ABSTRACT
Stable associations between plants and microbes are critical to promoting host health and productivity. The objective of this work was to test the hypothesis that restructuring of the core microbiota may be associated with the progression of huanglongbing (HLB), the devastating citrus disease caused by Liberibacter asiaticus, Liberibacter americanus, and Liberibacter africanus. The microbial communities of leaves (n = 94) and roots (n = 79) from citrus trees that varied by HLB symptom severity, cultivar, location, and season/time were characterized with Illumina sequencing of 16S rRNA genes. The taxonomically rich communities contained abundant core members (i.e., detected in at least 95% of the respective leaf or root samples), some overrepresented site-specific members, and a diverse community of low-abundance variable taxa. The composition and diversity of the leaf and root microbiota were strongly associated with HLB symptom severity and location; there was also an association with host cultivar. The relative abundance of Liberibacter spp. among leaf microbiota positively correlated with HLB symptom severity and negatively correlated with alpha diversity, suggesting that community diversity decreases as symptoms progress. Network analysis of the microbial community time series identified a mutually exclusive relationship between Liberibacter spp. and members of the Burkholderiaceae, Micromonosporaceae, and Xanthomonadaceae. This work confirmed several previously described plant disease-associated bacteria, as well as identified new potential implications for biological control. Our findings advance the understanding of (i) plant microbiota selection across multiple variables and (ii) changes in (core) community structure that may be a precondition to disease establishment and/or may be associated with symptom progression.
IMPORTANCE This study provides a comprehensive overview of the core microbial community within the microbiomes of plant hosts that vary in extent of disease symptom progression. With 16S Illumina sequencing analyses, we not only confirmed previously described bacterial associations with plant health (e.g., potentially beneficial bacteria) but also identified new associations and potential interactions between certain bacteria and an economically important phytopathogen. The importance of core taxa within broader plant-associated microbial communities is discussed.
KEYWORDS: core microbiome, plant-associated microbiota, Liberibacter, pathogen-microbe interactions
INTRODUCTION
The coevolution of microorganisms with plant hosts has given rise to finely tuned symbiotic and parasitic relationships that are central to plant growth and health. In addition to tightly coevolved relationships between individual partners, recent studies have demonstrated that plant-associated microbial communities play critical roles in plant development. Under the right circumstances, the microbiome can bolster plant productivity by providing protection against pathogens, among other mechanisms (1–4). Several studies have indicated that managing microbial taxonomic/functional diversity at the field scale can have positive effects on crop production (5–8).
Commonly occurring organisms across similar microbiomes comprise a core microbial community that is hypothesized to play key roles in ecosystem functioning within that type of microbial habitat (6, 9). While numerous deep sequencing studies have revealed thousands of bacterial operational taxonomic units (OTUs) to comprise plant microbiomes, there is typically a small number of taxa that dominate the broader community (10–18). Some of the highly abundant taxa within these studies are noticeably conserved across microbiomes of similar plant species, even under variable experimental conditions. This suggests that a core microbial community forms stable associations with particular hosts across temporal and geographic scales. Nonetheless, the structure of plant microbiota is known to be associated with a suite of biotic factors (e.g., plant developmental stage and phytopathogens) and abiotic factors (e.g., soil type, climate, and season) (11, 13, 15, 16, 19). Given the limited number of studies that have discerned the core members of plant-associated microbial communities (11, 13, 15), much remains unknown about the structure of the core community among all microbiota and its importance with regard to plant health.
A growing number of studies have focused on understanding how the plant microbiome changes during disease development (15, 17, 20–26). While results from these studies suggest that plant-associated microbial communities undergo perturbation during environmental change or disease progression, one may alternatively argue that the disease itself is a consequence of complex changes to the communities. For example, infections of tomato plants with Rhizoctonia solanacearum led to changes in relative proportions of several bacterial classes that were consistently dominant among all classes detected in the rhizosphere (i.e., changes in abundances of core classes) (21). Similar trends were observed with another Rhizoctonia species in beets; however, the presence of a complex rhizosphere community that had included potentially beneficial bacterial groups (i.e., Pseudomonadaceae, Burkholderiaceae, Xanthomonadales, and Lactobacillaceae) was linked to suppressing the development of beet root rot caused by the pathogen (27). In addition, several plant diseases are caused, or at least enhanced, by synergistic interactions among multiple bacterial pathogens (e.g., tomato pith necrosis, mulberry wilt, and broccoli head rot) (28). Collectively, these studies suggest that plant disease establishment may be associated with changes in the microbiota that involve suppressing key, possibly core, members of the native community. The interactions between native bacteria that may mediate the potential for microbiome-related pathogen suppression are still not well documented. These characterizations are needed for better understanding of the role of (core) native bacteria in disease progression and, ultimately, for formulating novel strategies to manipulate plant-associated microbial communities to mitigate disease.
While plant biocontrol bacteria have shown great promise in the lab (29–31), their efficacy under field conditions has been only moderately successful (32, 33). However, harnessing the beneficial potential of native microbiota is still one of the few logistically and economically feasible solutions for controlling certain phytopathogens. This is likely the case for the devastating citrus greening disease, or huanglongbing (HLB), which is caused by the phloem-limited alphaproteobacteria Liberibacter asiaticus, Liberibacter americanus, and Liberibacter africanus (34). Infection by any one of these organisms causes host callose depositions and inhibition of nutrient uptake and transport, which lead to a series of symptoms that culminate in tree death (34). The severity of HLB is underscored by the billions of dollars in economic damage and thousands of jobs lost throughout Florida, where L. asiaticus is widespread, that occurred during the first few years of its documented emergence (35). Interestingly, clone library sequencing and PhyloChip surveys have shown citrus-associated microbiota restructure during HLB symptom development, and it has been suggested that the population dynamics of native bacteria can affect the titer of L. asiaticus (17, 36–38). In the present study, the microbial communities of citrus trees were defined across a number of variables, including HLB symptom severity, geographic location, citrus cultivar, season, and time. We used citrus and HLB as a plant host/disease model to investigate (i) the relationship between leaf- and root-associated microbial communities and host plant, (ii) the interactions between native microbiota and a phytopathogen, and (iii) the core microbiota, their prevalence within the broader microbial communities, and correlations between community structure and plant disease, among other factors.
RESULTS
Citrus-associated microbial community structure and core microbiota.
Approximately 13.77% ± 1.18% and 91.22% ± 1.22% (mean ± standard error [SE]) of all quality-filtered reads from leaf and root samples, respectively, were classified as 16S rRNA genes of bacteria/archaea. We used peptide nucleic acid (PNA) clamps (39) during library preparation in order to limit the amplification of contaminant sequences (i.e., 16S rRNA of mitochondria and chloroplasts). In a preliminary study without the use of PNA, libraries prepared from leaf samples (n = 9) and root samples (n = 3) yielded 0.90% ± 0.15% and 16.78% ± 6.58% (mean ± SE) of reads corresponding to bacteria/archaea (data not shown). Thus, the PNA clamps allowed for substantial enrichment of desired sequences. Moreover, after removing the residual contaminant sequences from the quality-filtered libraries, there were 94 microbial communities of leaves and 79 microbial communities of roots with more than 1,000 16S rRNA gene sequences assigned to 16S rRNA of bacteria/archaea (Table 1). From these 173 microbial communities, the 7,767,069 assigned reads, with an average of 13,725 ± 730 sequences per leaf sample and 81,986 ± 5,168 sequences per root sample (mean ± SE), were included in further analyses.
TABLE 1.
Characterization of the microbial communities of leaf and root samples from 73 citrus trees across Floridaa
| Location | Citrus tree species and cultivar | Date(s) surveyed (mo/day/yr) | No. of: |
HLB symptom appearance | ||
|---|---|---|---|---|---|---|
| Trees | Leaf communities | Root communities | ||||
| Gainesville, FL | Citrus sinensis L. Osbeck cv. Valencia | 4/1/15, 6/1/15, 9/22/15, 3/29/16 | 8b | 8/date | 8/datec | IV |
| Fort Pierce, FL | Citrus sinensis L. Osbeck cv. Valencia | 10/13/15 | 15 | 14 | 9 | III |
| Immokalee, FL | Citrus sinensis L. Osbeck cv. Valencia | 11/3/15 | 5 | 5 | 0 | III |
| Citrus paradisi Macfadyen cv. Ray Ruby | 11/3/15 | 5 | 5 | 5 | IV | |
| Citrus sinensis (L.) Osbeck cv. Navel | 11/3/15 | 5 | 5 | 5 | II | |
| Quincy, FL | Citrus sinensis (L.) Osbeck cv. Navel | 10/20/15 | 10 | 9 | 10 | I |
| Citrus unshiu Marcovitch cv. Owari | 10/20/15 | 5 | 5 | 5 | I | |
| Citrus × tangelo cv. Honeybell | 10/20/15 | 3 | 3 | 3 | I | |
| Vero Beach, FL | Citrus sinensis L. Osbeck cv. Valencia | 11/2/15 | 5 | 4 | 2 | II |
| Citrus paradisi Macfadyen cv. Ray Ruby | 11/2/15 | 12 | 12 | 9 | I | |
For each sample set, the location, citrus cultivar, sampling date, number of trees sampled, numbers of leaf and root microbial communities included in data analyses (i.e., the subset of total samples that passed quality control and contained >1,000 16S sequences with assigned taxonomy), and huanglongbing (HLB) symptom appearance are listed. For the latter, the categories I, II, III, and IV correspond to asymptomatic, symptomatic—mild, symptomatic—moderate, or symptomatic—severe, respectively. (See Fig. S2 for detailed description.)
The same 8 trees were sampled on each date.
On 6/1/15, only 7 replicates were included in the analysis.
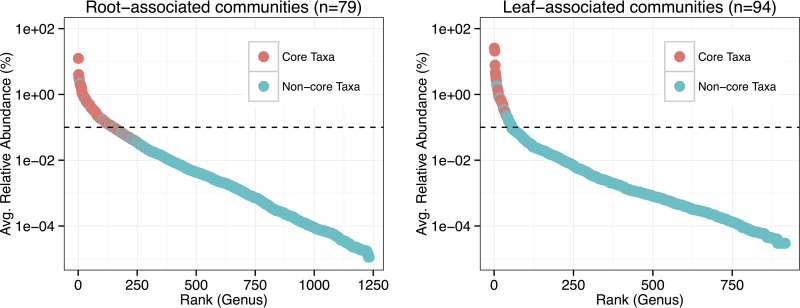
Several hundred genera were detected in the leaf- and root-associated microbial communities. Approximately 6% of the leaf microbiota and 12% of the root microbiota accounted for 90% of microbial abundance within the respective communities (Fig. 1). There was a larger consortium of core bacteria associated with roots (183 genera, 137 families, 83 orders, 45 classes, and 8 phyla) than leaves (28 genera, 28 families, 21 orders, 13 classes, and 8 phyla), and most of the relatively dominant members within the microbial communities were core bacteria (Fig. 1). The ratios of core members to all members within the microbiota consistently decreased as the taxonomic rank became finer (see Fig. S3 in the supplemental material). Accordingly, certain non-core taxa at lower taxonomic ranks comprised broader core groups. For example, there were more core members at the family level in leaf microbiota (n = 28) than there were total families of core genera in leaf microbiota (n = 21; see Table S1 in the supplemental material) (i.e., any non-core genera within the other 7 core families must have been present across samples). With regard to the non-core citrus-associated microbiota, most taxa appeared to be sample specific rather than well replicated (Fig. S3). In summary, the microbial communities were comprised of the key abundant core members and some overrepresented site-specific members, with a diverse community of low-abundance variable taxa.
FIG 1.
Rank abundance curves for the genera assigned to 16S sequences detected in the root- and leaf-associated microbial communities. Color indicates whether the genus was a member of the core microbiota (i.e., present in >95% of respective samples [58]). The dotted line represents the threshold for the rare biosphere (i.e., 0.1% relative abundance [40]).
Spatiotemporal variability of HLB.
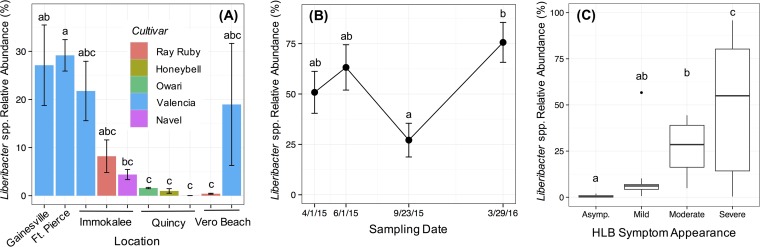
In leaf-associated communities (n = 94), the relative abundance of Liberibacter spp. was 25.60% ± 3.14% (mean ± SE), which was, on average, the most abundant out of all genera detected. Alternatively, in root-associated communities (n = 79), the relative abundance of Liberibacter spp. was 0.038% ± 0.012% (mean ± SE). Analysis of variance (ANOVA) indicated that the relative abundance of the pathogen among leaf microbiota significantly differed across locations (P < 0.001) and among citrus cultivars (P < 0.001). Figure 2A illustrates pairwise comparisons between specific treatment groups. Also, as expected, there was a positive correlation between the severity of HLB symptoms displayed by citrus trees and the relative abundance of Liberibacter spp. among leaf microbiota (Fig. 2C).
FIG 2.
Relative abundance of Liberibacter spp. (i.e., the HLB pathogen) in citrus leaf samples by location and cultivar (A), by sampling date for Valencia trees at the Gainesville site (B), and by severity of HLB symptoms (C). See Fig. S2 for a description of symptom severity categories. Letters a, b, and c indicate significant difference based on Tukey's post hoc test following ANOVA (α set at 0.05).
To test the hypothesis that the prevalence of the HLB pathogen within the microbiomes of HLB-symptomatic trees varies by season and increases over time, we compared the relative abundances of Liberibacter spp. within leaves from Valencia trees sampled in Gainesville in spring 2015 (1 April 2015), summer 2015 (1 June 2015), fall 2015 (23 September 15), and spring 2016 (29 March 2016). Over the course of the year, the abundance of the pathogen fluctuated, with a drop during the fall season. There was still a net increase between spring 2015 and spring 2016 (Fig. 2B). Interestingly, HLB symptoms also progressed between spring 2015 and spring 2016; that is, the chlorosis-like appearance of leaves, as well as branch dieback, became more widespread throughout the canopy (see Fig. S4 in the supplemental material). Thus, while seasonal effects contributed to changes in the abundance of the HLB pathogen, there were increases over time that concurred with disease symptom progression.
Biotic and abiotic factors associated with community structure.
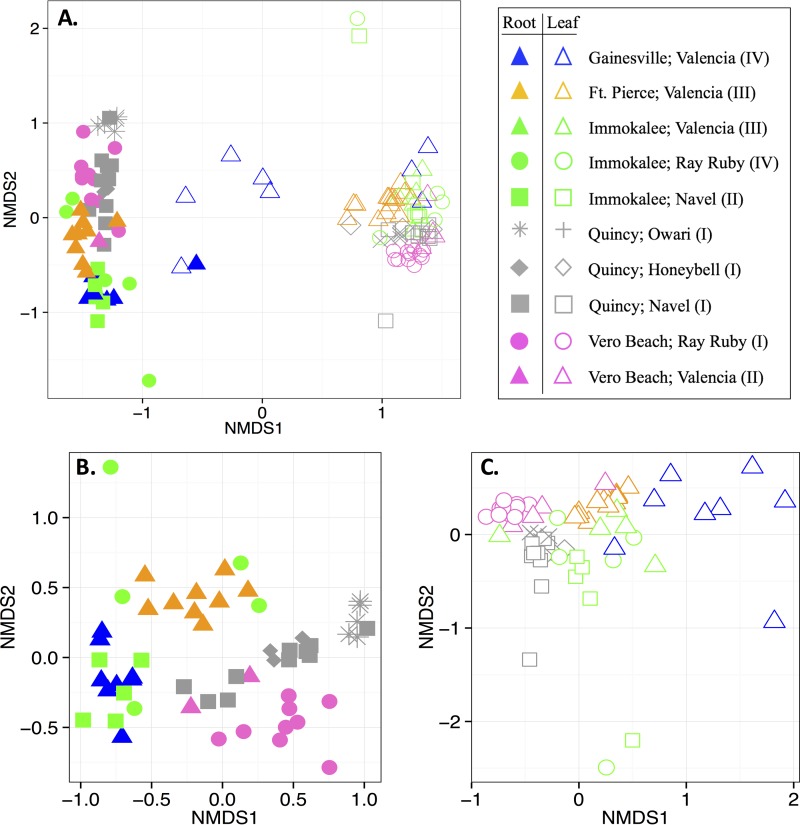
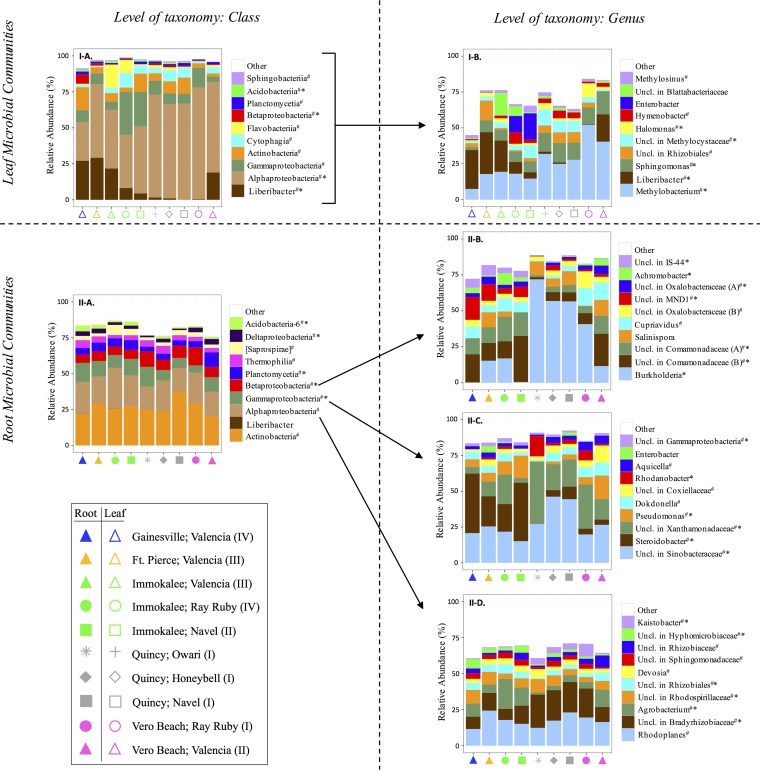
There were fundamental differences in the structures of leaf and root microbiota (analysis of similarity [ANOSIM] R statistic, 0.950; significance based on 999 permutations, 0.001), which was illustrated by clustering in nonmetric multidimensional scaling (NMDS) plots (Fig. 3). Interestingly, while the proportions of taxa within broader phylogenetic groups (i.e., at the class level) of respective leaf- and root-associated communities were generally conserved across locations and cultivars, there were trends for differences in relative abundances of dominant microbiota at the genus level (Fig. 4). Accordingly, the composition and diversity of leaf and root microbiota were substantially associated with location (leaves, ANOSIM R statistic, 0.655, and significance based on 999 permutations, 0.001; roots, ANOSIM R statistic, 0.577, and significance based on 999 permutations, 0.001); there was also an association with citrus cultivar (leaves, ANOSIM R statistic, 0.318, and significance based on 999 permutations, 0.001; roots, ANOSIM R statistic, 0.289, and significance based on 999 permutations, 0.001) (Fig. 3). Additional ANOSIMs were performed for subsets of data in order to determine (i) whether the microbial communities of specific cultivars collected at different sites varied and (ii) whether the microbial communities of different cultivars varied within the sites where multiple cultivars were sampled. Interestingly, the structures of leaf microbiota from each cultivar that had multiple collection sites (i.e., Valencia, Navel, and Ray Ruby) substantially varied by location (Table 2). Alternatively, the differences in root microbiota structure across locations were substantial for Navel trees, but only moderate for Valencia trees (Table 2). Furthermore, the differences in microbial community structure by cultivar were location specific. At Vero Beach, the microbiota of the different different cultivars (i.e., Valencia, Ray Ruby) substantially varied, whereas, at Immokalee and Quincy, only moderate differences were detected among the cultivars sampled (Table 2).
FIG 3.
NMDS plots for differences in the structure of microbial communities that were associated with the citrus trees sampled in fall 2015. (A) All samples; (B) only root samples; (C) only leaf samples. Solid and open symbols correspond to root and leaf microbiota, respectively. Symbols correspond to tree location (color) and cultivar (shape). The category for HLB symptom severity (I, asymptomatic; II, symptomatic—mild; III, symptomatic—moderate; IV, symptomatic—severe) is indicated.
FIG 4.
Relative abundances of dominant taxa assigned to 16S sequences detected in leaves (I) and roots (II) of the citrus trees sampled in fall 2015. (A) Classes, with Liberibacter spp. separate from other Alphaproteobacteria; (B to D) genera. Symbols correspond to tree location (color) and cultivar (shape). The category for HLB symptom severity (I, asymptomatic; II, symptomatic—mild; III, symptomatic—moderate; IV, symptomatic—severe) is indicated. # indicates a member of core microbiota, and * indicates significant difference, with a statistical power of >0.9, from Student's t test for differences in relative abundance in asymptomatic trees (category I) and HLB-symptomatic trees (categories II to IV) (α set at 0.05). Uncl., unclassified.
TABLE 2.
Differences in microbial community structure across locations for cultivars that were sampled at multiple sites and across cultivars within a location where multiple cultivars were sampleda
| Factor | Microbial community | Cultivar(s) | Location(s) | ANOSIM R | Significance |
|---|---|---|---|---|---|
| Location | Leaf | Navel | Immokalee and Quincy, FL | 0.606 | 0.002 |
| Ray Ruby | Vero Beach and Immokalee, FL | 0.897 | 0.001 | ||
| Valencia | Ft. Pierce, Gainesville, Immokalee, and Vero Beach, FL | 0.663 | 0.001 | ||
| Root | Navel | Immokalee and Quincy, FL | 0.766 | 0.001 | |
| Valencia | Ft. Pierce, Gainesville, and Vero Beach, FL | 0.226 | 0.015 | ||
| Cultivar | Leaf | Navel, Ray Ruby, Valencia | Immokalee, FL | 0.296 | 0.034 |
| Ray Ruby, Valencia | Vero Beach, FL | 0.625 | 0.046 | ||
| Honeybell, Navel, Owari | Quincy, FL | 0.365 | 0.006 | ||
| Root | Navel, Ray Ruby, Valencia | Immokalee, FL | 0.308 | 0.033 | |
| Ray Ruby, Valencia | Vero Beach, FL | 0.758 | 0.003 | ||
| Honeybell, Navel, Owari | Quincy, FL | 0.319 | 0.016 |
The ANOSIM R statistic and the significance based on 999 permutations are listed for each ANOSIM test. An R statistic of 0 means the communities are identical, whereas an R value of 1 means the communities have no overlap. R values greater than 0.6 with significance less than 0.05 indicate substantial difference and are listed in boldface.
In order to determine effects of location and cultivar on microbial community structure that were free from confounding effects of HLB, subsets of data from trees within the same HLB symptom severity category were compared. Regarding location, the leaf microbiota of the moderately symptomatic Valencia trees in both Ft. Pierce and Immokalee still substantially differed (ANOSIM R statistic, 0.749; significance based on 999 permutations, 0.001) (see Table S2 in the supplemental material). In addition, for the three asymptomatic varieties sampled in Quincy, there were substantial differences between the community structures of Owari and Honeybell leaf microbiota (ANOSIM R statistic, 0.959; significance based on 999 permutations, 0.023) and root microbiota (ANOSIM R statistic, 0.867; significance based on 999 permutations, 0.016). Alternatively, little or no differences were detected between the microbiota of Navel trees with that of either Owari or Honeybell trees. Overall, the structure of the leaf and root microbiota strongly correlated with location and moderately correlated with cultivar. Even though these associations were partially attributable to HLB symptom severity, examples of correlations between microbiota and location, as well as microbiota and cultivar, free from confounding effects of each other and HLB were evident.
Most importantly, according to ANOSIM, HLB symptom severity of trees (i.e., asymptomatic, mildly symptomatic, moderately symptomatic, and severely symptomatic) was also strongly associated with community structure of leaf microbiota (ANOSIM R statistic, 0.572; significance based on 999 permutations, 0.001) and root microbiota (ANOSIM R statistic, 0.592; significance based on 999 permutations, 0.001), regardless of the other variables being considered. At the genus level, the leaf-associated communities of asymptomatic trees had relatively low abundances of Liberibacter and relatively high abundances of Enterobacter, Hymenobacter, and/or Methylobacterium, compared to those from HLB-symptomatic trees (Fig. 4I-B). Regression analysis showed a negative correlation between the alpha diversity of leaf microbiota and the relative abundance of Liberibacter spp. (see Fig. S5 in the supplemental material). Furthermore, the root-associated microbial communities of HLB-symptomatic trees had (i) higher abundances of Bradyrhizobiaceae and lower abundances of Agrobacterium within Alphaproteobacteria, (ii) higher abundances of Burkholderia and lower abundances of Comamonadaceae within Betaproteobacteria, and (iii) higher abundances of Rhodanobacter, Sinobacteraceae, or Xanthamonadaceae and lower abundances of Comamonadaceae or Pseudomonas within Gammaproteobacteria, compared to that of asymptomatic trees (Fig. 4II-B to -D). Many of the taxa whose relative abundances differed based on HLB symptoms were members of the core citrus microbial community (Fig. 4; Table S1).
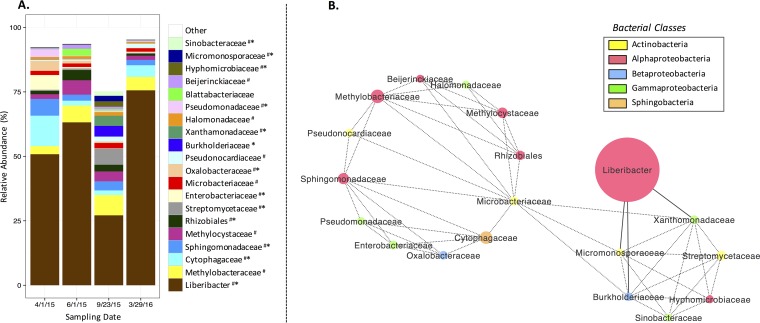
Moreover, to test the hypothesis that season and time were associated with community structure, we compared microbiota from Valencia trees sampled in Gainesville on 1 April 2015, 1 June 2015, 23 September 2015, and 29 March 2016. By comparing the microbiota sampled in 2015, we determined that season correlated with the structure of the leaf-associated communities (ANOSIM R statistic, 0.282; significance based on 999 permutations, 0.001), but not with that of root-associated communities (ANOSIM R statistic, 0.064; significance based on 999 permutations, 0.091). By comparing the samples from spring 2015 and spring 2016, we determined that there were also differences in leaf microbiota based on time (ANOSIM R statistic, 0.218; significance based on 999 permutations, 0.012), which were smaller than those by season, and there were no differences in root microbiota (ANOSIM R statistic, 0.012; significance based on 999 permutations, 0.337) based on time. These results are illustrated by the sample date-specific trends observed for proportions of dominant taxa within the leaf microbiota (Fig. 5A) but not root microbiota (see Fig. S6 in the supplemental material).
FIG 5.
(A) Relative abundances of the 20 most abundant families, with Liberibacter spp. separate from other members of Rhizobiaceae, in leaf-associated microbial communities of Valencia trees sampled in Gainesville, FL, on 1 April 2015, 1 June 2015, 23 September 2015, and 29 March 2016. # indicates the taxon was a member of the core microbiota, and * indicates significant difference (P < 0.05) for different dates based on ANOVA. (B) Correlation-based network analysis of the relative abundances of these top 20 taxa reveals significant interactions (i.e., dotted lines indicate cooccurrence, and solid lines indicate mutual exclusion) (P < 0.05). The size of each circle corresponds to the average relative abundance, and the color corresponds to the bacterial class represented.
Putative interactions between native bacteria and Liberibacter spp.
To test the hypothesis that microbe-microbe interactions could potentially influence the progression or suppression of the HLB pathogen, we examined cooccurrence patterns of the 20 most abundant bacterial families (with Liberibacter spp. separate from other members of Rhizobiaceae), most of which were core microbiota (Fig. 5A), over the course of the year. Family level was chosen because it was the lowest taxonomic rank where almost all of the top 20 taxa had full classification (i.e., only 1/20 families was unclassified within an assigned bacterial order). A total of 51 significant interactions (P < 0.05) were detected between 19 of these taxa, one of which was Liberibacter spp. All taxa in the network, except for the pathogen, had at least one positive interaction (cooccurrence) (P < 0.05) with another taxon, and two cluster groups were observed (Fig. 5B). The smaller cluster was linked to negative interactions (mutual exclusion) (P < 0.05) with Liberibacter spp. (Fig. 5B). The negative interactions between Liberibacter spp. and Burkholderiaceae, Micromonosporaceae, and Xanthomonadaceae appeared to mediate the negative relationship between the HLB pathogen and the majority of native bacteria, since the top 20 families constituted >75% of all family-level members of the microbial community at any time point (Fig. 5A) during HLB disease progression.
DISCUSSION
Harnessing the beneficial potential of the plant microbiome to mitigate major crop diseases, such as HLB, is gaining interest as a sustainable approach to improve agricultural production (5, 6, 32). It has been suggested that the abundances of core members within plant-associated microbial communities, which may stably associate with particular hosts (11, 13, 15), are a determinant factor in whether the community is functioning in a “healthy” state (9). Understanding the importance of the core group with regard to its implications for plant health and productivity is still at a stage of infancy. Plant diseases are known to be associated with changes in the microbiome (15, 17, 20–26), but much remains to be learned about potential interactions between phytopathogens and native bacteria, notably core microbiota, that could play key roles in disease progression. The present study investigated these associations and interactions through a comprehensive analysis of the core leaf- and root-associated communities of citrus hosts that varied in the extent of HLB disease progression.
The core compositional members of the microbial communities were defined at each taxonomic rank, and as rank became more specific (i.e., from phylum to genus), the ratios of the numbers of core taxa to all taxa consistently decreased. Certain non-core members at finer levels of taxonomy collectively comprised some of the taxa that were defined as core at higher taxonomic ranks. This could be attributable to plant microbiome selection being dependent on microbial resource requirements and functional genes that are redundant within broader phylogenetic groups (9). Moreover, the vast majority of all genera detected in the communities were members of the rare biosphere (40) and were highly variable in terms of occurrence across samples, even among replicates. Although the numbers of core members comprised a small fraction of all citrus-associated taxa, these organisms made up the majority of the relatively dominant microbiota.
Microbial community structure was strongly associated with HLB symptom severity. The positive correlation between the relative abundance of the Liberibacter spp. and symptom severity, as well as its negative correlation with alpha diversity, suggests that the diversity of leaf microbiota decreases as HLB progresses. While Liberibacter spp. were significantly more abundant within leaves of HLB-symptomatic trees, other Alphaproteobacteria (Methylobacterium, Sphingomonas, and Methylocystaceae) were present in greater proportions in those of asymptomatic trees. Since niche similarities can adversely impact coexistence (41), there may have been resource-related competition between the pathogen and other members of its bacterial class. Previous studies have also indicated Methylobacterium and Sphingomonas to be involved in protecting host plants from various pathogens (42–45). Furthermore, by examining the year-long cooccurrence patterns for the 20 most abundant families and Liberibacter spp., we uncovered negative correlations between the pathogen and three bacterial families (Burkholderiaceae, Xanthomonadaceae, and Micromonosporaceae) that have previously been described to have plant-beneficial properties (18, 23, 27, 46). These taxa may have, to an extent, been involved in mediating the suppressive relationship between the pathogen and the stable microbial community. Future work is needed to validate such microbe-pathogen interactions.
With regard to root microbiota, even though the abundance of Liberibacter spp. was miniscule, there were significant differences in the structures of communities of asymptomatic and HLB-symptomatic trees. The low detection of the pathogen in root-associated communities was probably due to it being phloem limited (i.e., an endophyte) (34), while the majority of bacteria in the root samples were likely present as epiphytes or on soil particles from the surrounding rhizosphere that had been in contact with the root surface (13). Several dominant core genera of citrus roots (Kaistobacter and unclassified genera in Bradyrhizobiaceae and Xanthomonadaceae) were found to be associated with asymptomatic trees. Bradyrhizobiaceae are known soil bacteria and root endophytes involved in symbiotic relationships with plants as nitrogen fixers (47, 48). While some members of Xanthomonadaceae are citrus pathogens (e.g., citrus canker is caused by Xanthomonas axonopodis) (49), others have been implicated in biological control (27). Alternatively, the relative abundances of other core genera, such as Steroidobacter and unclassified genera in Comamonadaceae, Hyphomicrobiaceae, MND1, IS-44, and Rhizobiales, were significantly greater in their respective classes among the root microbiota of HLB-symptomatic trees. It is unclear whether these disease-associated taxa may have synergistically impacted HLB progression or if they were opportunistic colonizers as the microbiomes were restructuring in response to disease progression having caused changes in the local microenvironments. In addition to the core taxa associated with roots, several dominant non-core/variable members were also linked to host health. For example, Rhodanobacter and Burkholderia were common within their bacterial classes in the root communities of asymptomatic trees, yet absent in those of diseased trees. The former has previously been shown to exhibit biological control activity toward phytopathogens both in vitro and in vivo (50), while the latter is known to colonize the root surface, gain entry into the internal tissue, and translocate in xylem and is capable of inducing host defense signaling (46). Interestingly, there were consistencies between some potentially beneficial taxa associated with both leaves and roots (e.g., Burkholderiaceae and Xanthomonadaceae), which may have implications for biological control of Liberibacter spp. Development of novel approaches to treat or manage HLB by bolstering the stability of the native microbial community or by using treatments that are target specific to Liberibacter spp. so that the native community is not disrupted warrants further investigation.
Separate from the correlations between microbial community structure and HLB symptom expression, there were unique associations between the plant microbiota and location, cultivar, season, and time, which have been previously discussed (11, 13, 15, 16, 19). A limitation to this study was that the analyses were performed at the taxonomic level of genus or above, so the strain specificity of potential microbial interactions or associations was not able to be determined. Although strain-level diversity of L. asiaticus (i.e., the HLB pathogen in Florida) is largely unexplored, perhaps due to the fact that the pathogen is not culturable (17), it cannot be ruled out that the differences in HLB symptom severity and microbial community structure that were noted for the different sampling locations could have been related to differences in pathogenicity of the Liberibacter strains that were present at the site. It is possible that, compared to the more severely symptomatic trees, the asymptomatic trees that were positive for the pathogen or the mildly symptomatic trees may have been colonized by benign or at least relatively less-virulent Liberibacter spp.
We emphasize that the findings from this work indicate correlation, not causation, between microbial community structure and HLB progression, among other variables. Although the directional associations between pathogen, microbiota, and host remain unclear, several hypotheses can be posited. For example, the differences in microbial community structures between healthy and diseased trees may have been a response to changes in host-associated microhabitats that were affected by disease progression. HLB is known to cause carbohydrate partitioning imbalances and deficiencies of essential micronutrients (e.g., Fe, Mn, and Zn) throughout the tree canopy (51, 52). These factors could have impacted the leaf-associated microbial communities. As for root microbiota, the declining health state of the trees may have altered the composition and amounts of root exudates produced. Trivedi et al. (24) reported HLB infection significantly alters the abundances of functional genes in the citrus rhizosphere that are involved in carbon fixation, nitrogen cycling, phosphorus utilization, and metal homeostasis, which reflected a state of environmental change. On the other hand, since the relative abundance of Liberibacter spp. within leaf-associated microbial communities was found to correlate with disease symptom severity, which was in agreement with a previously made suggestion that a minimal titer of the pathogen is required for HLB symptoms to develop (17), certain changes within the microbial communities that could have involved a pathway for proliferation of Liberibacter spp. among native bacteria may have been a critical factor for disease establishment. After all, the pathogen was still present in the leaf microbiota of almost all asymptomatic trees (i.e., in 27/29 leaf samples from asymptomatic trees) at very low relative abundances. Potential plant-beneficial bacteria that were differentially abundant within the trees that had varied in HLB symptom severity could have been involved in (i) competing with Liberibacter for nutrient resources, limiting its ability to grow and spread, (ii) antibiosis toward the pathogen or other disease-associated bacteria, (iii) assistance with host nutrient acquisition, which could help alleviate disease symptom-related nutrient deficiencies, or (iv) induction of host plant signaling pathways that stimulated plant defense responses. Distinguishing the changes within core plant-associated microbial communities that are associated with disease establishment from those that are associated with consequences of disease symptoms is an interesting avenue for future research.
Overall, our results advance the understanding of (i) plant microbiota selection across multiple variables and (ii) changes in community structure that are associated with disease establishment or symptom progression, having implications for biological control. We not only confirmed previously described bacterial associations with plant health (e.g., potentially beneficial bacteria) but also demonstrated the importance of core taxa within the broader community, as well as identified new associations and potential interactions between certain bacteria and the economically important phytopathogens Liberibacter spp.
MATERIALS AND METHODS
Sample collection and processing.
Leaves and roots were collected from citrus trees across Florida (Gainesville, Ft. Pierce, Immokalee, Quincy, Vero Beach) (see Fig. S1 in the supplemental material), where huanglongbing (HLB) disease symptoms ranged from asymptomatic to severely symptomatic during 2015 to 2016. Valencia (Citrus sinensis L. Osbeck), Navel [Citrus sinensis (L.) Osbeck], Honeybell (Citrus × tangelo), Owari (Citrus unshiu Marcovitch), and Ray Ruby (Citrus paradisi Macfadyen) were sampled in the study (Table 1). The HLB symptom appearance for each set of replicate trees was classified by 4 categories (I, II, III, or IV) corresponding to asymptomatic, symptomatic—mild, symptomatic—moderate, or symptomatic—severe, respectively. Figure S2 in the supplemental material provides a detailed description of each category.
To sample leaves, the canopy of each tree was divided into four quadrants and 10 leaves from each quadrant were randomly selected. The leaves were collected aseptically by being cut at the stem and placed into sterile Whirl-Pak stomacher bags (Nasco, Fort Atkinson, WI). To sample roots, 3 soil cores (5-cm diameter, 25-cm depth) were taken within the root zone of each tree and the fibrous roots were screened from the soil and combined. A composite root sample for each tree was collected in a sterile 50-ml polypropylene tube. The leaf and root samples were transported to the lab in a cooler on dry ice.
All samples were frozen at −80°C and lyophilized. The leaf samples were crushed into <1-cm fragments in their respective stomacher bags. Each set of leaf samples that was taken from the same tree was combined at equal volume in sterile 15-ml polypropylene tubes. The root samples were crushed into <1-cm fragments by adding a sterile steel grinding ball (9.5-mm diameter) to each tube and shaking vigorously by hand. The crushed leaf and root samples, along with a sterile steel grinding ball (9.5-mm diameter), were placed in 5-ml polyethylene vials (Spex, Metuchen, NJ) and homogenized via bead beating with three 1-min bursts in the GenoGrinder 2000 (Spex, Metuchen, NJ).
16S Illumina sequencing.
Genomic DNA for Illumina sequencing was extracted from each homogenized sample with the Isolate II plant DNA kit (Bioline, Taunton, MA). Amplicon libraries were prepared for the V4 region of 16S rRNA genes with PCRs that incorporated Illumina-compatible universal bacteria/archaeal primers 515f/806r, with a 12-bp indexing barcode added to the forward primer (53, 54). Samples were amplified in 25-μl reaction mixtures that contained 0.5 U of Phusion high-fidelity (HF) DNA polymerase (M0530S; New England BioLabs), 1× Phusion HF reaction buffer, 3% dimethyl sulfoxide (DMSO), 200 μM deoxynucleoside triphosphates (dNTPs), 0.25 μM each primer, and 1 μl of template DNA (approximately 45 ng). PNA clamps (0.75 μM pPNA or 0.75 μM mPNA) (39) were included in each reaction in order to limit amplification of contaminant 16S plastid and 16S mitochondrial sequences. PCRs were performed using a SimpliAmp thermal cycler (Applied Biosystems, Foster City, CA) with an initial denaturation stage at 94°C for 3 min, followed by 35 cycles of denaturation at 94°C for 45 s, annealing of PNA clamps at 78°C for 10 s, annealing of primers at 50°C for 60 s, and elongation at 72°C for 90 s, and completed with a final elongation stage at 72°C for 10 min (39). Triplicate amplifications for each sample were pooled and cleaned with the MinElute PCR purification kit (Qiagen, Valencia, CA). The amplicons were quantified for DNA concentration and the 260-nm/280-nm ratio using a NanoDrop 1000 (ThermoFisher Scientific, Waltham, MA). Final pools containing 500 ng of DNA from each amplicon library were prepared and submitted to the NextGen DNA sequencing core at the Interdisciplinary Center for Biotechnology Research at the University of Florida. The samples were quantified at the sequencing center by quantitative PCR (qPCR) and QUBIT for quality control and size selected with ELF to produce fragment sizes in the desired range for the amplicon sequencing. Paired-end sequencing (2× 150 cycles) was performed on an Illumina MiSeq platform.
Data analysis.
Sequenced reads were demultiplexed based on the indexing barcodes at the sequencing center. We processed the reads with Cutadapt (https://github.com/marcelm/cutadapt) and Sickle (https://github.com/najoshi/sickle) to remove any residual Illumina adapters and primer sequences, truncate sequences at the first N position, trim sequences at a base pair with a phred score below 30, and remove reads that were shorter than 120 bp. Paired-end reads were joined with Eautils (https://github.com/ExpressionAnalysis/ea-utils) with the requirements of having a minimum overlap of 30 bp and a 3% maximum difference in the overlap region. Sample names were added to the definition lines of sequencing reads using the sed command and concatenated into one fasta file in order to make them compatible for analysis in QIIME v.1.8 (55). Clustering of OTUs at 97% similarity, with no removal of singletons, was performed in QIIME by the open-reference OTU picking method (56), and taxonomy assignments were made by mapping to the Greengenes reference database version 13.8 (57). Unassigned OTUs and those that were identified as 16S mitochondrial or plastid DNA were removed from further analyses. The total counts of OTUs and assigned taxa for each taxonomic rank were transformed to relative abundance values.
The core members of the citrus leaf and root microbiota were defined as those that had at least 1 sequencing read in at least 95% of the respective samples (58). Rank abundance curves were generated to inspect the distribution of core members and non-core/variable members (i.e., those found in less than 95% of samples). The ratios of core members to non-core/variable members were compared at the phylum, class, order, family, and genus levels.
In order to evaluate the spatiotemporal variability of the relative abundance of Liberibacter spp. and how it correlated with HLB disease symptom severity, the statistical software PAST (59) was used to perform ANOVA and Tukey's post hoc tests to determine the differences in the relative abundance of the pathogen in microbiota of citrus trees based on (i) location and citrus cultivar (trees sampled in fall 2015), (ii) season/time (trees sampled in Gainesville during 2015 to 2016), and (iii) HLB symptom severity (all trees).
Microbial community structure was analyzed with phyloseq (60) and plotted with ggplot2 (61) in R v.3.2.1. Regression analysis was used to determine the relationship between microbiota alpha diversity and Liberibacter species relative abundance. The associations of microbial community structure with plant organ (i.e., leaf and root), location, cultivar, HLB symptom severity, season, and time were evaluated with nonmetric multidimensional scaling (NMDS) plots and/or analysis of similarity (ANOSIM) (significance based on 999 permutations) in Vegan v.2.3.2 (62). Note that an ANOSIM R statistic of 0 means the communities are identical, whereas an R value of 1 means the communities have no overlap. The Student t test and power calculation were applied to determine the differences in relative abundances of core bacterial genera that were associated with asymptomatic trees (i.e., category I) and HLB symptomatic trees (i.e., categories II to IV).
Network analysis of interactions between the 20 most abundant families, with Liberibacter spp. separate from other members of their family (Rhizobiaceae), of leaf microbiota from the Valencia trees sampled in Gainesville during 2015 to 2016 (n = 32) was performed using the CoNet app (63) in Cytoscape v.3.0.2 (64). Significant positive and negative correlations between taxa were determined by support of three separate measures: Pearson's correlation, Spearman's correlation, or Bray-Curtis dissimilarity. Networks from the three measures were merged by intersection, keeping only significant interactions (α set at 0.05) with support from all methods. In addition, an ANOVA was applied to determine differences in the relative abundances of these taxa at the four different time points.
Accession number(s).
Parsed raw sequencing reads are publicly available through NCBI's Sequence Read Archive under BioProject accession no. PRJNA362723.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institute of Food and Agriculture, U.S. Department of Agriculture grant (award no. 2015-70016-23029) to G.L.L., C.F.G., and M.T. R.A.B. gratefully acknowledges the University of Florida Graduate School Fellowship for support. The content is solely the responsibility of the authors and does not necessarily represent the official views of the granting agencies.
We are indebted to Christopher Gardner for help with sample collection and providing photographs of the trees and to Javier Silfa-Cifuentes and Sarah Stavros for technical assistance with sample processing. We thank Robert Adair, José Chaparro, Nick Comerford, Yong-Ping Duan, and Kelly Morgan for access to the various field sites that were sampled in this study.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00210-17.
REFERENCES
- 1.Andreote FD, Gumiere T, Durrer A. 2014. Exploring interactions of plant microbiomes. Sci Agric 71:528–539. doi: 10.1590/0103-9016-2014-0195. [DOI] [Google Scholar]
- 2.Lebeis SL. 2014. The potential for give and take in plant-microbiome relationships. Front Plant Sci 5:287. doi: 10.3389/fpls.2014.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mendes R, Garbeva P, Raaijmakers JM. 2013. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37:634–663. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- 4.Schlaeppi K, Bulgarelli D. 2015. The plant microbiome at work. Mol Plant Microbe Interact 28:212–217. doi: 10.1094/MPMI-10-14-0334-FI. [DOI] [PubMed] [Google Scholar]
- 5.Berg G, Zachow C, Müller H, Philipps J, Tilcher R. 2013. Next-generation bio-products sowing the seeds of success for sustainable agriculture. Agronomy 3:648–656. doi: 10.3390/agronomy3040648. [DOI] [Google Scholar]
- 6.Gopal M, Gupta A, Thomas GV. 2013. Bespoke microbiome therapy to manage plant diseases. Front Microbiol 4:10–13. doi: 10.3389/fmicb.2013.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein E, Katan J, Minz D, Gamliel A. 2013. Soil suppressiveness to Fusarium disease: shifts in root microbiome associated with reduction of pathogen root colonization. Phytopathology 103:23–33. doi: 10.1094/PHYTO-12-11-0349. [DOI] [PubMed] [Google Scholar]
- 8.Mavrodi OV, Mavrodi DV, Parejko JA, Thomashow LS, Weller DM. 2012. Irrigation differentially impacts populations of indigenous antibiotic-producing Pseudomonas spp. in the rhizosphere of wheat. Appl Environ Microbiol 78:3214–3220. doi: 10.1128/AEM.07968-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shade A, Handelsman J. 2012. Beyond the Venn diagram: the hunt for a core microbiome. Environ Microbiol 14:4–12. doi: 10.1111/j.1462-2920.2011.02585.x. [DOI] [PubMed] [Google Scholar]
- 10.Bodenhausen N, Horton MW, Bergelson J. 2013. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS One 8:e56329. doi: 10.1371/journal.pone.0056329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P. 2012. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488:91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- 12.Gottel NR, Castro HF, Kerley M, Yang Z, Pelletier DA, Podar M, Karpinets T, Uberbacher ED, Tuskan GA, Vilgalys R, Doktycz MJ, Schadt CW. 2011. Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Appl Environ Microbiol 77:5934–5944. doi: 10.1128/AEM.05255-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL. 2012. Defining the core Arabidopsis thaliana root microbiome. Nature 488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE. 2013. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci U S A 110:6548–6553. doi: 10.1073/pnas.1302837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rastogi G, Sbodio A, Tech JJ, Suslow TV, Coaker GL, Leveau JHJ. 2012. Leaf microbiota in an agroecosystem: spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J 6:1812–1822. doi: 10.1038/ismej.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Redford AJ, Fierer N. 2009. Bacterial succession on the leaf surface: a novel system for studying successional dynamics. Microb Ecol 58:189–198. doi: 10.1007/s00248-009-9495-y. [DOI] [PubMed] [Google Scholar]
- 17.Sagaram US, Deangelis KM, Trivedi P, Andersen GL, Lu SE, Wang N. 2009. Bacterial diversity analysis of Huanglongbing pathogen-infected citrus, using phyloChip arrays and 16S rRNA gene clone library sequencing. Appl Environ Microbiol 75:1566–1574. doi: 10.1128/AEM.02404-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weinert N, Piceno Y, Ding GC, Meincke R, Heuer H, Berg G, Schloter M, Andersen G, Smalla K. 2011. PhyloChip hybridization uncovered an enormous bacterial diversity in the rhizosphere of different potato cultivars: many common and few cultivar-dependent taxa. FEMS Microbiol Ecol 75:497–506. doi: 10.1111/j.1574-6941.2010.01025.x. [DOI] [PubMed] [Google Scholar]
- 19.Berg G, Smalla K. 2009. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol 68:1–13. doi: 10.1111/j.1574-6941.2009.00654.x. [DOI] [PubMed] [Google Scholar]
- 20.Bulgari D, Casati P, Crepaldi P, Daffonchio D, Quaglino F, Brusetti L, Bianco PA. 2011. Restructuring of endophytic bacterial communities in grapevine yellows-diseased and recovered Vitis vinifera L plants. Appl Environ Microbiol 77:5018–5022. doi: 10.1128/AEM.00051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J-G, Ren G-D, Jia Z-J, Dong Y-H. 2014. Composition and activity of rhizosphere microbial communities associated with healthy and diseased greenhouse tomatoes. Plant Soil 380:337–347. doi: 10.1007/s11104-014-2097-6. [DOI] [Google Scholar]
- 22.Schreiner K, Hagn A, Kyselková M, Moënne-Loccoz Y, Welzl G, Munch JC, Schloter M. 2010. Comparison of barley succession and take-all disease as environmental factors shaping the rhizobacterial community during take-all decline. Appl Environ Microbiol 76:4703–4712. doi: 10.1128/AEM.00481-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trivedi P, Duan Y, Wang N. 2010. Huanglongbing, a systemic disease, restructures the bacterial community associated with citrus roots. Appl Environ Microbiol 76:3427–3436. doi: 10.1128/AEM.02901-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trivedi P, He Z, Van Nostrand JD, Albrigo G, Zhou J, Wang N. 2012. Huanglongbing alters the structure and functional diversity of microbial communities associated with citrus rhizosphere. ISME J 6:363–383. doi: 10.1038/ismej.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu L, Ravnskov S, Larsen J, Nicolaisen M. 2012. Linking fungal communities in roots, rhizosphere, and soil to the health status of Pisum sativum. FEMS Microbiol Ecol 82:736–745. doi: 10.1111/j.1574-6941.2012.01445.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Du B-H, Jin Z, Li Z, Song H, Ding Y-Q. 2011. Analysis of bacterial communities in rhizosphere soil of healthy and diseased cotton (Gossypium sp.) at different plant growth stages. Plant Soil 339:447–455. doi: 10.1007/s11104-010-0600-2. [DOI] [Google Scholar]
- 27.Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, Piceno YM, DeSantis TZ, Andersen GL, Bakker PA, Raaijmakers JM. 2011. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 28.Lamichhane JR, Venturi V. 2015. Synergisms between microbial pathogens in plant disease complexes: a growing trend. Front Plant Sci 6:1–12. doi: 10.3389/fpls.2015.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andreote FD, Rocha UN, Araújo WL, Azevedo JL, van Overbeek LS. 2010. Effect of bacterial inoculation, plant genotype and developmental stage on root-associated and endophytic bacterial communities in potato (Solanum tuberosum). Antonie Van Leeuwenhoek 97:389–399. doi: 10.1007/s10482-010-9421-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dematheis F, Kurtz B, Vidal S, Smalla K. 2013. Multitrophic interactions among western corn rootworm, Glomus intraradices and microbial communities in the rhizosphere and endorhiza of maize. Front Microbiol 4:1–12. doi: 10.3389/fmicb.2013.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herschkovitz Y, Lerner A, Davidov Y, Rothballer M, Hartmann A, Okon Y, Jurkevitch E. 2005. Inoculation with the plant-growth-promoting rhizobacterium Azospirillum brasilense causes little disturbance in the rhizosphere and rhizoplane of maize (Zea mays). Microb Ecol 50:277–288. doi: 10.1007/s00248-004-0148-x. [DOI] [PubMed] [Google Scholar]
- 32.Bakker MG, Manter DK, Sheflin AM, Weir TL, Vivanco JM. 2012. Harnessing the rhizosphere microbiome through plant breeding and agricultural management. Plant Soil 360:1–13. doi: 10.1007/s11104-012-1361-x. [DOI] [Google Scholar]
- 33.Schreiter S, Sandmann M, Smalla K, Grosch R. 2014. Soil type dependent rhizosphere competence and biocontrol of two bacterial inoculant strains and their effects on the rhizosphere microbial community of field-grown lettuce. PLoS One 9:1–11. doi: 10.1371/journal.pone.0103726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bové JM. 2006. Huanglongbing: a destructive, newly-emerging, century-old disease of citrus. J Plant Pathol 88:7–37. [Google Scholar]
- 35.Hodges AW, Spreen TH. 2012. Economic impacts of citrus greening (HLB) in Florida. University of Florida IFAS Extension document FE903. University of Florida-IFAS, Gainesville, FL. [Google Scholar]
- 36.Zhang M, Powell CA, Benyon LS, Zhou H, Duan Y. 2013. Deciphering the bacterial microbiome of citrus plants in response to “Candidatus Liberibacter asiaticus”-infection and antibiotic treatments. PLoS One 8:e76331. doi: 10.1371/journal.pone.0076331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang M, Powell CA, Guo Y, Benyon L, Duan Y. 2013. Characterization of the microbial community structure in Candidatus Liberibacter asiaticus-infected citrus plants treated with antibiotics in the field. BMC Microbiol 13:112. doi: 10.1186/1471-2180-13-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang MQ, Guo Y, Powell CA, Doud MS, Yang CY, Zhou H, Duan YP. 2016. Zinc treatment increases the titre of “Candidatus Liberibacter asiaticus” in huanglongbing-affected citrus plants while affecting the bacterial microbiomes. J Appl Microbiol 120:1616–1628. doi: 10.1111/jam.13102. [DOI] [PubMed] [Google Scholar]
- 39.Lundberg DS, Yourstone S, Mieczkowski P, Jones CD, Dangl JL. 2013. Practical innovations for high-throughput amplicon sequencing. Nat Methods 10:999–1002. doi: 10.1038/nmeth.2634. [DOI] [PubMed] [Google Scholar]
- 40.Lynch MDJ, Neufeld JD. 2015. Ecology and exploration of the rare biosphere. Nat Rev Microbiol 13:217–229. doi: 10.1038/nrmicro3400. [DOI] [PubMed] [Google Scholar]
- 41.Mordecai EA. 2011. Pathogen impacts on plant communities: unifying theory, concepts, and empirical work. Ecol Monogr 81:429–441. doi: 10.1890/10-2241.1. [DOI] [Google Scholar]
- 42.Ardanov P, Sessitsch A, Häggman H, Kozyrovska N, Pirttilä AM. 2012. Methylobacterium-induced endophyte community changes correspond with protection of plants against pathogen attack. PLoS One 7:e46802. doi: 10.1371/journal.pone.0046802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Enya J, Shinohara H, Yoshida S, Tsukiboshi T, Negishi H, Suyama K, Tsushima S. 2007. Culturable leaf-associated bacteria on tomato plants and their potential as biological control agents. Microb Ecol 53:524–536. doi: 10.1007/s00248-006-9085-1. [DOI] [PubMed] [Google Scholar]
- 44.Innerebner G, Knief C, Vorholt JA. 2011. Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl Environ Microbiol 77:3202–3210. doi: 10.1128/AEM.00133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madhaiyan M, Suresh Reddy BV, Anandham R, Senthilkumar M, Poonguzhali S, Sundaram SP, Sa T. 2006. Plant growth-promoting Methylobacterium induces defense responses in groundnut (Arachis hypogaea L.) compared with rot pathogens. Curr Microbiol 53:270–276. doi: 10.1007/s00284-005-0452-9. [DOI] [PubMed] [Google Scholar]
- 46.Compant S, Reiter B, Sessitsch A, Clément C, Ait Barka E, Nowak J. 2005. Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Appl Environ Microbiol 71:1685–1693. doi: 10.1128/AEM.71.4.1685-1693.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lugtenberg B, Kamilova F. 2009. Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 48.Verma JP, Yadav J, Tiwari KN, Singh L, Singh V. 2010. Impact of plant growth promoting rhizobacteria on crop production. Int J Agric Res 5:954–983. doi: 10.3923/ijar.2010.954.983. [DOI] [Google Scholar]
- 49.Das AK. 2003. Citrus canker—a review. J Appl Hortic 5:52–60. [Google Scholar]
- 50.De Clercq D, Van Trappen S, Cleenwerck I, Ceustermans A, Swings J, Coosemans J, Ryckeboer J. 2006. Rhodanobacter spathiphylli sp. nov., a gammaproteobacterium isolated from the roots of Spathiphyllum plants grown in a compost-amended potting mix. Int J Syst Evol Microbiol 56:1755–1759. doi: 10.1099/ijs.0.64131-0. [DOI] [PubMed] [Google Scholar]
- 51.Etxeberria E, Gonzalez P, Achor D, Albrigo G. 2009. Anatomical distribution of abnormally high levels of starch in HLB-affected Valencia orange trees. Physiol Mol Plant Pathol 74:76–83. doi: 10.1016/j.pmpp.2009.09.004. [DOI] [Google Scholar]
- 52.Tian S, Lu L, Labavitch JM, Webb SM, Yang X, Brown PH, He Z. 2014. Spatial imaging of Zn and other elements in Huanglongbing-affected grapefruit by synchrotron-based micro X-ray fluorescence investigation. J Exp Bot 65:953–964. doi: 10.1093/jxb/ert450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Apprill A, McNally S, Parsons R, Weber L. 2015. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 75:129–137. doi: 10.3354/ame01753. [DOI] [Google Scholar]
- 54.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rideout JR, He Y, Navas-Molina JA, Walters WA, Ursell LK, Gibbons SM, Chase J, McDonald D, Gonzalez A, Robbins-Pianka A, Clemente JC, Gilbert JA, Huse SM, Zhou H-W, Knight R, Caporaso JG. 2014. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ 2:e545. doi: 10.7717/peerj.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huse SM, Ye Y, Zhou Y, Fodor AA. 2012. A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS One 7:e34242. doi: 10.1371/journal.pone.0034242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hammer Ø, Harper DAT, Ryan PD. 2001. PAST: PAleontological STatistics software package for education and data analysis. Palaeontol Electron 4:1–9. [Google Scholar]
- 60.McMurdie PJ, Holmes S. 2013. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 62.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 63.Faust K, Raes J. 2012. Microbial interactions: from networks to models. Nat Rev Microbiol 10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 64.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.