, two SNPs (PZE0166290049 and PZE0166357949) were selected to represent qhir11 and two SNPs (SYN26730 and PZE101085336) to represent qhir12 in the genotyping of the F2 seeds. Based on the results, 428 recombinant F2 seeds in the four F2 genotype classes described above were selected and planted in the field. Leaf DNA of these plants was extracted at the four-leaf stage following CIMMYT’s laboratory protocols (CIMMYT 2001). Because the two SNPs of each sub-region in the seeds did not cover the respective physical interval entirely, we analyzed additionally four SNPs for qhir11 and 10 SNPs for qhir12, which were part of the SNPs on the MaizeSNP50 BeadChip polymorphic between the two parents. This assay was also used to ascertain the classification of the recombinant F2 plants; plants showing any discrepancy were discarded. Moreover, some F2 plants did not survive or failed to produce selfed seed. Thus, selfed ears were harvested from 21 AABb, 72 aaBb, 56 AaBB, and 44 Aabb genotypes in the F2 generation, adding up to a total of 193 ears.
Genotyping and phenotypic analysis of F2:3 families from recombinants
Ten ears were randomly selected from each of the four afore-mentioned F2 genotype classes for raising F2:3 families. DNA was extracted from ~45 individual seeds from each of the 40 F2:3 families and genotyped with three SNP markers for both qhir11 and qhir12, covering the entire physical interval of the sub-regions as identified by Hu et al. (2016). From each family, only the seeds homozygous for the qhir11 and qhir12 sub-regions were selected as male parents for pollination of liguleless tester PDH3 × PDH8.
Among the 756 F3 plants that were test-crossed, 83.7% resulted in more than 1000 seeds, 12% resulted between 500 and 999 seeds, and 4.2% resulted in less than 500 seeds. For each F3 plant with more than 1000 testcross seeds, 1000 seeds were germinated in styrofoam trays in a shade house at the Agua Fria experimental station. Each tray accommodated 100 seeds. After 14 days of germination, each tray was evaluated for the number of germinated seedlings and the number of seedlings with and without ligule. For the F3 plants with less than 1000 testcross seeds, all seeds were germinated. The HIR was calculated as HIR = N L/(N L+N NL), where N L and N NL refer to the number of plants with and without ligule, respectively.
Phenotyping of kernel abortion
We refer here to endosperm abortion as kernel abortion, because most endosperm aborted seeds in our study lacked an embryo similar to the observation by Xu et al. (2013a). Selfed ears obtained from the F3 plants in each of the four genotype classes were visually rated for a kernel abortion score (KAS) on a scale of 1–5, where 1 represents no aborted seed visible on the ear, and 5 represents complete abortion with no seed set on the ear. To measure the extent of kernel abortion quantitatively, the number of normal seeds and number of kernel aborted seeds were counted on each ear from the AAbb and aaBB genotype classes as suggested by Xu et al. (2013a). Kernel abortion rate (KAR) for each entry was calculated as KAR = N a/(N a + N n), where N a refers to the number of aborted seeds and N n to the number of normal seeds.
Statistical analyses
The HIR for each F3 genotype within each F2 genotype class was calculated as the least-squares means in the following generalized linear model assuming a binomial distribution:
where Y ijk is the ith observation in the jth genotype class for the kth F2:3 family, µ is the general mean, g j is the effect of the jth genotype, f k is the effect of the kth family and e ijk the residual error. The model was fitted using the glm function in the R software package, version 3.3.0. Least-squares means and corresponding confidence intervals were calculated with the lsmeans package, version 2.23–5, and compact letters displays were produced with the multcompView package, version 0.1–7, at significance level α = 5%. We used an over-dispersion factor to account for variance in the data in excess of the binomial sampling variance that may result in an inflation of the standard errors.
For each of the four recombinant genotype classes selected from the F2, we tested the following hypotheses: (i) H0: aabb = AAbb vs. HA: aabb ≠ AAbb (from F2 genotype class Aabb); (ii) H0: aabb = aaBB vs. HA: aabb ≠ aaBB (from F2 genotype class aaBb); (iii) H0: AABB = aaBB vs. HA: AABB ≠ aaBB (from F2 genotype class AaBB); and (iv) H0: AABB = AAbb vs. HA: AABB ≠ AAbb (from F2 genotype class AABb).
F3 plants heterozygous for either of the sub-regions were not tested in this experiment. Significant differences in these tests determine whether the qhir11 or qhir12 sub-region alone is sufficient to exhibit HIR equivalent to qhir11 and qhir12 together.
KAS for each F3 genotype was calculated with the same generalized linear model as for HIR except that a Poisson distribution was assumed and KAS was used as response variable. KAR for the two F3 genotype classes AAbb and aaBB was calculated with the same generalized linear model but without the family term because of confounding between family and genotype.
Segregation distortion (SD) in the F2 generation was investigated with a G-test for goodness-of-fit to the segregation ratios expected under Mendelian inheritance and applying a significance level α = 5%. The G-test of goodness of fit to expected segregation ratios and the expected allele frequencies was carried out with the R software function GTest from the DescTools package, version 0.99.17.
Gene annotations
The gene annotations by the MAKER gene annotation pipeline (Cantarel et al. 2008) in the physical interval of qhir11 in the B73 genome sequence (V2) available in http://ensembl.gramene.org/Zea_mays was used to search for putative candidate genes in the studied interval.
Results
Recombination and segregation in the F2 and F3 generations
A total of 475 recombinants in the F2 generation falling into different genotype classes were identified between qhir11 and qhir12 based on the segregation analysis of 7154 F2 seeds (Table 1). No recombination was observed between the two sub-regions in most of the F2 seeds (93.4%), which had the same genotype as the F1 cross or the parent lines. Single recombination events between qhir11 and qhir12 were observed in 6% of the F2 seeds, and double recombination events between qhir11 and qhir12 were observed in 0.1% of F2 seeds. In addition, 0.6% of F2 seeds had recombination events which occurred within either of the sub-regions. Based on the recombination observed between the distal SNP of qhir11 and the proximal SNP of qhir12, the recombination rate between the qhir11 and qhir12 sub-regions was 3.1%. From the 428 single recombinant F2 plants between qhir11 and qhir12, a total of 193 plants remained for further analyses, with the following numbers in the four F2 genotype classes: 72 aaBb, 56 AaBB, 44 Aabb and 21 AABb. Among F3 plants, highly significant (P < 0.001) segregation distortion against the homozygous inducer genotype was observed for the qhir11 sub-region (Table 2). The segregation distortion observed for the qhir12 sub-region was also significant (P < 0.01) but against the non-inducer genotype. The same trends were observed for the allele frequencies at both sub-regions.
Table 1.
Recombination in parental gametes observed in the F2 generation between the sub-regions qhir11 and qhir12
| No. of recombinations | Genotypea | Counts | Frequency (%) |
|---|---|---|---|
| 0 | aabb | 2340 | 32.7 |
| AABB | 1020 | 14.25 | |
| AaBb | 3319 | 46.38 | |
| 1 (∑ = 428) | AABb | 59 | 0.82 |
| aaBb | 174 | 2.43 | |
| AaBB | 106 | 1.48 | |
| Aabb | 89 | 1.24 | |
| 2 (∑ = 6) | AAbb | 2 | 0.03 |
| aaBB | 4 | 0.06 |
aaa and bb (homozygous for the non-inducer CML269 in sub-region qhir11 and qhir12, respectively, based on two SNPs assayed in each sub-region); AA and BB (homozygous for the inducer TAIL8); Aa and Bb heterozygous
Table 2.
Segregation, allele counts and allele frequencies observed for sub-regions qhir11 and qhir12 in F3 seeds from the four F2 genotype classes segregating only at the respective locus. P values are shown for a G-test of goodness of fit of observed counts to the expected segregation ratio of 0.25:0.50:0.25, and for expected allele frequencies of 0.5
| Sub-region | Genotypes in F3 | Alleles in F3 | |||
|---|---|---|---|---|---|
| qhir11 | AA | Aa | aa | A | a |
| Counts | 150 | 455 | 323 | 755 | 1101 |
| % | 0.16 | 0.49 | 0.35 | 0.41 | 0.59 |
| G-test | P = 7.42 × 10− 15 | P = 8.03 × 10− 16 | |||
| qhir12 | BB | Bb | aa | B | b |
|---|---|---|---|---|---|
| Counts | 212 | 464 | 174 | 888 | 812 |
| % | 0.25 | 0.55 | 0.20 | 0.52 | 0.48 |
| G-test | P = 0.00428 | P = 0.0652 | |||
Effects of the qhir11 and qhir12 sub-regions on haploid induction rate
F3 plants with genotype AAbb, derived from F2 plants in genotype class Aabb, revealed on average a significantly (P < 0.01) higher HIR (6.45%) than aabb plants having a mean HIR = 0.12% (Table 3). Thus, the AA genotype showed a strong positive effect on HIR. In F3 plants of F2 genotype class AABb, HIR was significantly (P < 0.01) higher in AAbb plants (7.16%) than in AABB plants (5.92%). Thus, a relatively small negative effect on HIR was observed for the BB genotype in the presence of the AA genotype. This negative effect was not observed in the absence of the AA genotype, because in F3 plants from F2 genotype class aaBb, the mean HIR of the aaBB genotypes (0.12%) was not significantly different from the mean HIR of the aabb genotypes (0.09%). In F3 plants of the F2 genotype class AaBB, the HIR of AABB genotypes was also significantly (P < 0.01) higher than the HIR of aaBB genotypes.
Table 3.
Least-squares means for HIR for each F3 genotype within each of the four F2 genotype classes
| F2 genotype class | F3 genotype | No. seedlings | HIR%a | |
|---|---|---|---|---|
| Haploid | Diploid | |||
| Aabb | aabb | 156 | 132,126 | 0.12a |
| AAbb | 3770 | 52,658 | 6.45b | |
| AaBB | aaBB | 262 | 106,820 | 0.24a |
| AABB | 2889 | 46,736 | 5.94b | |
| aaBb | aabb | 87 | 87,819 | 0.09a |
| aaBB | 125 | 98,396 | 0.12a | |
| AABb | AAbb | 3933 | 51,616 | 7.16a |
| AABB | 4016 | 61,841 | 5.92b | |
aDifferent letters (a and b) indicate a significant difference in least-squares means of HIR between genotypes within each F2 genotype class at an overall significance level α = 5%, with a Bonferroni correction for multiple comparisons
Regarding the HIR of all F3 plants irrespective of their origin from the four F2 genotype classes, the highest HIR (5.96%) was observed for genotype AAbb, followed by a significantly (P < 0.05) smaller value (HIR = 5.02%) for genotype AABB (Table 4). A large decrease in HIR was found in the aaBB genotype (HIR = 0.19%) and a further significant (P < 0.05) decrease in the aabb genotype (HIR = 0.12%). Thus, in the presence of AA at qhir11, BB had a reducing effect on HIR but in the presence of aa, it had an increasing effect on HIR, whereas no significant effect was observed in the analysis of means in F3 genotypes derived from individual F2 genotype classes.
Table 4.
Least-squares means for haploid induction rate (HIR), kernel abortion score (KAS) and kernel abortion rate (KAR) for each F3 genotype
| Genotype | HIRa | KASab | KARab |
|---|---|---|---|
| aabb | 0.12a | 1.25a | – |
| aaBB | 0.19b | 1.34a | 4.26a |
| AAbb | 5.96d | 2.99b | 29.62b |
| AABB | 5.02c | 2.84b | – |
aDifferent letters indicate significant differences at an overall significance level α = 5%, using the Tukey method for comparing a group of four estimates for HIR and KAS and no adjustment for KAR, because only one comparison could be made
bKAS is shown on the original score scale, KAR is shown in percent
Effects of the qhir11 and qhir12 sub-regions on kernel abortion
Most ears harvested from AAbb and AABB genotypic class F3 plants showed some level of kernel abortion while most ears of aaBB and aabb classes did not record any abortion (Suppl. Figure 1a and 1b). Regardless of the genotype at the other sub-region, F3 plants of genotype AA had a significantly (P < 0.01) higher KAS than the aa genotype. Quantitative evaluation of kernel abortion in the F3 generation showed that genotype AAbb had a six-fold higher KAR than the genotype aaBB (Table 4).
Discussion
Strategy for genetic delineation of qhir1 influencing haploid induction
Genetic delineation of the qhir11 and qhir12 sub-regions required large population sizes in the F2 generation considering that they are physically located very close to each other on chromosome 1.04 (Hu et al. 2016). Regarding the incomplete penetrance of qhir1 for HIR (Prigge et al. 2012b), the choice of the parents for this study was critical to guarantee sufficient variation in HIR of progenies recombinant for the qhir11 and qhir12 sub-regions. The non-inducer parent CML269 had shown highly significant difference in HIR values between progeny selected for qhir1 in combination with multiple haploid inducers (CIMMYT, unpublished data). Therefore, CML269 was chosen as non-inducer parent to develop a large F2 population with the selected tropicalized haploid inducer TAIL8. The 14 SNP markers selected for our analyses provided good coverage of the qhir1 region and were sufficient to delineate the sub-regions qhir11 and qhir12. The recombinants observed in the F2 generation showed a genetic distance of 3.1 cM between them, which is consistent with the estimate for qhir11 and qhir12 reported by Hu et al. (2016). We did not study the effect of qhir11 and qhir12 in homozygous recombinants (AAbb or aaBB) of the F2 generation because they were too few to make valid inferences. Given the huge efforts required in phenotyping for HIR, we had to restrict the number of individuals analyzed from each F2 genotype class to 10 F2 plants, resulting in 40 F2:3 families which could be analyzed within and among the four genotype classes. Seed DNA was genotyped for each of these F3 families to eliminate heterozygotes before planting and conducting testcrosses and selfings with the F3 plants. A liguless tester was used in testcrosses for measuring HIR because this method was recommended for accurate measurement of HIR in comparison to other methods (Melchinger et al. 2016) and has been reliably used for determining the HIR in previous studies (Prigge et al. 2012a; Melchinger et al. 2013; Chaikam et al. 2016). Staggered planting of the liguleless tester multiple times allowed achieving synchrony in flowering with the majority of the F3 plants differing widely in anthesis date (data not shown). For the majority of F3 plants (83.7%), we could evaluate HIR based on the recommended number of testcross seed (1000) and for only less than 1% of the F3 plants we had to measure HIR with fewer than 200 testcross seeds, which was the lower limit suggested by Prigge et al. (2012a).
Effects of qhir11 and qhir12 on maternal haploid induction rate
The F3 progenies, which were homozygous recombinants for the qhir11 and qhir12 sub-region, showed unambiguous differences in HIR (Table 3). HIR is known to be a trait with incomplete penetrance and hence, has a tendency to show highly variable expression in different genetic backgrounds (Prigge et al. 2012b). In the population studied here, there appeared to be no alleles masking the HIR trait, because HIR ranged from normal inducer levels to non-inducer levels. In contrast to the hypothesis put forward by Hu et al. (2016), the 535 kb segment of the qhir11 sub-region was in our study the only sub-region of qhir1 mandatory for HI ability. The inducer qhir11 allele (A) increased the HIR significantly in the presence of inducer (B) or non-inducer (b) alleles at the sub-region qhir12. The inducer qhir12 allele alone, in the absence of the inducer qhir11 allele did not cause a HIR higher than the spontaneous occurrence of haploids observed in normal non-inducer maize lines (Chase 1969). Actually, qhir12 significantly decreased HIR in the presence of the inducer allele at qhir11 but significantly increased HIR in the presence of the non-inducer allele at qhir11. In both cases, the significant differences due to the qhir12 allele were not strong enough to change the overall expression of HI due to the qhir11 allele, but merely modified the HIR.
A genome-wide study on 53 haploid inducers publicly available and 1,482 normal maize lines provided strong evidence that qhir11 and qhir12 were fixed in all the inducers and this was exclusively attributed to selection for HI (Hu et al. 2016). The qhir11 sub-region, also found significant in the study by Hu et al. (2016), revealed two haplotypes, where the minor haplotype was shared by two non-inducer lines, which did not have HI ability. Additionally, Hu et al. (2016) identified qhir12 as the most probable genomic segment carrying gene(s) responsible for HI, as this region had a single haplotype that was unchanged in all the inducers. In contrast, the results of our validation study clearly show that the major haplotype of qhir11 found by Hu et al. (2016) is mandatory for HI and that the presence or absence of inducer qhir12 did not affect the HIR significantly. Our study cannot make any inference on the effect of the minor haplotype of qhir11 that was present only among two publically available inducers analyzed. Also, our study cannot make any specific conclusion regarding the 243 kb fine-mapped genomic region for HIR (Dong et al. 2013), as we have not studied this region in particular, but rather a larger genomic region harboring this fine-mapped region.
Traits associated with maternal haploid induction
Various authors suggested investigating segregation distortion as a means to further fine-map the qhir11 sub-region influencing maternal haploid induction in maize (Barret et al. 2008; Prigge et al. 2012b; Dong et al. 2013). Strong segregation distortion was reported against the haploid inducer allele in many genetic studies (Barret et al. 2008; Prigge et al. 2012b; Dong et al. 2013). Xu et al. (2013) studied segregation distortion in regard to HI and mapped a major QTL associated with segregation distortion, sed1, on chromosome 1, overlapping with the fine-mapped qhir1 QTL. It is not clear yet, if segregation distortion is due to the same gene causing HI, or if another gene reducing fitness is closely linked to the gene(s) in qhir1 causing HI. It is also not clear exactly what type of reduction in fitness is linked to HI. Barret et al. (2008) suggested impediments in male gametic transmission associated with HI, while Xu et al. (2013) proved that there is both gametic and zygotic selection responsible for segregation distortion associated with HI. Our study did not aim to distinguish whether segregation distortion was caused by the same gene responsible for HI, or by another tightly linked gene. However, we observed in this study that both HI ability and strong segregation distortion against the inducer qhir11 allele, both of which were not observed for qhir12. For qhir12, the observed segregation distortion was significantly smaller, and in the opposite direction, favoring the inducer allele, while a much smaller effect was found on the HIR.
In addition to SD, high maternal HI also is strongly associated with the formation of defective kernels, including embryo and endosperm abortion (Xu et al. 2013) and reduced seed set (Satarova and Cherchel 2010). Similar to its effects on SD, the qhir11 sub-region in our study strongly increased kernel abortion while qhir12 had negligible effect on this. It is possible that the same gene(s) conditioning the HIR or another tightly linked gene within the qhir11 region can condition kernel abortion. One hypothesis for this relationship is that one of the sperm cells from the inducer pollen could be defective while the other sperm cell is normal (Geiger 2009). When the defective sperm cell fertilizes the central cell, endosperm abortion can result, and when the defective sperm cell fertilizes the egg cell, a haploid embryo or aborted embryo can result. This hypothesis was supported by the occurrence of morphologically different sperm cells (Bylich and Chalyk 1996), aneuploid microsporocytes which may produce aneuploid sperm cells (Chalyk et al. 2003), and an increase in heterofertilization when haploid inducer pollen is used (Kraptchev et al. 2003; Rotarenco and Eder 2003). Another hypothesis involves epigenetic, dosage-dependent modification of the chromosomes exerted by the sed1 locus which overlaps with the qhir1 locus resulting in incomplete penetrance of the sed1/qhir1 locus (Xu et al. 2013). It was proposed that expression of the sed1 locus can differ between the pollen grains resulting in some pollen grains having strong epigenetic modification while others are less modified. A strong modification of the sperm cell chromosomes may lead to kernel abortion or haploid formation while less epigenetically modified pollen leads to normal kernel formation. Further studies are required to understand the exact mechanism(s) behind kernel abortion associated with HI, for which cloning the gene(s) underlying these loci could be critical.
Putative candidate genes in the qhir11 physical interval
The physical interval of qhir11 in the B73 genome sequence (V2) has 13 protein-coding genes annotated by the MAKER gene annotation pipeline (Cantarel et al. 2008) as available in http://ensembl.gramene.org/Zea_mays (Suppl. Table 2). Out of these genes, 11 are predicted to have protein domains with known functions. Among these, gene Zm00001d029411 is predicted to have a protein which falls into the CULLIN family of ubiquitin ligases.
CULLIN-dependent ubiquitin ligases form a class of structurally related multi-subunit enzymes that control the rapid and selective degradation of important regulatory proteins involved in cell cycle progression and development (Thomann et al. 2005). In mice, knocking out a cullin-RING ubiquitin ligase leads to infertile male mice, due to fewer numbers of mature spermatozoa, most of which exhibit morphological defects, rendering them immotile and unable to fertilize eggs. In addition to the morphological abnormalities, chromosomal defects were also observed which may also contribute to infertility (Yin et al. 2011). The gene Zm00001d029411 in B73 had maximum similarity to AtCUL1 in Arabidopsis thaliana, based on a BLAST N alignment (E = 0.0012). CUL1 forms part of the SCF (SKP1-CUL1-F-box) complex in plants and animals, where SCF-dependant ubiquitylation plays a critical role in the control of the cell cycle (Thomann et al. 2005). Consistent with such a role, Arabidopsis cul1 loss-of function mutants arrest early during embryogenesis at the zygote stage (Shen et al. 2002). Genetic analysis also indicated a reduction in transmission of the atcul1 mutation through both male and female gametes. Considering the specific roles the protein domain plays in cell cycle and gametophyte development and transmission, this gene could be an interesting putative candidate gene for HI ability. Several recent studies indicate that manipulation of Centromere Histone CENH3 could lead to in vivo haploid induction in Arabidopsis (Ravi and Chan 2010; Seymour et al. 2012; Ravi et al. 2014), and in maize—(Kelliher et al. 2016). However, native CENH3 may not have any role in in vivo HI using maternal haploid inducers in maize. CENH3 is localized on chromosome 6.06 (Prigge et al. 2012b) and no mapping study has so far detected a major QTL for HI in this region. Also, study by Kelliher et al. (2016) showed that altered CENH3 when introduced into maize showed a maximum of 3.6% HIR, which is significantly lower than the high HIR (~10% or more) obtained using the improved maternal haploid inducers (Röber et al. 2005; Prigge et al. 2012a; Chaikam et al. 2016). Our study also showed that none of the annotated genes at qhir11 are related to CENH3. Therefore, cloning of the gene(s) responsible for maternal haploid induction, underlying qhir11, may provide a better insight into the genetic mechanism underlying gynogenesis in maize. It also needs to be explored whether CENH3-mediated HI can be synergistic to the qhir1 mediated HI in maize.
Conclusions
In this study, the qhir1 region was genetically delineated, and the haploid induction ability of qhir11 and qhir12 sub-regions was dissected through analysis of recombinants from a large F2 population derived from a non-inducer x haploid inducer cross. The study clearly revealed that qhir11 is the only sub-region with a strong effect on HIR, whereas qhir12 had a negligible effect on HIR, in contrast to the hypothesis of Hu et al. (2016) based on a selective sweep based GWAS approach. Furthermore, our study proved that qhir11 is more strongly associated than qhir12 with segregation distortion and kernel abortion, two traits that are associated with maternal haploid induction. The results of this study give direction in further fine mapping and cloning of the gene/s underlying qhir1. The molecular markers delineating qhir11 can be used for more efficient development of new inducer lines adapted to diverse agro climatic zones using marker assisted selection.
Author’s note
When this publication was in production, three articles (Kelliher et al. 2017; Gilles et al. 2017; Liu et al. 2017) were published about cloning the gene underlying qhir1 QTL that codes for a sperm specific phospholipase and triggers haploid induction.
Author contribution statement
AEM, SKN, VC and PMB designed the experiments. VC, ML and LLA coordinated the field trials and phenotyping. SKN and VC coordinated the sample collection, DNA extraction and genotyping. WM, SKN, VC and AEM analyzed the data. SKN, VC and WM wrote the manuscript. AEM and PMB edited the manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Suppl. Fig 1: Effects of AAbb and aaBB genotypes (a) and AABB and aabb (b) on kernel abortion in selfed F3 ears (TIF 2581 KB)
Acknowledgements
The authors gratefully acknowledge the financial support received from the Bill and Melinda Gates foundation to CIMMYT and University of Hohenheim through a project “Double Haploid Facility for Strengthening Maize Breeding Programs in Africa”. CIMMYT also acknowledges the financial support for research on DH technology from CGIAR Research Program (CRP) MAIZE and the MASAGRO Project funded by the Secretariat of Agriculture, Livestock, Rural Development, Fisheries and Food (SAGARPA), Mexico. The authors wish to thank Haixiao Hu, formerly with University of Hohenheim for his contributions in the planning of the study. The authors also thank Gordon Huestis for facilitating DNA extraction and Alberto Vergara Alva for DNA extraction.
Abbreviations
- DH
Doubled haploid
- KAR
Kernel abortion rate
- KAS
Kernel abortion score
- HI
Haploid induction
- HIR
Haploid induction rate
- SD
Segregation distortion
- GWAS
Genome wide association study
Compliance with ethical standards
The authors declare that the experiments comply with the laws of Mexico.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
S. K. Nair and V. Chaikam contributed equally to the work.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-017-2873-9) contains supplementary material, which is available to authorized users.
References
- Barret P, Brinkmann M, Beckert M. A major locus expressed in the male gametophyte with incomplete penetrance is responsible for in situ gynogenesis in maize. Theor Appl Genet. 2008;117:581–594. doi: 10.1007/s00122-008-0803-6. [DOI] [PubMed] [Google Scholar]
- Bylich VG, Chalyk ST. Existence of pollen grains with a pair of morphologically different sperm nuclei as a possible cause of the haploid-inducing capacity in ZMS line. Maize Genet Coop Newsl. 1996;70:33. [Google Scholar]
- CIMMYT . Laboratory protocols: CIMMYT applied molecular genetics laboratory protocols. Mexico D.F: CIMMYT; 2001. [Google Scholar]
- Cantarel BL, Korf I, Robb SMC, et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaikam V, Martinez L, Melchinger AE, et al. Development and validation of red root marker-based haploid inducers in maize. Crop Sci. 2016;56:1–11. doi: 10.2135/cropsci2015.10.0653. [DOI] [Google Scholar]
- Chaikam V. In vivo maternal haploid induction in maize. In: Prasanna B, Chaikam V, Mahuku G, editors. Doubled haploid technology in maize breeding: theory and practice. Mexico, DF: CIMMYT; 2012. pp. 14–19. [Google Scholar]
- Chalyk S, Baumann A, Daniel G, Eder J. Aneuploidy as a possible cause of haploid-induction in maize. Maize Genet Coop Newsl. 2003;77:29. [Google Scholar]
- Chase SS. Monoploids and monoploid-derivatives of maize (Zea mays L.) Bot Rev. 1969;35:117–168. doi: 10.1007/BF02858912. [DOI] [Google Scholar]
- Coe EH. A line of maize with high haploid frequency. Am Nat. 1959;93:381–382. doi: 10.1086/282098. [DOI] [Google Scholar]
- Deimling S, Röber F, Geiger HH. Methodik und Genetik der in-vivo-Haploideninduktion bei Mais. Vor Pflanzenzüchtg. 1997;38:203–224. [Google Scholar]
- Dong X, Xu X, Miao J, et al. Fine mapping of qhir1 influencing in vivo haploid induction in maize. Theor Appl Genet. 2013;126:1713–1720. doi: 10.1007/s00122-013-2086-9. [DOI] [PubMed] [Google Scholar]
- Ganal MW, Durstewitz G, Polley A, et al. A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS One. 2011;6:e28334. doi: 10.1371/journal.pone.0028334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Martinez C, Skinner DJ, et al. Development of a seed DNA-based genotyping system for marker-assisted selection in maize. Mol Breed. 2008;22:477–494. doi: 10.1007/s11032-008-9192-4. [DOI] [Google Scholar]
- Geiger HH. Doubled haploids. In: Bennetzen JL, Hake S, editors. Handbook of Maize. Berlin: Springer; 2009. pp. 641–657. [Google Scholar]
- Gilles LM, Khaled A, Laffaire J, et al. Loss of pollen-specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. EMBO J. 2017 doi: 10.15252/embj.201796603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Schrag TA, Peis R, et al. The genetic basis of haploid induction in maize identified with a novel Genome-Wide Association Method. Genetics. 2016;202:1267–1276. doi: 10.1534/genetics.115.184234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher T, Starr D, Wang W, et al. Maternal haploids are preferentially induced by CENH3-tailswap transgenic complementation in maize. Front Plant Sci. 2016;7:414. doi: 10.3389/fpls.2016.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher T, Starr D, Richbourg L, Chintamanani S, Delzer B, Nuccio ML, Green J, Chen Z, McCuiston J, Wang W, Liebler T, Bullock P, Martin B. MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature. 2017;542(7639):105–109. doi: 10.1038/nature20827. [DOI] [PubMed] [Google Scholar]
- Kraptchev B, Kruleva M, Dankov T. Induced heterofertilization in maize (Zea mays L.) Maydica. 2003;48:271–274. [Google Scholar]
- Lashermes P, Beckert M. Genetic control of maternal haploidy in maize (Zea mays L.) and selection of haploid inducing lines. Theor Appl Genet. 1988;76:405–410. doi: 10.1007/BF00265341. [DOI] [PubMed] [Google Scholar]
- Li L, Xu X, Jin W, Chen S. Morphological and molecular evidences for DNA introgression in haploid induction via a high oil inducer CAUHOI in maize. Planta. 2009;230:367–376. doi: 10.1007/s00425-009-0943-1. [DOI] [PubMed] [Google Scholar]
- Liu C, Li W, Zhong Y, et al. Fine mapping of qhir8 affecting in vivo haploid induction in maize. Theor Appl Genet. 2015;128:2507–2515. doi: 10.1007/s00122-015-2605-y. [DOI] [PubMed] [Google Scholar]
- Liu C, Li X, Meng D, Zhong Y, Chen C, Li W, Li L, Tian X, Zhao H, Song W, Luo H, Zhang Q, Lai J, Jin W, Yan J, Chen S. A 4-bp Insertion at ZmPLA1 Encoding a Putative Phospholipase A Generates Haploid Induction in Maize. Mol Plant. 2017;10(3):520–522. doi: 10.1016/j.molp.2017.01.011. [DOI] [PubMed] [Google Scholar]
- Melchinger AE, Brauner PC, Böhm J, Schipprack W. In vivo haploid induction in maize: comparison of different testing regimes for measuring haploid induction rates. Crop Sci. 2016;56:1127–1135. doi: 10.2135/cropsci2015.11.0668. [DOI] [Google Scholar]
- Melchinger AE, Schipprack W, Würschum T, et al. Rapid and accurate identification of in vivo-induced haploid seeds based on oil content in maize. Sci Rep. 2013;3:2129. doi: 10.1038/srep02129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge V, Schipprack W, Mahuku G, et al. Development of in vivo haploid inducers for tropical maize breeding programs. Euphytica. 2012;185:481–490. doi: 10.1007/s10681-012-0657-5. [DOI] [Google Scholar]
- Prigge V, Sánchez C, Dhillon BS, et al. Doubled haploids in tropical maize: I. Effects of inducers and source germplasm on in vivo haploid induction rates. Crop Sci. 2011;51:1498–1506. doi: 10.2135/cropsci2010.10.0568. [DOI] [Google Scholar]
- Prigge V, Xu X, Li L, et al. New insights into the genetics of in vivo induction of maternal haploids, the backbone of doubled haploid technology in maize. Genetics. 2012;190:781–793. doi: 10.1534/genetics.111.133066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge V, Melchinger AE. Production of haploids and doubled haploids in maize. In: Loyola-Vargas VM, Ochoa-Alejo N, editors. Plant cell culture protocols. New Jersey: Springer; 2012. pp. 161–172. [Google Scholar]
- Qiu F, Liang Y, Li Y, et al. Morphological, cellular and molecular evidences of chromosome random elimination in vivo upon haploid induction in maize. Curr Plant Biol. 2014;1:83–90. doi: 10.1016/j.cpb.2014.04.001. [DOI] [Google Scholar]
- Ravi M, Chan SWL. Haploid plants produced by centromere-mediated genome elimination. Nature. 2010;464:615–618. doi: 10.1038/nature08842. [DOI] [PubMed] [Google Scholar]
- Ravi M, Marimuthu MPA, Tan EH, et al. A haploid genetics toolbox for Arabidopsis thaliana. Nat Commun. 2014;5:5334. doi: 10.1038/ncomms6334. [DOI] [PubMed] [Google Scholar]
- Röber FK, Gordillo GA, Geiger HH. In vivo haploid induction in maize-performance of new inducers and significance of doubled haploid lines in hybrid breeding. Maydica. 2005;50:275. [Google Scholar]
- Rotarenco V, Eder J. Possible effects of heterofertilization on the induction of maternal haploids in maize. Maize Genet Coop Newsl. 2003;77:30. [Google Scholar]
- Sarkar KR, Coe EH., Jr A genetic analysis of the origin of maternal haploids in maize. Genetics. 1966;54:453. doi: 10.1093/genetics/54.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satarova TN, Cherchel VY. Inheritance of matroclinal haploidy in maize. Tsitol Genet. 2010;44(3):35–40. [PubMed] [Google Scholar]
- Seymour DK, Filiault DL, Henry IM, et al. Rapid creation of Arabidopsis doubled haploid lines for quantitative trait locus mapping. Proc Natl Acad Sci. 2012;109:4227–4232. doi: 10.1073/pnas.1117277109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W-H, Parmentier Y, Hellmann H, et al. Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol Biol Cell. 2002;13:1916–1928. doi: 10.1091/mbc.E02-02-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swapna M, Sarkar KR. Anomalous fertilization in haploidy inducer lines in maize (Zea mays L.) Maydica. 2012;56:221–225. [Google Scholar]
- Thomann A, Brukhin V, Dieterle M, et al. Arabidopsis CUL3A and CUL3B genes are essential for normal embryogenesis. Plant J. 2005;43:437–448. doi: 10.1111/j.1365-313X.2005.02467.x. [DOI] [PubMed] [Google Scholar]
- Xu X, Li L, Dong X, et al. Gametophytic and zygotic selection leads to segregation distortion through in vivo induction of a maternal haploid in maize. J Exp Bot. 2013;64:1083–1096. doi: 10.1093/jxb/ers393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Lin C, Kim ST, et al. The E3 ubiquitin ligase Cullin 4 A regulates meiotic progression in mouse spermatogenesis. Dev Biol. 2011;356:51–62. doi: 10.1016/j.ydbio.2011.05.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Qiu F, Liu Y, et al. Chromosome elimination and in vivo haploid production induced by Stock 6-derived inducer line in maize (Zea mays L.) Plant Cell Rep. 2008;27:1851–1860. doi: 10.1007/s00299-008-0601-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Fig 1: Effects of AAbb and aaBB genotypes (a) and AABB and aabb (b) on kernel abortion in selfed F3 ears (TIF 2581 KB)


