Abstract
Metabolic alteration constitutes a hallmark of cancer. Glycolysis and antioxidant pathways in kidney cancer are elevated, with frequent mutation of the VHL gene. Intratumor genetic heterogeneity has been recently demonstrated in kidney cancer. However, intratumor metabolic heterogeneity has not been investigated. Here, we used global metabolomics analysis and tissue slice tracer studies to demonstrate that different portions of a human primary kidney tumor possess different metabolic characteristics and drug sensitivity. Pyruvate levels were elevated and pyruvate metabolism was altered in some tumor sections. These observations indicated that pyruvate metabolism may constitute a possible vulnerability of kidney cancer; indeed, pyruvate stimulated the growth of primary kidney cancer cells and pharmacological inhibition of pyruvate transporters slowed the growth of patient-derived kidney tumors in mice. These findings deepen our understanding of the intratumor metabolic heterogeneity of kidney cancer and may inform novel therapeutic approaches in human kidney cancer.
Keywords: Kidney cancer, Metabolism, Metabolomics, Lipidomics, Heterogeneity, Mutation
Highlights
-
•
Human primary kidney tumors harbor intratumor metabolic heterogeneity.
-
•
Some tumor portions have high pyruvate levels with low glycolysis metabolites.
-
•
Different pyruvate metabolic pathways are present in a tumor and represent a potential vulnerability
Understanding the features of cancer can lead to effective cancer therapy. One of the reasons why anti-cancer drugs have limited efficacy is the existence of intratumor heterogeneity. Intratumor heterogeneity is usually revealed by diverse genetic alterations. In this study, we studied metabolic features to comprehend intratumor heterogeneity. In human primary kidney tumors, some portions contained high levels of upper-glycolysis metabolites; however, other portions contained high pyruvate levels. Difference in pyruvate metabolism was observed in a tumor. Additional studies suggested that pyruvate metabolism represented a vulnerability in kidney cancer.
1. Introduction
Aerobic glycolysis in cancer tissue, the Warburg effect (Warburg, 1956), has been known for a long time. Recent metabolic analyses revealed that the metabolism of cancer cells is altered to allow them to maintain high proliferation rates in spite of fluctuating nutrient availability (Vander Heiden et al., 2009, Cairns et al., 2011). However, to date, metabolic hallmarks of human cancer have not been fully clarified by metabolomics analyses.
Renal cell carcinoma (RCC) is considered to be a suitable model for studying altered metabolism in cancer as inherited predisposition to RCC is reportedly associated with genes responsible for cellular metabolism (Linehan et al., 2010). Genes, such as VHL, PBRM1, SETD2, MET, and NF2, are frequently mutated in RCC (The Cancer Genome Atlas Research Network, 2013, Durinck et al., 2015). Recently, exome sequencing of spatially separated portions of primary RCC revealed extensive intratumor genetic heterogeneity (Gerlinger et al., 2012). Altered metabolism, including enhanced glycolysis and antioxidant response pathways, has also been reported in clear cell RCC (Hakimi et al., 2016), but to date, no study has explored intratumor metabolic heterogeneity in kidney cancer.
In this study, we addressed intratumor metabolic heterogeneity by global metabolomics analyses of surgically resected primary kidney tumors, some of which were obtained from spatially-separated different sites within the same tumor. In addition, we investigated the roles of pyruvate—the featured metabolite in our metabolic profiling—in kidney cancer growth to identify potential clinical vulnerability in human kidney cancer.
2. Materials and Methods
2.1. Tissue Collection
Our institutional committees approved the study experiments, and informed consent was obtained from all participating patients. Kidney tumors and adjacent normal kidney tissues were obtained from patients after radical nephrectomy for RCC at the Kyorin University Hospital (Tokyo Japan). Clinical and pathological characteristics of patients are described in Table S1. Samples were collected from macroscopically identified normal or tumor tissue. For global metabolomics analysis, 32 kidney tumor samples and 18 normal adjacent kidney tissue samples were obtained from 18 patients (patients no. 20–38); multiple spatially separated samples were obtained from tumors from eight patients (patients no. 21, 24, 30, 33–35, 37, and 38). For tissue slice tracer studies, 30 kidney tumor samples and 12 adjacent normal tissue samples were obtained from two patients (patients no. 42 and 45). A patient-derived kidney cancer cell line, 5K, and its paired normal kidney cell line, 5KN, were established from the tissues of patient no. 5 and PDX tumors were established from the tissues of patients no. 11, 12, 24, and 34. For global metabolomic and genomic analyses, tissue samples were soaked in liquid nitrogen immediately after resection and stored in liquid nitrogen until analysis.
2.2. Global Metabolomics Analysis
Global metabolomics profiles were obtained by combining data derived from three platforms: gas chromatography (GC) MS-MS, ion-pair liquid chromatography (IP-LC) MS-MS, and lipidomics. Details are included in the Supplemental Experimental Procedures.
2.3. Cell Culture and Growth Assays
Preparation of patient-derived cell cultures was performed according to an existing protocol (Valente et al., 2011), with modifications. Patient tissues were cut into small pieces and treated with 1 mg/mL collagenase in culture medium, DMEM-F-12 (Gibco) supplemented with 10% fetal bovine serum (FBS) (Hyclone) and human transferrin (holo) (BBI Solutions). After 20 min incubation at 37 °C with gentle stirring, the cells were washed with Hank's balanced salt solution, and seeded on a culture dish. The cells were maintained at 37 °C in 5% CO2 atmosphere.
For growth assays, the established 5K patient-derived RCC cells or the respective paired normal 5KN cells were plated at a density of 2500 cells/well in 96-well plates, and cultured overnight. Next day, sodium pyruvate (Gibco), l-cystine (Sigma-Aldrich), 2-oxobutyrate (Wako), or UK-5099 (Sigma-Aldrich), were added to the wells. After 5 days, cell viability was assessed using the Cell Titer-Glo Luminescent Cell Viability Assay (Promega). RPMI-1640 medium containing 205 μM glutamine and 10% FBS was used in these experiments. Growth assays with UK-5099 were conducted in the presence of 1 mM sodium pyruvate.
2.4. Tissue Slice Stable Isotopic Tracer Experiments
The ex vivo studies were performed using tissue slices according to the modified method of Sellers et al. (2015). Briefly, the tumor and the adjacent normal kidney tissue were resected and cut with a scalpel into ca. slices (1-mm thick) within 2 h of surgery. The tissue slices were then incubated for 24 h at 37 °C in a 5% CO2 atmosphere in DMEM supplemented with 10% FBS (dialyzed) and either 10 mM [U-13C]-glucose (Cambridge Isotope Laboratories, Inc.) or 2 mM [U-13C,15N]-glutamine (Cambridge Isotope Laboratories, Inc.). The tissue fragments were then washed in saline, frozen in liquid nitrogen, homogenized in methanol using the TissueLyser II, and extracted for GC–MS analysis.
2.5. Stable Isotopic Tracer Experiments in Cell Culture
The 5K and 5KN cells were plated at a density of 1 × 105 cells/10 mL in a 10-cm dish in RPMI-1640 (Wako, 189-02025) supplemented with 10% FBS. The next day, the medium was replaced with glucose- and glutamine-free RPMI-1640 (Wako, 185-02865 and 183-02165, respectively) supplemented with 10% FBS and 2 g/L [U-13C]-glucose (Cambridge Isotope Laboratories, Inc., CLM-1396) or 300 mg/L [U-13C,15N]-glutamine (Cambridge Isotope Laboratories, Inc., 520-75,044). Cells were incubated for 0.5, 1, or 4 h. Metabolites were extracted with methanol for GC–MS analysis as described above. The RPMI-1640 media in this study did not contain pyruvate or lactate.
2.6. Animal Experiments
The use of animals for this study was approved by Takeda Pharmaceutical Company Shonan Research Center Animal Ethics Committee (Kanagawa, Japan), and all procedures were performed according to protocols approved by the Institutional Animal Care and Use Committee of the Pharmaceutical Research Division. Female C.B-17-Icr-scid-scidJcl mice were purchased from CLEA Japan Inc. (Tokyo, Japan). Patient-derived grafts were implanted subcutaneously into one flank of each 6–8-week-old mouse. For the UK-5099 study, daily treatment of mice with the vehicle (0.5% methylcellulose) or UK-5099 (10 mg/kg twice daily or 100 mg/kg/day daily, po) was initiated 3 weeks after implantation of tumor grafts from patients no. 24 and 34. For the temsirolimus (Pfizer Japan Inc., Tokyo, Japan) study, weekly treatment of mice with the vehicle (saline) or temsirolimus (10 mg/kg/day, qw, iv) was initiated 3–4 weeks after the implantation of tumor grafts from patients no. 11 and 12. Tumor sizes were measured using calipers twice a week. The tumor volume was expressed in mm3 using the formula 0.5 × a × b2, where a is the largest diameter and b is largest diameter perpendicular to a.
2.7. Statistical Analysis
Hierarchical clustering and heat map analysis were performed using Partek Genomics Suite 6.6 (Partek, Inc.). Differences between the control and treatment groups in cell growth assays were analyzed by Williams' test. Differences between the control and treatment groups in PDX assays were analyzed by Welch's t or Dunnett's test.
3. Results
3.1. A Metabolic Cluster in Kidney Cancer Characterized by High Pyruvate Levels
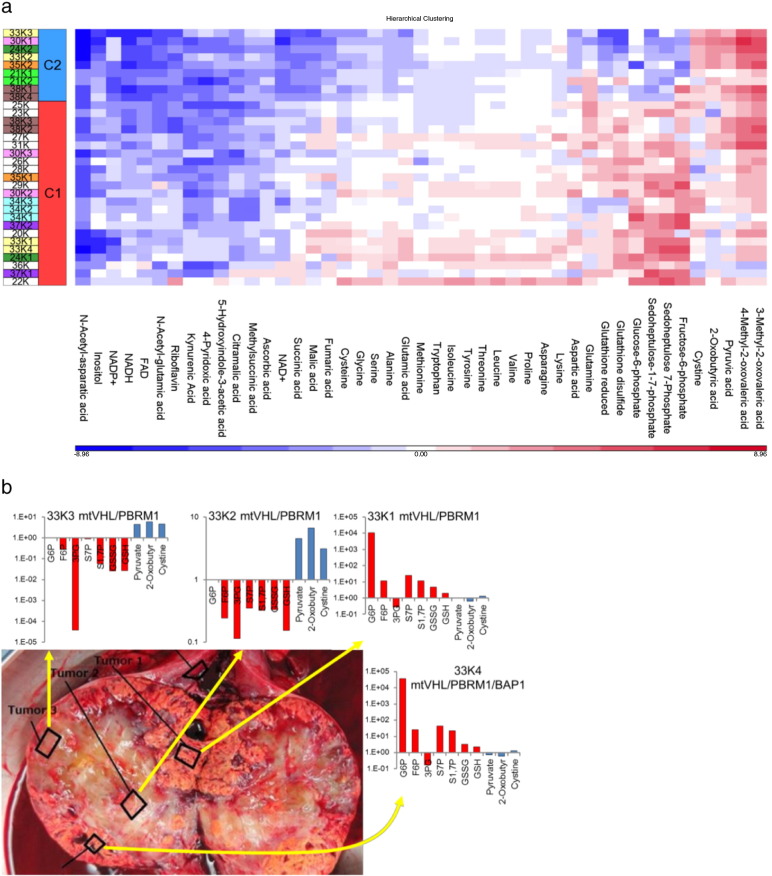
The metabolic patterns of 32 tumor tissues were separated into two major clusters based on the metabolomics data (Table S1, Fig. S1a). Metabolic cluster 1 (MC1) was characterized by elevated levels of metabolites of the glycolysis-pentose phosphate pathway (PPP), glutathione, and amino acids; metabolic cluster 2 (MC2) was characterized by elevated levels of metabolites such as pyruvate, cystine, and 2-oxobutyric acid, with reduced levels of glycolysis-PPP metabolites and of the reduced form of glutathione (Fig. 1a).
Fig. 1.
Intratumor heterogeneity in kidney cancer revealed by metabolomics analysis (a) Heat map of representative metabolites, with MC1- and MC2-specific features. The log2 (tumor/normal) values of metabolites were used for the construction of the heat map. Sample names are expressed as patient number and K (tumor) followed by sample site designated in Fig. 1b and S1d. (b) Regions harvested from a nephrectomy specimen from patient no. 33, together with the tissue levels of representative metabolites and mutation status. The metabolite levels are expressed as the tumor/normal ratio.
Sequencing of 23 RCC-associated genes did not reveal any clear correlation between the mutation status and the metabolic clusters (Table S2). Similarly, clustering tumors according to gene expression did not match the metabolic cluster classification (Fig. S1b); however, the gene expression levels of lactate dehydrogenase A (LDHA) and pyruvate dehydrogenase A1 (PDHA1), which convert pyruvate to lactate and acetyl-CoA, respectively, were decreased in MC2 tumors (Fig. S1c).
3.2. Intratumor Heterogeneity in Kidney Cancer Revealed by Metabolomics Analysis
Multiple spatially separated samples were obtained from tumors from eight patients. The metabolic clustering data in Fig. S1a include the multi-sampled tumor tissue. Metabolic patterns at different tumor sites from five patients (no. 24, 30, 33, 35, and 38) were classified into different metabolic clusters (Fig. S1a). Macroscopic observation of the sampled sites and determination of levels of representative metabolites clearly indicated that the tumor from patient no. 33 harbored both MC1 (high level of glycolysis metabolites in 33K1 and 33K4) and MC2 (high pyruvate levels in 33K2 and 33K3) domains (Fig. 1b). Metabolomics analysis of other multi-sampled tumors demonstrated similar intratumor metabolic heterogeneity (Fig. S1d). Intratumor genetic heterogeneity was confirmed, but genetic mutations were not associated with metabolic patterns (Fig. 1b, Fig. S1d).
In addition to hydrophilic metabolites, we also analyzed lipid profiles in tumor samples, by lipidomics; the profiles were relatively uniform and did not highlight metabolic heterogeneity (Fig. S1e). Several unfamiliar lipid species were detected specifically in tumor tissues; these included cholesteryl esters (CEs) and triglycerols (TGs) which harbor longer chain and more highly desaturated fatty acids (Fig. S1f).
3.3. Intratumor Heterogeneity Revealed by Stable Isotopic Tracer Studies
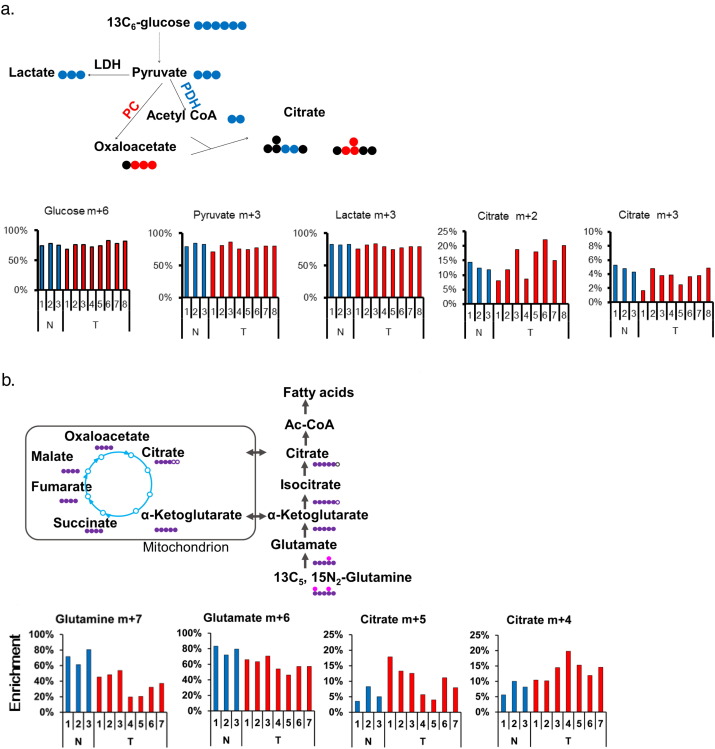
Metabolomics data provide a snapshot of metabolite levels. Here, a question was raised about what metabolic flow is like in tumors; metabolic flow better reflects the biological function and may explain the presence of high-level pyruvate sites in tumors. Hence, we addressed the intratumor metabolic heterogeneity by another approach, a stable isotopic-tracer study using tissue slices obtained from multiple sites within a tumor from patient no. 42. Incubation of the tumor and the adjacent normal tissue slices with 13C6-glucose resulted in an equal fractional enrichment of pyruvate (m + 3) in each tissue slice, but the subsequent metabolic flow was not consistent in the tumor tissue (Fig. 2a). The fractional enrichment of citrate (m + 2), the expected product if 13C6-glucose–derived pyruvate would have been converted to citrate by PDH, was higher in some tumor slices than in normal tissue (Fig. 2a). Citrate (m + 2) enrichment below or equal to that in the normal tissue was observed in other slices within the same tumor (Fig. 2a). This indicated different degrees of PDH activity at different sites within a single tumor. Regarding the fractional enrichment of citrate (m + 3), the expected product if 13C6-glucose–derived pyruvate would have been metabolized by pyruvate carboxylase (PC), it was lower than or the same as in the normal tissue in some parts of the tumor (Fig. 2a). This suggested different degrees of PC activity at different sites within a single tumor. Fractional enrichment of lactate (m + 3) was comparable in normal and tumor tissues (Fig. 2a). However, we did not measure the lactate (m + 3) in the conditioned medium. Therefore, LDH activity in these tissues was unclear.
Fig. 2.
Intratumor heterogeneity revealed by stable isotopic tracer studies. Tumor slices and the adjacent normal tissue from patient no. 42 were incubated for 24 h in the presence of tracers, as described in Materials and Methods. (a) Eight tumor tissue slices and three normal kidney tissue slices were incubated with 13C6-glucose. The schematic illustrates the production of 13C isotopologues of citrate by PC and PDH. Black circles denote 12C; blue and red circles denote 13C. (b) Seven tumor tissue slices and three normal kidney tissue were incubated with 13C5, 15N2-glutamine. The schematic illustrates the fates of 13C and 15N isotopologues in glutamine metabolism. White, purple, and pink circles denote 12C, 13C, and 15N, respectively. The y-axis represents the percent enrichment.
In addition, tissue slice tracer experiments with 13C5, 15N2-glutamine revealed intratumor heterogeneity, with a reciprocal relationship between the oxidative (citrate m + 4) and reductive (citrate m + 5) carboxylation pathways (Fig. 2b). Intratumor difference in metabolite flow was also confirmed in analogous labeled glucose or glutamine experiments in samples from patient no. 45 (Fig. S2a and 2b).
These results clearly demonstrated the existence of intratumor metabolic heterogeneity in human kidney cancer. They also suggested that reduced pyruvate metabolism may contribute to high pyruvate levels in some portions of the tumor. These observations raised two questions: (1) does pyruvate metabolic pathway constitute vulnerability in kidney tumors? (2) does metabolic heterogeneity correlate with cancer sensitivity to a currently available anti-cancer drug?
3.4. Pyruvate Metabolism as a Vulnerability of Kidney Cancer
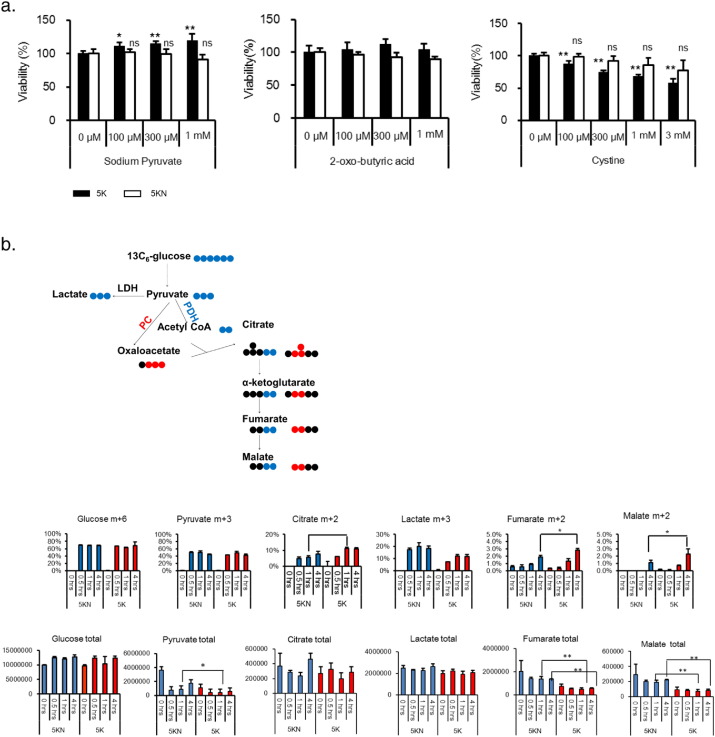
We proceeded to analyze the possible vulnerabilities of kidney cancer. Metabolomics data revealed that pyruvate, cystine, and 2-oxobutyrate levels were clearly elevated in some tumor portions (Fig. 1). In addition, tissue slice tracer studies indicated that pyruvate fueled the tricarboxylic acid (TCA) cycle in some tumor portions (Fig. 2). We therefore focused on the role of the three metabolites in cell growth, using 5K and 5KN cells derived from kidney cancer and normal kidney tissues, respectively, from patient no. 5. Pyruvate, not cystine or 2-oxobutyrate, stimulated the growth of 5K cells but not 5KN cells (Fig. 3a), suggesting that kidney cancer cells may be more dependent on pyruvate than normal kidney cells.
Fig. 3.
Pyruvate fuels the TCA cycle and stimulates growth in kidney cancer cells. (a) The effects of pyruvate, 2-oxo-butyric acid, and cystine on the growth of 5K (cancer) and 5KN (normal) cells. Cells were seeded in 96-well plates at a density of 2500 cells/well (n = 4). The metabolites were added the next day, and cell viability was assessed after 5 d. Values are normalized to the control group (100%). Data represent the mean ± SD. *p < 0.025, **p < 0.005 in Williams' test. (b) 5K and 5KN cells (n = 3) were treated with 13C6-glucose, and the amount of metabolites were measured, as described in Materials and Methods. Total metabolite levels were calculated by summing up each labeled metabolites. The schematic illustrates the fates of 13C isotopologues in glycolytic pathway and TCA cycle. Black circles denote 12C; blue and red circles denote 13C. Data are presented as the mean ± SD; p-values between 5K and 5KN were calculated by Bonferroni's corrected t-test at 1 h and 4 h; *p < 0.05, **p < 0.01.
Next, we performed tracer experiments with labeled glucose to investigate which pyruvate metabolic pathway was active in these cells. 5K and 5KN cells were treated with 13C6-glucose. The total amounts of glucose in 5K and 5KN cells were not different (Fig. 3b), whereas the total amounts of TCA metabolites such as pyruvate, fumarate, and malate were lower in 5K cells than in 5KN cells (Fig. 3b). The fractional enrichments of glucose (m + 6) and pyruvate (m + 3) did not differ between 5K and 5KN cells (Fig. 3b), whereas the fractional enrichments of citrate (m + 2), fumarate (m + 2), and malate (m + 2) in 5K cells were higher than those in 5KN cells (Fig. 3b). These data suggested that cataplerosis of TCA cycle metabolites might occur more efficiently in 5K cells than in 5KN cells. Citrate (m + 3) was not detected in both cell types (data not shown). Less lactate (M + 3) enrichment occurred in 5K cells than in 5KN cells (Fig. 3b). The lactate level in conditioned medium of 5K cells after adding non-labeled glucose was also less than that of 5KN cells (Fig. S2c), indicating that the LDH activity in 5K cells was lower than that in 5KN cells. Taken together, these data suggested that pyruvate fuels the TCA cycle through enhanced PDH-mediated flux in cancer cells.
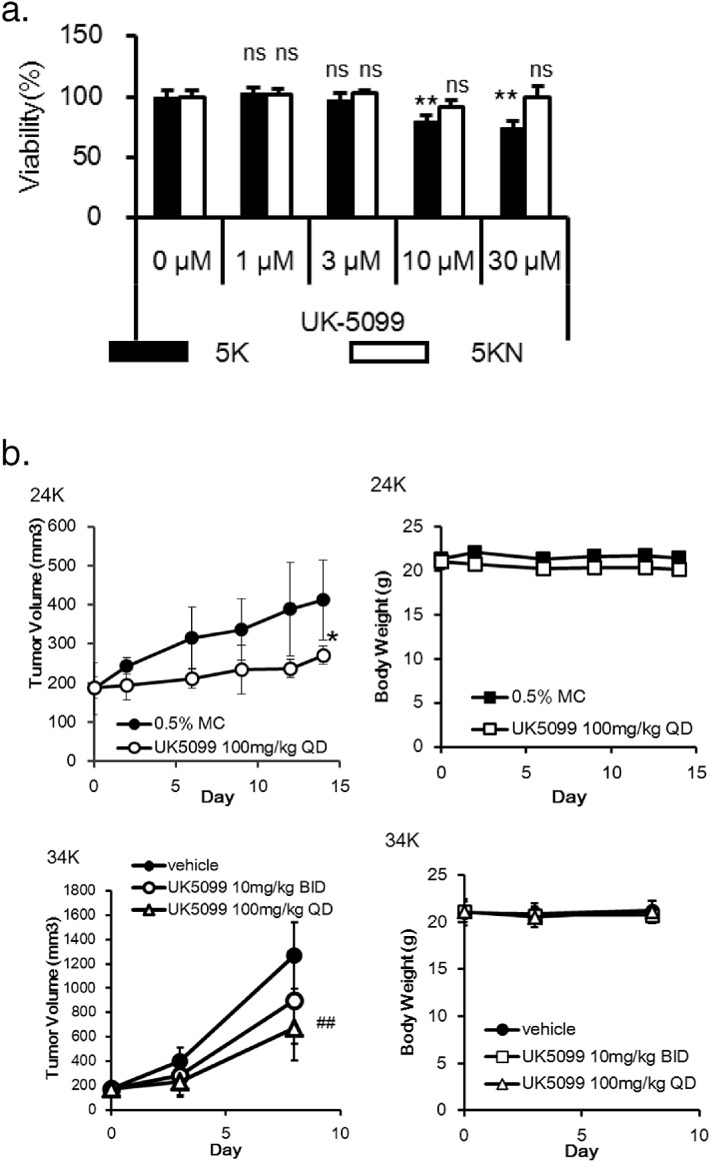
Because pyruvate is transported into mitochondria via a mitochondrial pyruvate carrier (MPC) (Bricker et al., 2012, Herzig et al., 2012) to fuel the TCA cycle, we next investigated whether inhibition of pyruvate transport might be an effective therapeutic option for RCC. UK-5099, an MPC inhibitor (Halestrap, 1975), suppressed the growth of 5K cells in the presence of pyruvate, but not the growth of 5KN cells (Fig. 4a). Moreover, UK-5099 slowed the growth of patient-derived xenograft (PDX) tumors in mice without severe loss of animal body weight (Fig. 4b). Taken together, these results indicate that kidney cancer cells may be more dependent on pyruvate than normal kidney cells, and suggest that pyruvate metabolism may be an exploitable vulnerability in human kidney cancer.
Fig. 4.
An MPC inhibitor suppresses cancer cell growth. (a) 5K and 5KN cells (n = 3) were seeded in 96-well plates at the density of 2500 cells/well. UK-5099 was added the next day, and cell viability was assessed after 5 d. Values are normalized to the control group (100%). Data represent the mean ± SD. **p < 0.005, by Williams' test. (b) Mice (n = 5) bearing tumors established from patient no. 24 were treated daily with 100 mg/kg of UK-5099. Mice (n = 6) bearing tumors established from patient no. 34 were treated twice daily with 10 mg/kg or daily with 100 mg/kg of UK-5099. Left, tumor volume; right, body weight. Data represent the mean ± SD. *p < 0.05 by Welch's t-test; ##p < 0.01 by Dunnett's test.
3.5. Metabolic Pattern and Drug Sensitivity
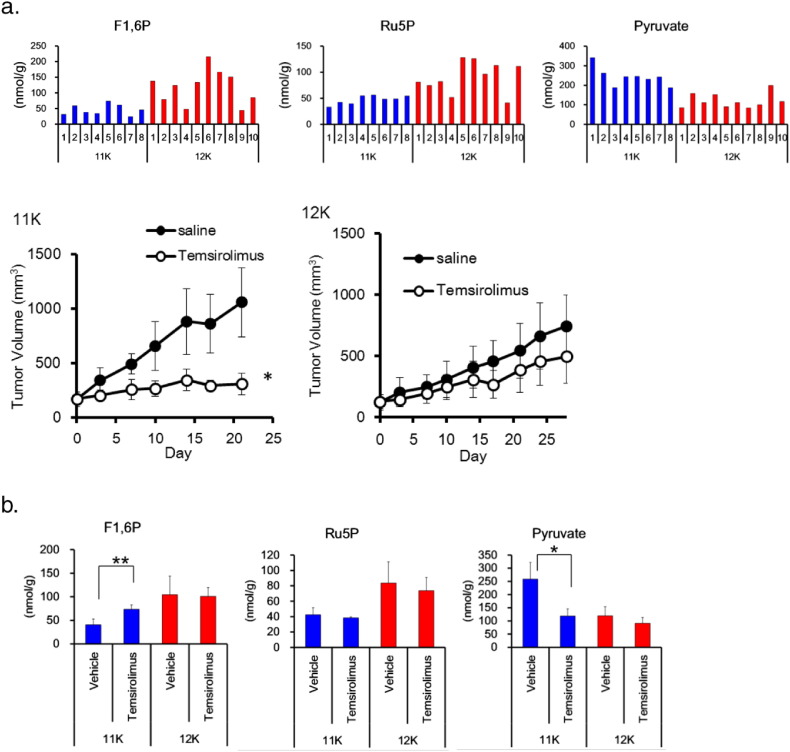
Finally, we addressed the question whether metabolic heterogeneity correlates with drug sensitivity. We tested the response of PDX tumors with different metabolic patterns to temsirolimus, a specific inhibitor of mTOR (Cai et al., 2007), used for RCC treatment in clinical practice. Pyruvate levels were higher and fructose-1,6-phosphate and ribulose-5-phosphate levels were lower in patient no. 11 PDX tumors in mice (tumor 11K) than in patient no. 12 PDX tumors (tumor 12K) (Fig. 5a). Tumor 11K responded better to temsirolimus than tumor 12K (Fig. 5a); further, temsirolimus treatment resulted in reduced pyruvate levels in tumor 11K, but not in tumor 12K (Fig. 5b). These findings suggest that tumors with high pyruvate content may respond well to temsirolimus, and that the tumor metabolic pattern and drug sensitivity may be associated.
Fig. 5.
Metabolic pattern and drug sensitivity. (a) Metabolite levels and anti-tumor activity of temsirolimus in patient-derived xenograft tumors from patients no. 11 (n = 4) and 12 (n = 5). Metabolite levels were measured in tumors without drug treatment (top). Mice bearing tumors were treated weekly with 10 mg/kg of temsirolimus (bottom). Data represent the mean ± SD. *p < 0.05 by Welch's t-test. F1,6P, Fructose 1,6-bisphosphate; Ru5P, ribulose-5-phosphate. (b) Metabolite levels in patient-derived xenograft tumors from patients no. 11 (n = 4) and 12 (n = 5) after once-weekly treatment with 10 mg/kg of temsirolimus. Tumor samples were taken 24 h after the last administration. Data represent the mean ± SD. *p < 0.05, **p < 0.01 by Welch's t-test.
4. Discussion
The existence of intratumor genetic heterogeneity in kidney cancer is becoming increasingly clear, but metabolic heterogeneity within a tumor had not been fully delineated until now. We demonstrated that different parts of a primary kidney tumor are characterized by different metabolic patterns and flows.
Currently, the cause of intratumor metabolic heterogeneity is unclear. Our metabolomics analysis indicated that RCCs exhibit the Warburg-like effect regardless of the status of the VHL gene; further, we did not find any clear correlations between gene mutations and metabolic patterns. Further, metabolic clustering could not be explained by gene expression. This is consistent with a recent report that metabolic alteration in clear cell RCC is not necessarily correlated with an altered expression of genes encoding metabolic enzymes (Hakimi et al., 2016). This may be caused by a non-canonical metabolic flux, mismatch between gene and protein expression levels, or the modulation of enzyme activity by cofactors. Indeed, our metabolomics analysis revealed decreased levels of cofactors (NAD+, FAD, and pyridoxic acid) in tumors. Nevertheless, some metabolic changes did correlate with gene expression of metabolic enzymes, e.g., pyruvate levels and LDHA and PDHA1 gene expression. Elevated pyruvate levels in MC2 tumors may be associated with reduced levels of LDHA and PDHA1, which metabolize pyruvate. Similarly, tissue slice glucose tracer experiments revealed different degrees of PDH activity in the tumor, supporting the notion that decreased PDH activity might lead to high pyruvate levels. Further studies are required to clarify the link between PDH activity and pyruvate levels. Another possible cause of metabolic heterogeneity that should be investigated in the future is histology and different tumor microenvironments.
We discovered that the metabolic pattern of primary kidney cancers might be divided into two major clusters. MC1 was consistent with previous reports (Catchpole et al., 2011, Li et al., 2014, Hakimi et al., 2016), whereas the other metabolomics cluster, MC2, had not been described until now. When considering the roles of metabolites in MC2, specifically pyruvate, cystine, and 2-oxobutyric acid, they may be required for energy metabolism or an anti-oxidative stress response within the cell. Equally possible, they may play a role after flowing to another portion inside the tumor, in the process known as metabolic symbiosis (Pisarsky et al., 2016, Sonveaux et al., 2008). MC2 domains may act as an energy reservoir of kidney tumors.
We demonstrated that pyruvate stimulates the growth of kidney cancer cells, but not normal kidney cells, both derived from patient samples. Pyruvate is a multifaceted metabolite associated with the production of energy, NADH, and a building block, via conversion into acetyl-coenzyme A, lactate, oxaloacetate, alanine, or carbohydrate. Our isotope studies demonstrated higher PDH activity in kidney cancer cells than in paired normal cells, and also in some tumor portions when compared to that in the paired normal tissues. Further study is required to elucidate how pyruvate exerts its growth-promoting effect in kidney cancer.
Studying how intratumor metabolic heterogeneity influences drug sensitivity is challenging. We used two different PDX models (MC1, tumor 12K; and MC2, tumor 11K) with different intratumor metabolic patterns to investigate the response of cancer cells to temsirolimus. Tumor 11K was more sensitive to temsirolimus than tumor 12K, suggesting that a tumor site with high pyruvate levels may respond better to temsirolimus. Since a PTEN mutation that activates mTOR was present in tumor 12K but not in tumor 11K (data not shown), metabolic patterns may exert a dominant effect over gene mutations in determining the sensitivity to temsirolimus. Interestingly, intratumor pyruvate levels were reduced by temsirolimus treatment only in tumor 11K. This may contribute, at least partially, to the antitumor activity of temsirolimus, although the mechanism for suppression of pyruvate by temsirolimus is currently unclear. Taken together, it is tempting to speculate that the metabolic pattern is associated with tumor sensitivity to temsirolimus; however, we were unable to unambiguously determine that because of the limited sample (model) size. Continued efforts are needed to clarify the relationship between the metabolic heterogeneity and drug sensitivity in cancer.
In contrast with hydrophilic metabolites, the lipid profile was relatively uniform among samples and did not highlight their heterogeneity. The levels CEs with long-chain highly desaturated fatty acids were elevated in tumors; this was consistent with the notion that clear cell RCCs are rich in CEs and TGs (Gebhard et al., 1987, Drabkin and Gemmill, 2012, Saito et al., 2016). Interestingly, our lipidomics platform identified rare lipid species that were specific to tumor tissues and that have not been reported before; these were typically CEs with very long chains or TGs with ether links. Although the function of these lipid molecules remains to be elucidated, they have the potential to be exploited as biomarkers for diagnosis or drug targeting.
Our metabolic analysis of kidney cancer samples revealed the existence of intratumor metabolic heterogeneity. The results led to the identification of pyruvate metabolism as a vulnerability of kidney cancer. Hensley et al. most recently showed metabolic heterogeneity by preoperative multimodality imaging combined with intraoperative 13C glucose in human lung cancer, and tied this heterogeneity to differences in PDH flux (Hensley et al., 2016). We must continue to identify exploitable vulnerabilities by further studying metabolic features of cancer in patients.
Funding Source
This work was wholly supported by Takeda Pharmaceutical Company Limited.
Conflicts of Interest
The authors declare no conflicts of interest.
Author Contributions
T.O. and T.H. designed and supervised the research. K.N. collected clinical samples. M.M., Y.S., A.A., and K.H. performed metabolomics studies. T.T., H.A., S.K., S.N., M.M., and T.H. performed the in vitro and in vivo experiments. K.H. and S.N. performed the bioinformatics analyses. S.N. and T.H. wrote the manuscript.
Acknowledgments
We thank Drs. Teresa W.-M. Fan, Andrew N. Lane, and Richard M. Higashi of Markey Cancer Center (University of Kentucky) for their technical advice regarding the tissue slice tracer experiments. We also thank Mayumi Deki, a lab technician in Kyorin University School of Medicine, for technical assistance, and the following employees of Takeda Pharmaceutical Company Limited: Shunsuke Ebara, Ayako Okamoto, Megumi Hirayama, Chisato Takahara, Ryuuichi Nishigaki, Kuniko Kikuchi, Ryo Dairiki, Yoshinori Ishikawa, Hirokazu Tozaki, and Koji Yamamoto for providing technical assistance.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, two figures, and two tables, and can be found with this article online.
Appendix A. Supplementary data
Table S1, S2, Fig S1, S2, Supplemental Experimental Procedures.
References
- Bricker D.K., Taylor E.B., Schell J.C., Orsak T., Boutron A., Chen Y.C., Cox J.E., Cardon C.M., Van Vranken J.G., Dephoure N. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science. 2012;337:96–100. doi: 10.1126/science.1218099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai P., Tsao R., Ruppen M.E. In vitro metabolic study of Temsirolimus: preparation, isolation, and identification of the metabolites. Drug Metab. Dispos. 2007;35:1554–1563. doi: 10.1124/dmd.107.014746. [DOI] [PubMed] [Google Scholar]
- Cairns R.A., Harris I.S., Mak T.W. Regulation of cancer cell metabolism. Nat. Rev. Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Catchpole G., Platzer A., Weikert C., Kempkensteffen C., Johannsen M., Krause H., Jung K., Miller K., Willmitzer L., Selbig J. Metabolic profiling reveals key metabolic features of renal cell carcinoma. J. Cell. Mol. Med. 2011;15:109–118. doi: 10.1111/j.1582-4934.2009.00939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabkin H.A., Gemmill R.M. Cholesterol and the development of clear-cell renal carcinoma. Curr. Opin. Pharmacol. 2012;12:742–750. doi: 10.1016/j.coph.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Durinck S., Stawiski E.W., Pavía-Jiménez A., Modrusan Z., Kapur P., Jaiswal B.S., Zhang N., Toffessi-Tcheuyap V., Nguyen T.T., Pahuja K.B. Spectrum of diverse genomic alterations define non-clear cell renal carcinoma subtypes. Nat. Genet. 2015;47:13–21. doi: 10.1038/ng.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhard R.L., Clayman R.V., Prigge W.F., Figenshau R., Staley N.A., Reesey C., Bear A. Abnormal cholesterol metabolism in renal clear cell carcinoma. J. Lipid Res. 1987;28:1177–1184. [PubMed] [Google Scholar]
- Gerlinger M., Rowan A.J., Horswell S., Larkin J., Endesfelder D., Gronroos E., Martinez P., Matthews N., Stewart A., Tarpey P. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi A.A., Reznik E., Lee C.H., Creighton C.J., Brannon A.R., Luna A., Aksoy B.A., Liu E.M., Shen R., Lee W. An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell. 2016;29:104–116. doi: 10.1016/j.ccell.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halestrap A.P. The mitochondrial pyruvate carrier. Kinetics and specificity for substrates and inhibitors. Biochem. J. 1975;148:85–96. doi: 10.1042/bj1480085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley C.T., Faubert B., Yuan Q., Lev-Cohain N., Jin E., Kim J., Jiang L., Ko B., Skelton R., Loudat L. Metabolic heterogeneity in human lung tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S., Raemy E., Montessuit S., Veuthey J.L., Zamboni N., Westermann B., Kunji E.R., Martinou J.C. Identification and functional expression of the mitochondrial pyruvate carrier. Science. 2012;337:93–96. doi: 10.1126/science.1218530. [DOI] [PubMed] [Google Scholar]
- Li B., Qiu B., Lee D.S., Walton Z.E., Ochocki J.D., Mathew L.K., Mancuso A., Gade T.P., Keith B., Nissim I. Fructose-1, 6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513:251–255. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linehan W.M., Srinivasan R., Schmidt L.S. The genetic basis of kidney cancer: a metabolic disease. Nat. Rev. Urol. 2010;7:277–285. doi: 10.1038/nrurol.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarsky L., Bill R., Fagiani E., Dimeloe S., Goosen R.W., Hagmann J., Hess C., Christofori G. Targeting metabolic symbiosis to overcome resistance to anti-angiogenic therapy. Cell Rep. 2016;15:1161–1174. doi: 10.1016/j.celrep.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Arai E., Maekawa K., Ishikawa M., Fujimoto H., Taguchi R., Matsumoto K., Kanai Y., Saito Y. Lipidomic signatures and associated transcriptomic profiles of clear cell renal cell carcinoma. Sci. Rep. 2016;6:28932. doi: 10.1038/srep28932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers K., Fox M.P., Bousamra M., 2nd, Slone S.P., Higashi R.M., Miller D.M., Wang Y., Yan J., Yuneva M.O., Deshpande R. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J. Clin. Invest. 2015;125:687–698. doi: 10.1172/JCI72873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonveaux P., Végran F., Schroeder T., Wergin M.C., Verrax J., Rabbani Z.N., De Saedeleer C.J., Kennedy K.M., Diepart C., Jordan B.F. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J. Clin. Invest. 2008;118:3930–3942. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente M.J., Henrique R., Costa V.L., Jerónimo C., Carvalho F., Bastos M.L., de Pinho P.G., Carvalho M. A rapid and simple procedure for the establishment of human normal and cancer renal primary cell cultures from surgical specimens. PLoS One. 2011;6:e19337. doi: 10.1371/journal.pone.0019337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, S2, Fig S1, S2, Supplemental Experimental Procedures.