Abstract
Escherichia coli rrnB terminator t1 contains an RNA hairpin-dependent (class I) and a sequence-specific (class II) termination signal. The latter consists of an 8-bp conserved sequence (CS), TATCTGTT, immediately followed by an 8-bp T rich sequence. In this study, elongation complexes of T7 RNA polymerase at various positions of the class II signal and several mutant signals were obtained by stepwise walking on immobilized DNA templates free of the class I signal. Multiple CS-associated conformational changes were observed, starting at the beginning of the signal and occurring sequentially. When the complexes reach the first base pair of the CS–DNA duplex, which is downstream of the RNA–DNA heteroduplex, their stability, as measured by time-course retention of radiolabeled transcripts, markedly decreases. Further elongation leads to an abrupt change in polymerase–RNA interaction. Cross-linking of the polymerase to a 4-thio-UMP incorporated into RNA 8 nucleotides upstream of the 3′ end and just upstream of the heteroduplex is initially strong but diminishes when the polymerase reaches the fourth base pair of the CS. After a further 7-nt elongation, the exposed single-stranded region of nontemplate strand is contracted; RNA in the upstream half of the heteroduplex becomes dissociated, and the CS–DNA duplex is reformed. During the next 5-nt elongation before termination, the CS duplex is prevented from translocation, and the contracted transcription bubble expands only downstream. These findings suggest that the CS duplex plays essential roles by successively binding to polymerase both downstream and upstream of the heteroduplex.
Keywords: photocross-linking, class II termination, stepwise walking of RNA polymerase, RNA–protein interaction, elongation complex stability
Bacterial RNA polymerases terminate transcription by forming an RNA hairpin followed by a U rich sequence, encoded by a class I termination signal. In contrast, bacteriophage T7 and SP6 RNA polymerases terminate at both class I signals and class II termination signals, the latter consisting of a conserved sequence (CS), HATCTGTT (H designating A, C, or T), followed by a T rich sequence. The class II termination signals found in the Escherichia coli rrnB operon (1), the human preproparathyroid hormone gene (2), and vesicular stomatitis virus DNA (3) terminate phage RNA polymerase transcription, but a CS alone, without a T rich sequence, in the concatamer junction of replicating T7 DNA only causes RNA polymerase to pause (4).
The class II sequence-specific termination differs from the class I hairpin-forming termination in several aspects. (i) Class II termination does not depend on formation of an RNA secondary structure; incorporation of IMP in place of GMP into RNA does not affect termination (1, 5). (ii) Class II termination is highly specific in sequence, requiring a perfect CS duplex; introduction of mutations and mismatches abolishes termination (1, 5, 6). (iii) Introduction of a nick into T7 or SP6 RNA polymerase resulting in N-terminal 20-kDa and C-terminal 80-kDa fragments abolishes class II termination activity but does not affect class I activity (5, 7). (iv) Addition of the denaturing agent guanidine hydrochloride into the reaction increases class II termination efficiency but does not affect class I efficiency (8). (v)Mn2+, when substituted for Mg2+, has different effects on the two classes of termination (9). (vi) Introduction of a disulfide link (D660C-R379C) into T7 RNA polymerase abrogates class II termination but does not affect class I termination (10). Thus, the two classes of termination operate via nonequivalent mechanisms. A hypertranslocation (11) model suggested for class I termination is not compatible with class II termination (8).
The t1 terminator of the rrnB operon has both classes of termination signal for phage T7 and SP6 RNA polymerases (1, 5). The class II signal of rrnB t1 contains a CS module, TATCTGTT, and a T rich module facilitating RNA release (5). The CS module functions as a duplex, causing the polymerase to pause and change conformation (5, 6). The exposed single-stranded region in the transcription bubble is shortened at several base pairs before the termination site, and elongation complexes pause at 2 bp before the termination site (8). Termination occurs through a slow extension from the pause state, but not through a conformational change at the termination site (8). During the extension just before termination, the thumb and fingers subdomains of polymerase should move apart (10). Thus, the earliest conformational change known to be associated with class II termination is the bubble contraction, but it is not known how this occurs.
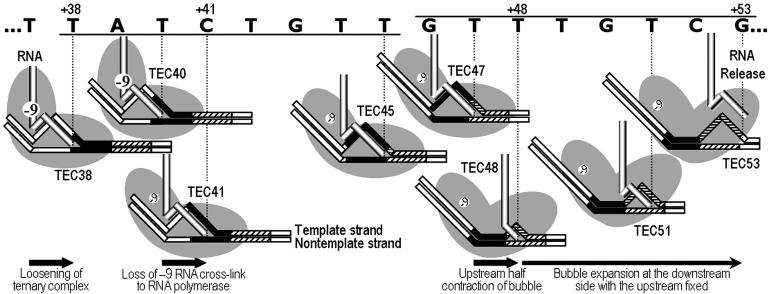
In investigating the mechanism of class II termination, we have found that signal-associated conformational changes start at the beginning of the 16-bp class II termination signal of rrnB t1. Drastic changes in complex stability, RNA cross-linking to polymerase and transcription bubble, occurred sequentially, as shown schematically in Fig. 1, and these were associated with the CS. When the CS–DNA duplex bound to polymerase downstream of the active site, the elongation complexes were drastically destabilized. This was followed by cessation of the strong RNA cross-linking to the elongating polymerase upstream of the DNA–RNA heteroduplex, leading to dissociation of RNA in the upstream half of the heteroduplex after a further 7-nt extension. The CS–DNA duplex subsequently formed in the collapsed upstream half of the transcription bubble, possibly binding to the polymerase upstream of the heteroduplex. These results suggest that the two successive bindings of the CS duplex and polymerase both play important roles in class II termination.
Fig. 1.
Sequential multiple changes occurring before class II termination. The CS (underlined) and T rich (overlined) modules of class II rrnB t1 termination signal from +38 to +53 are indicated by filled and shaded boxes, respectively, in the illustrations of elongation complexes of T7 RNA polymerase (two overlapping gray ovals). During elongation, two DNA strands move to the left as the RNA strand exits upward. Ternary complexes are destabilized at +38 when CS binds to polymerase downstream of the active site. At +41, the –9 RNA residue (circled –9) ceases cross-linking to polymerase. At +48, the transcription bubble contracts to 3–4 bp as CS strands form a duplex in the collapsed upstream half of the bubble. The CS duplex could bind to polymerase upstream of the RNA–DNA heteroduplex. During the last 5-nt extension before termination at +53, the bubble expands only downstream, whereas the CS duplex is prevented from translocation.
Materials and Methods
DNA Templates. The plasmid pBT1, containing a T7 promoter and the class II termination signal of E. coli rrnB t1 terminator but free of the class I termination signal (8), was used to prepare a biotinylated linear DNA template t1b of 186 bp by PCR amplification, using reverse primer 5′-GCGGATAACAATTTCACACAGGA-3′ biotinylated at its 5′ end and forward primer 5′-GTAAAACGACGGCCAGT-3′. The sequence from nucleotide +33 to nucleotide +58 (where the transcription start site is nucleotide +1) was derived from the rrnB t1 terminator (Fig. 2A). Several plasmids bearing mutant forms of the signal were constructed by inserting synthetic oligonucleotides into the BamHI/HindIII site of pBT1.
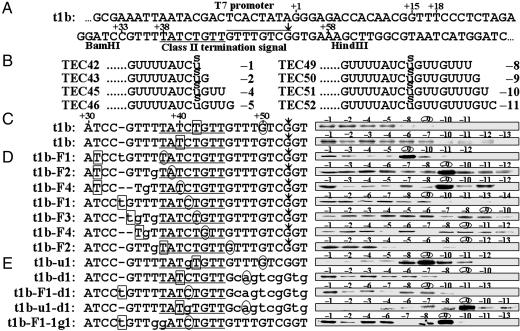
Fig. 2.
Photocross-linking of RNA and RNA polymerase. (A) The template t1b, containing the E. coli rrnB t1 class II termination signal, terminates T7 RNA polymerase transcription at +53 (with respect to the 5′ end of the RNA), indicated by an arrow. The CS module is underlined, and the T rich module overlined. (B) RNA sequences of t1b elongation complexes obtained by stepwise walking after an sU was incorporated into radiolabeled RNA at +42. The positions of sU with respect to the 3′ end of RNA are indicated in negative numbers. (C) The complexes in B were irradiated for photocross-linking (first line). Another series of t1b elongation complexes were obtained after an sU was incorporated into RNA at boxed position +40, followed by irradiation (second line). (D) Template variants t1b-F1 through -F4 differ from t1b only at the positions indicated by lowercase letters, and all are active in termination. Seven series of elongation complexes were obtained after sU was incorporated at the boxed positions in RNA. Complexes that will have an sU at -9 RNA residue are indicated in the sequences by the circled positions of their 3′ ends. (E) Five mutants differing from t1b at the positions indicated by lowercase letters. All are inactive in termination. The efficiencies of the “strong” cross-linking (in the six complexes mentioned in the text) averaged over five experiments range from 14% to 21% (an average of 17%), whereas those of the “weak” cross-linking are all <6.0%.
Stepwise Polymerase Walking. Elongation complexes of biotinylated templates were obtained by stepwise walking, as described (12). The initial 80-μl reaction mixture containing 40 mM Tris·HCl, pH 7.9, 6 mM MgCl2, 100 mM KCl, 10 mM DTT, 10 pmol biotinylated DNA template, 10 pmol T7 RNA polymerase (Amersham Pharmacia Biosciences), 50 μM ATP, 50 μM GTP, and 5 μM CTP was incubated at room temperature for 20 min. The product, a ternary elongation complex of 15-mer RNA, here called TEC15, was then subjected to a walking reaction using 80 μCi [α-32P]UTP (3,000 Ci/mmol; 1 Ci = 37 GBq) for 1 min. The resulting radiolabeled TEC18, containing an 18-mer RNA, was further walked by repetition of resuspension and 1 min incubation in 80-μl transcription buffer containing the 0.5 μM NTP required for the next step. To determine the length and amount of RNA in complexes, a 5-μl aliquot of each complex was mixed with two volumes of gel-loading solution A (12 M urea/10 mM EDTA/0.1% bromophenol blue) prewarmed to 55°C, heated at 95°C for 2 min, and electrophoresed on 8 M urea/10% polyacrylamide gels.
Photocross-Linking. Addition of 1 μM 4-thio-UTP (pH 7.2) at particular stages of the polymerase walking procedure results in incorporation of 4-thio-UMP (sU) at specific sites in the transcripts (12, 13). A 5-μl aliquot of each complex was assayed for the quantity of expected size RNA as above, and a second aliquot (<10 μl) of each complex containing the same amount of RNA was placed on ice and irradiated with 360-nm UV light for 20 min. Streptavidin beads were dissociated from biotinylated DNA by boiling in 0.1% SDS for 5 min and removed by a magnet. The cross-linked and uncross-linked polymerases were separated by 0.1% SDS/8% polyacrylamide gel electrophoresis. Radioactivity of cross-linked complexes was measured by using a Storm 860 scanner (Amersham Pharmacia Biosciences).
Mapping of Exposed Single-Stranded DNA Region. DNA templates were radiolabeled at the 5′ end of the nontemplate strand and biotinylated at the 5′ end of the template strand. The KMnO4 footprinting assay was performed as described (8) for various ternary complexes (0.5 pmol) obtained by stepwise walking, and the products were analyzed by electrophoresis on 8 M urea/12% polyacrylamide gels.
Results
Absence of Strong Cross-Linking of RNA to T7 RNA Polymerase Near the Termination Site. Previously reported photocross-linking results indicated that elongating T7 RNA polymerase can be efficiently cross-linked to the transcript at 8 nucleotides upstream (nucleotide -9) of the 3′ end, nucleotide –1 (13, 14). To determine whether there were changes in the interaction between RNA and elongating polymerase near the class II termination site, we repeated the cross-linking experiments using DNA templates containing the class II, but not the class I, signal of E. coli rrnB terminator t1.
Template t1b contains the 16-bp class II signal from nucleotide +38 to nucleotide +53 (Fig. 2 A), and transcription by T7 RNA polymerase under standard conditions has been shown to produce 53-mer termination and 111-mer runoff products (8). Using stepwise walking on biotinylated templates (12, 13), we isolated various active ternary elongation complexes with RNA lengths up to 52 nucleotides.
The RNA residues interacting with the polymerase were determined by incorporating an sU into radiolabeled RNA and photocross-linking this residue to elongating polymerases. An elongation complex of 41-mer RNA (TEC41), referred to as template position +41, was obtained by stepwise walking on t1b. Then, an sU residue was added to the 3′-end of the RNA in place of +42U to form TEC42 with the sU residue located at position -1 of the RNA (Fig. 2B). [The positions of RNA residues are identified by positive or negative numbers depending on whether the 5′-end (+1) or 3′-end (-1) is used as the reference.] The resulting product was walked to TEC43, in which the +42sU was located at residue –2 of the RNA, and further walked to every sequential position, up to 1 bp before the termination site. The products TEC45, TEC46, TEC49, TEC50, TEC51, and TEC52 contain a +42sU residue located at positions –4, –5, –8, –9, –10, and –11, respectively, with respect to the 3′-end of the RNA (Fig. 2B).
When these complexes were irradiated (Fig. 2C), we found that all had a low level of polymerase–RNA cross-linking (Fig. 2C, first line). Because strong polymerase cross-linking of sU at residue –9 had been observed in various elongation complexes of other templates (13, 14), it was especially interesting that our cross-linking of TEC50 with –9 sU was weak. (We were not able to assay cross-linking of sU at –3, –6, and –7, because the consecutive Ts at positions +44 to +45 and +47 to +49 did not permit the formation of these complexes.)
To determine whether strong cross-linking had moved from –9 to a position untested in the above experiments, we incorporated sU into RNA at +40 and advanced complexes to all possible template positions (Fig. 2C, second line), to produce complexes with sU at every RNA position from –1 to –13, except for –5, –8, and –9. All cross-linking levels were still low. These two sets of experimental results indicate that polymerase cross-linking of RNA residues from –1 to –13 is low in ternary complexes near the termination site.
Polymerase Cross-Linking with the –9 Residue Ceases from the Fourth Base Pair of the CS. Because the loss of strong –9 cross-linking may be a common characteristic of complexes with a class II signal sequence (as in TEC50 of +42sU), we investigated other complexes at different positions of the CS. To obtain all complexes from TEC38 through TEC43 and TEC46 containing a –9 sU, the sequence upstream of the class II signal was altered (lowercase letters in Fig. 2), resulting in template variants t1b-F1 through -F4 (Fig. 2D). For example, t1b-F1 had an insertion of t between nucleotides +33 and +34 (template position numbering refers to t1b rather than the variant template). After incorporation of sU into radiolabeled RNA at nucleotide +31 of t1b-F1, TEC31 was walked to various positions, such that TEC38 had the sU at –9 (Fig. 2D, first line). Likewise, when an sU was incorporated at the inserted t of t1b-F1, we obtained TEC41 with a –9 sU (Fig. 2D, fourth line).
After incorporation of sU into RNA at various positions using the variant templates, we obtained seven series of complexes (Fig. 2D). When these were irradiated, polymerase–RNA cross-linking was weak in all of the complexes, except for TEC38 of t1b-F1, TEC39 of t1b-F2, and TEC40 of t1b-F4, all of which had sU at –9, and all of which exhibited strong cross-linking (Fig. 2D, lines 1–3). The other –9 sU complexes, TEC41 of t1b-F1, TEC42 of t1b-F3, TEC43 of t1b-F4, and TEC46 of t1b-F2, however, did not exhibit strong cross-linking (Fig. 2D, lines 4–7).
These results indicate that –9 sU is strongly cross-linked to polymerases at positions equivalent only to +38, +39, and +40 but not to +41, +42, +43, +46, and +50, and that no other RNA residues up to –14 are strongly cross-linked. Thus, the –9 RNA residue is continually cross-linked strongly to polymerase until the third position (+40) of the CS, but not at the fourth position (+41) or beyond.
The variant templates t1b-F1 through -F4 are active in termination and the termination site is not shifted. Although a double substitution of +37T and +38T for Gs in t1b-1g1 was previously reported to inactivate the signal (5), we found that +37T→g alone in t1b-F2 has no effect, indicating that only +38T→g inactivates the signal. Moreover, the signal is not inactivated by +36 (t1b-F4) and +35 (t1b-F3) substitutions, thus confirming that the class II signal sequence starts from +38.
The –9 RNA Cross-Link Is Maintained on Conserved-Sequence Mutants. When the fourth position of the CS module was changed from +41C to g (in t1b-u1), cross-linking was strong in TEC50 with sU at –9 (Fig. 2E), but termination did not occur (5). A similar pattern was observed in two other mutant templates: t1b-u1-d1, which contains mutations in both modules, and t1b-F1-1g1, which contains a substitution in the CS module (Fig. 2E). In contrast, when the T rich module was changed in t1b-d1 and t1b-F1-d1, –9 cross-linking was weak in TEC48 or TEC41, respectively (Fig. 2E), although termination did not occur. Thus, the cessation of strong –9 RNA cross-linking is associated with an intact CS rather than an intact T rich module.
Marked Decrease in Complex Stability at the Start of the Signal. The cessation of strong –9 RNA cross-linking to polymerase at location +41 of intact CS led us to examine the stability of t1b complexes on +31 through +51; after each complex was obtained by using stepwise walking at room temperature, aliquots were incubated at 37°C for 0–120 min. Remaining complexes containing biotinylated DNA were captured by streptavidin-coated magnetic beads, and their radioactive transcripts were measured by gel electrophoresis (Fig. 3A). A decrease in radioactivity will be seen when RNA is released from the ternary complex or when the binary complex of RNA–polymerase dissociates from DNA during the incubation. Thus, this assay measures the stability of ternary complexes.
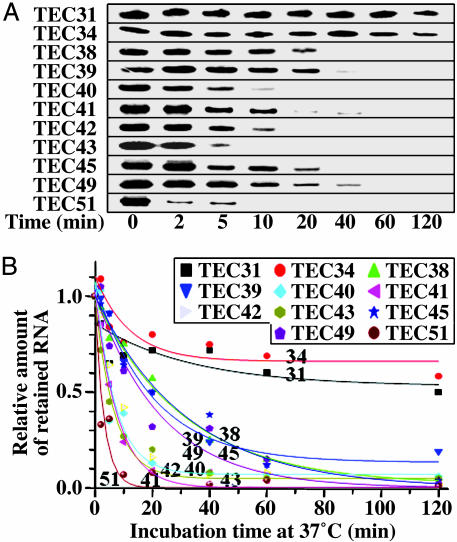
Fig. 3.
Ternary complex stability. (A) Complexes at +31 through +51 were separately obtained by using stepwise walking on biotinylated t1b at room temperature, with the last step for 2 min. Each complex was divided into 30-μl aliquots for incubation at 37°C. After 0–120 min, the complexes were isolated on streptavidin-coated magnetic microbeads and radioactive transcripts retained after washing were analyzed by gel electrophoresis. (B) Amounts of radioactive RNA measured in three separate time courses for each complex. Values relative to the zero time point are shown, indicating the fraction of remaining ternary complexes; each point represents the average of three samples. Decay half-times were estimated from first-order curves except for TEC31 and TEC34 with coefficients of determination r2 of 0.98 or 0.99, except for TEC40 (0.91), TEC49 (0.95), and TEC51 (0.91). The plateau levels were <5%, except for TEC31 (53%), TEC34 (66%), and TEC39 (13%).
In TEC31 and TEC34, more than half the transcripts remained as complexes even after 2 h of incubation. Their decay curves approached plateau levels of 53% and 66%, respectively (Fig. 3B), suggesting that only a portion of the complex, referred to as loose complex, could lose RNA and/or DNA, and the decay half-time of the remaining tight complexes was too long to measure. In contrast, loose complexes dominated in TEC38 through TEC51, because most of the transcripts disappeared within 2 h (Fig. 3A). The half-times of these apparent first-order decay kinetics were estimated to be 1–23 min, and <5% of complexes remained intact, except for TEC39 (13%).
TEC35, TEC36, and TEC37 could not be obtained with t1b due to the TTTT at +35 to +38. Instead, these complexes and TEC38 were obtained from an active variant template t1b-F3, and their stabilities were measured in the same way. The decay curves of TEC35, TEC36, and TEC37 of t1b-F3 (63–71% remaining as complex) were similar to those of TEC31 and TEC34 of t1b. On the other hand, the decay curve of TEC38 of t1b-F3 approached the plateau level of 22%, and most of TEC40, TEC45, and TEC51 of t1b-F3 were dissociated within 2 h (Fig. 5, which is published as supporting information on the PNAS web site). Thus, complex stability markedly decreases from the first position (+38) of the signal sequence and thereafter.
The stretch of four Ts at +35 to +38 in t1b is not responsible for this decrease in complex stability, because a G substitution at +37 in t1b-F2 or a double substitution at +35 and +37 in t1b-F3 did not increase the plateau level and decay half-time of TEC40, TEC45, and TEC51. In contrast, when we used t1b-1g1, which carries mutations at +37 and +38, the plateau levels of TEC40, TEC45, and TEC51 were raised to 62–68% with a half-time of 100–120 min. Thus, mutation at the first position (+38) of CS, but not at +37 or +35, restores the stability of the complex, suggesting that CS is important for the destabilization at +38 and beyond. In contrast, when we used t1b-d1, which is devoid of the T rich module downstream of +46, the plateau level and half-time were still small in TEC40 and TEC45 but were increased to 75% and 200 min, respectively, in TEC51. (Data for all of the mutant template complexes are shown in Fig. 5). Thus, the stability of the complexes around +51 is also affected by the T rich module sequence.
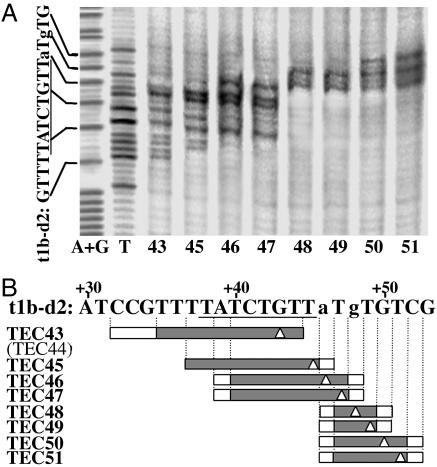
The Exposed Single-Stranded Region Contracts at 5 bp Upstream of the Termination Site. We previously observed (8) that the exposed single-stranded region in the transcription bubble of template t1b contracted from ≈10 to 4–5 bp during extension from TEC46 to TEC49 when transcribing the T rich sequence. This contraction was not observed with a CS mutant, suggesting its association with CS (8). The TTT sequence at +47 to +49 of t1b prevents a determination of the starting position of the contraction. Here, we used the template variant, t1b-d2, which carries substitutions at +46 and +48 to determine the starting position of the contraction and to examine the effect of mutations in the T rich module. We obtained the complexes TEC43 through TEC51 from t1b-d2 by stepwise walking and mapped unpaired exposed Ts in the nontemplate strand of each complex by KMnO4 footprinting (Fig. 4).
Fig. 4.
Mapping of exposed unpaired Ts in the nontemplate strands of ternary complexes. (A) Template t1b-d2 was radiolabeled at the 5′ end of the nontemplate strand and biotinylated at the 5′ end of the template strand. Ternary complexes halted at +43 through +51 were obtained by stepwise walking and subjected to KMnO4 footprinting. The products were analyzed by gel electrophoresis. (B) Exposed single-strand DNA regions in the complexes are shown by filled boxes and uncertain regions by empty ones. The band intensities were averaged over more than five experiments, and when average band intensities were below 25% of the most intense band in each lane, the corresponding residues were regarded as not reactive. The 3′ ends of RNA are marked by empty triangles.
The exposed single-stranded region contracted to 3–4 bp in TEC48, where only the Ts at +47 and +49 were unpaired and exposed, although t1b-d2 was inactive in termination. Other features were the same as for the t1b complexes (8). After the contraction, the single-stranded region expanded on the downstream side without any changes in the upstream side. The leading boundary of the bubble was 1 bp downstream of the 3′ end of RNA, as in TEC43, TEC46, and TEC50. The size of the bubble was normally 10 bp, as in TEC45. Thus, bubble contraction starts in TEC48 at a position in the T rich module but is not associated with this module.
Discussion
Sequential Events Starting at the Beginning of the Class II Signal. The t1b template contains a class II termination signal, consisting of a CS module, TATCTGTT, at +38 to +45, and a T rich module at +46 to +53. Its transcription by T7 RNA polymerase terminates at +53 under standard conditions (1, 8). In the present study, we observed multiple distinct changes in the properties of elongation complexes throughout the 16-bp termination signal of t1b (Fig. 1). (i) The stability of ternary complexes markedly decreases at +38. (ii) Strong cross-linking of the –9 RNA residue to the elongating polymerase ceases in TEC41. (iii) The exposed single-stranded DNA region in the transcription bubble is abruptly contracted in TEC48. (iv) The half-contracted region expands only on the downstream side from TEC49 until termination at +53.
If elongation complexes get into termination mode only at or very near the termination site, their general properties including high stability, -9 RNA cross-linking to the polymerase and normal-size transcription bubble (≈10 bp) should persist until then. We found, however, that these properties change substantially much earlier, and all these changes are specifically associated with the CS module of the class II signal. Thus, the propensity for termination starts 16 bp before the termination site. The changes occur sequentially, and a change at one position may set up the next change at a later position.
Elongation complexes isolated through the immobilized template DNA were ternary when examined by using radiolabeled DNA and RNA (data not shown), as reported (15). This is in contrast to elongation complexes of immobilized polymerase isolated near the termination site (16), which include some binary complexes of polymerase–RNA and polymerase–DNA (15).
Multiple Conformational Changes Along the Class II Termination Signal. The earliest conformational change, observed as a marked decrease in complex stability, occurs at +38. The length of the DNA duplex interacting with the polymerase downstream of the active site was estimated to be 5–8 bp by footprinting (17–20), consistent with the crystal structure of the T7 elongation complex (21). Thus, in TEC38, more than half of the 8-bp CS would bind to the polymerase as a duplex. A mutation at +38 substantially restores the stability of TEC40, TEC45, and TEC51, suggesting that this position is important, and that the CS duplex may be involved in decreasing complex stability.
Complex destabilization may be due to the DNA and/or RNA losing some interactions with the polymerase at sites other than the –9 RNA residue or to the RNA–DNA heteroduplex being shortened or weakened. The –1 and –14 RNA residues can be cross-linked to elongating polymerase but not as efficiently as the –9 residue (13, 22). Cross-linking of sU at –1 and –14 appeared to be weaker in some of the complexes we studied than seen previously, but this effect was not as large or consistent as seen for the –9 residue. If destabilization in TEC38 arises from a substantially shortened heteroduplex, one would still expect most, if not all, of the four consecutive Us encoded by Ts at +35 to +38 and located at the 3′ end of RNA in TEC38 to participate in a heteroduplex. We found, however, that substitutions of Gs at +35 and +37 in t1b-F2 and -F3 did not have a substantial stabilizing effect.
The second conformational change involves the –9 RNA interaction with polymerase during the transition from TEC40 to TEC41. The strong –9 cross-linking ceases abruptly at that point, suggesting the occurrence of a conformational change. The –9 residue is located just upstream of the RNA–DNA heteroduplex and is cross-linked to a part of the specificity loop (amino acids 744–750) in the elongating polymerase (13, 14). This part is presumably located near the RNA exit path in elongation complexes. The loss of the –9 cross-link may reflect a conformational change in this region of the polymerase. In TEC41, this change could release a structural strain that accumulated during RNA extension beyond TEC38.
The third conformational change is the half contraction of the exposed unpaired DNA region in TEC48. It is unlikely that the nontemplate strand alone, or two separate DNA strands simultaneously, bind to polymerase, based on the effects of various mutations in the CS (8). Thus, the entire CS (+38 to +45) appears to form a duplex in TEC48. The contraction is not likely due to the difference in stability between the DNA duplex and the RNA–DNA heteroduplex of the CS, because it does not occur earlier than TEC48. It seems more likely that the RNA is displaced in the upstream half of the bubble after the RNA path has been displaced in TEC41.
During the last 5-nt extension, the upstream boundary of the bubble remains fixed, whereas the downstream DNA is unwound, and the bubble size increases until termination at +53. The pattern of bubble expansion is similar to the abortive initiation cycling phase in that the upstream DNA is held by polymerase as the active site moves downstream on the DNA (8). This finding is supported by results obtained using a chemical nuclease, suggesting that the leading domain of the polymerase moves further along the DNA than the trailing domain (10), and that the CS duplex formed in TEC48 could bind to polymerase. Tyrosine 385 of T7 RNA polymerase has been suggested as a candidate for binding to the CS in a paused state just before the termination site (10).
A Termination Model Involving Roles of the CS (Fig. 1). All of the conformational changes observed in this study are associated with the CS module. The interaction of this module with the polymerase is thought to involve duplex CS (5, 6). Here, we determined the progression of CS duplex binding to the elongating polymerase, and we suggest that the CS plays crucial roles during duplex binding to the polymerase at two different points. When downstream of the active site, in TEC38, the CS duplex presumably binds to polymerase, resulting in destabilization of complexes and later displacement of RNA (-9 residue) upstream of the heteroduplex. When the RNA is further displaced, as the upstream half of the heteroduplex is unwound in TEC48, the DNA strands of the CS again form a duplex that binds to polymerase. This binding thus prevents the CS duplex from translocation.
It remains to be determined what regions of RNA polymerase bind to the CS duplex when it is located downstream and upstream of the heteroduplex (e.g., in TEC38 and TEC48, respectively). This might be determined by DNA–polymerase cross-linking experiments or by the elucidation of crystal structures of these complexes. It would be interesting to see whether the CS duplex-binding regions are the same in TEC38 and TEC48, because this would suggest a major conformational change as the region moves from downstream to upstream.
Further extension from TEC48 results in termination at different sites, depending on the T rich module sequence (5), the polymerase, and the transcription conditions. For example, the T7 RNA polymerase terminates at +53 in 100–500 μM NTP, but at +51 in 0.5–50 μM NTP, whereas SP6 RNA polymerase terminates at +51 even at 500 μM NTP (5). In the presence of Mn2+ in place of Mg2+, RNA is extended by a few nucleotides before termination by SP6 polymerase but not by T7 polymerase (9).
During the extension from TEC48, the upstream boundary of the transcription bubble is fixed, and the downstream boundary and the active site advance together downstream on the DNA. If the active-site domain moves away from the fixed upstream domain, the polymerase would be stretched to accommodate the extension. If the active-site domain does not move, the nontemplate strand and the heteroduplex would be compressed or looped out. In either case, there would be a spatial limit, and further translocation and nucleotide incorporation would be blocked in a pretranslocated state, facilitating termination. The size of the spatial limit could vary for different polymerases under different conditions, explaining the shifts in termination sites.
We favor the former possibility, polymerase stretching, not just because termination efficiency is increased by addition of a denaturing agent (8), but also because termination requires that the thumb subdomain move away from the fingers subdomain of T7 RNA polymerase. When the two subdomains are linked by a disulfide bridge, the polymerase fails to terminate, although it pauses normally just before the termination site (10).
Supplementary Material
Acknowledgments
We thank Professor Charles R. Cantor for critical reading of this manuscript as well as for constructive criticisms and suggestions. This work was partially supported by grants from the Korea Science and Engineering Foundation (R01-1999-00111) and the 21C Frontier Project (M1-03KB010006). Y.S. is a recipient of a postdoctoral fellowship (M02-2004-000-11191-0), and C.K. is a participant of the Brain Korea 21 Project.
Author contributions: Y.S. and C.K. designed research; Y.S. performed research; Y.S. and C.K. analyzed data; and Y.S. and C.K. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CS, conserved sequence; sU, 4-thio-UMP.
References
- 1.Hartvig, L. & Christiansen, J. (1996) EMBO J. 15, 4767-4774. [PMC free article] [PubMed] [Google Scholar]
- 2.Macdonald, L. E., Durbin, R. K., Dunn, J. J. & McAllister, W. T. (1994) J. Mol. Biol. 238, 145-158. [DOI] [PubMed] [Google Scholar]
- 3.Whelan, S. P., Ball, L. A., Barr, J. N. & Wertz, G. T. (1995) Proc. Natl. Acad. Sci. USA 92, 8388-8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyakhov, D. L., He, B., Zhang, X., Studier, F. W., Dunn, J. J. & McAllister, W. T. (1997) J. Mol. Biol. 269, 28-40. [DOI] [PubMed] [Google Scholar]
- 5.Kwon, Y. S. & Kang, C. (1999) J. Biol. Chem. 274, 29149-29155. [DOI] [PubMed] [Google Scholar]
- 6.He, B., Kukarin, A., Temiakov, D., Chin-Bow, S. T., Lyakhov, D. L., Rong, M., Durbin, R. K. & McAllister, W. T. (1998) J. Biol. Chem. 273, 18802-10811. [DOI] [PubMed] [Google Scholar]
- 7.Macdonald, L. E., Zhou, Y. & McAllister, W. T. (1993) J. Mol. Biol. 232, 1030-1047. [DOI] [PubMed] [Google Scholar]
- 8.Song, H. & Kang, C. (2001) Genes Cells 6, 291-301. [DOI] [PubMed] [Google Scholar]
- 9.Boudvillain, M., Schwartz, A. & Rahmouni, A. R. (2002) Biochemistry 41, 3137-3146. [DOI] [PubMed] [Google Scholar]
- 10.Mukherjee, S., Brieba, L. G. & Sousa, R. (2003) EMBO J. 22, 6483-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Artsimovitch, I. & Landick, R. (1998) Genes Dev. 12, 3110-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sohn, Y., Shen, H. & Kang, C. (2003) Methods Enzymol. 371, 170-179. [DOI] [PubMed] [Google Scholar]
- 13.Shen, H. & Kang, C. (2001) J. Biol. Chem. 276, 4080-4084. [DOI] [PubMed] [Google Scholar]
- 14.Temiakov, D., Mentesana, P. E., Ma, K., Mustaev, A., Borukhov, S. & McAllister, W. T. (2000) Proc. Natl. Acad. Sci. USA 97, 14109-14114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kashlev, M. & Komissarova, N. (2002) J. Biol. Chem. 277, 14501-14508. [DOI] [PubMed] [Google Scholar]
- 16.Gusarov, I. & Nudler, E. (1999) Mol. Cell 3, 495-504. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda, R. A. & Richardson, C. C. (1986) Proc. Natl. Acad. Sci. USA 83, 3614-3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mookhtiar, K. A., Peluso, P. S., Muller, D. K., Dunn, J. J. & Coleman, J. E. (1991) Biochemistry 30, 6305-6313. [DOI] [PubMed] [Google Scholar]
- 19.Basu, S. & Maitra, U. (1986) J. Mol. Biol. 190, 425-437. [DOI] [PubMed] [Google Scholar]
- 20.Huang, J. & Sousa, R. (2000) J. Mol. Biol. 303, 347-358. [DOI] [PubMed] [Google Scholar]
- 21.Tahirov, T. H., Temiakov, D., Anikin, M., Patlan, V., McAllister, W. T., Vassylyev, D. G. & Yokoyama, S. (2002) Nature 420, 43-50. [DOI] [PubMed] [Google Scholar]
- 22.Ma, K., Temiakov, D., Jiang, M., Anikin, M. & McAllister, W. T. (2002) J. Biol. Chem. 277, 43206-43215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.