ABSTRACT
Photobacterium damselae subsp. damselae is a pathogen of marine animals, including fish of importance in aquaculture. The virulence plasmid pPHDD1, characteristic of highly hemolytic isolates, encodes the hemolysins damselysin (Dly) and phobalysin (PhlyP). Strains lacking pPHDD1 constitute the vast majority of the isolates from fish outbreaks, but genetic studies to identify virulence factors in plasmidless strains are scarce. Here, we show that the chromosome I-encoded hemolysin PhlyC plays roles in virulence and cell toxicity in pPHDD1-negative isolates of this pathogen. By combining the analyses of whole genomes and of gene deletion mutants, we identified two hitherto uncharacterized chromosomal loci encoding a phospholipase (PlpV) and a collagenase (ColP). PlpV was ubiquitous in the subspecies and exerted hemolytic activity against fish erythrocytes, which was enhanced in the presence of lecithin. ColP was restricted to a fraction of the isolates and was responsible for the collagen-degrading activity in this subspecies. Consistent with the presence of signal peptides in PlpV and ColP sequences, mutants for the type II secretion system (T2SS) genes epsL and pilD exhibited impairments in phospholipase and collagenase activities. Sea bass virulence experiments and cell culture assays demonstrated major contributions of PhlyC and PlpV to virulence and toxicity.
IMPORTANCE This study constitutes genetic and genomic analyses of plasmidless strains of an emerging pathogen in marine aquaculture, Photobacterium damselae subsp. damselae. To date, studies on the genetic basis of virulence were restricted to the pPHDD1 plasmid-encoded toxins Dly and PhlyP. However, the vast majority of the recent isolates of this pathogen from fish farm outbreaks lack this plasmid. Here we demonstrate that the plasmidless strains produce two hitherto uncharacterized ubiquitous toxins encoded in chromosome I, namely, the hemolysin PhlyC and the phospholipase PlpV. We report the main roles of these two toxins in fish virulence and in cell toxicity. Our results constitute the basis for a better understanding of the virulence of a widespread marine pathogen.
KEYWORDS: Photobacterium damselae, phospholipase, lecithinase, hemolysin, damselysin, hlyA, collagenase, PhlyC, PhlyP, phobalysin
INTRODUCTION
Photobacterium damselae subsp. damselae is a marine bacterium of the family Vibrionaceae that causes infections in a variety of marine animals and also in humans (1). It is a primary pathogen of fish species of economic importance in aquaculture, causing wound infections and hemorrhagic septicemia (2, 3). Affected species include, among others, turbot (Scophthalmus maximus) (4), sea bream (Sparus aurata) (5), and sea bass (Dicentrarchus labrax) (6, 7). The recent reports on the isolation of this pathogen from newly cultured species led to its consideration as an emerging pathogen in marine aquaculture (8).
Highly hemolytic strains harbor a 153-kb virulence plasmid named pPHDD1, which contains the hemolysin genes dly and hlyApl (9). In addition, all the hemolytic strains have a third, chromosome I-borne hemolysin gene, hlyAch (10, 11). Damselysin (Dly), the product of dly, is a phospholipase D (12). Phobalysin P (PhlyP, “photobacterial lysin encoded on a plasmid”), the product of hlyApl, is a small β-pore-forming toxin (13). The amino acid sequence of phobalysin C (PhlyC), the product of hlyAch, shares 92% identity with that of PhlyP. Dly produces a synergistic effect in combination with either PhlyP or PhlyC against erythrocytes, and this synergy is responsible for maximum virulence for fish. Mutation of both dly and hlyApl genes in a pPHDD1-harboring strain causes a hemolytic phenotype on sheep blood agar that resembles that of naturally plasmidless strains, which is due to the activity of hlyAch on chromosome I (10).
To date, virulence studies with specific gene mutants of P. damselae subsp. damselae have been conducted exclusively with pPHDD1-harboring strains using turbot as an animal model of infection (9, 10). In addition, the role of hlyAch in virulence has been evaluated exclusively in strains that also produce Dly and PhlyP. However, the vast majority of the P. damselae subsp. damselae isolates from recent outbreaks in aquaculture farms do not produce Dly and PhlyP toxins and have been isolated mainly from sparid fish and from sea bass (6, 7, 11). Therefore, there is a need to investigate the genetic basis of virulence in strains that do not produce Dly and PhlyP. Previous studies detected phospholipase activity (but not protease production) in pPHDD1-harboring and plasmidless P. damselae subsp. damselae strains (4, 14, 15), and other studies reported that some strains exhibited collagenase activity (16, 17), but the genetic basis of these activities remains uncharacterized.
These previous observations suggest that the array of virulence genes of P. damselae subsp. damselae is not completely characterized. Recently, we described a type II secretion system (T2SS) in this pathogen and demonstrated its role in the secretion of Dly, PhlyP, and PhlyC (18), but to date, no additional proteins of the T2SS secretome have been shown. In this study, we identified the genetic basis of P. damselae subsp. damselae phospholipase and protease activities and their roles in virulence for fish using a sea bass animal model and in toxicity for fish and human cell lines. Here, we demonstrate that the T2SS secretome of P. damselae subsp. damselae also includes a novel phospholipase (PlpV) and a gelatinase/collagenase (ColP). Our results revealed main roles of the chromosomal hemolysin gene hlyAch and the phospholipase gene plpV in fish virulence and cell toxicity. In addition, we provide the complete genome sequences of two plasmidless strains of this widespread marine bacterial pathogen.
RESULTS AND DISCUSSION
Virulence of plasmidless strains and pPHDD1-harboring strains for sea bass.
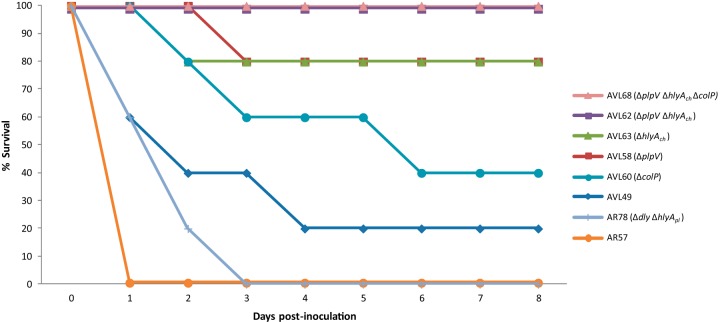
P. damselae subsp. damselae is being isolated as a causative agent of disease in novel species of cultivated fish, as well as in fish from areas where it had not been previously reported, such as Egypt (19), Tunisia (20), and the Black Sea (7). The majority of P. damselae subsp. damselae strains isolated from outbreaks in aquaculture farms lack pPHDD1 (6, 7, 11), a virulence plasmid present in highly pathogenic P. damselae subsp. damselae isolates (9). To assess the differences in virulence between a plasmidless strain and a pPHDD1-carrying strain, we performed virulence assays in sea bass, selecting the rifampin-resistant derivatives of the plasmidless strain LD-07 (AVL49) and of the pPHDD1-harboring strain RM-71 (AR57) because they are susceptible to genetic modification and facilitate counterselection after conjugation experiments (9, 10). At a dose of 1 × 105 CFU/fish, the plasmidless strain AVL49 did not cause fish death after 8 days while the pPHDD1 strain AR57 killed 100% of the animals within the first 24 h (data not shown). However, inoculation of 2.5 × 106 CFU/fish of AVL49 killed 80% of the fish after 8 days, while at the same dose, AR57 caused the fulminant death of 100% of the fish within 24 h postinoculation (Fig. 1). These observations suggest that the plasmidless strain AVL49 is less virulent than strain AR57. The hemolytic phospholipase damselysin (Dly) and the pore-forming toxin phobalysin (PhlyP) are the main known virulence factors encoded on pPHDD1 (9). To determine whether differences in virulence between AR57 and AVL49 were caused, at least in part, by Dly and PhlyP, we carried out virulence assays with a Δdly ΔhlyApl mutant of AR57 (AR78), inoculating at a dose of 2.5 × 106 CFU/fish. Although it showed a temporal delay in the mortality pattern with respect to the parental strain, this double mutant killed 100% of the fish, and it was more virulent than strain AVL49 at the same dose (Fig. 1). For each virulence experiment, the inoculated strain was isolated from the kidneys of all dead fish, and colonies tested positive in a colony PCR for the subspecies-specific ureC gene (data not shown). Next, we carried out ATP assays with extracellular products (ECP) from both strains and showed that strain AR78 decreased cellular ATP 10-fold in zebrafish AB.9 cells compared with strain AVL49 (Fig. 2). This result suggests that P. damselae subsp. damselae cell toxicity relies mainly on secreted virulence factors.
FIG 1.
Survival (%) of sea bass intraperitoneally challenged with different P. damselae subsp. damselae parental strains and deletion mutants (see Table 3) for a series of candidate virulence genes (n = 10 fish per strain). AR57 and AR78 are derivatives of RM-71, a highly virulent pPHDD1-harboring isolate. The rest of the strains are derivatives of the plasmidless isolate, LD-07.
FIG 2.
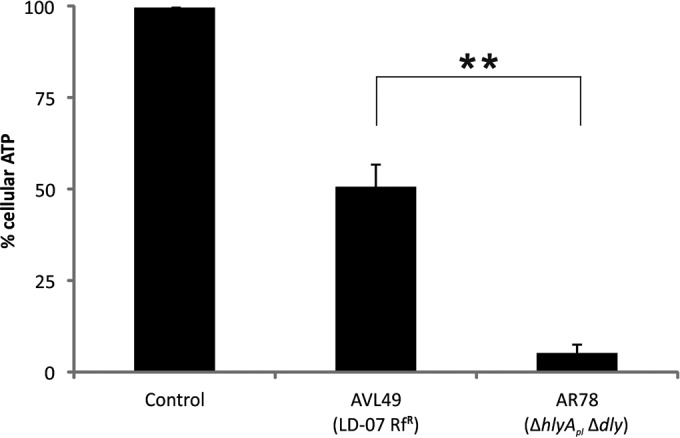
Loss of ATP in cells treated with P. damselae subsp. damselae ECP. Percentages of cellular ATP were measured after incubating AB.9 cells at 28°C with extracellular products (ECP) from indicated strains (400 μg/ml total protein) for 90 min. Control, medium only. Mean values ± SE; n = 3; **, P ≤ 0.01. Statistical significance was determined by an unpaired two-tailed Student's t test.
Analysis of four P. damselae subsp. damselae genomes identifies potential virulence factors in plasmidless strains.
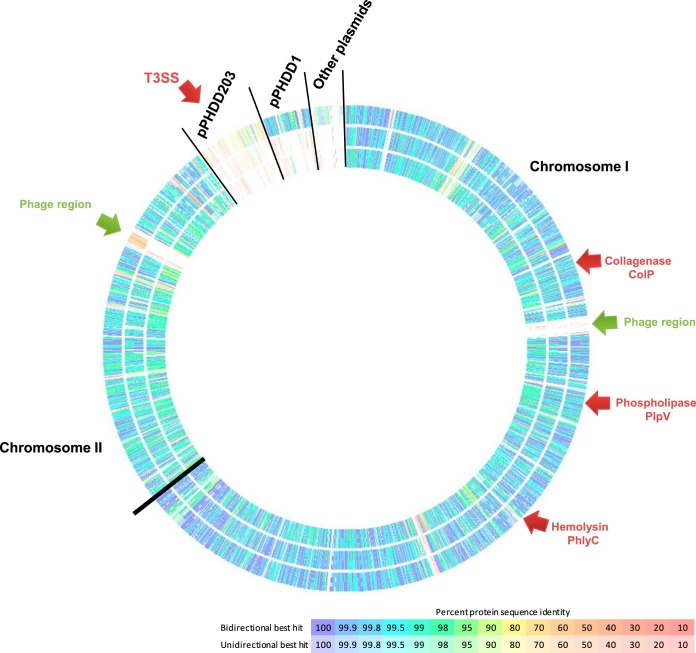
To identify potential secreted virulence factors and to gain insight into the genome differences between plasmidless and pPHDD1-harboring strains, we sequenced and assembled the genomes of two plasmidless strains (LD-07 and A-162) and one pPHDD1-harboring strain (RM-71) in a comparative analysis using the genome of the type strain CIP 102761 as the framework (GenBank accession no. ADBS00000000). The general features of the four genomes are summarized in Table 1. The core genome of the four strains included 2,583 protein-coding genes, and the pangenome was calculated to comprise 4,769 genes. Notably, each strain contained a number of strain-specific genes (Fig. 3; Table 1). Overall, the assembly of the three de novo sequenced genomes showed a high degree of synteny in chromosomes I and II (Fig. 4). We confirmed that 5 scaffolds of the type strain CIP 102761 contained typical plasmid-related genes (therefore constituting putative plasmids). In addition, two gene regions encoding potential prophages (one in chromosome I and one in chromosome II) were shown to be specific to the type strain CIP 102761 (Fig. 4). These observations explain the larger genome size of this strain (Table 1). Although some sequences of RM-71, LD-07, and A-162 strains showed some similarity to these CIP 102761-unique regions, this was mainly due to the presence of insertion sequence elements, phage integrases, partitioning proteins, and mating pair formation proteins sharing low identity among strains (10 to 20% identity) (data not shown). One of the CIP 102761-specific plasmids (dubbed here pPHDD203) corresponds to a scaffold of 203 kb (GenBank accession. no. ADBS01000003), and it encodes a putative type III secretion system (T3SS) which is absent from the other three genomes (Table 1). Although T3SS is a candidate virulence factor, we ruled it out from our study because it is absent from the pPHDD1 strain RM-71 and from the two plasmidless strains. One hundred twelve genes shared by RM-71 and CIP 102761 strains, which included most of the genes in the pPHDD1 plasmid, were absent from the two plasmidless strains (LD-07 and A-162) (Fig. 3). This suggests that the lower virulence of LD-07 compared with that of RM-71 may rely on the existence of additional virulence factors specific to pPHDD1. In this regard, the recent evidence that pPHDD1 contains a homologue of the Vibrio vulnificus virulence gene vep20, which encodes a transferrin receptor (21), suggests that the array of virulence genes in pPHDD1 might remain incompletely understood. In addition, it is feasible that the accumulation of point mutations within the sequence of the chromosomal hlyAch gene constitutes an additional explanation for the differences in virulence between LD-07 and AR78 (Δdly ΔhlyApl). In this regard, we have previously reported that point mutations in hlyAch affect the hemolytic activity (11).
TABLE 1.
General features of the Photobacterium damselae subsp. damselae genomes analyzed in this study
| Characteristic | Strain |
|||
|---|---|---|---|---|
| CIP 102761 | RM-71 | A-162 | LD-07 | |
| Genome feature | ||||
| Genome size (bp) | 5,046,328 | 4,485,055 | 4,272,996 | 4,321,079 |
| GC % | 40.7 | 40.6 | 40.8 | 40.6 |
| No. of contigs | 8 | 127 | 203 | 80 |
| Total no. of genes | 4,572 | 4,022 | 3,859 | 3,787 |
| Total no. of CDSa | 4,267 | 3,864 | 3,701 | 3,680 |
| No. of unique CDS | 241 | 262 | 34 | 207 |
| Virulence factor | ||||
| pPHDD1 plasmid | + | + | − | − |
| T3SS | + (VDA_000181 to VDA_000203) | − | − | − |
| hlyAch | + (VDA_002420) | + (A0J47_02585) | + (A9D46_00870) | + (A0J46_02060) |
| plpV | + (VDA_002242) | + (A0J47_05605) | + (A9D46_03080) | + (A0J46_07665) |
| colP | − | − | + (A9D46_13210) | + (A0J46_15580) |
CDS, coding DNA sequences.
FIG 3.
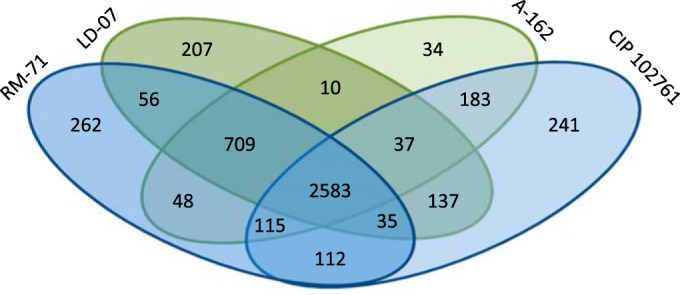
Venn diagram of the orthologous and specific proteins in each of the four P. damselae subsp. damselae genomes analyzed. Plasmidless strains (A-162 and LD-07) are depicted in green, and pPHDD1 strains (RM-71 and CIP 102761) are depicted in blue. Putative orthologous genes were defined as reciprocal best-hit proteins with a minimum of 90% identity.
FIG 4.
Comparison of P. damselae subsp. damselae genomes. From the outer ring to the inner ring: RM-71, A-162, and LD-07. The genome of the type strain CIP 102761 was used as the reference for the comparison and is not drawn as it would constitute a continuous circle. Black lines are drawn to separate chromosomes and plasmids. Note that the CIP 102761 genome includes a large putative plasmid (pPHDD203), several smaller putative plasmids, and two phage regions, all of which are absent in the other three strains.
We identified virulence-related genes that were common between the four analyzed strains and included the above-mentioned chromosomal hemolysin gene, hlyAch, and a hitherto uncharacterized phospholipase gene, which we termed plpV (Table 1; Fig. 4). Most pertinent to this study, the number of proteins shared by the two plasmidless strains at a minimum of 90% identity and that were absent in the two pPHDD1 strains accounted for only 10 open reading frames (ORFs). Among them, we identified a toxin-antitoxin system and a putative collagenase gene, which we termed colP (see Table S1 in the supplemental material).
Chromosome-encoded PhlyC of plasmidless strains contributes to hemolysis and virulence for sea bass.
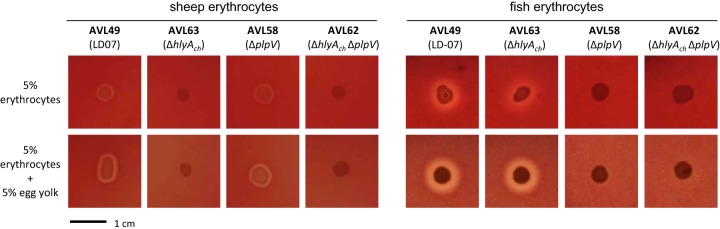
The comparative genome analysis described above suggests that some of the virulence factors of plasmidless strains might be shared with the pPHDD1 strains. Here, we confirmed that hlyAch is ubiquitous in the four genomes (Table 1), but its role in virulence for fish has not been evaluated in plasmidless isolates. We constructed an in-frame deletion mutant of hlyAch in strain AVL49 (LD-07 Rfr) and confirmed that this mutant (AVL63) lost the ability to cause hemolysis of sheep erythrocytes in a blood agar plate test (Fig. 5). We also found that the extracellular products (ECP) of AVL49, but not of AVL63, exhibited hemolytic activity, and this activity was completely lost after heating at 60°C for 10 min (data not shown), indicating that the hlyAch product, PhlyC, is thermosensitive.
FIG 5.
Hemolytic activity of P. damselae subsp. damselae plasmidless strain AVL49 and mutant derivatives on TSA-1 plates supplemented with 5% sheep or fish (trout) erythrocyte suspensions, either in the presence or absence of lecithin (supplemented with 5% egg yolk extract). hlyAch and plpV are the chromosome I-borne genes encoding PhlyC hemolysin and PlpV phospholipase, respectively. Bar, 1 cm.
The activity of certain bacterial hemolysins is known to be enhanced after supplementation with exogenous lecithin (22, 23). We found that the addition of lecithin to sheep erythrocytes caused a slight increase in the translucency of the hemolytic halo, whereas the presence of lecithin clearly enhanced the hemolysis of fish erythrocytes, not only in terms of halo translucency but also in terms of its diameter (Fig. 5). Surprisingly, mutation of hlyAch did not abolish hemolysis of fish erythrocytes (Fig. 5). This indicates a differential activity of PhlyC depending on the species source of erythrocytes and clearly suggests the existence of at least an additional hemolysin which is active against fish erythrocytes. Finally, when we inoculated sea bass with the hlyAch mutant at 2.5 × 106 CFU/fish, we found a decrease in virulence with respect to the parental strain, from 80% to 20% mortality (Fig. 1). Although previous studies reported that plasmidless strains show virulence for a variety of fish species (7, 17), our study constitutes evidence that hlyAch (the gene that encodes PhlyC) is a major contributor to virulence of plasmidless strains.
Chromosome-encoded phospholipase PlpV plays a role in virulence of plasmidless strains and exerts hemolytic activity against fish erythrocytes in a lecithin-dependent manner.
The ΔhlyAch mutant exhibited some degree of virulence for fish, albeit attenuated, as well as hemolytic activity against fish erythrocytes. This suggests that the plasmidless strain AVL49 produces additional virulence factors. Previous studies reported that the ECP of P. damselae subsp. damselae possess phospholipase activity, but the genetic basis remains unknown (14, 15). After an in silico analysis, we found that the translated product of the locus VDA_002242 in chromosome I of the type strain CIP 102761 showed homology to phospholipases described in other species of the family Vibrionaceae. VDA_002242 is located within the same genetic context in the four P. damselae subsp. damselae genomes analyzed here (Fig. 6A). Moreover, a PCR test demonstrated that VDA_002242 is ubiquitous in the subspecies (Table 2). We dubbed this gene plpV, which stands for phospholipase of Vibrionaceae. Then, we evaluated the phospholipase activities of our collection of 34 P. damselae subsp. damselae strains on tryptic soy agar (TSA) supplemented with 1% NaCl and 3% egg yolk. All the isolates yielded opaque precipitation halos in the egg yolk plates, but the pPHDD1-harboring strains (Dly-producing strains) produced 5-fold-wider halos (Table 2; Fig. 6B). Deletion of dly in AR57 caused a strong decrease in phospholipase activity, yielding a halo similar to that of the plasmidless strain AVL49 (Fig. 6B). These results provide clear evidence that the phospholipase activity of AR57 is not exclusively due to the activity of Dly. Next, we confirmed that deletion of plpV (VDA_002242) in the plasmidless strain AVL49 caused a complete suppression of the phospholipase activity in egg yolk plates (Fig. 6B). This activity was abolished in ECP from parental strain AVL49 after heating to 60°C for 10 min, suggesting that PlpV is thermolabile (data not shown). Single deletion of plpV in AR57 also affected the width of the halo in the phospholipase test (1.44 ± 0.2 cm in the mutant versus 2.06 ± 0.2 cm in the parental strain), and its deletion in combination with the deletion of dly completely abolished phospholipase activity (Fig. 6B). Altogether, these results indicate that phospholipase activity in the pPHDD1-harboring strains is due to both Dly and PlpV, with a major contribution of Dly. In plasmidless strains, PlpV is solely responsible for the phospholipase activity detectable in the egg yolk plate assay.
FIG 6.
(A) Genomic context of VDA_002242 (shown in black) encoding PlpV phospholipase. Gene numbers refer to the annotation of the type strain CIP 102761. (B) Phospholipase activity of P. damselae subsp. damselae parental strains and mutants on TSA-1 supplemented with 3% egg yolk. dly and plpV are the genes for the plasmid and chromosome-encoded phospholipases, respectively, and epsL and pilD encode components of the type II secretion system. Bar, 1 cm.
TABLE 2.
Results of the PCR screening for gene markers and of phenotypic tests for the 34 Photobacterium damselae subsp. damselae strains used in this study
| Strain | Source | pPHDD1a | hlyAch | Caseinase | Gelatinase |
Phospholipase |
||
|---|---|---|---|---|---|---|---|---|
| Testb | colP | Testc | plpV | |||||
| RM-71 | Turbot, Spain | + | + | − | − | − | ++ | + |
| RG-153 | Turbot, Spain | + | + | − | − | − | ++ | + |
| RG-91 | Turbot, Spain | + | + | − | − | − | ++ | + |
| RG-214 | Turbot, Spain | + | + | − | − | − | ++ | + |
| AZ 245.1 | Turbot, Spain | + | + | − | − | − | ++ | + |
| CDC-2227-81 | Human, United States | + | + | − | − | − | ++ | + |
| CIP 102761 | Damselfish, United States | + | + | − | − | − | ++ | + |
| ATCC 35083 | Brown shark, United States | − | + | − | − | − | + | + |
| 9FT1M-3 | Shark, United States | − | + | − | + | + | + | + |
| 9FT2B-2 | Shark, United States | − | + | − | + | + | + | + |
| RS80L1V1 | Red snapper, United States | − | + | − | − | − | + | + |
| RS78SPL1 | Red snapper, United States | − | + | − | − | − | + | + |
| ST-1 | Sea trout, United States | − | + | − | − | − | + | + |
| A-162 | Eel, Belgium | − | + | − | + | + | + | + |
| 158 | Eel, Belgium | − | + | − | + | + | + | + |
| PG-801 | Shrimp, Taiwan | − | + | − | − | − | + | + |
| J3G-801 | Shrimp, Taiwan | − | − | − | − | − | + | + |
| 309 | Mussel, Spain | − | + | − | − | − | + | + |
| 192 | Dolphin, United States | − | + | − | − | − | + | + |
| 238 | Dolphin, United States | − | + | − | − | − | + | + |
| AZ 247.1 | Turbot, Spain | − | + | − | − | − | + | + |
| ACR 208.1 | Turbot, Spain | − | + | − | − | − | + | + |
| USC-Viro-1 | Turbot, Spain | − | + | − | − | − | + | + |
| ACRP-72.1 | Turbot, Spain | − | − | − | − | − | + | + |
| LD-07 | Gilthead seabream, Spain | − | + | − | + | + | + | + |
| TW 294 L2 | European sea bass, Spain | − | + | − | − | + | + | |
| TW 250/03 | Gilthead seabream, Spain | − | + | − | + | + | + | + |
| TW 462/02.1 | Gilthead seabream, Spain | − | + | − | − | − | + | + |
| DCL 1.2 | Seabream, Spain | − | + | − | − | − | + | + |
| DCL 4.1 | Seabream, Spain | + | + | − | − | − | ++ | + |
| DCL 7.1 | Seabream, Spain | + | + | − | + | + | ++ | + |
| DCL 7.3 | Seabream, Spain | + | + | − | − | − | ++ | + |
| DCL 8.1 | Seabream, Spain | − | + | − | + | + | + | + |
| DCL 9.1 | Seabream, Spain | − | + | − | − | − | + | + |
pPHDD1 presence was assessed by PCR targeted to the damselysin (dly) gene.
Gelatinase agar plate test with 1% gelatin.
Phospholipase agar plate test with 3% egg yolk. +, medium halo (strains that only produce PlpV); ++, large halo (strains that produce Dly and PlpV).
Subsequently, we observed that deletion of plpV abolished hemolysis of fish erythrocytes, suggesting that PlpV is responsible for this hemolytic activity. Furthermore, the addition of lecithin enhanced the hemolysis of fish erythrocytes by plpV-positive strains, and this enhancement disappeared after the deletion of plpV (Fig. 5). However, the hemolysis of sheep erythrocytes was not affected by the plpV deletion (Fig. 5). As expected, the plpV hlyAch double mutant did not produce hemolysis of sheep or fish erythrocytes (Fig. 5). Thus, our results suggest that PhlyC is mainly responsible for the hemolysis of sheep erythrocytes and that PlpV is mainly responsible for hemolysis of fish erythrocytes. This erythrocyte-type selectivity might be explained by differences in the lipid compositions of the cell membranes (24).
PlpV showed high sequence similarity to phospholipases in other pathogenic Vibrio and Photobacterium species, including phospholipases of P. damselae subsp. piscicida (98% identity), Vibrio alginolyticus (61% identity), and V. cholerae (53% identity), among others (Fig. S1A). The plpV ORF consisted of 1,218 bp encoding a predicted 405-amino-acid protein with a lipase domain of the SGNH hydrolase family. In accordance with this, we found that PlpV contained four conserved amino acid positions in four sequence blocks characteristic of the SGNH family members (25) (Fig. S1B). Since phospholipase activity remains in ECP, it is likely that PlpV is secreted. SignalP 4.1 prediction showed a signal peptide sequence in PlpV (Fig. S1C), suggesting that this enzyme is secreted via the type II secretion system (T2SS). In support of this, we observed that the phospholipase phenotype was abolished in epsL and pilD mutants of strain AVL49 (Fig. 6B), involving two genes that encode components of the T2SS of P. damselae subsp. damselae (18), thereby confirming that the T2SS plays a role in PlpV secretion. A well-characterized homologue of PlpV is the Vibrio anguillarum Plp (26). Deletion of plp in V. anguillarum did not reduce virulence in a rainbow trout model (27). In our study, we show that deletion of plpV in the P. damselae subsp. damselae plasmidless strain AVL49 reduced its virulence for sea bass (20% mortality rate) to a level comparable to that of the hlyAch single mutant (Fig. 1). This demonstrates that plpV is a virulence determinant in the P. damselae subsp. damselae plasmidless strain AVL49. Importantly, a plpV hlyAch double mutant showed a drastic reduction of its virulence for sea bass, as it did not cause any fish death within 8 days postinoculation. This indicates that PlpV and PhlyC are the main chromosomally encoded virulence factors in P. damselae subsp. damselae. Besides, virulence is notably increased when both toxins are present (80% mortality) compared with mortalities reached in the presence of the individual toxins (20% mortality) and resembles the synergistic effects we previously reported for PhlyP and Dly (10).
Proteolytic activity of P. damselae subsp. damselae: identification of a collagenase gene.
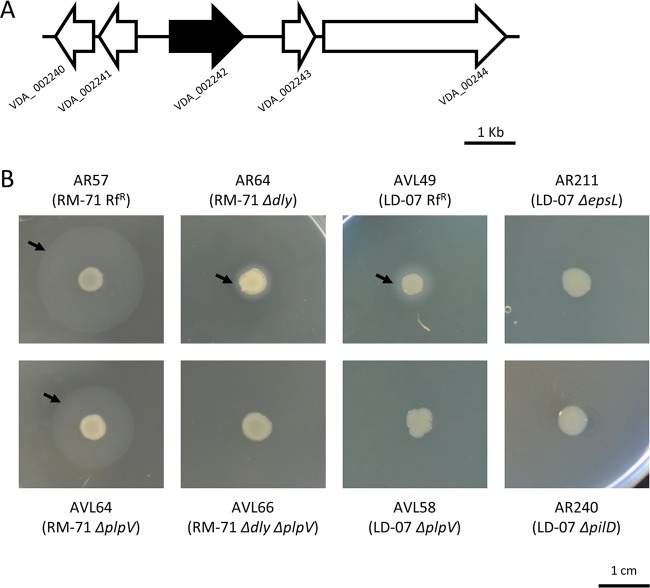
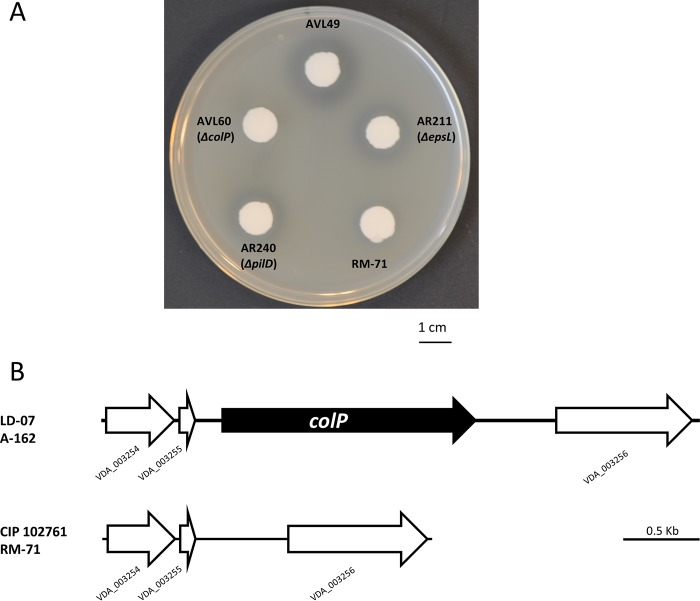
Proteolytic enzymes with a role in virulence have been previously described in marine bacterial pathogens. However, protease activity in P. damselae subsp. damselae remains unsubstantiated (4, 14, 15). We examined protease activities in our collection of P. damselae subsp. damselae strains and found that none of the 34 isolates tested were able to degrade casein in agar plate assays (Table 2), which strongly suggests that P. damselae subsp. damselae is a noncaseinolytic bacterium. Furthermore, plasmidless strain AVL49 was able to degrade gelatin in agar plate assays (Fig. 7A), as did 7 additional isolates (including strain A-162) (Table 2). In this regard, only a few studies detected gelatinolytic activity in a small fraction of P. damselae subsp. damselae isolates (16, 17), while other studies reported negative results for this trait (14). No correlation could be established between gelatin degradation and the isolation source or geographical origin. Indeed, gelatinase-positive isolates were represented among both the pPHDD1-harboring and the plasmidless groups of strains (Table 2). As shown in Table 1, the comparative genome analysis identified a putative collagenase gene only in plasmidless strains LD-07 and A-162, and we dubbed this gene colP (for collagenase of Photobacterium). colP is located between two genes annotated as VDA_003255 and VDA_003256 in the type strain (Fig. 7B). By PCR analysis, we confirmed that the presence of colP correlates with the ability to degrade gelatin among the subspecies isolates (Table 2) and that the same genetic context of colP is conserved in all P. damselae subsp. damselae strains (data not shown). The lack of a gelatin degradation halo in a colP mutant of AVL49 confirmed that the capability for gelatin degradation is conferred by colP (Fig. 7A). We also demonstrated that AVL49 ECP hydrolyzed the synthetic peptide 2-furanacryloyl-Leu-Gly-Pro-Ala (FALGPA), a substrate of collagenolytic proteases, whereas the colP mutant yielded zero units in the FALGPA hydrolysis test (Table S2). A heat treatment at 60°C for 10 min abolished the ability to hydrolyze FALGPA, indicating that the collagenase/gelatinase produced by AVL49 is thermolabile. colP consists of 2,406 bp and encodes a predicted protein of 801 amino acids with 47 to 44% identity to with microbial collagenases of different Vibrio spp. (Fig. S2A). According to the MEROPS database, ColP is a class II member of the M9 family. This family comprises bacterial metalloproteinases (predicted to be zinc dependent) from Vibrio and Clostridium with presumable collagenolytic activity. It possesses a catalytic domain (positions 25 to 552) and a prepeptidase C-terminal domain (positions 589 to 665) (Fig. S2B). An exception among the class II proteases, this enzyme does not have a C-terminal PKD-like domain and thus cannot hydrolyze casein (28), explaining the lack of casein degradation in our P. damselae subsp. damselae collection. We also predicted a signal peptide in the ColP sequence (amino acid positions 1 to 21) (Fig. S2B), suggesting that ColP is secreted via the T2SS. We observed that gelatin degradation was reduced, but not abolished, in a strain with a deletion of either epsL or pilD, two genes of the P. damselae subsp. damselae T2SS, which indicates that the T2SS contributes to ColP secretion but is not essential (Fig. 7A). Proteolytic enzymes can play a variety of roles in pathogenic bacteria. They can contribute directly to virulence through tissue damage, and they can provide a source of amino acids by degrading host proteins. In this regard, proteolytic enzymes of Vibrio anguillarum (29) and of Aeromonas salmonicida (30) are considered true virulence factors since they degrade fish tissues. However, there are examples where proteolytic enzymes do not play a role in virulence (31, 32). We found that a single deletion of colP in the AVL49 strain reduced virulence with respect to the parental strain, albeit this reduction was lower than that observed after hlyAch and plpV mutations (Fig. 1). This result shows that when injected intraperitoneally, colP has a low effect on virulence for sea bass. Nevertheless, it cannot be excluded that the route of infection we have used in this study may bypass host defense mechanisms as well as natural barriers. Therefore, a potential role of ColP cannot be ruled out in fish colonization, where gelatinase activity might provide a selective advantage during infection under natural conditions. As expected, a hlyAch plpV colP triple mutant was nonvirulent under the conditions and doses tested (Fig. 1). Altogether, these results indicate that these three genes contribute, although at different degrees, to shape the virulence of P. damselae subsp. damselae for sea bass.
FIG 7.
(A) Gelatinase activity on TSA-1 supplemented with 1% gelatin of P. damselae subsp. damselae parental strains and mutants. colP is the gene encoding the predicted gelatinase ColP, and epsL and pilD encode components of the type II secretion system. Bar, 1 cm. (B) Genomic context of colP in the colP-positive strains LD-07 and A-162 and its comparison with the same genome region in the colP-negative strains RM-71 and CIP 102761. Gene numbers refer to the annotation of the type strain CIP 102761.
hlyAch and plpV play roles in toxicity for fish cell cultures.
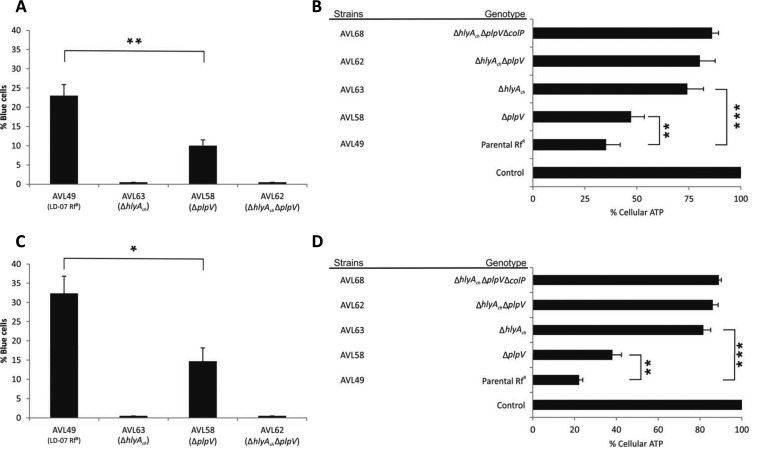
To date, the genetic basis of the cytotoxicity of plasmidless strains had not been evaluated. Here, we assayed the effect of the parental strain AVL49 and different mutant derivatives for their in vitro toxicity against the fish fibroblast AB.9 cell line. First, we incubated AB.9 fish cells with AVL49 and its mutant derivatives (multiplicity of infection [MOI], 1:50) for 90 min and evaluated membrane integrity by counting the cells permeable to the vital dye trypan blue (TB) (Fig. 8A). As a result, we observed entry of TB only when cells were challenged with P. damselae subsp. damselae strains producing PhlyC (Fig. 8A). On the contrary, consistent with the lack of blue cells, strains producing only PlpV did not affect membrane permeability (Fig. 8A). This implies that the destruction of membrane integrity depends on hlyAch. Since PhlyC shares 92% identity at the protein level with PhlyP (13), it is likely that PhlyC is also a small β-pore-forming toxin. Interestingly, the number of permeable cells increased significantly when both toxins, PlpV and PhlyC, were present. In line with our results in virulence assays, TB experiments demonstrated that PhlyC alone is not responsible for the full toxicity of the parental strain AVL49 and that PlpV is required for full toxicity and virulence. PlpV is believed to act as a phosphatidylcholine-specific phospholipase, and its interaction with PhlyC resembles the synergistic effects observed with Dly and PhlyP (10). Our results suggest that the combination PlpV/PhlyC in P. damselae subsp. damselae can constitute another example in which a phospholipase enhances the virulence and toxicity caused by a pore-forming toxin. At present, we do not have information on the molecular mechanisms that explain the role of PlpV in virulence and cell toxicity, but our results open the door to future studies aimed at unraveling the mode of action of PlpV.
FIG 8.
(A and C) Analysis of membrane integrity of AB.9 and HaCat cells coinfected with P. damselae subsp. damselae. Cells were infected with the indicated strains at an MOI of 1:50, were incubated for 90 min at 28ºC (AB.9; A) or 37ºC (HaCat; C), and were washed with PBS and subsequently stained for 10 min with trypan blue (TB) (final concentration, 0.2%). TB-positive and -negative cells were counted in each assayed well as groups of 100 cells (four quadrants), and a mean value was obtained per individual tested well. Final data are mean values ± SE; n = 3. *, P < 0.05; **, P ≤ 0.01. Statistical significance was determined by an unpaired, parametric two-tailed Student's t test. (B and D) Loss of ATP in cells treated with P. damselae subsp. damselae ECP. AB.9 (B) and HaCat (D) cells were incubated at 28ºC and 37ºC, respectively, for 90 min with ECP from indicated strains (400 μg/ml total protein). Control, medium only. Mean values ± SE; n = 3; **, P ≤ 0.01; ***, P ≤ 0.001. Statistical significance was determined by an unpaired two-tailed Student's t test.
Next, we determined cellular ATP production in AB.9 cells treated with ECP from strain AVL49 and its mutant derivatives. ECP (400 μg/ml total protein) from parental strain AVL49 decreased cellular ATP to 35% (Fig. 8B). This effect was reduced in cells treated with ECP from the ΔplpV mutant strain (AVL58), in which cellular ATP was decreased to 47%. As expected, ECP from AVL63 (ΔhlyAch) diminished cellular ATP to 74% (Fig. 8B), confirming the main role of PhlyC in the toxicity of P. damselae subsp. damselae plasmidless strains against fish. While the double mutant strain AVL62 (ΔhlyAch ΔplpV) was more toxic than the triple mutant strain AVL68 (ΔhlyAch ΔplpV ΔcolP) with regard to depletion of cellular ATP (Fig. 8B), the differences were not statistically significant. Similar results on toxicity were observed with various fish cell lines where bacteria induced cell rounding and detachment at 12 h postinfection (Fig. S3).
Most of the human infections caused by P. damselae subsp. damselae occur after wounded skin is exposed to contaminated seawater (1). To elucidate whether plasmidless strains are also toxic to human cells and have the potential to cause skin infections, we performed trypan blue and ATP assays in nonvirally transformed human keratinocytes (HaCaT) (Fig. 8C and D). Interestingly, bacteria and ECP caused levels of toxicity similar to those observed in AB.9 cells, and ECP from strain AVL49 proved to be more toxic to HaCaT cells than to AB.9 fish cells (78% versus 65% cellular ATP decrease, respectively) (Fig. 8B and D). That these toxins are able to produce similar toxic effects in fish and human cell lines raises an interesting question about how broad the spectrum of cells susceptible to them is.
Altogether, our results indicate that PhlyC is the only toxin with the ability to disrupt membrane integrity and that PlpV is not only toxic but also enhances PhlyC activity against fish and human cells.
Conclusions.
We provide strong evidence that hlyAch and plpV can be considered ubiquitous chromosomal virulence gene markers in P. damselae subsp. damselae. We also provide evidence that ColP contributes to an increased virulence during the infection process. Therefore, we conclude from our study that virulence for fish and the cell toxicity of plasmidless strains of P. damselae subsp. damselae strongly correlate with the presence of hlyAch and plpV. These results represent a significant contribution in deciphering the genetic basis of virulence in this bacterial fish pathogen and open the door for future studies aimed at providing novel approaches for the prevention and control of vibriosis caused by P. damselae subsp. damselae in marine fish aquaculture.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The Escherichia coli strains used for cloning experiments and the plasmids and P. damselae subsp. damselae strains used and constructed in this study for the genetic and virulence experiments are listed in Table 3. P. damselae subsp. damselae strains were routinely grown at 25°C on tryptic soy broth and agar supplemented with up to 1% NaCl (TSB-1 and TSA-1, respectively). The antibiotics used were rifampin (Rf) at 50 μg · ml−1 and kanamycin (Km) at 50 μg · ml−1.
TABLE 3.
Strains and plasmid used and constructed in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| P. damselae subsp. damselae | ||
| RM-71 | Isolated from turbot; pPHDD1 | 4 |
| AR57 | RM-71 derivative, spontaneous rifampin-resistant mutant, Rfr | 9 |
| AR78 | AR57 with in-frame deletion of dly and hlyApl, Rfr | 9 |
| AR64 | AR57 with in-frame deletion of dly, Rfr | 9 |
| AVL64 | AR57 with in-frame deletion of plpV, Rfr | This study |
| AVL66 | AR57 with in-frame deletion of dly and plpV, Rfr | This study |
| A-162 | Isolated from eel (Anguilla anguilla) in Belgium, 1991 | Laboratory collection |
| LD-07 | Isolated from gilthead seabream, plasmidless strain | 5 |
| AVL49 | LD-07, spontaneous rifampin-resistant mutant, Rfr | This study |
| AVL63 | AVL49 with in-frame deletion of hlyAch, Rfr | This study |
| AVL58 | AVL49 with in-frame deletion of plpV, Rfr | This study |
| AVL60 | AVL49 with in-frame deletion of colP, Rfr | This study |
| AVL62 | AVL49 with in-frame deletion of hlyAch and plpV, Rfr | This study |
| AVL68 | AVL49 with in-frame deletion of hlyAch, plpV, and colP, Rfr | This study |
| AR211 | LD-07 derivative with in-frame deletion of epsL, Rfr | 17 |
| AR240 | LD-07 derivative with in-frame deletion of pilD, Rfr | 17 |
| E. coli | ||
| DH5α | Cloning strain | Laboratory stock |
| S17-1 λpir | RP4-2(Km::Tn7, Tc::Mu-1) pro-82 λpir recA1 endA1 thiE1 hsdR17 creC510 | 47 |
| Plasmid | ||
| pNidKan | Suicide vector derived from pCVD442, Kmr | 48 |
Rfr, rifampin resistance; Kmr, kanamycin resistance.
General DNA techniques and PCR assays.
Genomic DNA was routinely extracted with the Easy-DNA kit (Invitrogen). The relevant PCR primers used in this study are listed in Table 4. PCRs were routinely performed with Taq DNA polymerase (Kapa) using a gradient thermocycler (Bio-Rad) with the following thermal cycling conditions: 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 57°C for 30 s, and an elongation step at 72°C of 1 min per kb. DNA sequences of PCR amplicons and plasmid inserts were determined by Sanger sequencing and a capillary DNA sequencer (ABI 3730xl; Applied Biosystems).
TABLE 4.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′−3′)a | Amplicon size (bp) |
|---|---|---|
| ureC screening | ||
| Ure-5′ | TCCGGAATAGGTAAAGCGGG | 448 |
| Ure-3′ | CTTGAATATCCATCTCATCTGC | |
| dly screening | ||
| Dly-5′ | CCTATGGGACATGAATGG | 549 |
| Dly-3′ | GCTCTAGGCTAAATGAATC | |
| hlyAch deletion | ||
| HlyAch mut 1 | GCGGATCCGAAGACGTTAATGAATGCGT | 2,078 |
| HlyAch mut 2 | GCGAATTCTCTGCTATCGGAGTATACAT | |
| hlyAch mut 3 | GCGAATTCTACAAAGCATCTCCTAATGA | 2,007 |
| hlyAch mut 4 | GCCTCGAGGCTTCGAACATTTTCTTACA | |
| hlyAch screening | ||
| HlyAch-5′ | AATGTTTCTTTCCGTTGGGC | 340 |
| HlyAch-3′ | CCGGAGTTCCACCAGTAAAT | |
| colP deletion | ||
| ColP mut 1 | GCTCTAGATCTTGCAATATCTCCACAGA | 1,964 |
| ColP mut 2 | GCGGATCCGTAATGGCCTTGACGAGATA | |
| ColP mut 3 | GCGGATCCAGATAATCAAGGTAATGTTG | 2,002 |
| ColP mut 4 | GCGAATTCGTACCTGAATGTGCTGAAGC | |
| colP screening | ||
| ColP_F | CCTGCTACAGTTCGAGCTCA | 1,617 |
| ColP_R | TCAACCACTCGAGATATTCC | |
| plpV deletion | ||
| PlpV mut 1 | GCTCTAGACACACTGCCGCCACTGCTCT | 1,993 |
| PlpV mut 2 | GCGAATTCTATAATAACTACCATCACTG | |
| PlpV mut 3 | GCGAATTCTTCCATAACATTAAAGATGC | 2,208 |
| PlpV mut 4 | GCCTCGAGTCATTGAATATGGCCAAAGT | |
| plpV screening | ||
| PlpV_F | TCTCATAATAGCAGTAATCT | 1,618 |
| PlpV_R | TTACTAAGCAGAATCCAGCC |
Underlined sequences denote recognition sites for restriction enzymes.
Genome sequencing of strains LD-07, A-162, and RM-71.
Genomic DNA of isolates LD-07, A-162, and RM-71 was purified using the GNOME DNA kit (Qbiogene) and sequenced using an Illumina MiSeq sequencer with 100× coverage. Reads were trimmed for quality, adapters, and ambiguous nucleotides and were assembled using SPAdes 3.6 (33). The resulting draft genome sequences were annotated and compared with the Rapid Annotations using Subsystems Technology (RAST) server (34). For the comparative analysis and the identification of common versus specific genes among strains, putative orthologous genes were defined as reciprocal best-hit proteins with a minimum of 90% identity.
Allelic-exchange deletion mutant construction.
Nonpolar deletions of hlyAch, plpV, and colP were constructed using PCR amplification of the amino- and carboxy-terminal fragments of each gene, which, when fused together, would result in an in-frame deletion of more than 90% of the coding sequence. Amplification was carried out using Hi-Fidelity Kapa Taq (Kapa). Allelic exchange was performed using the Kmr suicide vector pNidKan containing the sacB gene, which confers sucrose sensitivity, and R6K ori, which requires the pir gene product for replication. The plasmid constructs containing the deleted alleles were transferred from E. coli S17-1-λpir into a spontaneous rifampin-resistant derivative of strain LD-07 (AVL49). After conjugation for 48 h on TSA plates prepared with seawater, cells were scraped off the plate and resuspended in TSB-1. Next, 100-μl aliquots of serial decimal dilutions were spread on TSA-1 plates, selecting for rifampin and for kanamycin resistance for plasmid integration and subsequently selecting for sucrose resistance (15% [wt/vol]) for a second recombination event. This led to the P. damselae subsp. damselae mutant strains described in Table 3. The absence of the deleted alleles was confirmed in each case by PCR, and the genome region involved in the deletion was sequenced to verify that the deletion was nonpolar.
Cells and culture conditions.
HaCaT cells (nonvirally transformed human keratinocytes) (35) were cultured in Dulbecco modified Eagle medium (DMEM)/F-12 GlutaMAX-I medium with 10% fetal calf serum, 1% HEPES buffer, and 1% penicillin-streptomycin in a humidified incubator with 5% CO2 at 37°C. AB.9 zebrafish cells (36) were cultured in DMEM GlutaMAX-I medium with 15% fetal calf serum, 1% HEPES buffer, and 1% penicillin-streptomycin in a humidified incubator with 5% CO2 at 28°C. All media and additives were obtained from Life Technologies. Cytotoxicity assays on the carp (Cyprinus carpio) epithelioma papilosum cyprini (EPC) cell line were basically carried out as previously described (37). Cells were cultured in Eagle minimal essential medium (EMEM) with Earle's salts supplemented with 10% (vol/vol) fetal bovine serum (FBS; Lonza) and penicillin (100 IU · ml−1). For the cytotoxicity assay, 12-well plates of EPC cell cultures were prepared and incubated at 20°C to reach cell confluence. Each bacterial strain to be tested was grown on TSA-1 plates, and cells were resuspended in saline solution (NaCl [0.85%]) to reach an optical density at 600 nm (OD600) of 1.5. A 0.2-ml volume of each suspension was added to the EPC cells. Saline solution was used as a negative control. Each experiment was conducted in triplicate. The development of cytotoxic effects was observed 6, 24, and 48 h postinoculation using a phase-contrast inverted microscope (Nikon) at ×200 magnification.
Assays for hemolysis, phospholipase, and gelatinase activities.
For hemolysis assays on agar plates, a single colony of each strain grown on TSA-1 plates was picked with the tip of a rounded wooden pick and seeded on sheep blood agar plates (Oxoid). Pictures were taken after 24 h of culture at 25°C. Hemolysis of fish erythrocytes was evaluated on agar plates supplemented with 5% (vol/vol) trout blood. The phospholipase/lecithinase activities were determined using agar plates supplemented with an egg yolk emulsion as a lecithin source, where hydrolysis of lecithin by the phospholipase enzyme produces water-insoluble diglycerides that form an opaque precipitate. A 10-μl volume of TSB-1 overnight culture was spotted onto TSA-1 plates supplemented with 3% egg yolk extract (Oxoid), and results were evaluated after 24 h of culture at 25°C. The gelatinase activity assay was conducted by spotting 10 μl of a TSB-1 overnight culture onto TSA-1 plates supplemented with 1% gelatin (Oxoid), and results were developed after 48 h of incubation at 25°C by covering the agar plate surface with a 12.5% (wt/vol) HgCl2 solution. In addition, a quantitative measure of collagenase activity was achieved by determining the hydrolysis of the synthetic peptide FALGPA. The reaction mixture consisted of 1 mM FALGPA (Sigma) in 50 mM Tricine, 400 mM NaCl, and 10 mM CaCl2 (pH 7.5), and hydrolysis was measured at 345 nm as previously described (38). All of these tests were repeated three times to ensure that the results were reproducible.
Extraction of ECP and heat stability tests of hemolytic, phospholipase, and collagenase activities in strain LD-07.
To obtain the extracellular products (ECP), 100 μl of P. damselae subsp. damselae TSB-1 cultures adjusted to an OD600 of 1 were spread over cellophane-covered TSA-1 plates with a sterile cotton swab as previously described (39). After incubating for 48 h at 25°C, bacterial cells were washed off the cellophane with a minimum volume of saline solution (0.85% NaCl [wt/vol]). The cell suspensions were centrifuged at 13,000 rpm for 5 min, and the supernatants were filtered through 0.22-μm-pore membranes and stored at −20°C until needed. The total protein concentrations were determined according to the Bradford method (40), and protein concentrations were standardized after being measured. Aliquots of 5 μl of ECP samples were spotted onto sheep blood agar plates (for hemolytic activity) and onto TSA-1 supplemented with 3% (vol/vol) egg yolk (for phospholipase activity). In parallel, aliquots of the ECP samples were heated at 60 and 100°C for 10 min, and the hemolytic, phospholipase, and collagenase activities were determined as described above.
ATP measurements.
HaCaT and AB.9 cells were seeded into 96-well plates at a density of 2 × 104 cells/well. After 24 h, the medium was replaced with diluted ECP (400 μg/ml) or medium controls and the plates were incubated for 90 min. The total amount of ATP in cell lysates was measured luminometrically using a firefly luciferase-based assay as described elsewhere (41).
Staining with trypan blue.
HaCaT and AB.9 cells were seeded into 96-well plates at a density of 1 × 104 cells/well. After 24 h, the medium was replaced with medium containing the strains (MOI, 1:50) indicated in the figures. The infections were performed at 37°C (HaCaT) or 28°C (AB.9) for 90 min. Subsequently, the medium was replaced with fresh medium and trypan blue (proportion, 1/2) and the plates were incubated for 10 min. Next, cells were washed with phosphate-buffered saline (PBS), and stained and unstained cells were counted visually. Each assayed well was divided into quadrants, and mean values were obtained from four individual counts of 100 cells. Experiments were performed in triplicate.
Sea bass experimental infection.
Sea bass (Dicentrarchus labrax) were obtained from IGAFA (Illa de Arousa, Galicia, Spain). A random sample of individuals was analyzed to ensure the absence of P. damselae subsp. damselae by plate culture and further by PCR for the subspecies-specific ureC gene (42). Groups of 10 fish (6 ± 1.2 g) per strain tested and per dose were acclimated in 100-liter flowthrough aquaria at 24°C for 1 week before the challenges were performed. The virulence tests were conducted by intraperitoneal injection of bacterial suspensions. Fish were challenged with 0.1 ml of bacterial suspensions in 0.85% NaCl solution at two different doses of 1 × 105 CFU/fish and 2.5 × 106 CFU/fish depending on the experiment. The bacterial inocula were first adjusted by OD600 values and further verified by plate counts. A control group was inoculated with the same volume of sterile 0.85% NaCl solution. Fish mortality was recorded daily for 8 days postchallenge. Reisolation on TSA-1 and thiosulfate-citrate-bile salts-sucrose (TCBS) agar plates and identification of the bacteria from the kidneys of dead fish were performed. Colonies were confirmed by the subsp. damselae-specific ureC gene PCR test as previously described (42). All protocols for animal experimentation used in this study have been reviewed and approved by the animal ethics committee of the Universidade de Santiago de Compostela.
Signal peptide prediction and molecular phylogenetics.
The presence of signal peptide sequences was predicted by SignalP 4.1 (43). The MEROPS database (http://merops.sanger.ac.uk) (44) was consulted for the in silico analysis of ColP gelatinase. The phylogenetic relationships among PlpV and ColP amino acid sequences in different species of the Vibrionaceae family were inferred using the neighbor-joining method (45). In this analysis, the numbers next to the branches denoted the percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates). The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances (number of base substitutions per site) were computed using the maximum composite likelihood method. Evolutionary analyses were conducted in MEGA6 (46).
Accession number(s).
DNA sequences have been deposited in the GenBank database under accession numbers LYBT00000000 (RM-71), LYBU00000000 (LD-07), and LZFN00000000 (A-162).
Supplementary Material
ACKNOWLEDGMENTS
We thank the Instituto Galego de Formación en Acuicultura (IGAFA) (Illa de Arousa, Galicia, Spain) for the valuable support in providing the sea bass for the virulence experiments. We thank Alicia Ana Silva Martínez for her valuable help with cell culture preparation and maintenance.
This work was supported by grants AGL2013-48353-R and AGL2016-79738-R from the Ministry of Economy and Competitiveness (MINECO) of Spain (both cofunded by the FEDER Programme of the European Union) to C.R.O. A.V. was supported by a predoctoral FPI fellowship (number BES-2013-064281) assigned to grant AGL2012-39274-C02-01 from the MINECO of Spain. A.J.R was supported by a postdoctoral grant awarded by Xunta de Galicia, Spain (Galician Plan of Research, Innovation and Growth 2011-2015 [Plan I2C]).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00401-17.
REFERENCES
- 1.Rivas AJ, Lemos ML, Osorio CR. 2013. Photobacterium damselae subsp. damselae, a bacterium pathogenic for marine animals and humans. Front Microbiol 4:283. doi: 10.3389/fmicb.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Love M, Teebkenfisher D, Hose JE, Farmer JJ, Hickman FW, Fanning GR. 1981. Vibrio damsela, a marine bacterium, causes skin ulcers on the damselfish Chromis punctipinnis. Science 214:1139–1140. doi: 10.1126/science.214.4525.1139. [DOI] [PubMed] [Google Scholar]
- 3.Fouz B, Novoa B, Toranzo AE, Figueras A. 1995. Histopathological lesions caused by Vibrio damsela in cultured turbot, Scophthalmus maximus (L): inoculations with live cells and extracellular products. J Fish Dis 18:357–364. doi: 10.1111/j.1365-2761.1995.tb00312.x. [DOI] [Google Scholar]
- 4.Fouz B, Larsen JL, Nielsen B, Barja JL, Toranzo AE. 1992. Characterization of Vibrio damsela strains isolated from turbot Scophthalmus maximus in Spain. Dis Aquat Organ 12:155–166. doi: 10.3354/dao012155. [DOI] [Google Scholar]
- 5.Vera P, Navas JI, Fouz B. 1991. First isolation of Vibrio damsela from seabream (Sparus aurata). Bull Eur Assoc Fish Pathol 11:112–113. [Google Scholar]
- 6.Labella A, Manchado M, Alonso MC, Castro D, Romalde JL, Borrego JJ. 2010. Molecular intraspecific characterization of Photobacterium damselae ssp. damselae strains affecting cultured marine fish. J Appl Microbiol 108:2122–2132. doi: 10.1111/j.1365-2672.2009.04614.x. [DOI] [PubMed] [Google Scholar]
- 7.Terceti MS, Ogut H, Osorio CR. 2016. Photobacterium damselae subsp. damselae, an emerging fish pathogen in the Black Sea: evidences of a multiclonal origin. Appl Environ Microbiol 82:3736–3745. doi: 10.1128/AEM.00781-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Labella A, Berbel C, Manchado M, Castro D, Borrego JJ. 2011. Photobacterium damselae subsp. damselae, an emerging pathogen affecting new cultured marine fish species in southern Spain, p 135–152. In Aral F. (ed), Recent advances in fish farms. InTech, Rijeka, Croatia. [Google Scholar]
- 9.Rivas AJ, Balado M, Lemos ML, Osorio CR. 2011. The Photobacterium damselae subsp. damselae hemolysins damselysin and HlyA are encoded within a new virulence plasmid. Infect Immun 79:4617–4627. doi: 10.1128/IAI.05436-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rivas AJ, Balado M, Lemos ML, Osorio CR. 2013. Synergistic and additive effects of chromosomal and plasmid-encoded hemolysins contribute to hemolysis and virulence in Photobacterium damselae subsp. damselae. Infect Immun 81:3287–3299. doi: 10.1128/IAI.00155-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivas AJ, Labella A, Borrego JJ, Lemos ML, Osorio CR. 2014. Evidences for horizontal gene transfer, gene duplication and genetic variation as driving forces of the diversity of haemolytic phenotypes in Photobacterium damselae subsp. damselae. FEMS Microbiol Lett 355:152–162. doi: 10.1111/1574-6968.12464. [DOI] [PubMed] [Google Scholar]
- 12.Kreger AS, Bernheimer AW, Etkin LA, Daniel LW. 1987. Phospholipase D activity of Vibrio damsela cytolysin and its interaction with sheep erythrocytes. Infect Immun 55:3209–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rivas AJ, Von Hoven G, Neukirch C, Meyenburg M, Qin Q, Füser S, Boller K, Lemos ML, Osorio CR, Husmann M. 2015. Phobalysin, a small β-pore-forming toxin of Photobacterium damselae subsp. damselae. Infect Immun 83:4335–4348. doi: 10.1128/IAI.00277-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Labella A, Sanchez-Montes N, Berbel C, Aparicio M, Castro D, Manchado M, Borrego JJ. 2010. Toxicity of Photobacterium damselae subsp. damselae strains isolated from new cultured marine fish. Dis Aquat Organ 92:31–40. [DOI] [PubMed] [Google Scholar]
- 15.Fouz B, Barja JL, Amaro C, Rivas C, Toranzo AE. 1993. Toxicity of the extracellular products of Vibrio damsela isolated from diseased fish. Curr Microbiol 27:341–347. doi: 10.1007/BF01568958. [DOI] [Google Scholar]
- 16.Pedersen K, Dalsgaard I, Larsen JL. 1997. Vibrio damsela associated with diseased fish in Denmark. Appl Environ Microbiol 63:3711–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pedersen K, Skall HF, Lassen-Nielsen AM, Bjerrum L, Olesen NJ. 2009. Photobacterium damselae subsp. damselae, an emerging pathogen in Danish rainbow trout, Oncorhynchus mykiss (Walbaum), mariculture. J Fish Dis 32:465–472. doi: 10.1111/j.1365-2761.2009.01041.x. [DOI] [PubMed] [Google Scholar]
- 18.Rivas AJ, Vences A, Husmann M, Lemos ML, Osorio CR. 2015. Photobacterium damselae subsp. damselae major virulence factors Dly, plasmid-encoded HlyA, and chromosome-encoded HlyA are secreted via the type II secretion system. Infect Immun 83:1246–1256. doi: 10.1128/IAI.02608-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdel-Aziz M, Eissa AE, Hanna M, Okada MA. 2013. Identifying some pathogenic Vibrio/Photobacterium species during mass mortalities of cultured Gilthead seabream (Sparus aurata) and European seabass (Dicentrarchus labrax) from some Egyptian coastal provinces. Int J Vet Sci Med 1:87–95. doi: 10.1016/j.ijvsm.2013.10.004. [DOI] [Google Scholar]
- 20.Khouadja S, Lamari F, Bakhrouf A, Gaddour K. 2014. Virulence properties, biofilm formation and random amplified polymorphic DNA analysis of Photobacterium damselae subsp. damselae isolates from cultured sea bream (Sparus aurata) and sea bass (Dicentrarchus labrax). Microb Pathog 69–70:13–19. [DOI] [PubMed] [Google Scholar]
- 21.Pajuelo D, Lee CT, Roig FJ, Hor LI, Amaro C. 2015. Novel host-specific iron acquisition system in the zoonotic pathogen Vibrio vulnificus. Environ Microbiol 17:2076–2089. doi: 10.1111/1462-2920.12782. [DOI] [PubMed] [Google Scholar]
- 22.Shinoda S, Matsuoaka H, Tsuchie T, Miyoshi S, Yamamoto S, Taniguchi H, Mizuguchi Y. 1991. Purification and characterization of a lecithin-dependent haemolysin from Escherichia coli transformed by a Vibrio parahaemolyticus gene. J Gen Microbiol 137:2705–2711. doi: 10.1099/00221287-137-12-2705. [DOI] [PubMed] [Google Scholar]
- 23.Naka H, Hirono I, Aoki T. 2007. Cloning and characterization of Photobacterium damselae ssp. piscicida phospholipase: an enzyme that shows haemolytic activity. J Fish Dis 30:681–690. doi: 10.1111/j.1365-2761.2007.00861.x. [DOI] [PubMed] [Google Scholar]
- 24.Ivanov IT. 2007. Allometric dependence of the life span of mammal erythrocytes on thermal stability and sphingomyelin content of plasma membranes. Comp Biochem Physiol A Mol Integr Physiol 147:876–884. doi: 10.1016/j.cbpa.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 25.Molgaard A, Kauppinen S, Larsen S. 2000. Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases. Structure 8:373–383. doi: 10.1016/S0969-2126(00)00118-0. [DOI] [PubMed] [Google Scholar]
- 26.Rock JL, Nelson DR. 2006. Identification and characterization of a hemolysin gene cluster in Vibrio anguillarum. Infect Immun 74:2777–2786. doi: 10.1128/IAI.74.5.2777-2786.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L, Mou X, Nelson DR. 2013. Characterization of Plp, a phosphatidylcholine-specific phospholipase and hemolysin of Vibrio anguillarum. BMC Microbiol 13:271. doi: 10.1186/1471-2180-13-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang YZ, Ran LY, Li CY, Chen XL. 2015. Diversity, structures, and collagen-degrading mechanisms of bacterial collagenolytic proteases. Appl Environ Microbiol 81:6098–6107. doi: 10.1128/AEM.00883-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denkin SM, Nelson DR. 2004. Regulation of Vibrio anguillarum empA metalloprotease expression and its role in virulence. Appl Environ Microbiol 70:4193–4204. doi: 10.1128/AEM.70.7.4193-4204.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunnlaugsdóttir B, Gudmundsdóttir BK. 1997. Pathogenicity of atypical Aeromonas salmonicida in Atlantic salmon compared with protease production. J Appl Microbiol 83:542–551. doi: 10.1046/j.1365-2672.1997.00247.x. [DOI] [PubMed] [Google Scholar]
- 31.Vipond R, Bricknell IR, Durant E, Bowden TJ, Ellis AE, Smith M, MacIntyre S. 1998. Defined deletion mutants demonstrate that the major secreted toxins are not essential for the virulence of Aeromonas salmonicida. Infect Immun 66:1990–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pérez-Pascual D, Gómez E, Álvarez B, Méndez J, Reimundo P, Navais R, Duchaud E, Guijarro JA. 2011. Comparative analysis and mutation effects of fpp2-fpp1 tandem genes encoding proteolytic extracellular enzymes of Flavobacterium psychrophilum. Microbiology 157:1196–1204. doi: 10.1099/mic.0.046938-0. [DOI] [PubMed] [Google Scholar]
- 33.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Stepanauskas R, Clingenpeel SR, Woyke T, McLean JS, Lasken R, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling genomes and mini-metagenomes from highly chimeric reads, vol 7821, p 158–170. In Deng M, Jiang R, Sun F, Zhang X (ed), Lecture notes in computer science. Springer–Verlag, Berlin, Germany. [Google Scholar]
- 34.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. 1988. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol 106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Badakov R, Jazwinska A. 2006. Efficient transfection of primary zebrafish fibroblasts by nucleofection. Cytotechnology 51:105–110. doi: 10.1007/s10616-006-9018-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang XH, Oon HL, Ho GWP, Wong WSF, Lim TM, Leung KY. 1998. Internalization and cytotoxicity are important virulence mechanisms in vibrio-fish epithelial cell interactions. Microbiology 144:2987–3002. doi: 10.1099/00221287-144-11-2987. [DOI] [PubMed] [Google Scholar]
- 38.Van Wart HE, Steinbrink DR. 1981. A continuous spectrophotometric assay for Clostridium histolyticum collagenase. Anal Biochem 113:356–365. doi: 10.1016/0003-2697(81)90089-0. [DOI] [PubMed] [Google Scholar]
- 39.Liu PV. 1957. Survey of hemolysin production among species of pseudomonads. J Bacteriol 74:718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 41.Haugwitz U, Bobkiewicz W, Han SR, Beckmann E, Veerachato G, Shaid S, Biehl S, Dersch K, Bhakdi S, Husmann M. 2006. Pore-forming Staphylococcus aureus alpha-toxin triggers epidermal growth factor receptor-dependent proliferation. Cell Microbiol 8:1591–1600. doi: 10.1111/j.1462-5822.2006.00733.x. [DOI] [PubMed] [Google Scholar]
- 42.Osorio CR, Toranzo AE, Romalde JL, Barja JL. 2000. Multiplex PCR assay for ureC and 16S rRNA genes clearly discriminates between both subspecies of Photobacterium damselae. Dis Aquat Organ 40:177–183. doi: 10.3354/dao040177. [DOI] [PubMed] [Google Scholar]
- 43.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 44.Rawlings ND, Barrett AJ, Finn R. 2016. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 44:D343–D350. doi: 10.1093/nar/gkv1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saitou N, Nei M. 1987. The neighbor-joining method:a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 46.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herrero M, De Lorenzo V, Timmis KN. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in Gram-negative bacteria. J Bacteriol 172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mouriño S, Osorio CR, Lemos ML. 2004. Characterization of heme uptake cluster genes in the fish pathogen Vibrio anguillarum. J Bacteriol 186:6159–6167. doi: 10.1128/JB.186.18.6159-6167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.