ABSTRACT
Chromosomal copy number variation (CCNV) plays a key role in evolution and health of eukaryotes. The unicellular yeast Saccharomyces cerevisiae is an important model for studying the generation, physiological impact, and evolutionary significance of CCNV. Fundamental studies of this yeast have contributed to an extensive set of methods for analyzing and introducing CCNV. Moreover, these studies provided insight into the balance between negative and positive impacts of CCNV in evolutionary contexts. A growing body of evidence indicates that CCNV not only frequently occurs in industrial strains of Saccharomyces yeasts but also is a key contributor to the diversity of industrially relevant traits. This notion is further supported by the frequent involvement of CCNV in industrially relevant traits acquired during evolutionary engineering. This review describes recent developments in genome sequencing and genome editing techniques and discusses how these offer opportunities to unravel contributions of CCNV in industrial Saccharomyces strains as well as to rationally engineer yeast chromosomal copy numbers and karyotypes.
KEYWORDS: aneuploidy, evolutionary adaptation, strain improvement, genome engineering, industrial yeast fermentation, fermentation, industrial yeast
INTRODUCTION
Saccharomyces yeasts are applied in a large and expanding number of industrial processes (1), ranging from traditional applications such as dough leavening (2) and beer (3) and wine fermentation (4) to modern processes such as the production of first- and second-generation fuel ethanol (5, 6), other low-molecular-weight compounds (7), and heterologous proteins (8). Selection and improvement of yeast strains remain essential to meet the complex, diverse, and continually changing performance criteria for industrial applications of Saccharomyces yeasts (9). Improving and extending yeast strain applications can be pursued by exploration of biodiversity, mating, interspecies hybridization, random mutagenesis and selection, evolutionary engineering, targeted genetic modification, or a combination of these approaches (10).
Understanding the genetic basis for industrial performance is invaluable for focusing and accelerating microbial strain improvement. In prokaryotes, genetic variation among related strains and species predominantly encompasses the presence or absence of protein-encoding and regulatory sequences, as well as mutations in these sequences. In eukaryotes, including the Saccharomyces yeasts, differences in ploidy, i.e., variations in copy number of chromosomes, provide an important additional source of genetic diversity (11).
While most eukaryotic cells are euploid, i.e., their chromosomes all have the same copy number, aneuploidy is encountered in nature as well as in manmade contexts. In aneuploid cells, the copy number of one or more chromosomes differs from that of the remainder of the genome. The existence of stable aneuploidy cells implies that chromosomal copy number variation (CCNV) contributes to genetic and physiological diversity within eukaryotic species and, in multicellular eukaryotes, within organisms. The biological significance of CCNV is powerfully illustrated by its impacts on human health. Effects of CCNV of human X and Y chromosomes range from infertility (XXY) to mental retardation (XXXXY), while trisomies of other chromosomes can cause decreased life span, mental retardation, and premature fetal death (12, 13). Spectacular CCNV occurs in most human cancer cell lines, leading to chromosome numbers of up to 90, and has been linked to the cancer hallmark of increased genome instability (14). Targeting of aneuploid cells is therefore considered a potential strategy for cancer therapy (15). Use of polyploid plants and animals in agriculture is related to their increased size and infertility (16, 17), while allopolyploid plants additionally combine industrially relevant traits from two parental genomes (18, 19). As will be discussed in this paper, CCNV is also an important phenomenon in industrial strains of Saccharomyces yeasts, whose history often involves prolonged domestication and/or industrial strain improvement.
Saccharomyces cerevisiae is an important model for studying how aneuploidy arises during mitotic and meiotic cell division, how it affects growth, and how it influences evolution of eukaryotes. These research fields are discussed in recent specialized review papers (20–22). The present paper specifically aims to review current knowledge on the analysis, occurrence, and significance of CCNV in Saccharomyces yeasts in industrial contexts. To this end, we review methods for analyzing CCNV in yeast strains, the mechanisms by which CCNV can arise spontaneously or be induced in the laboratory, and the mechanisms by which CCNV can negatively affect fitness of yeast cells. Subsequently, we discuss the occurrence and significance of CCNV for domestication and development of industrial strains of Saccharomyces yeasts and its relevance in evolutionary engineering.
METHODS FOR CCNV ANALYSIS IN YEASTS
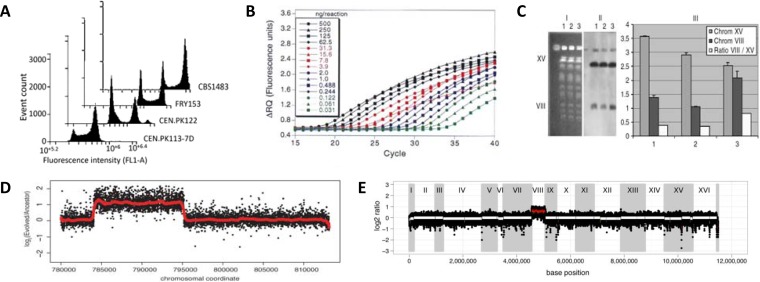
Analysis of chromosomal copy numbers in yeasts predominantly relies on five, largely complementary methods (Fig. 1). Flow cytometry analysis of cells stained with fluorescent DNA-intercalating dyes, using reference strains for calibration, enables absolute quantification of cellular DNA content and overall ploidy (23). The choice of fluorescent dyes should consider excitation/emission spectra, RNA/DNA specificity, mutagenicity, effects on viability, and the required accuracy (24). When the fluorescent dye does not compromise viability, fluorescence-activated cell sorting (FACS) can be used to select cells with a deviating DNA content. FACS-based selection has enabled selection of mutants whose DNA content differed by less than 2% from that of the parent population (25). While this FACS approach cannot select cells with specific chromosome amplifications or deletions, it can preselect cells with a deviating overall DNA content.
FIG 1.
Methods to analyze chromosome copy number and DNA content in yeast cells. (A) Absolute quantification of the DNA content of strain CBS1483 by flow cytometry using the DNA-intercalating dye Sytox Green and calibration with three strains of known ploidy. (Adapted from reference 23.) (B) qPCR fluorescence profiles for different initial concentrations of a template DNA sequence can be used to infer the amount of initial template in a reaction and to calculate relative copy numbers of different parts of the template DNA. (Republished from reference 157.) (C) Chromosome copy number determination of S. cerevisiae variants using contour-clamped homogeneous electric field electrophoresis and Southern blotting. I, stained CHEF gel; II, Southern blot hybridization; III, quantification of the hybridization bands. Lanes 1 and 2 show two disomic knockout strains that have only a single copy of chromosome VIII, while lane 3 shows a diploid control strain. (Modified from reference 28 with permission [copyright 2005 John Wiley & Sons Ltd.].) (D) Copy number estimation of chromosome II by array comparative genomic hybridization of an evolved strain relative to its unevolved parental strain. Deviating copy number can be detected by significant deviations of the measured signal and has been accentuated by a red line. (Republished from reference 108.) (E) Copy number estimation of the genome of the wine production strain VL3, based on whole-genome sequencing and read depth analysis. A marked increase of the read depth for chromosome VIII indicates a gain of copy of that chromosome. (Adapted from reference 117.)
Contour-clamped homogeneous electric field (CHEF) electrophoresis separates yeast chromosomes on agarose gels and is used to analyze chromosome complements (karyotypes) of yeast strains (26, 27). Southern hybridization of CHEF gels can reveal copy numbers of individual chromosomes by comparison of hybridization intensity with reference strains (Fig. 1C) (28). However, the accuracy of CCNV estimates obtained by this method is limited.
Copy numbers of individual yeast chromosomes can be analyzed by quantitative real-time PCR (qPCR, Fig. 1B), using primers that amplify chromosome-specific genomic sequences (29). Accuracy of PCR-based copy number estimates can be boosted by digital droplet PCR (ddPCR), which uses microfluidics to generate thousands of replicate PCRs in water-in-oil emulsions (30, 31). Since qPCR analysis estimates copy numbers of only the amplified region(s), additional methods are required to assess whether these reflect copy number variations of entire chromosomes or of specific chromosomal regions (segmental aneuploidy).
Array comparative genomic hybridization (aCGH) compares local copy number differences by hybridizing genomic DNA from related yeast strains to oligonucleotide arrays (Fig. 1D) (32). Depending on oligonucleotide size and genome coverage of the arrays, copy number variations can be analyzed across entire genomes at resolutions down to 20 bp (33).
High-resolution, accurate analysis of CCNV in yeast increasingly depends on next-generation sequencing (NGS) of entire yeast genomes (34). NGS enables ploidy estimation from allele frequency in the whole genome and in specific regions (35). Moreover, when sequence bias in DNA isolation and/or sequencing (36) is prevented, the number of reads generated for any particular sequence (i.e., its read depth) directly reflects its copy number relative to the remainder of the genome (Fig. 1E) (37). Computational tools assist CCNV identification via read depth, either by mapping of NGS reads to a preassembled genome sequence or via de novo genome assembly (38). With both approaches, the accuracy of copy number estimates increases with increasing sequencing coverage. When many copies of a chromosome are present in a yeast strain, (dis)appearance of a single copy causes only a small relative change. Accurate analysis of aneuploid yeast genomes with large variations in chromosomal copy numbers therefore requires high sequencing coverage. Short-read-length NGS methods currently provide the most cost-effective access to high sequencing depth (>100× coverage at read lengths from 75 to 400 bp can be obtained routinely with, for example Illumina and Ion Torrent platforms). Sequencing reads can be mapped to a preassembled, accurate reference genome similar to that of the sequenced strain, yielding accurate CCNV estimates. If no such reference genome is available, de novo assembly of the genome and subsequent copy number analysis can provide unbiased and more accurate results (23). However, short-read-length NGS does not allow assembly of repetitive regions whose length exceeds the read length, such as TY, subtelomeres, and ribosomal DNA (rDNA) sequences in Saccharomyces genomes. De novo genome assembly is strongly facilitated by long-read-length sequencing platforms (e.g., Pacific Biosystems and Oxford Nanopore Technologies), either alone or combined with short-read-length data. Moreover, when genes are present in multiple nonidentical copies, it can be difficult to perform full reconstruction of duplicated alleles (“phasing”) (39). Indeed, when two single nucleotide polymorphisms (SNPs) occur in only one copy of a gene, nucleotides can be assigned to a specific allele only if individual reads that cover both variable positions are available. Allelic reconstruction, and by extension reconstruction of (parts of) chromosome copies, is enhanced by the use of long-read or mate-pair sequencing data (39). Long-read sequencing technologies still have higher error rates than short-read platforms. Fast developments in real-time, single-molecule methods for replication (Pacific Biosystems) or nanopore (Oxford Nanopore Technologies) sequencing enable generation of extremely long reads with increasing accuracy (40–43) and are likely to transform whole-genome resequencing (44). The potential of long-read sequencing to capture entire chromosome arms or even entire chromosomes within a single read offers unique possibilities to unravel chromosome structure, translocation breakpoints, and allelic variation among duplicate chromosomes and chromosomal fragments (41).
INDUCTION OF CHROMOSOME MISSEGREGATION
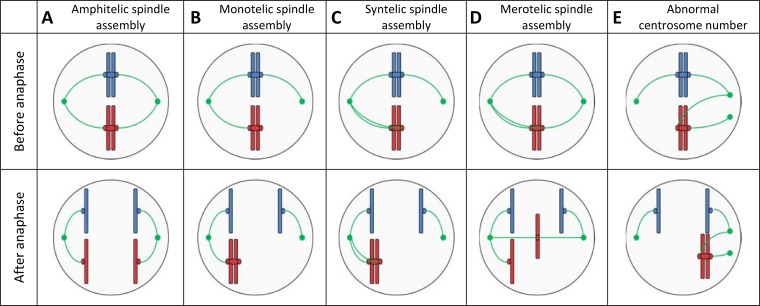
The anaphase of the eukaryotic cell cycle has evolved to conserve chromosomal copy number during cell division. Its crucial steps include chromatid cohesion, centrosome formation at opposite cell poles, kinetochore-microtubule attachment, and quality control at the spindle assembly checkpoint (45). Imperfections in any of these steps can cause chromosome missegregation and, thereby, CCNV in eukaryotic cell populations, tissues, and tumors (45–47). Even in cell lines without predisposing defects, chromosome missegregation occurs, albeit at very low frequencies (21, 48). In yeast, chromosome missegregation can occur during mitosis (48) and, with a higher incidence, during the meiotic process of sporulation (49). Figure 2 provides a schematic overview of mechanisms by which missegregation of chromosomes can occur.
FIG 2.
Schematic representation of chromosome segregation and of the common mechanisms leading to chromosome missegregation. Two chromatids of two different chromosomes are shown in red and blue, with their centromeres and kinetochores. In green, the centrosomes are shown with the assembled microtubule attached to the kinetochores of the chromatids. For each case, the microtubule-kinetochore assembly is shown before and after the anaphase. (A) Correct chromosome segregation is achieved by amphitelic spindle assembly, where microtubules connect each chromatid to a different centrosome, resulting in separation to opposite cellular poles during anaphase and maintaining a stable karyotype in the daughter cells (45). (B and C) If only one of the chromatids is attached to a centrosome or both chromatids are attached to the same kinetochore, referred to as monotely and syntely, respectively, proceeding to anaphase would result in the missegregation of both chromatids to that centrosome. However, monotelic and syntelic spindle assemblies are detected at the spindle assembly checkpoint and therefore rarely cause chromosome missegregation. (D) In the case of a merotelic spindle assembly, a chromatid is attached to both centrosomes and, as a result, cannot migrate to a cellular pole. The resulting random segregation of the lagging chromosome can cause missegregation, damage, and micronucleus formation (158). (E) When more than two centrosomes are formed, random attachment of chromatids can result in chromosome missegregation due to chromosome lagging or unequal chromosome segregation (159).
A wide range of chemical and physical stress factors increase the incidence of chromosome missegregation in growing cultures. Stimuli that increase occurrence of CCNV in mitotic yeast cultures include nutrient-limited growth (50), heat shock (51), UV or X-ray irradiation (52), and chemical stress. Chemical compounds such as nocodazole, fumaronitrile, and methyl benzimidazole-2-yl-carbamate induce a high incidence of chromosome missegregation in S. cerevisiae (53–55). Polar aprotic solvents, including ethanol esters, are other known inducers of CCNV (56), and high concentrations of ethanol itself have also been reported to enhance chromosome missegregation in fungal cells (57). Exposure to high ethanol concentrations may therefore contribute to the frequent occurrence of CCNV in industrial yeast strains used for production of alcoholic beverages and fuel ethanol (see below).
Chromosome missegregation can also be stimulated by genetic factors. Increased ploidy strongly enhances chromosome missegregation (58), in particular when uneven numbers of chromosome sets preclude equal distribution of chromosomes during meiosis (59). Strongly increased chromosome missegregation rates have also been observed in allopolyploid Saccharomyces yeasts, which carry chromosomes from different parental species and show a high incidence of aneuploidy (60). Since aneuploidy itself, including segmental aneuploidy, also stimulates chromosome missegregation, aneuploid cells are more prone to acquire further CCNV (61).
In contrast to chemical, physical, and genetic stresses, which affect segregation of all chromosomes, targeted molecular genetic approaches enable elimination or amplification of specific chromosomes. In S. cerevisiae, copy gain and loss of specific chromosomes have been achieved by cloning a strong inducible promoter upstream of the centromere of the targeted chromosome (62, 63). When induced, transcription from the promoter interferes with centromere function, thus causing missegregation during mitosis. Aneuploid daughter cells that have lost or gained a copy of the targeted chromosome can then be isolated from the resulting culture. Alternatively, by crossing with kar1 null mutants, mating is prematurely aborted but chromosome transfer between nuclei can still occur, yielding aneuploid cells. Aneuploidy of specific chromosomes can be easily selected for when they carry marker sequences (64).
NEGATIVE IMPACTS OF CCNV ON FITNESS
Aneuploid yeasts typically show a reduced fitness relative to congenic euploid strains (64). The molecular basis of generic transcriptional responses to aneuploidy remains to be fully elucidated. Reported transcriptional responses in aneuploid strains include downregulation of genes involved in cell growth and proliferation and upregulation of genes involved in the environmental stress response (ESR) (64, 65). Studies on the impact of gain or loss of chromosomes in otherwise euploid yeast strains showed that the aneuploidy-associated stress response (AASR) includes increased genome instability, low sporulation efficiency, reduced growth rate, increased nutrient uptake rates, and reduced replicative life span (21, 66, 67). Phenotypic consequences of chromosome gain and those of chromosome loss are similar, suggesting that the responsible cellular mechanisms overlap (68). AASR intensity is positively correlated with the length of the affected chromosome(s) and with the number of affected genes (20, 64, 69). A much less pronounced AASR in polyploid strains has been attributed to a smaller relative impact on chromosome number (64, 70). The absence of AASR-related phenotypes upon introduction of a yeast artificial chromosome harboring nontranscribed mammalian genes indicates that AASR is not due to increased DNA content per se (64).
Genome instability of aneuploid yeasts has been linked to the missegregation events that cause aneuploidy and, in particular, to “lagging” (Fig. 2D) of chromosomes during anaphase. DNA damage and imperfect repair of lagging chromosomes cause mutations, deletions, and translocations (71, 72). Additionally, formation of transient micronuclei by lagging chromosomes increases the mutation rate during subsequent mitosis (73, 74). At a longer time scale, aneuploidy promotes generation of CCNV by enhancing chromosome missegregation and mitotic recombination as well as by impairing DNA repair (61, 75, 76). Impaired sporulation of aneuploid strains has been linked to disruption of homologous chromosome pairing during meiosis (77). AASR-related cell cycle defects involve slow accumulation of G1 cyclins, causing an abnormal delay in the G1 phase (78, 79).
CCNV-associated changes in gene dosage can directly affect expression levels of the affected genes. Typically, gain or loss of a chromosome coincides with an increased or decreased expression level, respectively, of the large majority of expressed genes that it carries (80). Correct subunit folding and assembly of multiprotein complexes (29, 64, 81, 82), which strongly depend on subunit stoichiometry (83), can be disturbed when one or more subunits are encoded by aneuploid chromosomes. A resulting “overload” of the cellular protein folding machinery can cause accumulation of un- and misfolded proteins and proteotoxic stress (67, 70). Indeed, some aneuploid strains show increased sensitivity to inhibitors of protein folding and degradation (84) and impaired functionality of the proteasome, the chaperone Hsp90, or endocytosis-mediated protein degradation (66, 70). Energy costs of protein misfolding and protein overproduction have been implicated in the increased nutrient consumption and slow growth of aneuploid yeast strains (85). The correlation between protein level and gene copy number is not always straightforward (29, 64), and situations have even been described in which the transcript level of individual genes decreased with increasing copy number (86–88). Signaling cascades and transcriptional regulation are among the core cellular systems that can be affected by aneuploidy (89). The impact of gene-dosage-related changes in gene expression on AASR (29) can be further intensified or attenuated by mutations in genes on nonaneuploid chromosomes (90). Such in trans effects can, for example, be related to stoichiometric imbalances in protein complexes or pathways, unspecific protein interactions, protein folding, and degradation (81).
Sensitivity to AASR is yeast strain dependent (91, 92). In tolerant strains, mutations that attenuate AASR, such as a loss-of-function mutation in the deubiquitinating enzyme Ubp6p, were identified (82). While not all mutations involved in AASR tolerance are known, its relevance is amply demonstrated by the frequent occurrence of aneuploidy in wild, clinical, and industrial isolates of Saccharomyces yeasts (35, 91, 93).
CCNV IN EVOLUTIONARY ENGINEERING
In addition to negative impacts on cellular fitness, chromosome-specific effects of CCNV can also confer fitness benefits in specific environmental or genetic contexts. Indeed, CCNV offers a fast way to modify gene copy number during natural evolution of eukaryotes and to increase evolvability by allowing neofunctionalization of amplified essential genes (51, 94–96). Under selective conditions, mutants with CCNV will outgrow the parental population whenever positive effects of CCNV on fitness outweigh any negative impacts of AASR, while further mutations that enhance positive effects or decrease AASR can further increase the initial fitness benefit. CCNV is therefore seen as a significant contributor to evolutionary adaptation in eukaryotes (51, 97).
Technically, adaptive laboratory evolution (ALE) encompasses prolonged cultivation of microorganisms under defined conditions, combined with an analysis of the phenotypic and/or genotypic changes that occur during evolutionary adaptation (98). ALE approaches that are specifically designed to select for industrially relevant traits are often referred to as evolutionary engineering (99, 100). Resequencing of evolved strains can provide insight into the genetic basis for industrially relevant traits and enable its reverse engineering into naive, nonevolved strains (101). Evolutionary engineering is particularly attractive for food and beverage applications, since it does not involve recombinant DNA techniques and associated consumer acceptance and regulatory issues (102).
While, on the time scales involved in natural evolution and speciation, CCNV is considered to be a transient adaptation mechanism that is usually replaced by more elegant and efficient mutations (103, 104), most ALE experiments with yeasts cover only 50 to 500 generations of selective growth. It is therefore not surprising that CCNV is frequently encountered during ALE of Saccharomyces yeasts, for example, for the selection of suppressor mutants (Table 1). Numerous evolutionary engineering studies have linked CCNV to industrially relevant traits, ranging from tolerance to products or inhibitors to improved kinetics of sugar fermentation or sedimentation behavior of yeast cultures (Table 1). In some cases, ALE even resulted in complete duplication of the genome of haploid S. cerevisiae strains, for instance, after selection for glucose-limited growth, high ethanol tolerance, and increased sedimentation (105–107). In the last case, increased ploidy played a major role in shaping an evolved, multicellular phenotype.
TABLE 1.
Examples of whole-chromosome copy number variations acquired during laboratory evolution experiments with Saccharomyces cerevisiae strainsa
| Selected phenotype | Aneuploid chromosome(s) | Confirmed causality | Contributing gene(s) | Reference |
|---|---|---|---|---|
| Biomass sedimentation | Whole-genome duplication | Yes | ACE2 | 105 |
| Glucose-limited growth | Whole-genome duplication | Yes | 107 | |
| High temp tolerance | III (+1) | Yes | 17 individual genes | 103 |
| High pH tolerance | V (+1) | Yes | 103 | |
| Glucose-limited growth | I (+1), III (+1), V (+1) | No | 108 | |
| Phosphate-limited growth | IV (+1), VI (+1), X (+1), XIII (+2), XVI (+1) | No | 108 | |
| Lactate utilization by jen1Δ strain | III (+1) | Yes | ADY2 | 112 |
| Xylose utilization | I (−1) | No | 160 | |
| p-Coumaric and ferulic acid tolerance | XIV (+1) | No | 160 | |
| Copper tolerance | II (+1), VIII (+1) | No | CUP1, SCO1, and SCO2 | 104 |
| Galactose tolerance | VIII (+1) | Yes | GAL80 | 161 |
| Ethanol tolerance | III (+1), VIII (+1) | No | 106 | |
| Radicicol resistance | XV (+1) | Yes | STI1 and PDR5 | 113 |
| Fluconazole resistance | VIII (+1) | No | ERG11 | 113 |
| Tunicamycin resistance | XVI (−1) | Yes | 113 | |
| Benomyl resistance | XII (−1) | No | 113 | |
| Suppressors of MEC1 deficiency | IV (+1) | Yes | RNR1 | 162 |
| Suppressors of MYO1 deletion | XIII (+1), XVI (+1) | Yes | HSP82, HSC82, RLM1, and MKK2 | 94 |
| Suppressors of RPS24A and RNR1 deletion | IX (+1) | No | RPS24B and RNR3 | 163 |
| Suppressors of telomerase insufficiency | VIII (−1) | No | PRP8, UTP9, KOG1, and SCH9 | 164 |
In the examples listed, the acquired CCNV was hypothesized to contribute to the selected phenotype. “Confirmed causality” indicates that a causal link between CCNV and the phenotype acquired during laboratory evolution was experimentally confirmed. In cases where the impact of a CCNV on phenotype was linked to specific genes, this is also indicated. Segmental aneuploidies observed in the cited studies are not included in the table.
In addition to whole-chromosome copy number variations, ALE frequently involves segmental aneuploidies (108–111). While both can be identified by analysis of high-coverage, short-read NGS data, precise definition of duplication and/or translocation events and karyotypes involved in segmental aneuploidy generally requires additional analysis by long-read sequencing or diagnostic PCR (110, 111).
Several methods can be applied to test if segmental or whole-chromosome aneuploidies do indeed contribute to phenotypes acquired in an ALE experiment. In some cases, hypothesis-based deletion or amplification of one or more genes on (an) affected chromosome(s) can directly confirm the relevance of a CCNV. For example, an increased copy number of chromosome III in jen1Δ mutants evolved for restoration of lactate transport could be rapidly linked to the ADY2 monocarboxylate-transporter gene on this chromosome (112). Overexpression or deletion studies were also successfully used to identify 17 genes that contributed to the benefit of a copy gain of chromosome III in an S. cerevisiae strain evolved for heat tolerance (103). Alternatively, the relevance of a CCNV in an evolved strain can be tested by introducing the deviating chromosome copy number in a euploid strain, e.g., via transcriptional interference with centromere function (103, 113). Similarly, the chromosome copy number variation can be reverted to wild type, e.g., by sporulation and analysis of segregants with wild-type karyotypes (103, 113). Although the method is not routinely applied, specific chromosomal regions that contribute to an acquired phenotype can be identified by targeted introduction of segmental aneuploidy of sets of tiled chromosomal regions (114). Two recently described PCR-based methods enable duplication or deletion of chromosome segment copies by introduction of telomere seed sequences and of an additional centromere to generate an additional autonomously replicating chromosome fragment. By introducing centromere and telomere seed sequences pointing outward of the region of interest, this region will be duplicated on an additional, independently replicating chromosome (115). Conversely, by introducing a centromere and telomere seed sequences pointing into the region of interest, the targeted chromosome is split into two autonomously replicating chromosomes that no longer contain the targeted region (116). This approach enables a nonbiased, systematic analysis of the positive and negative contributions of chromosomal regions and/or individual genes.
CCNV IN INDUSTRIAL SACCHAROMYCES YEASTS
Aneuploidy has been observed in Saccharomyces strains used in diverse industrial applications, including dough leavening, bioethanol production, beer brewing, spirit production, wine fermentation, and production of cacao and coffee (Table 2). In industrial strains, CCNV may have occurred during centuries-long domestication processes and/or during strain improvement programs that involved CCNV-inducing mutagenesis procedures such as UV irradiation (52).
TABLE 2.
Examples of CCNV in industrial Saccharomyces strainsa
| Strain | Species | Industrial product | Approximate overall ploidy | Aneuploid chromosome(s) | Reference |
|---|---|---|---|---|---|
| BR001 | S. cerevisiae | Bread | 4n | IX (+1) | 93 |
| BR004 | S. cerevisiae | Bread | 4n | IX (+1) | 93 |
| E-IM3 | S. cerevisiae | Cacao | 3n | VII | 165 |
| AY529517 | S. cerevisiae | Cacao | 2n | IV, XII | 165 |
| YE 2-2 | S. cerevisiae | Coffee | 3n | I, XV, XVI | 165 |
| JV2 | S. cerevisiae | Coffee | 4n | Extensive aneuploidy | 165 |
| Y-393 | S. cerevisiae | Kefir | 3n | I, III, IX | 165 |
| YJM1356 | S. cerevisiae | Cider | 2n | I (+2) | 147 |
| YJM1439 | S. cerevisiae | Ginger beer | 2n | VIII (+2) | 147 |
| FostersO | S. cerevisiae | Ale beer | >2n | III (+1), XIV (−1) | 117 |
| FostersB | S. cerevisiae | Ale beer | >2n | III (+1), V (+1), XV (+1) | 117 |
| CBS1483 | S. cerevisiae × eubayanus | Lager beer | >2n | Extensive aneuploidy | 23 |
| CBS1270 | S. cerevisiae × eubayanus | Lager beer | >2n | Extensive aneuploidy | 23 |
| AWRI796 | S. cerevisiae | Wine | 2n | I (+1) | 117 |
| VL3 | S. cerevisiae | Wine | 2n | VIII (+1) | 117 |
| F-12 | S. cerevisiae | Flor wine | 2n | VII (+1), XIII (+2) | 130 |
| SA001 | S. cerevisiae | Sake | 2–3n | V (+1) | 93 |
| SA003 | S. cerevisiae | Sake | 2–3n | I (+1) | 93 |
| SP011 | S. cerevisiae | Spirits | 2n | I (−1), III (−1), VI (−1), IX (−1), XII (−1) | 93 |
| SP001 | S. cerevisiae | Spirits | 2n | I (−1), VI (−1) | 93 |
| Y-999 | S. cerevisiae | Bioethanol from starch | 3n | III | 165 |
| CBS7960 | S. cerevisiae | Bioethanol from sugarcane | 2n | VIII | 165 |
| ZTW1 | S. cerevisiae | Bioethanol from corn mash | 3n | IX (+1) | 166 |
The overall ploidy of the strains and identified aneuploid chromosomes are indicated. For strains in which the copy number deviation from euploidy has been determined, this is reported between parentheses. Extensive aneuploidy refers to strains with more than 10 aneuploid chromosomes. Segmental aneuploidies that occur in many of these strains are not indicated in the table.
Currently available information suggests that aneuploidy is not prevalent among S. cerevisiae strains used in dough leavening, bioethanol production, ale-type beer fermentation, and distilled-beverage production. In these strains, aneuploidy typically involves small deviations in copy number of one or a few chromosomes (117–119). Since accurate information is available for only a few of the many hundreds of such strains stored in culture collections, the incidence of CCNV may well be underestimated. Indeed, a recent whole-genome sequencing study revealed extensive CCNV among several beer-related S. cerevisiae strains that were previously assumed to be mostly euploid (93).
There is ample evidence that copy numbers of individual genes or loci affect industrially relevant traits of S. cerevisiae strains. For example, rates of sucrose, maltose, and melibiose fermentation correlate with copy numbers of SUC, MAL, and MEL loci, respectively (120–122), while proline utilization rates correlate with the copy number of the PUT1 proline oxidase gene (123). So far, the industrial significance of CCNV in industrial S. cerevisiae strains has not been systematically explored. S. cerevisiae ZTW1, a strain isolated from corn mash used in a Chinese bioethanol factory, provides an interesting exception. In this strain, chromosomal and segmental aneuploidy were shown to directly contribute to industrially relevant traits, including copper tolerance and ethanol yield (124).
Consistent with the increased rate of chromosome missegregation in alloploid cells, aneuploidy is highly prevalent among wine and lager-type beer yeasts originating from domestication of natural hybrids of different Saccharomyces species. Despite its frequent occurrence, the impacts of aneuploidy in these genetic contexts have not been explored in depth, and it is unclear how AASR and chromosome-specific copy number effects compare to those observed in otherwise euploid S. cerevisiae strains. In general, these alloploid genomes tolerate aneuploidy well, with massive diversity in chromosome copy numbers across strains (23, 125, 126). Some aneuploid lager brewing yeasts even sporulate, albeit at low efficiency, by anomalous cell division (79). Wine yeasts include S. cerevisiae × Saccharomyces kudriavzevii, S. cerevisiae × Saccharomyces uvarum, and S. cerevisiae × S. kudriavzevii × S. uvarum hybrids (127, 128), many of which are alloaneuploids, with a large diversity in chromosome copy numbers (129). Aneuploidy has a strong impact on performance of “flor” wine yeast. An increased copy number of chromosome VII, which carries the alcohol dehydrogenase genes ADH2 and ADH3, correlated with increased ethanol oxidation capacity of the characteristic vellum formed by these yeasts during sherry wine fermentation (130).
Saccharomyces pastorianus lager beer brewing strains have long been assumed to originate from a hybridization event involving S. cerevisiae and another Saccharomyces species (131). The genome of the cold-tolerant species Saccharomyces eubayanus, first isolated in Patagonia in 2011 (132) and later also found in North America, Asia, and New Zealand (132–135), was shown to exhibit a 99.56% identity with the non-cerevisiae part of S. pastorianus genomes (136). It is postulated that, after one or more spontaneous hybridization events, centuries of domestication and selection of the resulting S. cerevisiae × S. eubayanus hybrid(s) in brewing environments generated the current diversity of lager brewing strains (137, 138). S. cerevisiae × S. eubayanus hybrids made in the laboratory combine at least two important brewing-related characteristics of their parents. The S. cerevisiae subgenome contributes the ability to ferment maltotriose, a major fermentable sugar in wort, while low-temperature performance, essential for the lager brewing process, is conferred by the S. eubayanus subgenome (139, 140).
Historically and mainly based on geographical origin, two groups of S. pastorianus strains were distinguished. Group I (Saaz-type) strains tend to ferment well at low temperatures but generally show poor maltotriose fermentation. Conversely, group II strains (Frohberg type) tend to have higher optimal growth temperatures and ferment maltotriose well (141). These phenotypic differences correlate with ploidy and with the contribution of genetic material from the two subgenomes. Consistent with their better performance at low temperature, group I strains contain more S. eubayanus DNA, while some S. cerevisiae chromosomes can even be absent (e.g., S. cerevisiae chromosome III is absent in all group I strains sequenced so far) (23, 32, 141–143); group II strains generally have a more balanced genome composition, with (multiple) chromosomes from both S. eubayanus and S. cerevisiae (23, 32, 141–143). These differences have been proposed to reflect different hybridization histories of the two groups (144). In this model, group I derives from an original hybridization event involving a haploid S. cerevisiae strain and a haploid or diploid S. eubayanus strain, while group II strains arose from hybridization of a diploid S. cerevisiae strain with a haploid S. eubayanus strain (23) or from two subsequent hybridization events (141). Different hybridization histories appear to be contradicted by conserved chromosome rearrangement breakpoints in group I and group II strains (32, 143). However, these might also have evolved independently due to fragility of the breakpoint and/or by conferring a selective advantage (145). The latter hypothesis is consistent with ALE studies with an S. uvarum × S. cerevisiae hybrid in nitrogen-limited cultures, which selected for recombination between alloploid chromosomes in the MEP2 ammonium permease gene (146).
Two key brewing-related properties of S. pastorianus strains have been correlated with CCNV. Production of diacetyl, an important off-flavor in lager beers that needs to be removed at the end of fermentation (“Ruh” phase), correlated with copy number of chromosomes III, VIII, X, XII, and XIV (23). These chromosomes harbor genes involved in the valine biosynthesis pathway, which generates α-acetolactate, the precursor for diacetyl production. Similarly, Ca2+-dependent flocculation, which is essential for yeast sedimentation during brewing, positively correlated with copy numbers of chromosomes I, VI, XI, and XII, all of which harbor flocculin genes (23).
OUTLOOK: UNDERSTANDING AND ENGINEERING CCNV IN INDUSTRIAL CONTEXTS
Whole-genome sequences of environmental and industrial isolates of Saccharomyces species, which are becoming available at a rapid and still accelerating pace (35, 93, 147), confirm the relevance of CCNV for the natural diversity, domestication, and industrial strain improvement of these yeasts. Experimental hybridization of strains from different Saccharomyces species is rapidly gaining popularity as a strategy for strain improvement and product diversification of wine and beer yeasts (139, 148, 149). Traits that have been improved by hybridization include fermentative vigor over wide temperature ranges and concentrations of minor fermentation products (150), flocculation capacity (151), and sugar uptake kinetics (152). Moreover, ploidy of laboratory-made hybrid strains correlates with fermentation rates, ethanol yield, and concentrations of aromatic esters (148). In view of the higher tendency of alloploid and allopolyploid genomes to develop aneuploidy, CCNV is likely to be a key factor in the stability and further diversification of the resulting strains.
Targeted introduction of CCNV, e.g., by using drugs that interfere with chromosome segregation, is rarely applied in industrial strain improvement (10). Use of the mitotic inhibitor methyl benzimidazole-2-yl-carbamate (MBC) to mutagenize the aneuploid bioethanol strain ZTW1 demonstrates the potential of this approach (153). Treatment of strain ZTW1 with MBC yielded strains with an improved fermentative capacity under industrial high-gravity conditions (119), enhanced viability after drying (154), and higher final ethanol titer (124). These observations and the frequent appearance of CCNV in ALE suggest that such interference with chromosome segregation may deserve reconsideration in industrial yeast strain improvement.
The relatively small number of cases in which molecular mechanisms by which CCNV contributes to industrial performance of Saccharomyces yeasts have been investigated in detail often identified gene dosage effects as a key contributor. Allelic variation of amplified genes can be an additional, as-yet-underexplored source of industrially relevant diversity within strains that carry CCNV, especially in alloploid strains with a long history of domestication and/or strain improvement. Novel long-read DNA-sequencing approaches (e.g., nanopore MinION sequencing [41]) should enable a much faster identification of such allelic variations and of their correlation with industrially relevant traits, including subtle differences in flavor and aroma production. Recent developments in genome editing, including the advent of CRISPR (clustered regularly interspaced short palindromic repeat)-based techniques (155, 156) and methods for experimentally introducing defined, segmental aneuploidies (115, 116), will accelerate the functional analysis of CCNV. Moreover, these techniques will enable rapid introduction of relevant mutations into strains that do not contain CCNV, without the potential disadvantages of AASR. The combination of these developments will enable a more thorough investigation of the importance of CCNV for the performance of industrial strains and is likely to open the way to using CCNV induction as a tool for strain improvement, either by direct generation of improved strains or by identification of chromosome fragments or genes whose copy number affects industrial performance.
ACKNOWLEDGMENTS
We thank Nick Brouwers, Alex Salazar, Jasper Diderich, Niels Kuijpers (Heineken Supply Chain B.V.), and Jan-Maarten Geertman (Heineken Supply Chain B.V.) for critically reading the manuscript.
This work was performed within the BE-Basic R&D Program (http://www.be-basic.org/), which was granted an FES subsidy from the Dutch Ministry of Economic Affairs, Agriculture and Innovation (EL&I).
REFERENCES
- 1.Mattanovich D, Sauer M, Gasser B. 2014. Yeast biotechnology: teaching the old dog new tricks. Microb Cell Fact 13:34. doi: 10.1186/1475-2859-13-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell PJL, Higgins VJ, Attfield PV. 2001. Comparison of fermentative capacities of industrial baking and wild-type yeasts of the species Saccharomyces cerevisiae in different sugar media. Lett Appl Microbiol 32:224–229. doi: 10.1046/j.1472-765X.2001.00894.x. [DOI] [PubMed] [Google Scholar]
- 3.Lodolo EJ, Kock JLF, Axcell BC, Brooks M. 2008. The yeast Saccharomyces cerevisiae—the main character in beer brewing. FEMS Yeast Res 8:1018–1036. doi: 10.1111/j.1567-1364.2008.00433.x. [DOI] [PubMed] [Google Scholar]
- 4.Pretorius IS. 2000. Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16:675–729. doi:. [DOI] [PubMed] [Google Scholar]
- 5.Argueso JL, Carazzolle MF, Mieczkowski PA, Duarte FM, Netto OVC, Missawa SK, Galzerani F, Costa GGL, Vidal RO, Noronha MF, Dominska M, Andrietta MGS, Andrietta SR, Cunha AF, Gomes LH, Tavares FCA, Alcarde AR, Dietrich FS, McCusker JH, Petes TD, Pereira GAG. 2009. Genome structure of a Saccharomyces cerevisiae strain widely used in bioethanol production. Genome Res 19:2258–2270. doi: 10.1101/gr.091777.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moysés DN, Reis VCB, de Almeida JRM, de Moraes LMP, Torres FAG. 2016. Xylose fermentation by Saccharomyces cerevisiae: challenges and prospects. Int J Mol Sci 17:207. doi: 10.3390/ijms17030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng A-P, Biebl H. 2002. Bulk chemicals from biotechnology: the case of 1,3-propanediol production and the new trends. Adv Biochem Eng Biotechnol 74:239–259. [DOI] [PubMed] [Google Scholar]
- 8.Mendoza-Vega O, Sabatie J, Brown SW. 1994. Industrial production of heterologous proteins by fed-batch cultures of the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 15:369–410. doi: 10.1111/j.1574-6976.1994.tb00146.x. [DOI] [PubMed] [Google Scholar]
- 9.Pretorius IS, du Toit M, van Rensburg P. 2003. Designer yeasts for the fermentation industry of the 21st century Food Technol Biotechnol 41:3–10. [Google Scholar]
- 10.Steensels J, Snoek T, Meersman E, Nicolino MP, Voordeckers K, Verstrepen KJ. 2014. Improving industrial yeast strains: exploiting natural and artificial diversity. FEMS Microbiol Rev 38:947–995. doi: 10.1111/1574-6976.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comai L. 2005. The advantages and disadvantages of being polyploid. Nat Rev Genet 6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- 12.Visootsak J, Graham JM Jr. 2006. Klinefelter syndrome and other sex chromosomal aneuploidies. Orphanet J Rare Dis 1:1–5. doi: 10.1186/1750-1172-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassold T, Hunt P. 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 14.Duesberg P, Rausch C, Rasnick D, Hehlmann R. 1998. Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc Natl Acad Sci U S A 95:13692–13697. doi: 10.1073/pnas.95.23.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon DJ, Resio B, Pellman D. 2012. Causes and consequences of aneuploidy in cancer. Nat Rev Genet 13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 16.D'Hont A. 2005. Unraveling the genome structure of polyploids using FISH and GISH; examples of sugarcane and banana. Cytogenet Genome Res 109:27–33. doi: 10.1159/000082378. [DOI] [PubMed] [Google Scholar]
- 17.Piferrer F, Beaumont A, Falguière J-C, Flajšhans M, Haffray P, Colombo L. 2009. Polyploid fish and shellfish: production, biology and applications to aquaculture for performance improvement and genetic containment. Aquaculture 293:125–156. doi: 10.1016/j.aquaculture.2009.04.036. [DOI] [Google Scholar]
- 18.Lee S-S, Lee S-A, Yang J, Kim J. 2011. Developing stable progenies of ×Brassicoraphanus, an intergeneric allopolyploid between Brassica rapa and Raphanus sativus, through induced mutation using microspore culture. Theor Appl Genet 122:885–891. doi: 10.1007/s00122-010-1494-3. [DOI] [PubMed] [Google Scholar]
- 19.Leitch AR, Leitch IJ. 2008. Genomic plasticity and the diversity of polyploid plants. Science 320:481–483. doi: 10.1126/science.1153585. [DOI] [PubMed] [Google Scholar]
- 20.Sheltzer JM, Amon A. 2011. The aneuploidy paradox: costs and benefits of an incorrect karyotype. Trends Genet 27:446–453. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santaguida S, Amon A. 2015. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol 16:473–485. doi: 10.1038/nrm4025. [DOI] [PubMed] [Google Scholar]
- 22.Mulla W, Zhu J, Li R. 2014. Yeast: a simple model system to study complex phenomena of aneuploidy. FEMS Microbiol Rev 38:201–212. doi: 10.1111/1574-6976.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van den Broek M, Bolat I, Nijkamp JF, Ramos E, Luttik MAH, Koopman F, Geertman JM, de Ridder D, Pronk JT, Daran JMG. 2015. Chromosomal copy number variation in Saccharomyces pastorianus is evidence for extensive genome dynamics in industrial lager brewing strains. Appl Environ Microbiol 81:6253–6267. doi: 10.1128/AEM.01263-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haase SB, Reed SI. 2002. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle 1:132–136. doi: 10.4161/cc.1.2.114. [DOI] [PubMed] [Google Scholar]
- 25.Pfosser M, Amon A, Lelley T, Heberle-Bors E. 1995. Evaluation of sensitivity of flow cytometry in detecting aneuploidy in wheat using disomic and ditelosomic wheatrye addition lines. Cytometry 21:387–393. doi: 10.1002/cyto.990210412. [DOI] [PubMed] [Google Scholar]
- 26.Chu G, Vollrath D, Davis RW. 1986. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science 234:1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- 27.Török T, Rockhold D, King AD. 1993. Use of electrophoretic karyotyping and DNA-DNA hybridization in yeast identification. Int J Food Microbiol 19:63–80. doi: 10.1016/0168-1605(93)90124-Y. [DOI] [PubMed] [Google Scholar]
- 28.Waghmare SK, Bruschi CV. 2005. Differential chromosome control of ploidy in the yeast Saccharomyces cerevisiae. Yeast 22:625–639. doi: 10.1002/yea.1226. [DOI] [PubMed] [Google Scholar]
- 29.Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. 2010. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bozdag GO, Greig D. 2014. The genetics of a putative social trait in natural populations of yeast. Mol Ecol 23:5061–5071. doi: 10.1111/mec.12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, Kitano TK, Hodel MR, Petersen JF, Wyatt PW, Steenblock ER, Shah PH, Bousse LJ, Troup CB, Mellen JC, Wittmann DK, Erndt NG, Cauley TH, Koehler RT, So AP, Dube S, Rose KA, Montesclaros L, Wang S, Stumbo DP, Hodges SP, Romine S, Milanovich FP, White HE, Regan JF, Karlin-Neumann GA, Hindson CM, Saxonov S, Colston BW. 2011. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunn B, Sherlock G. 2008. Reconstruction of the genome origins and evolution of the hybrid lager yeast Saccharomyces pastorianus. Genome Res 18:1610–1623. doi: 10.1101/gr.076075.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gresham D, Dunham MJ, Botstein D. 2008. Comparing whole genomes using DNA microarrays. Nat Rev Genet 9:291–302. doi: 10.1038/nrg2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie C, Tammi MT. 2009. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics 10:80. doi: 10.1186/1471-2105-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu YO, Sherlock GJ, Petrov DA. 2016. Whole genome analysis of 132 clinical Saccharomyces cerevisiae strains reveals extensive ploidy variation. bioRxiv doi: 10.1101/044958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Dijk EL, Jaszczyszyn Y, Thermes C. 2014. Library preparation methods for next-generation sequencing: tone down the bias. Exp Cell Res 322:12–20. doi: 10.1016/j.yexcr.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 37.Yoon S, Xuan Z, Makarov V, Ye K, Sebat J. 2009. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res 19:1586–1592. doi: 10.1101/gr.092981.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao M, Wang Q, Wang Q, Jia PL, Zhao Z. 2013. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: features and perspectives. BMC Bioinformatics 14:S1. doi: 10.1186/1471-2105-14-S11-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chin C-S, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O'Malley R, Figueroa-Balderas R, Morales-Cruz A. 2016. Phased diploid genome assembly with single molecule real-time sequencing. bioRxiv doi: 10.1101/056887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodwin S, Gurtowski J, Ethe-Sayers S, Deshpande P, Schatz MC, McCombie WR. 2015. Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome. Genome Res 25:1750–1756. doi: 10.1101/gr.191395.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Istace B, Friedrich A, d'Agata L, Faye S, Payen E, Beluche O, Caradec C, Davidas S, Cruaud C, Liti G, Lemainque A, Engelen S, Wincker P, Schacherer J, Aury J-M. 2016. De novo assembly and population genomic survey of natural yeast isolates with the Oxford Nanopore MinION sequencer. bioRxiv doi: 10.1101/066613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhoads A, Au KF. 2015. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics 13:278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McIlwain SJ, Peris D, Sardi M, Moskvin OV, Zhan F, Myers KS, Riley NM, Buzzell A, Parreiras LS, Ong IM. 2016. Genome sequence and analysis of a stress-tolerant, wild-derived strain of Saccharomyces cerevisiae used in biofuels research. G3 6:1757–1766. doi: 10.1534/g3.116.029389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, Jovanovich SB, Krstic PS, Lindsay S, Ling XS, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey JM, Riehn R, Soni GV, Tabard-Cossa V, Wanunu M, Wiggin M, Schloss JA. 2008. The potential and challenges of nanopore sequencing. Nat Biotechnol 26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson SL, Bakhoum SF, Compton DA. 2010. Mechanisms of chromosomal instability. Curr Biol 20:R285–295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parry JM, Sharp D, Parry EM. 1979. Detection of mitotic and meiotic aneuploidy in the yeast Saccharomyces cerevisiae. Environ Health Perspect 31:97–111. doi: 10.1289/ehp.793197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Compton DA. 2011. Mechanisms of aneuploidy. Curr Opin Cell Biol 23:109–113. doi: 10.1016/j.ceb.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu YO, Siegal ML, Hall DW, Petrov DA. 2014. Precise estimates of mutation rate and spectrum in yeast. Proc Natl Acad Sci U S A 111:E2310–E2318. doi: 10.1073/pnas.1323011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parry EM, Cox BS. 1970. The tolerance of aneuploidy in yeast. Genet Res 16:333–340. doi: 10.1017/S0016672300002597. [DOI] [PubMed] [Google Scholar]
- 50.Adams J, Puskas-Rozsa S, Simlar J, Wilke CM. 1992. Adaptation and major chromosomal changes in populations of Saccharomyces cerevisiae. Curr Genet 22:13–19. doi: 10.1007/BF00351736. [DOI] [PubMed] [Google Scholar]
- 51.Chen G, Rubinstein B, Li R. 2012. Whole chromosome aneuploidy: big mutations drive adaptation by phenotypic leap. Bioessays 34:893–900. doi: 10.1002/bies.201200069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parry JM, Sharp D, Tippins RS, Parry EM. 1979. Radiation-induced mitotic and meiotic aneuploidy in the yeast Saccharomyces cerevisiae. Mutat Res 61:37–55. doi: 10.1016/0027-5107(79)90005-8. [DOI] [PubMed] [Google Scholar]
- 53.Albertini S, Zimmermann FK. 1991. The detection of chemically induced chromosomal malsegregation in Saccharomyces cerevisiae D61.M: a literature survey (1984–1990). Mutat Res 258:237–258. doi: 10.1016/0165-1110(91)90011-J. [DOI] [PubMed] [Google Scholar]
- 54.Liu M, Grant SG, Macina OT, Klopman G, Rosenkranz HS. 1997. Structural and mechanistic bases for the induction of mitotic chromosomal loss and duplication (‘malsegregation') in the yeast Saccharomyces cerevisiae: relevance to human carcinogenesis and developmental toxicology. Mutat Res 374:209–231. doi: 10.1016/S0027-5107(96)00236-9. [DOI] [PubMed] [Google Scholar]
- 55.Wood JS. 1982. Genetic effects of methyl benzimidazole-2-yl-carbamate on Saccharomyces cerevisiae. Mol Cell Biol 2:1064–1079. doi: 10.1128/MCB.2.9.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zimmermann FK, Mayer VW, Scheel I, Resnick MA. 1985. Acetone, methyl ethyl ketone, ethyl acetate, acetonitrile and other polar aprotic solvents are strong inducers of aneuploidy in Saccharomyces cerevisiae. Mutat Res 149:339–351. doi: 10.1016/0027-5107(85)90150-2. [DOI] [PubMed] [Google Scholar]
- 57.Crebelli R, Conti G, Conti L, Carere A. 1989. A comparative study on ethanol and acetaldehyde as inducers of chromosome malsegregation in Aspergillus nidulans. Mutat Res Fund 215:187–195. doi: 10.1016/0027-5107(89)90183-8. [DOI] [PubMed] [Google Scholar]
- 58.Storchova Z. 2014. Ploidy changes and genome stability in yeast. Yeast 31:421–430. doi: 10.1002/yea.3037. [DOI] [PubMed] [Google Scholar]
- 59.Mayer VW, Aguilera A. 1990. High levels of chromosome instability in polyploids of Saccharomyces cerevisiae. Mutat Res 231:177–186. doi: 10.1016/0027-5107(90)90024-X. [DOI] [PubMed] [Google Scholar]
- 60.Delneri D, Colson I, Grammenoudi S, Roberts IN, Louis EJ, Oliver SG. 2003. Engineering evolution to study speciation in yeasts. Nature 422:68–72. doi: 10.1038/nature01418. [DOI] [PubMed] [Google Scholar]
- 61.Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, Amon A. 2011. Aneuploidy drives genomic instability in yeast. Science 333:1026–1030. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reid RJD, Sunjevaric I, Voth WP, Ciccone S, Du W, Olsen AE, Stillman DJ, Rothstein R. 2008. Chromosome-scale genetic mapping using a set of 16 conditionally stable Saccharomyces cerevisiae chromosomes. Genetics 180:1799–1808. doi: 10.1534/genetics.108.087999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anders KR, Kudrna JR, Keller KE, Kinghorn B, Miller EM, Pauw D, Peck AT, Shellooe CE, Strong IJ. 2009. A strategy for constructing aneuploid yeast strains by transient nondisjunction of a target chromosome. BMC Genet 10:36. doi: 10.1186/1471-2156-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 65.Sheltzer JM, Torres EM, Dunham MJ, Amon A. 2012. Transcriptional consequences of aneuploidy. Proc Natl Acad Sci U S A 109:12644–12649. doi: 10.1073/pnas.1209227109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sunshine AB, Ong GT, Nickerson DP, Carr D, Murakami CJ, Wasko BM, Shemorry A, Merz AJ, Kaeberlein M, Dunham MJ. 2016. Aneuploidy shortens replicative lifespan in Saccharomyces cerevisiae. Aging Cell 15:317–324. doi: 10.1111/acel.12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oromendia AB, Amon A. 2014. Aneuploidy: implications for protein homeostasis and disease. Dis Model Mech 7:15–20. doi: 10.1242/dmm.013391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beach RR. 2016. Insights into the consequences of chromosome gains and losses in S. cerevisiae. PhD thesis Massachusetts Institute of Technology, Cambridge, MA. [Google Scholar]
- 69.Tang Y-C, Amon A. 2013. Gene copy-number alterations: a cost-benefit analysis. Cell 152:394–405. doi: 10.1016/j.cell.2012.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oromendia AB, Dodgson SE, Amon A. 2012. Aneuploidy causes proteotoxic stress in yeast. Genes Dev 26:2696–2708. doi: 10.1101/gad.207407.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Janssen A, van der Burg M, Szuhai K, Kops GJPL, Medema RH. 2011. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 333:1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- 72.Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. 2012. DNA breaks and chromosome pulverization from errors in mitosis. Nature 482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hoffelder DR, Luo L, Burke NA, Watkins SC, Gollin SM, Saunders WS. 2004. Resolution of anaphase bridges in cancer cells. Chromosoma 112:389–397. doi: 10.1007/s00412-004-0284-6. [DOI] [PubMed] [Google Scholar]
- 74.Terradas M, Martín M, Tusell L, Genescà A. 2009. DNA lesions sequestered in micronuclei induce a local defective-damage response. DNA Repair 8:1225–1234. doi: 10.1016/j.dnarep.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 75.Blank HM, Sheltzer JM, Meehl CM, Amon A. 2015. Mitotic entry in the presence of DNA damage is a widespread property of aneuploidy in yeast. Mol Biol Cell 26:1440–1451. doi: 10.1091/mbc.E14-10-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Skoneczna A, Kaniak A, Skoneczny M. 2015. Genetic instability in budding and fission yeast-sources and mechanisms. FEMS Microbiol Rev 39:917–967. doi: 10.1093/femsre/fuv028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Anderson E, Martin PA. 1975. The sporulation and mating of brewing yeasts. J Inst Brew 81:242–247. doi: 10.1002/j.2050-0416.1975.tb03685.x. [DOI] [Google Scholar]
- 78.Thorburn RR, Gonzalez C, Brar GA, Christen S, Carlile TM, Ingolia NT, Sauer U, Weissman JS, Amon A. 2013. Aneuploid yeast strains exhibit defects in cell growth and passage through START. Mol Biol Cell 24:1274–1289. doi: 10.1091/mbc.E12-07-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Niwa O, Tange Y, Kurabayashi A. 2006. Growth arrest and chromosome instability in aneuploid yeast. Yeast 23:937–950. doi: 10.1002/yea.1411. [DOI] [PubMed] [Google Scholar]
- 80.Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer MR, Kobayashi S, Davis C, Dai H, He YD, Stephaniants SB, Cavet G, Walker WL, West A, Coffey E, Shoemaker DD, Stoughton R, Blanchard AP, Friend SH, Linsley PS. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol 19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 81.Dephoure N, Hwang S, O'Sullivan C, Dodgson SE, Gygi SP, Amon A, Torres EM. 2014. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. eLife 3:e03023. doi: 10.7554/eLife.03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A. 2010. Identification of aneuploidy-tolerating mutations. Cell 143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li GW, Burkhardt D, Gross C, Weissman JS. 2014. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tang YC, Williams BR, Siegel JJ, Amon A. 2011. Identification of aneuploidy-selective antiproliferation compounds. Cell 144:499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Geiler-Samerotte KA, Dion MF, Budnik BA, Wang SM, Hartl DL, Drummond DA. 2011. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc Natl Acad Sci U S A 108:680–685. doi: 10.1073/pnas.1017570108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Birchler JA, Bhadra U, Bhadra MP, Auger DL. 2001. Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev Biol 234:275–288. doi: 10.1006/dbio.2001.0262. [DOI] [PubMed] [Google Scholar]
- 87.Berman J. 2016. Ploidy plasticity: a rapid and reversible strategy for adaptation to stress. FEMS Yeast Res 16:fow020. doi: 10.1093/femsyr/fow020. [DOI] [PubMed] [Google Scholar]
- 88.Gasch AP, Hose J, Newton MA, Sardi M, Yong M, Wang Z. 2016. Further support for aneuploidy tolerance in wild yeast and effects of dosage compensation on gene copy-number evolution. eLife 5:e14409. doi: 10.7554/eLife.14409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Veitia RA, Bottani S, Birchler JA. 2008. Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet 24:390–397. doi: 10.1016/j.tig.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 90.Dodgson SE, Kim S, Costanzo M, Baryshnikova A, Morse DL, Kaiser CA, Boone C, Amon A. 2016. Chromosome-specific and global effects of aneuploidy in Saccharomyces cerevisiae. Genetics 202:1395–1409. doi: 10.1534/genetics.115.185660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hose J, Yong CM, Sardi M, Wang Z, Newton MA, Gasch AP. 2015. Dosage compensation can buffer copy-number variation in wild yeast. eLife 4:e05462. doi: 10.7554/eLife.05462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cromie GA, Dudley AM. 2015. Aneuploidy: tolerating tolerance. Curr Biol 25:R771–773. doi: 10.1016/j.cub.2015.06.056. [DOI] [PubMed] [Google Scholar]
- 93.Gallone B, Steensels J, Prahl T, Soriaga L, Saels V, Herrera-Malaver B, Merlevede A, Roncoroni M, Voordeckers K, Miraglia L. 2016. Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell 166:1397–1410. doi: 10.1016/j.cell.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rancati G, Pavelka N, Fleharty B, Noll A, Trimble R, Walton K, Perera A, Staehling-Hampton K, Seidel CW, Li R. 2008. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell 135:879–893. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rancati G, Pavelka N. 2013. Karyotypic changes as drivers and catalyzers of cellular evolvability: a perspective from non-pathogenic yeasts. Semin Cell Dev Biol 24:332–338. doi: 10.1016/j.semcdb.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 96.Voordeckers K, Verstrepen KJ. 2015. Experimental evolution of the model eukaryote Saccharomyces cerevisiae yields insight into the molecular mechanisms underlying adaptation. Curr Opin Microbiol 28:1–9. doi: 10.1016/j.mib.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 97.Kohn LM. 2005. Mechanisms of fungal speciation. Annu Rev Phytopathol 43:279–308. doi: 10.1146/annurev.phyto.43.040204.135958. [DOI] [PubMed] [Google Scholar]
- 98.Dragosits M, Mattanovich D. 2013. Adaptive laboratory evolution—principles and applications for biotechnology. Microb Cell Fact 12:1. doi: 10.1186/1475-2859-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sauer U. 2001. Evolutionary engineering of industrially important microbial phenotypes. Adv Biochem Eng Biotechnol 73:129–169. [DOI] [PubMed] [Google Scholar]
- 100.Çakar ZP, Turanlı-Yıldız B, Alkım C, Yılmaz Ü. 2012. Evolutionary engineering of Saccharomyces cerevisiae for improved industrially important properties. FEMS Yeast Res 12:171–182. doi: 10.1111/j.1567-1364.2011.00775.x. [DOI] [PubMed] [Google Scholar]
- 101.Oud B, van Maris AJA, Daran J-MG, Pronk JT. 2012. Genome-wide analytical approaches for reverse metabolic engineering of industrially relevant phenotypes in yeast. FEMS Yeast Res 12:183–196. doi: 10.1111/j.1567-1364.2011.00776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bachmann H, Pronk JT, Kleerebezem M, Teusink B. 2015. Evolutionary engineering to enhance starter culture performance in food fermentations. Curr Opin Biotechnol 32:1–7. doi: 10.1016/j.copbio.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 103.Yona AH, Manor YS, Herbst RH, Romano GH, Mitchell A, Kupiec M, Pilpel Y, Dahan O. 2012. Chromosomal duplication is a transient evolutionary solution to stress. Proc Natl Acad Sci U S A 109:21010–21015. doi: 10.1073/pnas.1211150109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gerstein AC, Ono J, Lo DS, Campbell ML, Kuzmin A, Otto SP. 2015. Too much of a good thing: the unique and repeated paths toward copper adaptation. Genetics 199:555–571. doi: 10.1534/genetics.114.171124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Oud B, Guadalupe-Medina V, Nijkamp JF, de Ridder D, Pronk JT, van Maris AJA, Daran J-MG. 2013. Genome duplication and mutations in ACE2 cause multicellular, fast-sedimenting phenotypes in evolved Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 110:E4223–E4231. doi: 10.1073/pnas.1305949110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Voordeckers K, Kominek J, Das A, Espinosa-Cantu A, De Maeyer D, Arslan A, Van Pee M, van der Zande E, Meert W, Yang Y, Zhu B, Marchal K, DeLuna A, Van Noort V, Jelier R, Verstrepen KJ. 2015. Adaptation to high ethanol reveals complex evolutionary pathways. PLoS Genet 11:e1005635. doi: 10.1371/journal.pgen.1005635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Venkataram S, Dunn B, Li Y, Agarwala A, Chang J, Ebel ER, Geiler-Samerotte K, Hérissant L, Blundell JR, Levy SF. 2016. Development of a comprehensive genotype-to-fitness map of adaptation-driving mutations in yeast. Cell 166:1585–1596.e22. doi: 10.1016/j.cell.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, DeSevo CG, Botstein D, Dunham MJ. 2008. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet 4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.González-Ramos D, Gorter de Vries AR, Grijseels SS, Berkum MC, Swinnen S, Broek M, Nevoigt E, Daran J-MG, Pronk JT, Maris AJA. 2016. A new laboratory evolution approach to select for constitutive acetic acid tolerance in Saccharomyces cerevisiae and identification of causal mutations. Biotechnol Biofuels 9:173. doi: 10.1186/s13068-016-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Payen C, Di Rienzi SC, Ong GT, Pogachar JL, Sanchez JC, Sunshine AB, Raghuraman M, Brewer BJ, Dunham MJ. 2014. The dynamics of diverse segmental amplifications in populations of Saccharomyces cerevisiae adapting to strong selection. G3 4:399–409. doi: 10.1534/g3.113.009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F, Botstein D. 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.de Kok S, Nijkamp JF, Oud B, Roque FC, de Ridder D, Daran JMG, Pronk JT, Maris AJA. 2012. Laboratory evolution of new lactate transporter genes in a jen1Δ mutant of Saccharomyces cerevisiae and their identification as ADY2 alleles by whole-genome resequencing and transcriptome analysis. FEMS Yeast Res 12:359–374. doi: 10.1111/j.1567-1364.2011.00787.x. [DOI] [PubMed] [Google Scholar]
- 113.Chen GB, Bradford WD, Seidel CW, Li R. 2012. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature 482:246–250. doi: 10.1038/nature10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sunshine AB, Payen C, Ong GT, Liachko I, Tan KM, Dunham MJ. 2015. The fitness consequences of aneuploidy are driven by condition-dependent gene effects. PLoS Biol 13:e1002155. doi: 10.1371/journal.pbio.1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Natesuntorn W, Iwami K, Matsubara Y, Sasano Y, Sugiyama M, Kaneko Y, Harashima S. 2015. Genome-wide construction of a series of designed segmental aneuploids in Saccharomyces cerevisiae. Sci Rep 5:12510. doi: 10.1038/srep12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sugiyama M, Nakazawa T, Murakami K, Sumiya T, Nakamura A, Kaneko Y, Nishizawa M, Harashima S. 2008. PCR-mediated one-step deletion of targeted chromosomal regions in haploid Saccharomyces cerevisiae. Appl Microbiol Biotechnol 80:545–553. doi: 10.1007/s00253-008-1609-9. [DOI] [PubMed] [Google Scholar]
- 117.Borneman AR, Desany BA, Riches D, Affourtit JP, Forgan AH, Pretorius IS, Egholm M, Chambers PJ. 2011. Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. PLoS Genet 7:e1001287. doi: 10.1371/journal.pgen.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Codon AC, Benitez T, Korhola M. 1998. Chromosomal polymorphism and adaptation to specific industrial environments of Saccharomyces strains. Appl Microbiol Biotechnol 49:154–163. doi: 10.1007/s002530051152. [DOI] [PubMed] [Google Scholar]
- 119.Zheng DQ, Chen J, Zhang K, Gao KH, Li O, Wang PM, Zhang XY, Du FG, Sun PY, Qu AM, Wu S, Wu XC. 2014. Genomic structural variations contribute to trait improvement during whole-genome shuffling of yeast. Appl Microbiol Biotechnol 98:3059–3070. doi: 10.1007/s00253-013-5423-7. [DOI] [PubMed] [Google Scholar]
- 120.Naumov GI, Naumova ES, Louis EJ. 1995. Genetic mapping of the α-galactosidase MEL gene family on right and left telomeres of Saccharomyces cerevisiae. Yeast 11:481–483. doi: 10.1002/yea.320110512. [DOI] [PubMed] [Google Scholar]
- 121.Carlson M, Celenza JL, Eng FJ. 1985. Evolution of the dispersed SUC gene family of Saccharomyces by rearrangements of chromosome telomeres. Mol Cell Biol 5:2894–2902. doi: 10.1128/MCB.5.11.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Brown CA, Murray AW, Verstrepen KJ. 2010. Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr Biol 20:895–903. doi: 10.1016/j.cub.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ibáñez C, Pérez-Torrado R, Chiva R, Guillamón JM, Barrio E, Querol A. 2014. Comparative genomic analysis of Saccharomyces cerevisiae yeasts isolated from fermentations of traditional beverages unveils different adaptive strategies. Int J Food Microbiol 171:129–135. doi: 10.1016/j.ijfoodmicro.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 124.Zhang K, Tong M, Gao K, Di Y, Wang P, Zhang C, Wu X, Zheng D. 2015. Genomic reconstruction to improve bioethanol and ergosterol production of industrial yeast Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 42:207–218. doi: 10.1007/s10295-014-1556-7. [DOI] [PubMed] [Google Scholar]
- 125.Querol A, Bond U. 2009. The complex and dynamic genomes of industrial yeasts. FEMS Microbiol Lett 293:1–10. doi: 10.1111/j.1574-6968.2008.01480.x. [DOI] [PubMed] [Google Scholar]
- 126.Tadami H, Shikata-Miyoshi M, Ogata T. 2014. Aneuploidy, copy number variation and unique chromosomal structures in bottom-fermenting yeast revealed by array-CGH. J Inst Brew 120:27–37. doi: 10.1002/jib.108. [DOI] [Google Scholar]
- 127.Gonzalez SS, Barrio E, Gafner J, Querol A. 2006. Natural hybrids from Saccharomyces cerevisiae, Saccharomyces bayanus and Saccharomyces kudriavzevii in wine fermentations. FEMS Yeast Res 6:1221–1234. doi: 10.1111/j.1567-1364.2006.00126.x. [DOI] [PubMed] [Google Scholar]
- 128.Peris D, Belloch C, Lopandic K, Alvarez-Perez JM, Querol A, Barrio E. 2012. The molecular characterization of new types of Saccharomyces cerevisiae × S. kudriavzevii hybrid yeasts unveils a high genetic diversity. Yeast 29:81–91. doi: 10.1002/yea.2891. [DOI] [PubMed] [Google Scholar]
- 129.Dunn B, Richter C, Kvitek DJ, Pugh T, Sherlock G. 2012. Analysis of the Saccharomyces cerevisiae pan-genome reveals a pool of copy number variants distributed in diverse yeast strains from differing industrial environments. Genome Res 22:908–924. doi: 10.1101/gr.130310.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Guijo S, Mauricio J, Salmon J, Ortega J. 1997. Determination of the relative ploidy in different Saccharomyces cerevisiae strains used for fermentation and ‘flor’ film ageing of dry sherry-type wines. Yeast 13:101–117. doi:. [DOI] [PubMed] [Google Scholar]
- 131.Nakao Y, Kanamori T, Itoh T, Kodama Y, Rainieri S, Nakamura N, Shimonaga T, Hattori M, Ashikari T. 2009. Genome sequence of the lager brewing yeast, an interspecies hybrid. DNA Res 16:115–129. doi: 10.1093/dnares/dsp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Libkind D, Hittinger CT, Valerio E, Goncalves C, Dover J, Johnston M, Goncalves P, Sampaio JP. 2011. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc Natl Acad Sci U S A 108:14539–14544. doi: 10.1073/pnas.1105430108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gayevskiy V, Goddard MR. 2016. Saccharomyces eubayanus and Saccharomyces arboricola reside in North Island native New Zealand forests. Environ Microbiol 18:1137–1147. doi: 10.1111/1462-2920.13107. [DOI] [PubMed] [Google Scholar]
- 134.Peris D, Sylvester K, Libkind D, Goncalves P, Sampaio JP, Alexander WG, Hittinger CT. 2014. Population structure and reticulate evolution of Saccharomyces eubayanus and its lager-brewing hybrids. Mol Ecol 23:2031–2045. doi: 10.1111/mec.12702. [DOI] [PubMed] [Google Scholar]
- 135.Bing J, Han P-J, Liu W-Q, Wang Q-M, Bai F-Y. 2014. Evidence for a Far East Asian origin of lager beer yeast. Curr Biol 24:R380–R381. doi: 10.1016/j.cub.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 136.Peris D, Langdon QK, Moriarty RV, Sylvester K, Bontrager M, Charron G, Leducq J-B, Landry CR, Libkind D, Hittinger CT. 2016. Complex ancestries of lager-brewing hybrids were shaped by standing variation in the wild yeast saccharomyces eubayanus. PLoS Genet 12:e1006155. doi: 10.1371/journal.pgen.1006155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gibson B, Liti G. 2015. Saccharomyces pastorianus: genomic insights inspiring innovation for industry. Yeast 32:17–27. doi: 10.1002/yea.3033. [DOI] [PubMed] [Google Scholar]
- 138.Wendland J. 2014. Lager yeast comes of age. Eukaryot Cell 13:1256–1265. doi: 10.1128/EC.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hebly M, Brickwedde A, Bolat I, Driessen MRM, de Hulster EAF, van den Broek M, Pronk JT, Geertman J-M, Daran J-MG, Daran-Lapujade P. 2015. S. cerevisiae × S. eubayanus interspecific hybrid, the best of both worlds and beyond. FEMS Yeast Res 15:fov005. doi: 10.1093/femsyr/fov005. [DOI] [PubMed] [Google Scholar]
- 140.Krogerus K, Magalhães F, Vidgren V, Gibson B. 2015. New lager yeast strains generated by interspecific hybridization. J Ind Microbiol Biotechnol 42:769–778. doi: 10.1007/s10295-015-1597-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Okuno M, Kajitani R, Ryusui R, Morimoto H, Kodama Y, Itoh T. 2016. Next-generation sequencing analysis of lager brewing yeast strains reveals the evolutionary history of interspecies hybridization. DNA Res 23:67–80. doi: 10.1093/dnares/dsv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Walther A, Hesselbart A, Wendland J. 2014. Genome sequence of Saccharomyces carlsbergensis, the world's first pure culture lager yeast. G3 4:783–793. doi: 10.1534/g3.113.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hewitt SK, Donaldson IJ, Lovell SC, Delneri D. 2014. Sequencing and characterisation of rearrangements in three S. pastorianus strains reveals the presence of chimeric genes and gives evidence of breakpoint reuse. PLoS One 9:e92203. doi: 10.1371/journal.pone.0092203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Baker EC, Wang B, Bellora N, Peris D, Hulfachor AB, Koshalek JA, Adams M, Libkind D, Hittinger CT. 2015. The genome sequence of Saccharomyces eubayanus and the domestication of lager-brewing yeasts. Mol Biol Evol 32:2818–2831. doi: 10.1093/molbev/msv168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gordon JL, Byrne KP, Wolfe KH. 2009. Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet 5:e1000485. doi: 10.1371/journal.pgen.1000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dunn B, Paulish T, Stanbery A, Piotrowski J, Koniges G, Kroll E, Louis EJ, Liti G, Sherlock G, Rosenzweig F. 2013. Recurrent rearrangement during adaptive evolution in an interspecific yeast hybrid suggests a model for rapid introgression. PLoS Genet 9:e1003366. doi: 10.1371/journal.pgen.1003366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Strope PK, Skelly DA, Kozmin SG, Mahadevan G, Stone EA, Magwene PM, Dietrich FS, McCusker JH. 2015. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res 25:762–774. doi: 10.1101/gr.185538.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Krogerus K, Arvas M, Chiara M, Magalhães F, Mattinen L, Oja M, Vidgren V, Yue J-X, Liti G, Gibson B. 2016. Ploidy influences the functional attributes of de novo lager yeast hybrids. Appl Microbiol Biotechnol 100:7203–7222. doi: 10.1007/s00253-016-7588-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bellon JR, Eglinton JM, Siebert TE, Pollnitz AP, Rose L, de Barros Lopes M, Chambers PJ. 2011. Newly generated interspecific wine yeast hybrids introduce flavour and aroma diversity to wines. Appl Microbiol Biotechnol 91:603–612. doi: 10.1007/s00253-011-3294-3. [DOI] [PubMed] [Google Scholar]
- 150.Zambonelli C, Passarelli P, Rainieri S, Bertolini L, Giudici P, Castellari L. 1997. Technological properties and temperature response of interspecific Saccharomyces hybrids. J Sci Food Agric 74:7–12. [Google Scholar]
- 151.Coloretti F, Zambonelli C, Tini V. 2006. Characterization of flocculent Saccharomyces interspecific hybrids for the production of sparkling wines. Food Microbiol 23:672–676. doi: 10.1016/j.fm.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 152.Tronchoni J, Gamero A, Arroyo-Lopez FN, Barrio E, Querol A. 2009. Differences in the glucose and fructose consumption profiles in diverse Saccharomyces wine species and their hybrids during grape juice fermentation. Int J Food Microbiol 134:237–243. doi: 10.1016/j.ijfoodmicro.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 153.Hou LH, Meng M, Guo L, He JY. 2015. A comparison of whole cell directed evolution approaches in breeding of industrial strain of Saccharomyces cerevisiae. Biotechnol Lett 37:1393–1398. doi: 10.1007/s10529-015-1812-6. [DOI] [PubMed] [Google Scholar]
- 154.Zheng D, Zhang K, Gao K, Liu Z, Zhang X, Li O, Sun J, Zhang X, Du F, Sun P, Qu A, Wu X. 2013. Construction of novel Saccharomyces cerevisiae strains for bioethanol active dry yeast (ADY) production. PLoS One 8:e85022. doi: 10.1371/journal.pone.0085022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mans R, van Rossum HM, Wijsman M, Backx A, Kuijpers NGA, van den Broek M, Daran-Lapujade P, Pronk JT, van Maris AJA, Daran J-MG. 2015. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res 15:fov004. doi: 10.1093/femsyr/fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. 2013. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Heid CA, Stevens J, Livak KJ, Williams PM. 1996. Real time quantitative PCR. Genome Res 6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 158.Musacchio A, Salmon ED. 2007. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 159.Ganem NJ, Godinho SA, Pellman D. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature 460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sato TK, Liu T, Parreiras LS, Williams DL, Wohlbach DJ, Bice BD, Ong IM, Breuer RJ, Qin L, Busalacchi D, Deshpande S, Daum C, Gasch AP, Hodge DB. 2014. Harnessing genetic diversity in Saccharomyces cerevisiae for fermentation of xylose in hydrolysates of alkaline hydrogen peroxide-pretreated biomass. Appl Environ Microbiol 80:540–554. doi: 10.1128/AEM.01885-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sirr A, Cromie GA, Jeffery EW, Gilbert TL, Ludlow CL, Scott AC, Dudley AM. 2015. Allelic variation, aneuploidy, and nongenetic mechanisms suppress a monogenic trait in yeast. Genetics 199:247–262. doi: 10.1534/genetics.114.170563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gasch AP, Huang M, Metzner S, Botstein D, Elledge SJ, Brown PO. 2001. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell 12:2987–3003. doi: 10.1091/mbc.12.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hughes TR, Roberts CJ, Dai H, Jones AR, Meyer MR, Slade D, Burchard J, Dow S, Ward TR, Kidd MJ, Friend SH, Marton MJ. 2000. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet 25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 164.Millet C, Makovets S. 2016. Aneuploidy as a mechanism of adaptation to telomerase insufficiency. Curr Genet 62:557–564. doi: 10.1007/s00294-015-0559-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ludlow CL, Cromie GA, Garmendia-Torres C, Sirr A, Hays M, Field C, Jeffery EW, Fay JC, Dudley AM. 2016. Independent origins of yeast associated with coffee and cacao fermentation. Curr Biol 26:965–971. doi: 10.1016/j.cub.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhang K, Zhang LJ, Fang YH, Jin XN, Qi L, Wu XC, Zheng DQ. 2016. Genomic structural variation contributes to phenotypic change of industrial bioethanol yeast Saccharomyces cerevisiae. FEMS Yeast Res 16:fov118. doi: 10.1093/femsyr/fov118. [DOI] [PubMed] [Google Scholar]