ABSTRACT
Pseudoalteromonas piscicida is a Gram-negative gammaproteobacterium found in the marine environment. Three strains of pigmented P. piscicida were isolated from seawater and partially characterized by inhibition studies, electron microscopy, and analysis for proteolytic enzymes. Growth inhibition and death occurred around colonies of P. piscicida on lawns of the naturally occurring marine pathogens Vibrio vulnificus, Vibrio parahaemolyticus, Vibrio cholerae, Photobacterium damselae, and Shewanella algae. Inhibition also occurred on lawns of Staphylococcus aureus but not on Escherichia coli O157:H7 or Salmonella enterica serovar Typhimurium. Inhibition was not pH associated, but it may have been related to the secretion of a cysteine protease with strong activity, as detected with a synthetic fluorogenic substrate. This diffusible enzyme was secreted from all three P. piscicida strains. Direct overlay of the Pseudoalteromonas colonies with synthetic fluorogenic substrates demonstrated the activity of two aminopeptidase Bs, a trypsin-like serine protease, and enzymes reactive against substrates for cathepsin G-like and caspase 1-like proteases. In seawater cultures, scanning electron microscopy revealed numerous vesicles tethered to the outer surface of P. piscicida and a novel mechanism of direct transfer of these vesicles to V. parahaemolyticus. Vesicles digested holes in V. parahaemolyticus cells, while the P. piscicida congregated around the vibrios in a predatory fashion. This transfer of vesicles and vesicle-associated digestion of holes were not observed in other bacteria, suggesting that vesicle binding may be mediated by host-specific receptors. In conclusion, we show two mechanisms by which P. piscicida inhibits and/or kills competing bacteria, involving the secretion of antimicrobial substances and the direct transfer of digestive vesicles to competing bacteria.
IMPORTANCE Pseudoalteromonas species are widespread in nature and reduce competing microflora by the production of antimicrobial compounds. We isolated three strains of P. piscicida and characterized secreted and cell-associated proteolytic enzymes, which may have antimicrobial properties. We identified a second method by which P. piscicida kills V. parahaemolyticus. It involves the direct transfer of apparently lytic vesicles from the surface of the Pseudoalteromonas strains to the surface of Vibrio cells, with subsequent digestion of holes in the Vibrio cell walls. Enzymes associated with these vesicles are likely responsible for the digestion of holes in the cell walls. Pseudoalteromonas piscicida has potential applications in aquaculture and food safety, in control of the formation of biofilms in the environment, and in food processing. These findings may facilitate the probiotic use of P. piscicida to inactivate pathogens and may lead to the isolation of enzymes and other antimicrobial compounds of pharmacological value.
KEYWORDS: Pseudoalteromonas piscicida, Vibrio cholerae, Vibrio parahaemolyticus, Vibrio vulnificus, bacterial inhibition, enzymes, lytic vesicles, predatory bacteria, probiotics, proteases
INTRODUCTION
Members of the genus Pseudoalteromonas are Gram-negative, motile, and rod-shaped gammaproteobacteria that are found in high abundance in marine waters. Many species have been identified, and some have been shown to produce antimicrobial compounds, including isovaleric acid and 2-methylbutyric acid (1); p-hydroxybenzoic acid, trans-cinnamic acid, 6-bromoindolyl-3-acetic acid, N-hydroxybenzoisoxazolone, and 2′-deoxyadenosine (2); and 3,3′,5,5′-tetrabromo-2-2′biphenyldiol (3). Offret et al. (4) reviewed the antimicrobial metabolites produced by Pseudoalteromonas spp., listing them as alkaloids, polyketides, and peptides. Only 16 of 41 Pseudoalteromonas spp. have been shown to produce these compounds (4). About one-half of the Pseudoalteromonas spp. are pigmented (5), and components of the pigment have been reported to have antibacterial properties (3–10). Extracellular serine proteases, metalloproteases, and other less-characterized proteolytic substances are also produced by some Pseudoalteromonas spp. (8, 11–17) and are, in most cases, believed to serve as antibacterial agents or to remove proteinaceous material from biofilms (18, 19). It has been suggested that Pseudoalteromonas and other bacteria that are associated with the surface of corals produce antimicrobial compounds that may serve as a mechanism to protect corals from pathogens (20, 21), although Choudhury et al. (15) reported on a strain of Pseudoalteromonas agarivorans that was a sponge pathogen. Together, Pseudoalteromonas spp. produce a host of compounds that suppress competing microorganisms in the marine environment. These qualities have allowed Pseudoalteromonas spp. to be used probiotically to prevent disease in cultured fish and shellfish (22–25). Their beneficial effects are likely related, at least in part, to their secretion of inhibitors to competing microflora, including fish and shellfish pathogens.
In this paper, we isolated three Pseudoalteromonas strains from seawater and identified them as P. piscicida. The isolates were tested for specificity against a variety of Gram-negative human and fish pathogens, as well as Gram-positive pathogens, to compare their ability to inhibit bacteria from natural and anthropogenic sources. Scanning electron microscopy (SEM) was performed to characterize the morphologies of these strains and to identify mechanisms by which P. piscicida attacks and kills competing bacteria. Substrate specificity tests were conducted on the P. piscicida strains to identify the proteolytic enzymes potentially responsible for the inhibition of competing bacteria.
RESULTS
Isolation, phylogenetic analysis, and pigment production.
Several bacteria were isolated from seawater and produced colonies surrounded by foci of clearing on lawns of Vibrio parahaemolyticus. They were designated strains DE1-A, DE2-A, and DE2-B. Pigmentation of the colonies was also assessed, resulting in the appearance of yellow colonies for DE1-A and orange colonies for DE2-A and DE2-B after 24 h. Pigmentation became increasingly intense over 96 h.
Sequencing of ∼1,400 bp of the 16S rRNA gene was performed and showed that the three strains were closely related to each other and were homologous to P. piscicida strains in the NCBI database, with over 99% sequence identity. We constructed a phylogenetic tree using the neighbor-joining method with 30 16S rRNA gene sequences from P. piscicida strains and eight additional Pseudoalteromonas species (Fig. 1). All P. piscicida strains branched together and were highly related to one another, indicating that DE1-A, DE2-A, and DE2-B are P. piscicida isolates. Sequences from two strains of Pseudoalteromonas flavipulchra also clustered with the 16S rRNA gene sequences from P. piscicida, suggesting that these two strains may be mislabeled. The seven additional Pseudoalteromonas species examined all branched divergently from P. piscicida strains (Fig. 1).
FIG 1.
Evolutionary relationships among Pseudoalteromonas piscicida strains and related species based on 16S rRNA gene sequence. The evolutionary relationships were inferred using the neighbor-joining method. The optimal tree with the sum of branch lengths of 0.10271041 is shown. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura 2-parameter method and are in the units of the number of base substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 5). The analysis involved 30 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 1,155 positions in the final data set. Evolutionary analyses were conducted in MEGA7.
Specificity of inhibition.
Nine genera of human- and fish-pathogenic bacteria encompassing 13 species were screened for sensitivity to antimicrobial substances produced by the three P. piscicida isolates. These assays involved the inoculation of plates with Pseudoalteromonas spp. before the lawns had formed to determine if the formation of the lawns (growth of the host cells) would be inhibited by the Pseudoalteromonas. The results are shown in Table 1. Vibrios and other bacteria that are indigenous to marine waters were inhibited by at least one of the three strains of P. piscicida. The largest zones of clearing (up to 3.5 mm) were produced in lawns of Vibrio vulnificus (MLT362). Somewhat smaller zones were observed in V. vulnificus MLT364, while considerably smaller zones were observed for V. vulnificus MLT1003 (Table 1). This indicates strain-related differences in inhibition among V. vulnificus. In V. parahaemolyticus O3:K6, the bacteria used in the initial screening of the seawater from which the isolates were obtained, inhibition was minimal (ranging from 0.5- to 1.5-mm zones of inhibition). Another member of the Vibrionaceae family, Photobacterium damselae, was strongly inhibited, with zones of clearing up to 2.5 mm, depending on the strain. Among the least inhibited bacteria were the coral and shellfish pathogen Vibrio coralliilyticus and the fish pathogen Vibrio alginolyticus. Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium DT-104 were resistant to all three P. piscicida strains, whereas only one strain (DE1-A) inhibited Shigella sonnei. All three strains inhibited Staphylococcus aureus similarly, with zones of clearing not exceeding 1 mm (Table 1).
TABLE 1.
Inhibition of human and/or fish pathogens by three Pseudoalteromonas piscicida strains, DE1-A, DE2-A, and DE2-B, in triplicate assays
| Pathogen | Zone of clearing (mm) around colonies of P. piscicidaa: |
||
|---|---|---|---|
| DE1-A | DE2-A | DE2-B | |
| Marine bacteria | |||
| Aeromonas hydrophila | 0.5 | <0.5 | 1.5 |
| 0.5 | 0 | 1 | |
| 0.5 | 0 | 0 | |
| Listonella anguillarum | 0.5 | 0.5 | 0.5 |
| 0.5 | 0.5 | 0.5 | |
| 0.5 | 0.5 | 0.5 | |
| Photobacterium damselae | 0.5 | 2 | 2.5 |
| 0.5 | 2 | 2.5 | |
| 0.5 | 1 | 2 | |
| Shewanella algae | 1 | 0.5 | 0.5 |
| 1 | 0.5 | 0.5 | |
| 1 | 0.5 | 0.5 | |
| Vibrio alginolyticus | 0.5 | 0 | 0 |
| <0.5b | 0 | 0 | |
| 0 | 0 | 0 | |
| Vibrio cholerae | 1 | 1 | 1 |
| 1 | 1 | 1 | |
| 1 | 1 | 1 | |
| Vibrio coralliilyticus | <0.5 | <0.5 | <0.5 |
| <0.5 | <0.5 | <0.5 | |
| <0.5 | <0.5 | <0.5 | |
| Vibrio parahaemolyticus O3:K6 | 1 | 1.5 | 1 |
| 1 | 1.5 | 0.5 | |
| 1 | 1 | 0.5 | |
| Vibrio vulnificus MLT362 | 3.5 | 3 | 3 |
| 3.5 | 3 | 3 | |
| 3.5 | 3 | 2.5 | |
| Vibrio vulnificus MLT364 | 2.5 | 2.5 | 2 |
| 2.5 | 2 | 2 | |
| 2 | 2 | 1.5 | |
| Vibrio vulnificus MLT1003 | 1.5 | 1 | 1.5 |
| 1.5 | 1 | 1.5 | |
| 1 | 1 | 1 | |
| Anthropogenic bacteria | |||
| Escherichia coli O157:H7 | 0 | 0 | 0 |
| 0 | 0 | 0 | |
| 0 | 0 | 0 | |
| Salmonella enterica serovar Typhimurium | 0 | 0 | 0 |
| 0 | 0 | 0 | |
| 0 | 0 | 0 | |
| Shigella sonnei | 1.5 | 0 | 0 |
| 1.5 | 0 | 0 | |
| 1.5 | 0 | 0 | |
| Staphylococcus aureus | 1 | 1 | 1 |
| 1 | 1 | 1 | |
| 0.5 | 1 | 0 | |
Zones of clearing were measured around 48-h-old colonies of three P. piscicida isolates grown at 24 to 26°C on lawns of human and/or fish pathogens. The results are shown for triplicate readings and were rounded to the nearest 0.5 mm. Distances were measured from the outer portion of the P. piscicida colony to the outer periphery of the zone of clearing.
A result of <0.5 indicates that an area of clearing was observed but was too small to accurately measure.
In a second set of assays, fully grown lawns of Aeromonas hydrophila, V. alginolyticus, V. parahaemolyticus, and V. vulnificus MLT362 and MLT1003 were stabbed with the three strains of P. piscicida and incubated for 48 h. Bands of clearing formed similarly to the bands that formed when lawns had been freshly seeded. Clearing of the lawns indicates lysis of the bacteria within those lawns. Formed and immature lawns both produced zones of clearing which increased in size over time, indicating that the substances secreted by P. piscicida were not only inhibitory to the growth of bacteria within the lawns but were actually lethal to the bacteria. This lethal effect occurred without direct contact with P. piscicida cells, indicating that it was from substances secreted from the colonies.
Morphology.
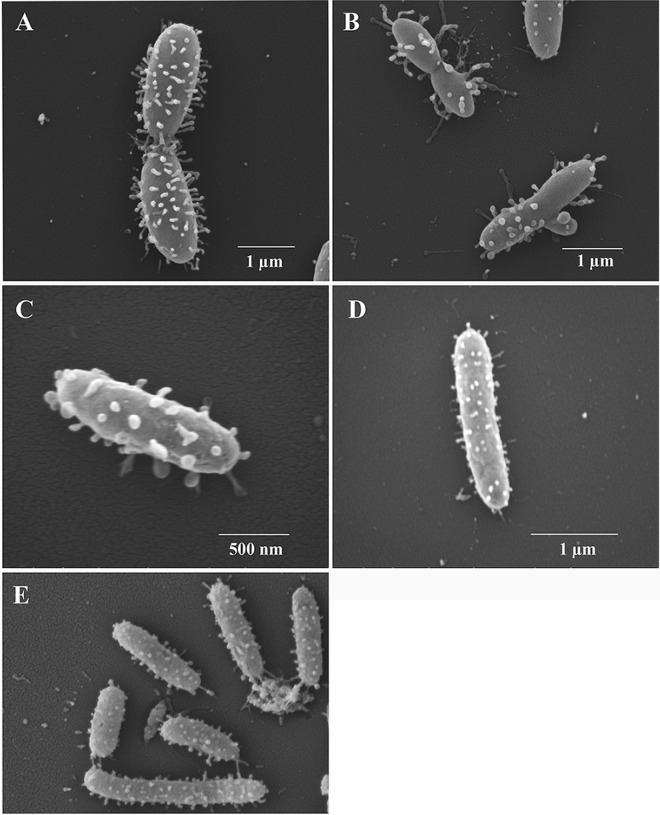
Scanning electron microscopy revealed that all three P. piscicida strains possessed surface vesicles of widely different shapes and sizes, ranging from under 50 nm to approximately 100 nm (Fig. 2). Each isolate often showed multiple vesicle morphologies, perhaps as a result of the age of the bacterium, possible induction of vesicle formation in the presence of either competing bacteria, changes in nutrients in the surrounding seawater, or combinations of those factors. For instance, isolate DE1-A displays different morphologies, with some vesicles attached to long stalks or pilus-like structures (Fig. 2A and B), which we refer to as bacterial “tethers.” Vesicles also varied in size and shape, as can be readily observed for isolate DE2-A in Fig. 2C. Isolate DE2-B was generally narrower than the other isolates (Fig. 2D). Its length also varied considerably, as shown in Fig. 2E. Overall, each strain was highly pleomorphic.
FIG 2.
Scanning electron micrographs of P. piscicida strains. (A and B) Multiple morphologies of P. piscicida strain DE1-A with notable vesicles. (C) Large flat vesicles extending outward from the surface of P. piscicida strain DE2-A. (D and E) Typical appearance of P. piscicida strain DE2-B with widely varied lengths.
Transfer of lytic vesicles.
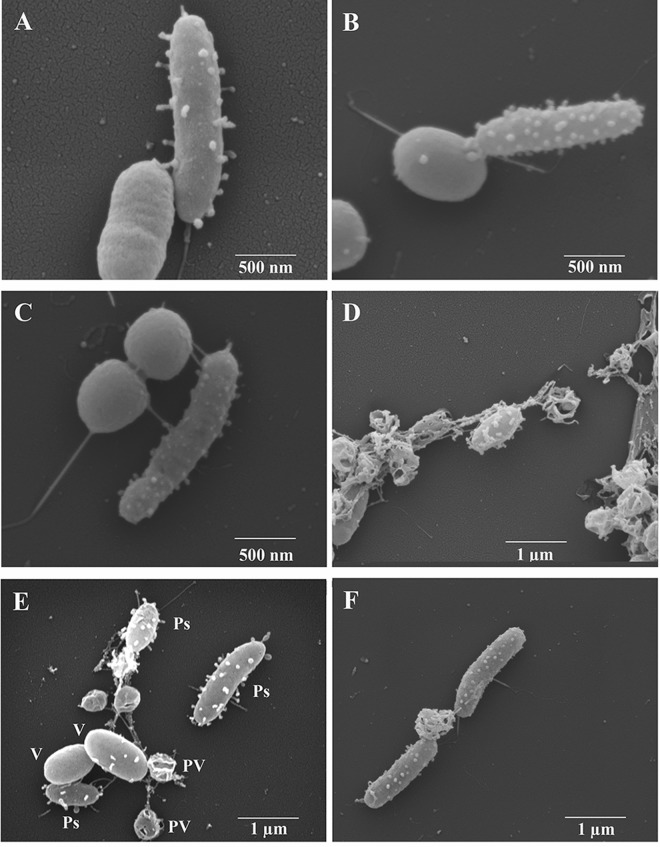
In the presence of V. parahaemolyticus O3:K6 in liquid culture (seawater), P. piscicida strains DE2-A (Fig. 3A and B) and DE1-A (Fig. 3C) appeared to transfer vesicles to the V. parahaemolyticus. In these micrographs, V. parahaemolyticus has a smooth surface prior to vesicle transfer. There appears to be some elasticity to the vesicles, as they stretched (Fig. 3C). Transferred vesicles produced characteristic holes in the V. parahaemolyticus cell walls (Fig. 3D). Note in Fig. 3D the intact V. parahaemolyticus in the center containing P. piscicida vesicles on its surface. A mixed culture containing P. piscicida DE2-B (Ps), V. parahaemolyticus (V), and permeabilized vibrios (PV) is shown in Fig. 3E. These holes indicate that the transferred vesicles contain one or more compounds capable of digesting holes through the bacterial cell walls. The appearance of bacteria containing multiple holes was seen only in cultures containing both P. piscicida and V. parahaemolyticus; thus, the appearance of such holes appears to be diagnostic for infection of V. parahaemolyticus O3:K6 by P. piscicida. In Fig. 3F, two P. piscicida bacteria may be feeding off the nutrients released by the lysed V. parahaemolyticus. This “feeding phenomenon” was commonly observed (also see Fig. 2E). Another isolate of Pseudoalteromonas, strain DE2-C, shows apparent scars where vesicles may have been pulled off the surface during transfer to the vibrios (Fig. 4). This pattern was frequently recorded in this particular strain, perhaps because the vesicles in this strain were often very large and bulbous. By measurement, the largest vesicles in this strain were approximately 125 nm in diameter (Fig. 4). This isolate was not characterized as extensively as the other three strains but was also isolated from the Delaware Bay. After viewing many micrographs of P. piscicida and its attached vesicles, it can be concluded that the vesicles are highly pleomorphic.
FIG 3.
Scanning electron micrographs of P. piscicida strains in coculture with V. parahaemolyticus O3:K6. (A to C) Apparent transfer of digestive vesicles from P. piscicida to the surfaces of V. parahaemolyticus in P. piscicida strain DE2-A (A and B) and strain DE1-A (C). (D) Vibrios containing vesicle-associated holes, except in center of micrograph, where an intact V. parahaemolyticus appears with vesicles on its surface. (E) P. piscicida (Ps), V. parahaemolyticus (V), and late-stage V. parahaemolyticus with vesicle-digested holes visible (permeabilized vibrios [PV]). (F) Two P. piscicida DE2-A bacteria suspected to be feeding (grazing) on nutrients released by a permeabilized V. parahaemolyticus.
FIG 4.

High magnification of P. piscicida strain DE2-C in a V. parahaemolyticus coculture showing large round bulbous vesicles and apparent scars on its surface, suspected to be from the transfer of vesicles to the vibrios. Scale bar signifies 500 nm.
Microscopic evaluation of zones of bacterial inhibition.
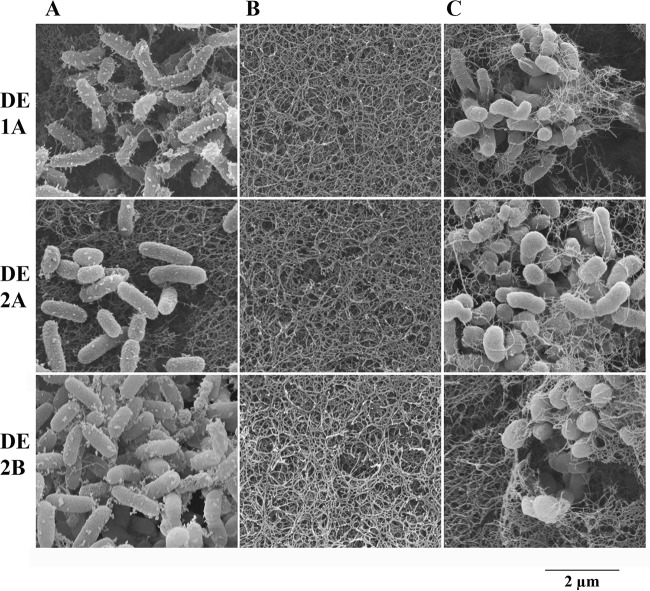
Pseudoalteromonas piscicida strains produced colonies surrounded by zones of clearing on a wide range of bacteria (Table 1). Scanning electron micrographs of agar from the outer edge of the Pseudoalteromonas colonies, when grown in the presence of V. vulnificus (Fig. 5A), showed only typical Pseudoalteromonas morphology and the general absence of vibrios. Within the zone of clearing around the colony (Fig. 5B) there was an absence of V. vulnificus and Pseudoalteromonas, indicating that the vibrios did not survive and the Pseudoalteromonas strains did not spread beyond the colony boundaries. Outside the zone of clearing, vibrios in the lawn were present at high density and did not show vesicles on their surface (Fig. 5C). Thus, the zones of clearing appear to be caused primarily by substances secreted from the Pseudoalteromonas colonies. Similar results were obtained with all Pseudoalteromonas strains examined (DE1-A, DE2-A, and DE2-B; Fig. 5). Additionally, V. parahaemolyticus gave similar results (not shown).
FIG 5.
Scanning electron micrographs of agar plugs excised from inhibition assays. Plates containing V. vulnificus in the agar were stabbed with three strains of P. piscicida and incubated at 22°C for 24 h until Pseudoalteromonas colonies and zones of clearing around the colonies became visible. The entire area (colonies, zones of clearing around the colonies, and the area just outside the clear zones) was excised, processed, and subjected to SEM. (A) The outer edges of three colonies of P. piscicida (strains DE1-A, DE2-A, and DE2-B) showing typical morphologies, including extracellular vesicles. (B) Areas within the zone of Vibrio inhibition showing the absence of bacteria. (C) Normal-appearing lawns of V. vulnificus just outside the zone of inhibition. Images taken at ×50,000 magnification; scale bar beneath images = 2.0 μm and applies to all micrographs.
Acid secretion.
The secretion of one or more diffusible compounds from colonies resulted in the formation of zones of inhibition in lawns of various host cells (Table 1). Literature suggests that these zones may be attributed to the secretion of acidic compounds (1, 2). We evaluated the possibility that changes in pH may have played a role in the formation of zones of inhibition around P. piscicida colonies grown on lawns of V. vulnificus. The results showed that the colonies, areas within the zones of inhibition, and the viable lawns of V. vulnificus host cells outside the zones of inhibition were approximately the same pH (8.1); thus, pH was not the cause of inhibition.
Enzyme activity.
Since protease secretion has been associated with some Pseudoalteromonas spp., we overlaid membranes containing six synthetic fluorogenic substrates onto colonies of the three P. piscicida strains, zones of clearing around the colonies, and the unaffected lawns of V. vulnificus to determine the possible presence of proteolytic enzymes (Table 2). Colonies from all strains of P. piscicida contained one or more enzymes which cleaved AFC-002 (l-Arg-AFC; AFC, 7-amino-4-trifluoromethylcoumarin), a substrate for aminopeptidase B, within 1 h, while strains DE2-A and DE2-B cleaved AFC-008 (l-Lys-AFC), another substrate for aminopeptidase B (Table 2). Substrate cleavage was observed only from the center of the colonies, suggesting that the older cells produced sufficient enzyme for detection. Aminopeptidase B typically cleaves arginyl and/or lysyl residues from the amino termini of proteins and peptides. Cleavage of AFC-002 was observed in the zone of clearing around strain DE2-A after 1 h, while the lawn of V. vulnificus did not show any comparable enzyme activity. In contrast, no enzyme activity was detected for any of the strains in the zones of inhibition using AFC-008 as the substrate. It should be noted that the lawns of V. vulnificus produced very strong lysyl-aminopeptidase activity toward AFC-008.
TABLE 2.
Enzyme activities of three P. piscicida isolates against six synthetic fluorogenic substrates 48 h postinoculation onto plates containing V. vulnificusa
| Fluorogenic substrateb | Substrate detects: | Enzyme activity from colony or zone of inhibition | Enzyme activity (15 min/1 h) from: |
|||
|---|---|---|---|---|---|---|
|
Pseudoalteromonas piscicida strains |
Lawns of Vibrio vulnificus | |||||
| DE1-A | DE2-A | DE2-B | ||||
| AFC-002 (l-Arg-AFC) | Aminopeptidase B | Colony | −/+ | −/+ | −/+ | −/− |
| Zone | −/− | −/+ | −/− | |||
| AFC-008 (l-Lys-AFC) | Aminopeptidase B | Colony | −/− | −/+ | −/+ | +/+ |
| Zone | −/− | −/− | −/− | |||
| AFC-059 (d-Pro-Phe-Arg-AFC) | Serine protease (trypsin-like) | Colony | +/+ | +/+ | −/+ | +/+ |
| Zone | −/− | +/+ | +/+ | |||
| AFC-094 (methoxysuccinyl-Ala-Ala-Pro-Val-AFC) | Serine protease (neutrophil elastase) | Colony | −/− | −/− | −/− | −/− |
| Zone | −/− | −/− | −/− | |||
| AFC-096 (methoxysuccinyl-Ala-Ala-Pro-Met-AFC) | Serine protease (chymotrypsin-like, cathepsin G-like) | Colony | +/+ | +/+ | −/+ | −/− |
| Zone | −/− | −/+ | −/+ | |||
| AFC-120 (N-acetyl-Tyr-Val-Ala-Asp-AFC) | Cysteine protease (caspase 1-like) | Colony | +/+ | +/+ | +/+ | −/− |
| Zone | +/+ | +/+ | +/+ | |||
Cellulose acetate membranes, containing substrates with different peptidyl side groups linked to fluorogenic 7-amino-4-trifluoromethylcoumarin (AFC) as the leaving group, were overlaid onto P. piscicida colonies, zones of clearing around the colonies, and lawns of healthy V. vulnificus cells outside the areas of inhibition. Fluorescence produced by (i) enzymatic activity from the colonies, (ii) the clear zones of Vibrio inhibition around the colonies, and (iii) lawns of Vibrio outside the zones of inhibition were recorded as positive (+) or negative (−) fluorescence after 15 min and 1 h.
Shown as the catalog number (Enzyme Systems Products, Livermore, CA), followed by the name of the substrate.
The centers of two of the three colonies of P. piscicida reacted strongly to AFC-059 (d-Pro-Phe-Arg-AFC) within 15 min, indicating the presence of a colony-associated serine protease in strains DE1-A and DE2-A and a weak proteolytic reactivity in DE2-B (Table 2). AFC-059 is an excellent substrate for trypsin and kallikrein-like proteases, since they typically cleave on the carboxyl side of arginyl and lysyl residues. The zone of clearing for DE1-A was negative even though the colony was positive for serine protease activity. In contrast, the zone of clearing was positive for strains DE2-A and DE2-B. Overall, these results show apparent differences in the production and secretion of serine protease(s), suggesting there could be multiple serine proteases produced by these strains. Lawns of V. vulnificus showed strong activity toward AFC-059. Another substrate, AFC-094 (methoxysuccinyl [MeoSuc]-Ala-Ala-Pro-Val-AFC), was not cleaved by enzymes in any of the Pseudoalteromonas strains, either by the colonies or within the zones of clearing (Table 2).
Colonies of DE1-A and DE2-A showed proteolytic activity toward the cathepsin substrate AFC-096 (MeoSuc-Ala-Ala-Pro-Met-AFC) within 15 min, while DE2-B was positive after 1 h (Table 2). Activity was not localized to the center of the colony but was diffuse over the entire colony. Only DE2-B displayed activity within the zone of clearing after membrane overlay for 1 h (Table 2).
Very strong activity was observed within 15 min from the colonies and from secreted enzymes within the zones of inhibition against the substrate AFC120 (AC-Tyr-Val-Ala-Asp-AFC), which is a substrate used to detect cysteine and aspartic acid proteases with high specificity for cleaving on the carboxyl side of aspartic acid (Table 2). The entire colony of each strain expressed strong activity, and a weaker activity was evident over the zones of clearing. The lawns of V. vulnificus did not show any signs of activity. Substrate AFC-120 is a substrate that has been marketed for the detection of caspases 1 and 4. None of the above-mentioned enzyme activities were inhibited by 10 mM EDTA, suggesting that they are not metalloproteases.
A photograph of DE2-A colonies and zones of clearing around the colonies is shown in Fig. 6A. Figure 6B shows the fluorescence produced from the various substrates after the overlay of P. piscicida DE2-A colonies, as well as the surrounding areas of clearing and the outer lawns of V. vulnificus using five different substrates (Fig. 6B). Substrate AFC-094 was omitted from this figure, since it was not cleaved by any of the P. piscicida-produced enzymes. Examples of fluorescence in the center of the colonies can be seen for substrates AFC-002, AFC-008, and AFC-059, as mentioned above. The outer lawns of V. vulnificus produced enzymes that cleaved only AFC-008 and AFC-059 from among the six substrates tested (Fig. 6B). It is well known from our previous work that membranes containing AFC-008 produce fluorescence when overlaid onto Vibrionaceae family members, and this has led to the development of a membrane-based assay for the quantification of total Vibrionaceae (26, 27). It should be noted that there is a halo of fluorescence around the colony of P. piscicida DE2-A when overlaid with AFC-059 (Fig. 6B). This fluorescence did not reach the outer levels of the zone of clearing, which remained dark blue (no enzyme activity), suggesting limited diffusion of this serine protease.
FIG 6.

Colonies and fluorogenic assays for enzyme activity. (A) Photographs of colonies of P. piscicida strain DE2-A, zones of clearing around the colonies, and surrounding healthy lawns of V. vulnificus strain MLT362. (B) Same areas as in panel A were overlaid with cellulose acetate membranes containing synthetic fluorogenic (7-amino-4-trifluoromethylcoumarin-linked [AFC]) substrates and photographed under UV illumination at 364 nm to identify proteolytic activity (light areas). Representative results are shown using substrates AFC-002 (l-Arg-AFC), AFC-008 (l-Lys-AFC), AFC-059 (d-Pro-Phe-Arg-AFC), AFC-096 (MeoSuc-Ala-Ala-Pro-Met-AFC), and AFC-120 (N-acetyl-Tyr-Val-Ala-Asp-AFC).
Bioinformatics analysis of available P. piscicida genome sequences for the presence of proteolytic enzymes.
In an effort to determine whether P. piscicida has the genetic components required for the enzymatic potential described above, we examined available genome sequences. In the NCBI genome database, four genome sequences are available for P. piscicida, of which P. piscicida ATCC 15057 (accession number NZ_ARMY00000000.1) is the most complete. There were 97 peptidases annotated within the 5.3-Mb genome sequence of P. piscicida ATCC 15057 and JCM 20779, 13 of which were annotated as aminopeptidases. To determine the number of extracellular peptidases, the 97 peptidases were examined for the presence of a signal peptide using SignalP 4.1 (28). Of these, 47 peptidases showed the presence of a signal sequence, with three peptidases containing a single transmembrane domain. There were 36 proteases annotated, of which seven were metalloproteases and nine were serine proteases. Thirteen of these proteases contained a signal peptide sequence, indicating they are likely extracellular proteins. The two additional P. piscicida genome sequences in the NCBI genome database had peptidases similar to those of ATCC 15057.
We also searched the P. piscicida ATCC 15057 genome for putative caspase 1-like proteins using the Pfam database (http://pfam.xfam.org/). This database contains a large collection of protein families represented by multiple sequence alignments. As of January 2017, there were 516 prokaryotic species that contained 1,416 caspase sequence homologs (protein family PF00656, peptidase C14). By using BLAST analysis, we examined all 1,416 sequences to identify possible homologs within P. piscicida but found no matches. Since our study and others have shown that there is significant variation among strains and species in terms of enzymatic activity, we broadened our search to examine all 42 Pseudoalteromonas representatives in the NCBI genome database. From this analysis, we discovered several caspase homologs among several Pseudoalteromonas species. Caspase domain proteins were identified in Pseudoalteromonas sp. strain P1-9 (accession no. LKBD01000006.1), Pseudoalteromonas prydzensis (accession no. WP_064665900), Pseudoalteromonas haloplanktis (accession no. ADOP01000022.1), Pseudoalteromonas elyakovii ATCC 700519 (accession no. JWIH01000017.1), and Pseudoalteromonas luteoviolacea DSM 6061 (accession no. AUYB01000147.1). By using the cathepsin G preproprotein from Mus musculus (accession no. NP_031826), we identified numerous serine protease homologs among Pseudoalteromonas spp. but not against P. piscicida.
DISCUSSION
The presence of various proteolytic enzymes in P. piscicida strains suggests a potential functional role in the inhibition of competing bacteria. All three strains of P. piscicida showed the strong presence of cysteine protease activity similar to that of caspase 1 from the colonies and within the zones of clearing. No comparable activity was observed in the lawns of V. vulnificus. In S. Typhimurium, caspase 1 has been shown to be involved in the formation of pores between 1.1 and 2.4 nm in diameter within the membrane of macrophages of infected host cells (29). Such digestive properties may help explain how a similar enzyme of the three P. piscicida strains may have contributed to the digestion of holes in the cell walls of V. parahaemolyticus. This enzyme is also a likely candidate for causing the formation of zones of inhibition in lawns of competing bacteria (Table 1).
The presence of trypsin-like serine protease activity associated with all three P. piscicida strains may have further contributed to vesicle-associated digestion of the cell walls of V. parahaemolyticus. Another serine protease, which hydrolyzed a substrate that is typically used for the detection of cathepsin G, was detected in the three P. piscicida strains. In humans, cathepsin G is a neutrophil lysosome-mediated enzyme with broad inhibitory activities toward Gram-positive and Gram-negative bacteria (30). It appears that our P. piscicida strains contain a homolog of cathepsin G, which might contribute to the inactivation of a broad range of bacteria. Inactivation may be through direct proteolytic processing of the bacterial cell walls to form holes, as observed in V. parahaemolyticus (Fig. 3D to F), or by the processing of proenzymes to activate them or make peptidases more easily secreted and diffusible. Either of the two aminopeptidase B-like proteases detected may also contribute toward zymogen activation or the processing of enzymes to make them more easily secreted and diffusible. Bioinformatic analyses demonstrated the presence of multiple aminopeptidases and serine proteases in P. piscicida, providing further evidence for our findings.
To date, methods have not been developed to isolate the vesicles from the surface of Pseudoalteromonas, due in part to their irregular sizes, apparent stickiness, and differences in their attachment to the P. piscicida strains. In fact, the structures by which the vesicles are tethered to the P. piscicida appear to be quite different (Fig. 2 and 4). Clearly, more work is needed to isolate and further characterize the enzymes detected in this study, especially if compounds of potential pharmacological value are to be identified.
This study demonstrated that secreted “inhibitors” are more than just inhibitory but are lethal to other bacteria, since they cleared (lysed) established lawns of vibrios and other bacterial genera. Inhibition assays demonstrated highly varied results, depending on the host bacterium and the strain of P. piscicida used. Marine bacteria and those that might be considered anthropogenic were inhibited by one or more P. piscicida strains. Staphylococcus aureus was the only Gram-positive bacterium tested for inhibition and was minimally inhibited by all three strains. From an evolutionary perspective, it would be expected that marine bacteria, like P. piscicida, would evolve to more effectively develop mechanisms to compete with organisms found in their native habitat; thus, it is a bit surprising that Shigella sonnei was inhibited to the degree it was by P. piscicida DE1-A (Table 1).
The second method to eliminate competing bacteria (e.g., V. parahaemolyticus) is through contact inhibition via lytic vesicles. Interestingly, electron micrographs showed the transfer of lytic vesicles (Fig. 3A to C) and the formation of vesicle-associated holes in the cell walls (Fig. 3D to F) of only V. parahaemolyticus; no such transfer of vesicles or formation of holes in the cell walls were observed in micrographs of the three P. piscicida strains in V. vulnificus, V. alginolyticus, Shewanella algae, or E. coli. It is unclear whether the transfer of lytic vesicles is receptor mediated, which could explain the differences between the transfer of vesicles to V. parahaemolyticus and the apparent failure to transfer vesicles to the other bacteria. After viewing hundreds of electron micrographs of P. piscicida in coculture with V. parahaemolyticus, it appears that the Pseudoalteromonas strains congregate around the permeabilized vibrios in a predatory fashion, where they may assimilate nutrients given off by their prey. We conclude that P. piscicida should be considered a predator of V. parahaemolyticus by virtue of its ability to identify target vibrios, perhaps through some receptor-mediated process; transfer lytic vesicles to the outer surface of the vibrios; digest holes in the Vibrio cell walls; congregate around the dying vibrios; and assimilate nutrients released by the vibrios in a true grazing manner. If we consider the modulation of marine bacteria by Pseudoalteromonas, by other marine predatory bacteria, like Halobacteriovorax spp., and by bacteriophages, we can begin to recognize the complexities of bacterial survival and death in the marine environment. Further studies are needed to explore vesicular transfer to determine if it is temperature, salinity, or nutrient dependent in V. parahaemolyticus and whether it occurs in other serotypes or species.
In the marine environment, which has currents and tides to help flush and dilute away secreted inhibitors, direct contact of P. piscicida with competing bacteria may be a more efficient approach for Pseudoalteromonas spp. to attack their prey, particularly in and around biofilms. Both secretion and lytic vesicles may take part in the modulation or destruction of pathogenic bacteria in biofilms where Pseudoalteromonas and other bacteria may remain in close proximity. Although some species of Pseudoalteromonas are widely reported to form biofilms, others have been reported to degrade and control biofilms (18, 19). We speculate that direct contact with lytic vesicles and the secretion of antimicrobial compounds act synergistically to reduce competing microbes and proteinaceous materials within biofilms. Proteinaceous materials may be highly susceptible to the proteolytic activities of the trypsin-like and chymotrypsin-like serine proteases and the cysteine protease of P. piscicida, thus giving P. piscicida a selective advantage in biofilm degradation.
We showed the presence of many vesicles of various sizes and shapes on the surfaces of all three P. piscicida strains, often connected to stalk-like tethers. The vesicles and tethers appear to be a natural part of these P. piscicida strains and are different from curli structures or flexible pili reported previously in other species of Pseudoalteromonas (31, 32). Likewise, these tethers are different from the stalks of stalked bacteria, like Caulobacter spp., or to the reproductive stalks that form during bacterial budding. The tethers of P. piscicida appear to have a unique role in the attachment and release of lytic vesicles to V. parahaemolyticus and perhaps to other organisms or to environmental surfaces, including biofilms.
There have been many reviews on the roles of bacterial outer membrane vesicles (OMVs) in regard to gene and protein transfer to host cells, the transfer of virulence factors, promotion of host cell destruction, cellular communications, elicitation of immune responses in host cells, etc. (33–38). Unlike typical OMVs, the vesicles of P. piscicida did not appear to be secreted and may not be considered extracellular, since they are tethered to the bacterium. The sizes of OMVs are generally listed as being between 10 and 300 nm in diameter (36, 37). Nevot et al. (39) identified small OMVs from Pseudoalteromonas antarctica that were only 25 to 70 nm in diameter. The largest vesicles in the current study were approximately 125 nm in diameter (Fig. 4). It is not certain if some of the vesicles from P. piscicida are spontaneously released or only transferred to competing bacteria by direct contact. The transfer mechanism is suspected to predominate, since vesicle-like structures within the extracellular milieu were not readily observed on micrographs. Shewanella oneidensis strain MR-1 also produces OMVs, but they appear uniform in size and are attached to the outer membrane by thin membranous structures referred to as nanowires, which conduct electrical impulses (40). The tethers and associated vesicles of P. piscicida appear to be highly pleomorphic, even within the same P. piscicida isolate (Fig. 2B). The full function of these appendages remains to be determined.
In conclusion, it appears that P. piscicida has the ability to target the destruction of natural competitors by two methods: the release of antimicrobial compounds and the direct transfer of lytic vesicles to competitors. Potential antimicrobial compounds identified in this study include several proteolytic enzymes. Further studies are needed to (i) characterize the digestive compounds that are secreted from P. piscicida and related Pseudoalteromonas spp., as well as those that are associated with lytic vesicles, with the goal of identifying new pharmacological compounds of potential therapeutic benefit; (ii) determine if the vesicles and tethers serve any other biological or mechanical functions; (iii) ascertain which bacteria can serve as hosts for this vesicular transfer or if different strains of P. piscicida exhibit different host specificities; (iv) determine if vesicular transfer is receptor mediated; (v) further investigate the predatory nature of P. piscicida and its modulation of overall bacterial levels in the marine environment; and (vi) identify potential probiotic uses for P. piscicida in aquaculture, seafood processing, and the reduction of biofilms.
MATERIALS AND METHODS
Bacteria.
Pseudoalteromonas isolates were obtained from seawater tested on lawns of V. parahaemolyticus O3:K6 (strain RIMD2210633), as described below. RIMD2210633 is a clinical pandemic strain originally obtained from an outbreak in Japan (41). The ability of the Pseudoalteromonas isolates to inhibit Gram-positive and Gram-negative bacteria was determined using the bacteria listed in Table 3.
TABLE 3.
Bacteria used in this study
| Bacterium | Designationa | Source or referenceb |
|---|---|---|
| Marine bacteria | ||
| Aeromonas hydrophila | K144 | USDA |
| Listonella (Vibrio) anguillarum | ATCC 43305 | ATCC |
| Photobacterium damselae | HSOA-9 | 50 |
| Shewanella algae | ATCC 51192 | ATCC |
| V. alginolyticus | ATCC 17749 | ATCC |
| V. cholerae O139 | CASHD | ICDDR-B |
| V. coralliilyticus | RE98 | 51 |
| V. parahaemolyticus O3:K6 | RIMD2210633, tdh+ | 41 |
| V. vulnificus | MLT362, biotype 1 | 52 |
| V. vulnificus | MLT364, biotype 1 | 52 |
| V. vulnificus | MLT1003, biotype 1 | 52 |
| Anthropogenic bacteria | ||
| Escherichia coli O157:H7 | ATCC 43889 | ATCC |
| Salmonella enterica serovar Typhimurium | DT104 | USDA |
| Shigella sonnei | BS514 | USUHS |
| Staphylococcus aureus 196E | ATCC 13565 | ATCC |
RIMD2210633, clinical isolate originally from Japan; tdh+, contains the thermostable direct hemolysin gene.
USDA, U.S. Department of Agriculture, Agricultural Research Service, Wyndmoor, PA; ATCC, American Type Culture Collection, Manassas, VA; ICDDR-B, International Centre for Diarrheal Disease Research-Bangladesh, Dhaka, Bangladesh; USUHS, Uniformed Services University of the Health Sciences, Bethesda, MD.
Isolation of inhibitory bacteria from seawater.
Seawater samples from three sites along the Delaware Bay (42, 43) were screened for bacteria that showed inhibitory properties toward V. parahaemolyticus. Screening was performed by use of a plaque assay method originally designed to detect Vibrio predatory bacteria, namely, Bdellovibrio and like organisms (BALOs), in seawater (42). In essence, 25 ml of bottom agar was added to 100-mm-diameter petri dishes and allowed to harden. The bottom agar consisted of Pp20 agar, made as follows: 1 g/liter Pp20 medium and 15 g/liter Bacto agar (both from Becton, Dickinson and Co., Sparks, MD), and, in place of distilled water, the medium was made with 30 ppt salinity seawater (seawater that had been autoclaved and passed through a 0.22-μm-pore filter prior to storage and use). Seawater for the medium was obtained at high tide off the dock of the Cape May-Lewes Ferry terminal in Lewes, DE, near the mouth of the Delaware Bay. A top agar (upper layer of agar) was prepared and maintained in a liquid state by combining 1 g/liter Pp20 medium and 7.5 g/liter Bacto agar made with seawater. Top agar was autoclaved and, while still hot, 7.5 ml was pipetted into a series of sterile 15-mm screw-cap glass culture tubes. The tubes were maintained in a water bath at 48°C. Vibrio parahaemolyticus O3:K6 was grown at 26°C and 250 rpm overnight in Luria-Bertani broth (Becton, Dickinson and Co.) made with sterile seawater that had been diluted to 20 ppt salinity to give a final salinity of 30 ppt (LB–3% broth). The following day, the enrichment was subcultured by reinoculation into fresh LB–3% broth until the optical density at 600 nm (OD600) reached ∼0.20, so that the culture was in exponential-growth phase. For each plaque assay isolation for Pseudoalteromonas in seawater, 7.5 ml of test seawater was added to a tube of warm top agar, along with 1 ml of the host Vibrio (at OD600, ∼0.20), and the tube was capped and gently inverted 3 times to mix. The mixture was overlaid onto a plate of bottom agar and allowed to harden with the lid partially opened. After solidification, the plates were inverted and incubated at 26°C. Since the top agar overlay was very soft, care was exercised to allow ample time (30 to 45 min) for the agar to solidify, and the plates were inverted gently. This procedure allowed the formation of lawns of V. parahaemolyticus and plaques or areas of clearing around visible colonies of presumptive Pseudoalteromonas spp. within 24 h.
Presumptive Pseudoalteromonas colonies were picked with a Pasteur pipette from the center of the colony to 15 μl of sterile seawater, which was streaked on Luria-Bertani agar made with 20 ppt seawater to give a final salinity of 3% (LB–3% agar). Isolated colonies were picked successively 3 to 4 times to obtain a pure culture. Purity was verified by plating for contaminating vibrios on Difco thiosulfate-citrate-bile salts-sucrose (TCBS) agar (Becton, Dickinson and Co.) with incubation overnight at 26°C. Only purified isolates without contaminating vibrios (e.g., plates that failed to produce colonies on TCBS) were evaluated. Pigment production by the colonies was also evaluated on LB–3% agar after 26°C incubation for 24 to 96 h.
Phylogenetic and bioinformatic analyses.
Purified isolates were grown in pure culture on LB–3% agar, and picks were submitted to Genewiz, Inc. (South Plainfield, NJ) for 16S rRNA gene sequencing using proprietary universal 16S rRNA primers. The 16S rRNA gene sequences from P. piscicida strains DE1-A, DE2-A, and DE2-B were compared to 16S rRNA gene sequences in the NCBI databases for P. piscicida and related species with the following NCBI accession numbers: NR_040946.1, KP757027.1, FJ457149.1, FJ457171.1, KF880965.1, KY382776.1, AHCC02000078.1, NR_119147.1, FJ457174.1, NR_025126.1, KC534410.1, HQ439521.1, AB681560.1, FJ457184.1, HQ439549.1, HQ439541.1, FJ457234.1, JQ250821.1, JQ250823.1, KT443875.1, NR_113299.1, KY073271.1, FR750950.1, KC534415.1, EU880522.2, NR_044837.2, and AB681915.1. The nucleotide sequences were aligned using CLUSTAL W using the program MEGA 7.0 (44). To infer evolutionary relationships, a phylogenetic tree was constructed using the neighbor-joining method using MEGA 7.0 (44, 45). The evolutionary distances were computed using the Kimura 2-parameter method and are in units of the number of nucleotide substitutions per site (46). The percentage of time the taxa clustered together, a test of robustness of branching patterns, was determined by the bootstrap replicate test (1,000 replicates) (47).
Four genome sequences of P. piscicida are present in the NCBI genome database. These four P. piscicida strains, recovered from the marine environment, were examined for the presence of peptidases, extracellular peptidases, and hydrolases. Sequence-based prediction of secreted peptidases was determined using SignalP 4.1 to identify putative signal peptides to differentiate peptidases and extracellular peptidases. All peptidases with signal sequences were then examined for transmembrane domains using the TMHMM server 2.0 to predict transmembrane helices (28, 48). The four genomes of P. piscicida examined were for strain ATCC 15057 (accession no. NZ_ARMY00000000.1), which has a genome size of 5.3 Mb with 4,467 annotated proteins; strain JCM 20779 (NZ_AHCC00000000.1), which has a genome size of 5.3 Mb with 4,437 annotated proteins; strain S2040 (NZ_JXXW00000000.1), which has a genome size of 5.0 Mb with 4,137 proteins; and strain S2724 (NZ_JXXX00000000.1), which has a genome size of 5.2 Mb with 4,395 annotated proteins. Caspase 1-like proteins were investigated using the Pfam database (http://pfam.xfam.org/) (49). BLASTn analysis was used to interrogate all sequenced Pseudoalteromonas spp. in the NCBI genome database for the presence of caspase 1-like proteins.
Specificity of inhibition.
The specificities of the Pseudoalteromonas strains to inhibit the naturally occurring marine bacteria Aeromonas hydrophila, Listonella anguillarum, Photobacterium damselae, Shewanella algae, V. alginolyticus, Vibrio cholerae, V. coralliilyticus, V. parahaemolyticus, and three strains of V. vulnificus were determined as follows. The host cells were grown to an OD600 of ∼0.20 in LB–3% broth at 26°C. Both the bottom and top agars were prepared as described above. The bottom agar was added to 100-mm-diameter petri dishes and allowed to solidify. Then, 1 ml of host cells was added to a tube of top agar, the tube was inverted to mix, and the agar was poured over the bottom layer. After solidification, each Pseudoalteromonas strain was picked from colonies grown on LB–3% agar plates, stabbed into the top agar in triplicate, and incubated for 48 h at 24 to 26°C. Assays for determining the inhibition caused by Pseudoalteromonas isolates on E. coli O157:H7, Salmonella enterica serovar Typhimurium, Shigella sonnei, and Staphylococcus aureus were similar, except the salt in the LB agar was reduced to 1.5% and the Pp20 medium was prepared with diluted seawater, such that the final salinity was 1.5%. This reduction in salinity was required for some of these nonmarine pathogens, which formed poor lawns at the higher (3%) salinity level. Under these conditions, bacterial lawns formed within 24 h. Zones of clearing were measured from the outer edge of each Pseudoalteromonas colony to the outer edge of the clear zone at ×1.5 magnification on a Quebec colony counter and recorded at 48 h postinoculation. To determine if Pseudoalteromonas spp. also formed zones of clearing on plates with established lawns, the lawns were poured the day before inoculation, and the surfaces of the lawns were dotted with Pseudoalteromonas spp. the following day. These plates were incubated at 24 to 26°C for 48 h, and the zones of inhibition around the colonies were measured as described above.
Acid production by Pseudoalteromonas strains.
To determine whether the P. piscicida strains secrete acidic compounds responsible for the zones of clearing around colonies grown on lawns of V. vulnificus, strains were dotted onto V. vulnificus plates, as described above (see “Specificity of inhibition,” above). After incubation at 26°C for 72 h, zones of clearing in the V. vulnificus lawns surrounding the Pseudoalteromonas colonies were tested with nonbleeding MColorpHast pH strips (pH, 6.5 to 10; EMD Millipore Corp., Billerica, MA). These colorimetric strips measure pH in 0.2 unit increments. Pseudoalteromonas piscicida colonies and lawns of V. vulnificus were also tested to compare pHs. Color development on the pH strips was recorded after 1 min.
Screening for proteolytic enzymes using fluorogenic substrates.
Petri dishes were inoculated with V. vulnificus MLT362 and dotted with the three P. piscicida strains (as described under “Specificity of inhibition,” above). Synthetic fluorogenic substrates were used to ascertain the proteolytic activity associated with the surfaces of the colonies, the zones of Vibrio inhibition around the colonies, and the unaffected lawns of vibrios. Synthetic substrates contained specific peptides covalently linked to 7-amino-4-trifluoromethylcoumarin (AFC) as the leaving group. The substrates used are listed in Table 2 and were obtained from Enzyme Systems Products, Livermore, CA. The AFC is naturally fluorescent, but its fluorescence is quenched by the peptidyl side groups; thus, only after enzymatic cleavage of the entire side group does the AFC regain its fluorescence. Substrates were dissolved in dimethyl sulfoxide and diluted to 20 mM in distilled water (dH2O). Cellulose acetate membranes were soaked in the diluted substrate for 15 s and then air-dried and stored in the dark for up to 1 week. On the day of use, membranes were cut into 1.5- to 2-cm squares, dipped for 2 s in dH2O, and overlaid onto 48-h-old colonies of the three strains of Pseudoalteromonas plus the zone of clearing around the colonies and onto the vibrios in the lawn outside the zone of clearing. The overlays were performed in triplicate. After 15 min and 1 h, the membranes were evaluated for enzyme activity by monitoring for fluorescence under a UV lamp at 364 nm. Membranes were then removed with forceps and allowed to dry under a hood, and both sides of the membranes were reevaluated under UV illumination. This process allowed enzyme activity, which was associated with the surface of the colonies, secreted enzyme in the zones of inhibition around the colonies and from the lawns of vibrios, to be determined. Fluorescence signaled the successful cleavage of the peptidyl side chain from the AFC, providing an indication of the family of enzyme present, based on the enzyme's specificity for cleavage.
To help characterize enzymes as possible metalloproteases, assays showing positive proteolytic activity were repeated, where substrate-containing membranes were wet in dH2O containing 10 mM EDTA immediately prior to overlay of the colonies, zones of inhibition around the colonies, and the healthy lawns of vibrios. The abolition of fluorescence would be an indication that the enzyme in question is a metalloprotease.
SEM.
Pseudoalteromonas strains were inoculated into sterile seawater containing V. parahaemolyticus, V. vulnificus, V. alginolyticus, Shewanella algae, and E. coli O157:H7. Cultures were grown for up to 5 h at room temperature (∼22°C), placed on coverslips for 1 h, glutaraldehyde fixed, and prepared for SEM, as previously described (42). Controls without P. piscicida were prepared similarly and viewed for their characteristic morphologies. Mixed cultures were viewed and observed for interactions between the bacteria and to identify any significant morphologies indicative of bacterial cell injury. In addition to liquid cultures, 48-h-old Pseudoalteromonas colonies surrounded by zones of clearing on lawns of V. vulnificus and V. parahaemolyticus were excised with a scalpel to capture the colony, the surrounding zone of clearing, and the lawn of host vibrios just outside the zone of clearing. The excised agar was processed for SEM as follows: the agar slices were placed in individual wells of 12-well plates and fixed by covering with 200 μl of 2.5% glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA) for 30 min. Wells were drained, the slices were rinsed twice for 30 min each with ∼2.5 ml of 0.1 M imidazole buffer (pH 7.0; Electron Microscopy Sciences) and dehydrated for 30-min intervals each in ∼2.5 ml of 50, 80, and 90% ethanol and 3 times for 30 min each time in ∼2.5 ml of 100% ethanol. The gel slices were stacked in a wire basket, separated by cloth, and placed in a critical point drying apparatus (Denton Vacuum, Inc., Cherry Hill, NJ), where they were dried for approximately 20 min using liquid carbon dioxide (Welco Co., Allentown, PA). The samples were mounted on stubs and sputter gold coated (EMS 150R ES; Electron Microscopy Sciences) for 1 min. Samples were then viewed with an FEI Quanta 200FEG scanning electron microscope (Hillsboro, OR) at an accelerating voltage of 10 kV in high-vacuum mode.
ACKNOWLEDGMENTS
We thank Douglas Soroka and Guoping Bao for assistance with electron microscopy.
This work was supported by in-house funding from the USDA, ARS under CRIS project 8072-42000-081-00D.
The use of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture (USDA).
REFERENCES
- 1.Hayashida-Soiza G, Uchida A, Mori N, Kuwahara Y, Ishida Y. 2008. Purification and characterization of antibacterial substances produced by a marine bacterium Pseudoalteromonas haloplanktis strain. J Appl Microbiol 105:1672–1677. doi: 10.1111/j.1365-2672.2008.03878.x. [DOI] [PubMed] [Google Scholar]
- 2.Yu M, Wang J, Tang K, Shi X, Wang S, Zhu W-M, Zhang X-H. 2012. Purification and characterization of antibacterial compounds of Pseudoalteromonas flavipulchra JG1. Microbiology 158:835–842. doi: 10.1099/mic.0.055970-0. [DOI] [PubMed] [Google Scholar]
- 3.Isnansetyo A, Kamei Y. 2003. MC21-A, a bactericidal antibiotic produced by a new marine bacterium, Pseudoalteromonas phenolica sp. nov. O-BC30(T), against methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 47:480–488. doi: 10.1128/AAC.47.2.480-488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Offret C, Desriac F, Le Chevalier P, Mounier J, Jégou C, Fleury Y. 2016. Spotlight on antimicrobial metabolites from the marine bacteria Pseudoalteromonas: chemodiversity and ecological significance. Mar Drugs 14:129. doi: 10.3390/md14070129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vynne NG, Må M, Nielsen KF, Gram L. 2011. Bioactivity, chemical profiling, and 16S rRNA-based phylogeny of Pseudoalteromonas strains collected on a global research cruise. Mar Biotechnol 13:1062–1073. doi: 10.1007/s10126-011-9369-4. [DOI] [PubMed] [Google Scholar]
- 6.Gauthier MJ. 1976. Morphological, physiological, and biochemical characteristics of some violet-pigmented bacteria isolated from seawater. Can J Microbiol 22:138–149. doi: 10.1139/m76-019. [DOI] [PubMed] [Google Scholar]
- 7.Gauthier MJ, Flatau GN. 1976. Antibacterial activity of marine violet-pigmented Alteromonas with special reference to the production of brominated compounds. Can J Microbiol 22:1612–1619. doi: 10.1139/m76-237. [DOI] [PubMed] [Google Scholar]
- 8.McCarthy SA, Johnson RM, Kakimoto D. 1994. Characterization of an antibiotic produced by Alteromonas luteoviolacea Gauthier 1982, 85 isolated from Kinko Bay, Japan. J Appl Bacteriol 77:426–432. doi: 10.1111/j.1365-2672.1994.tb03445.x. [DOI] [PubMed] [Google Scholar]
- 9.Isnansetyo A, Kamei Y. 2003. Pseudoalteromonas phenolica sp. nov., a novel marine bacterium that produces phenolic anti-methicillin-resistant Staphylococcus aureus substances. Int J Syst Evol Microbiol 53:583–588. [DOI] [PubMed] [Google Scholar]
- 10.Gram L, Melchiorsen J, Bruhn JB. 2010. Antibacterial activity of marine culturable bacteria collected from a global sampling of ocean surface waters and surface swabs of marine organisms. Mar Biotechnol (NY) 12:439–451. doi: 10.1007/s10126-009-9233-y. [DOI] [PubMed] [Google Scholar]
- 11.Huston AL, Deming JW. 2002. Relationships between microbial extracellular enzymatic activity and suspended and sinking particulate organic matter: seasonal transformations in the North Water Deep-Sea Res Pt II 49:5211–5225. doi: 10.1016/S0967-0645(02)00186-8. [DOI] [Google Scholar]
- 12.Lijima S, Washio K, Okahara R, Morikawa M. 2009. Biofilm formation and proteolytic activities of Pseudoalteromonas bacteria that were isolated from fish farm sediments. Microb Biotechnol 2:361–369. doi: 10.1111/j.1751-7915.2009.00097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou MY, Chen XL, Zhao HL, Dang HY, Luan XW, Zhang XY, He HL, Zhou BC, Zhang YZ. 2009. Diversity of both cultivable protease-producing bacteria and their extracellular proteases in the sediments of the South China Sea. Microb Ecol 58:582–590. doi: 10.1007/s00248-009-9506-z. [DOI] [PubMed] [Google Scholar]
- 14.Zhou M-Y, Wang G-L, Li D, Zhao D-L, Qin Q-L, Chen X-L, Chen B, Zhou B-C, Zhang X-Y, Zhang Y-Z. 2013. Diversity of both cultivable protease-producing bacteria and bacterial extracellular proteases in coastal sediments of King George Island, Antarctica. PLoS One 8:e79668. doi: 10.1371/journal.pone.0079668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choudhury JD, Pramanik A, Webster NS, Llewellyn LE, Gachhui R, Mukherjee J. 2015. The pathogen of the Great Barrier Reef sponge Rhopaloeides odorabile is a new strain of Pseudoalteromonas agarivorans containing abundant and diverse virulence-related genes. Mar Biotechnol (NY) 17:463–478. doi: 10.1007/s10126-015-9627-y. [DOI] [PubMed] [Google Scholar]
- 16.Han SJ, Park H, Kim S, Kim D, Park HJ, Yim JH. 2016. Enhanced production of protease by Pseudomonas arctica PAMC 21717 via statistical optimization of mineral components and fed-batch fermentation. Prep Biochem Biotechnol 46:328–335. doi: 10.1080/10826068.2015.1031390. [DOI] [PubMed] [Google Scholar]
- 17.He HL, Guo J, Chen XL, Xie BB, Zhang XY, Yu Y, Chen B, Zhou BC, Zhang YZ. 2012. Structural and functional characterization of marine forms of metalloprotease E495 from Arctic sea-ice bacterium Pseudoalteromonas sp. SM495. PLoS One 7:e35442. doi: 10.1371/journal.pone.0035442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein GL, Soum-Soutéra E, Guede Z, Bazire A, Compère C, Dufour A. 2011. The anti-biofilm activity secreted by a marine Pseudoalteromonas strain. Biofouling 27:931–940. doi: 10.1080/08927014.2011.611878. [DOI] [PubMed] [Google Scholar]
- 19.Mitra S, Sana B, Mukherjee J. 2014. Ecological roles and biotechnological applications of marine and intertidal microbial biofilms. Adv Biochem Eng Biotechnol 146:163–205. [DOI] [PubMed] [Google Scholar]
- 20.Nissimov J, Rosemberg E, Munn CB. 2009. Antimicrobial properties of resident coral mucus bacteria of Oculina patagonica. FEMS Microbiol Lett 292:210–215. doi: 10.1111/j.1574-6968.2009.01490.x. [DOI] [PubMed] [Google Scholar]
- 21.Shnit-Orland M, Kushmaro A. 2009. Coral mucus-associated bacteria: a possible first line of defense. FEMS Microbiol Ecol 67:371–380. doi: 10.1111/j.1574-6941.2008.00644.x. [DOI] [PubMed] [Google Scholar]
- 22.Fjellheim AJ, Klinkenberg G, Skjermo J, Aasen IM, Vadstein O. 2010. Selection of candidate probionts by two different screening strategies from Atlantic cod (Gadus morhua L.) larvae. Vet Microbiol 144:153–159. doi: 10.1016/j.vetmic.2009.12.032. [DOI] [PubMed] [Google Scholar]
- 23.Goulden EF, Hall MR, Pereg LL, Høj L. 2012. Identification of an antagonistic probiotic combination protecting ornate spiny lobster (Parulirus ornatus) larvae against Vibrio owensii infection. PLoS One 7:e39667. doi: 10.1371/journal.pone.0039667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morya VK, Choi W, Kim EK. 2014. Isolation and characterization of Pseudoalteromonas sp. from fermented Korean food, as an antagonist to Vibrio harveyi. Appl Microbiol Biotechnol 93:1389–1395. doi: 10.1007/s00253-013-4937-3. [DOI] [PubMed] [Google Scholar]
- 25.Rodrigues S, Paillard C, Dufour A, Bazire A. 2015. Antibiofilm activity of the marine bacterium Pseudoalteromonas sp. 3J6 against Vibrio tapetis, the causative agent of brown ring disease. Probiotics Antimicrob Proteins 7:45–51. doi: 10.1007/s12602-014-9173-3. [DOI] [PubMed] [Google Scholar]
- 26.Richards GP, Watson MA, Parveen S. 2005. Development of a simple and rapid fluorogenic procedure for identification of Vibrionaceae family members. Appl Environ Microbiol 71:3524–3527. doi: 10.1128/AEM.71.7.3524-3527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richards GP, Watson MA. 2006. A simple fluorogenic method to detect Vibrio cholerae and Aeromonas hydrophila in well water for areas impacted by catastrophic disasters. Am J Trop Med Hyg 75:516–521. [PubMed] [Google Scholar]
- 28.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 29.Fink SL, Cookson BT. 2006. Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell Microbiol 8:1812–1825. doi: 10.1111/j.1462-5822.2006.00751.x. [DOI] [PubMed] [Google Scholar]
- 30.Pham CT. 2006. Neutrophil serine proteases: specific regulators of inflammation. Nat Rev Immunol 6:541–550. doi: 10.1038/nri1841. [DOI] [PubMed] [Google Scholar]
- 31.Dalisay DS, Webb JS, Scheffel A, Svenson C, James S, Holmström C, Egan S, Kjelleberg S. 2006. A mannose-sensitive haemagglutinin (MSHA)-like pilus promotes attachment of Pseudoalteromonas tunicata cells to the surface of the green alga Ulva australis. Microbiology 152:2875–2883. doi: 10.1099/mic.0.29158-0. [DOI] [PubMed] [Google Scholar]
- 32.Thomas T, Evans FF, Schleheck D, Mai-Prochnow A, Burke C, Penesyan A, Dalisay DS, Steizer-Braid S, Saunders N, Johnson J, Ferriera S, Kjelleberg S, Egan S. 2008. Analysis of the Pseudoalteromonas tunicata genome reveals properties of a surface-associated life style in the marine environment. PLoS One 3:e3252. doi: 10.1371/journal.pone.0003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellis TN, Kuehn MJ. 2010. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev 74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manning AJ, Kuehn MJ. 2013. Functional advantages conferred by extracellular prokaryotic membrane vesicles. J Mol Microbiol Biotechnol 23:131–141. doi: 10.1159/000346548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schertzer JW, Whiteley M. 2013. Bacterial outer membrane vesicles in trafficking, communication and host-pathogen interaction. J Mol Microbiol Biotechnol 23:118–130. doi: 10.1159/000346770. [DOI] [PubMed] [Google Scholar]
- 36.Bonnington KE, Kuehn MJ. 2014. Protein selection and export via outer membrane vesicles. Biochim Biophys Acta 1843:1612–1619. doi: 10.1016/j.bbamcr.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kulkarni HM, Jagannadham MV. 2014. Biogenesis and multifaceted roles of outer membrane vesicles from Gram-negative bacteria. Microbiology 160:2109–2121. doi: 10.1099/mic.0.079400-0. [DOI] [PubMed] [Google Scholar]
- 38.Kim JH, Lee J, Park J, Gho YS. 2015. Gram-negative and Gram-positive bacterial extracellular vesicles. Semin Cell Dev Biol 40:97–104. doi: 10.1016/j.semcdb.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 39.Nevot M, Deroncelé V, Messner P, Guinea J, Mercadé E. 2006. Outer membrane vesicles released by Pseudoalteromonas antarctica NF3. Environ Microbiol 8:1523–1533. doi: 10.1111/j.1462-2920.2006.01043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gorby Y, McLean J, Korenevsky A, Rosso K, El-Naggar MY, Beveridge TJ. 2008. Redox-reactive membrane vesicles produced by Shewanella. Geobiology 6:232–241. doi: 10.1111/j.1472-4669.2008.00158.x. [DOI] [PubMed] [Google Scholar]
- 41.Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, Iijima Y, Najima M, Nakano M, Yamashita A, Kubota Y, Kimura S, Yasunaga T, Honda T, Shinagawa H, Hattori M, Iida T. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743–749. doi: 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- 42.Richards GP, Fay JP, Dickens KA, Parent MA, Soroka DS, Boyd EF. 2012. Predatory bacteria as natural modulators of Vibrio parahaemolyticus and Vibrio vulnificus in seawater and oysters. Appl Environ Microbiol 78:7455–7466. doi: 10.1128/AEM.01594-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richards GP, Watson MA, Boyd EF, Burkhardt W III, Lau R, Uknalis J, Fay JP. 2013. Seasonal levels of the Vibrio predator Bacteriovorax in Atlantic, Pacific and Gulf Coast seawater. Intl J Microbiol 2013:375371. doi: 10.1155/2013/375371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetic Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1884. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saitou N, Nel M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 46.Kimura M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 47.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 48.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 49.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richards GP, Watson MA, Crane EJ III, Burt IG, Bushek D. 2008. Shewanella and Photobacterium spp. in oysters and seawater from the Delaware Bay. Appl Environ Microbiol 74:3323–3327. doi: 10.1128/AEM.00060-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richards GP, Bono JL, Watson MA, Needleman DS. 2014. Complete genome sequence for the shellfish pathogen Vibrio coralliilyticus RE98 isolated from a shellfish hatchery. Genome Announc 2(6):e01253-14. doi: 10.1128/genomeA.01253-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richards GP, Hammer CH, Garfield MK, Parveen S. 2004. Characterization of a lysyl aminopeptidase activity associated with phosphoglucose isomerase of Vibrio vulnificus. Biochim Biophys Acta 1700:219–229. doi: 10.1016/j.bbapap.2004.05.005. [DOI] [PubMed] [Google Scholar]