ABSTRACT
Wetlands constitute the main natural source of methane on Earth due to their high content of natural organic matter (NOM), but key drivers, such as electron acceptors, supporting methanotrophic activities in these habitats are poorly understood. We performed anoxic incubations using freshly collected sediment, along with water samples harvested from a tropical wetland, amended with 13C-methane (0.67 atm) to test the capacity of its microbial community to perform anaerobic oxidation of methane (AOM) linked to the reduction of the humic fraction of its NOM. Collected evidence demonstrates that electron-accepting functional groups (e.g., quinones) present in NOM fueled AOM by serving as a terminal electron acceptor. Indeed, while sulfate reduction was the predominant process, accounting for up to 42.5% of the AOM activities, the microbial reduction of NOM concomitantly occurred. Furthermore, enrichment of wetland sediment with external NOM provided a complementary electron-accepting capacity, of which reduction accounted for ∼100 nmol 13CH4 oxidized · cm−3 · day−1. Spectroscopic evidence showed that quinone moieties were heterogeneously distributed in the wetland sediment, and their reduction occurred during the course of AOM. Moreover, an enrichment derived from wetland sediments performing AOM linked to NOM reduction stoichiometrically oxidized methane coupled to the reduction of the humic analogue anthraquinone-2,6-disulfonate. Microbial populations potentially involved in AOM coupled to microbial reduction of NOM were dominated by divergent biota from putative AOM-associated archaea. We estimate that this microbial process potentially contributes to the suppression of up to 114 teragrams (Tg) of CH4 · year−1 in coastal wetlands and more than 1,300 Tg · year−1, considering the global wetland area.
IMPORTANCE The identification of key processes governing methane emissions from natural systems is of major importance considering the global warming effects triggered by this greenhouse gas. Anaerobic oxidation of methane (AOM) coupled to the microbial reduction of distinct electron acceptors plays a pivotal role in mitigating methane emissions from ecosystems. Given their high organic content, wetlands constitute the largest natural source of atmospheric methane. Nevertheless, processes controlling methane emissions in these environments are poorly understood. Here, we provide tracer analysis with 13CH4 and spectroscopic evidence revealing that AOM linked to the microbial reduction of redox functional groups in natural organic matter (NOM) prevails in a tropical wetland. We suggest that microbial reduction of NOM may largely contribute to the suppression of methane emissions from tropical wetlands. This is a novel avenue within the carbon cycle in which slowly decaying NOM (e.g., humic fraction) in organotrophic environments fuels AOM by serving as a terminal electron acceptor.
KEYWORDS: anaerobic methane oxidation, humus, methanotrophy, wetlands
INTRODUCTION
Microbial processes produce and consume methane (CH4) in anoxic sediments, playing a crucial role in regulating Earth's climate. Virtually 90% of the CH4 produced from marine environments is oxidized by microorganisms, preventing its release into the atmosphere (1). Anaerobic oxidation of methane (AOM) associated with sulfate reduction was first discovered in marine environments (2). More recently, AOM has also been linked to the microbial reduction of nitrate (3, 4) and nitrite (5), as well as Fe(III) and Mn(IV) oxides (6–8), in freshwater and marine environments. Wetlands are the largest natural source of CH4 (9), contributing to about one-third of global emissions (10), but key drivers, such as electron acceptors fueling methanotrophic activities in these habitats, are poorly understood. CH4 emissions from wetlands have been strongly responsive to climate in the past and will likely continue to be responsive to anthropogenic-driven climate change in the future, predicting a large impact on global atmospheric CH4 concentration (10). The traditional assumption is that aerobic methanotrophy dominates CH4 cycling in wetlands by oxidizing an estimated 40 to 70% of the gross CH4 production in these ecosystems (11). Recent findings (12) challenged this conjecture by providing evidence that AOM may consume up to 200 Tg of CH4 · year−1, decreasing their potential CH4 emission by 50% in these habitats. Most AOM activities observed in wetlands have been related to sulfate reduction (12, 13), but other electron acceptors remain feasible. Natural organic matter (NOM), circumscribed to humic substances (HS) in many studies (14), occurs at high concentrations in wetlands in both soluble and solid phases (15). Recent evidence indicates that HS suppress methane production in different ecosystems (16, 17), yet the mechanisms involved are still enigmatic. HS can theoretically promote AOM, as they can serve as terminal electron acceptors for microbial respiration (18, 19) and have a higher redox potential than sulfate (20). However, compelling evidence demonstrating AOM driven by the microbial reduction of NOM present in anoxic environments remains elusive (21, 22).
We aimed to document 13CH4 anaerobic oxidation and the ongoing reduction of intrinsic electron acceptors, including the electron-accepting fraction of NOM, by the biota of freshly sampled sediment from a coastal tropical wetland. We provide 13CH4 tracer studies and spectroscopic evidence demonstrating that AOM is linked to the microbial reduction of redox functional groups present in the NOM of this tropical marsh. Furthermore, we found evidence, based on 16S rRNA gene sequences, indicating that microbial populations potentially involved in AOM coupled to microbial reduction of NOM were dominated by divergent biota from putative AOM-associated microorganisms.
RESULTS
Kinetics of 13C-methane oxidation and electron balances.
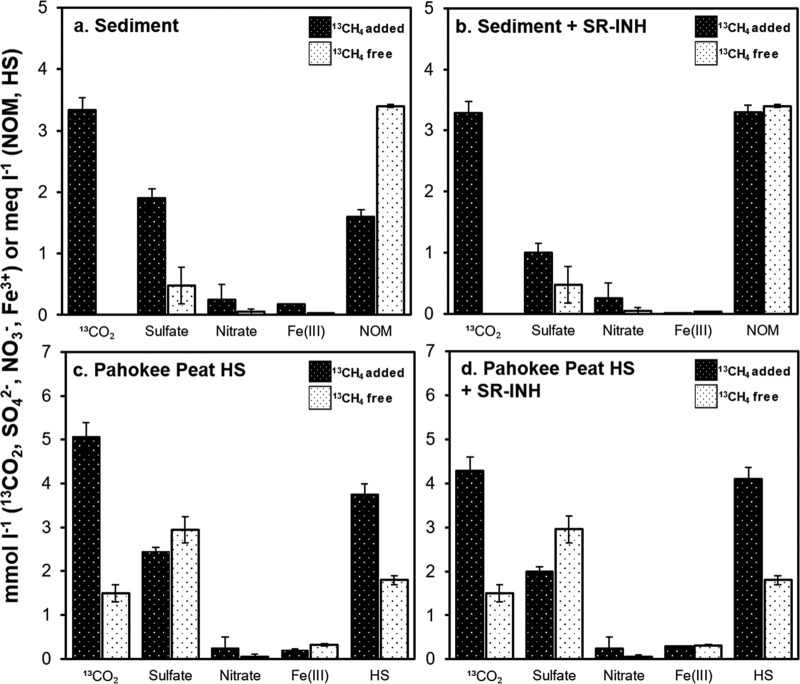
The exponential phase of AOM was observed in microcosms over the first 15 days of incubation in cases with unamended sediment (i.e., sediment without external NOM addition). The methanotrophic rate in this experimental treatment was ∼1.34 μmol 13C-methane oxidized · cm−3 · day−1 (Fig. 1). At the end of the exponential phase, sulfate and Fe(III) reduction accounted for 42.5% and 0.5% of the 13C-methane oxidized, respectively, while the role of nitrate was marginal (Fig. 2 and Table S2). These unamended sediment microcosms exhibited a reduction in intrinsic NOM during the course of AOM, which was expected due to the high concentration of organic carbon in the tropical wetland, with the capacity to accept electrons (Table S1 in the supplemental material and Fig. 2). Nevertheless, large perturbation caused by endogenous NOM reduction in experimental controls lacking 13C-methane obstructed an accurate assessment of AOM driven by this microbial process (Fig. 2). The large endogenous NOM reduction observed in these control experiments may be explained by the concomitant methane production (and subsequent consumption) observed (Fig. S2) and by oxidation of labile organic matter present in the sediment (Table S1). Supplementary incubations spiked with the sulfate reduction inhibitor molybdate (25 mmol liter−1) showed decreased sulfate-reducing activity (∼50%, Fig. 2), while AOM rates remained high compared against their noninhibited counterparts (Fig. 1). Remarkably, when sulfate reduction was inhibited, the reduction of intrinsic NOM was doubled (from 1.6 ± 0.11 to 3.4 ± 0.19 millielectron equivalents [meq] · liter−1), implying that the reduction of redox functional groups in NOM was promoted when the utilization of sulfate was impeded.
FIG 1.
Anaerobic methane oxidation measured as 13CO2 production in microcosms' headspace and 13C enrichment (inset) calculated as 13FCO2 = (13CO2/[13CO2 + 12CO2]). (a) Kinetics for incubations performed with unamended sediment. (b) Kinetics for incubations performed with sediment enriched with 2.5 g · liter−1 of external NOM in the form of Pahokee peat humic substances. Error bars represent the standard error among replicates (n = 4, or 3*). SR-INH stands for sediment incubations performed with molybdate (25 mmol liter−1) in order to inhibit sulfate reduction. 13CO2 production rates were based on the maximum slope observed on linear regressions considering at least three sampling points.
FIG 2.
Production of 13CO2 and reduction of intrinsic or added electron acceptors at the end of the exponential phase (20 days of incubation) in the absence (a and b) and in the presence (c and d) of external NOM as HS from Pahokee peat. SR-INH stands for controls amended with the sulfate reduction inhibitor molybdate (25 mmol liter−1). Error bars represent the standard error among replicates. 13CO2 produced was measured as described for Fig. 1. Quantification of sulfate and nitrate reduction implies a decrease in their concentration at this sampling time, whereas Fe(III) reduction was quantified in terms of the ferrous iron produced. Reduction of NOM and HS was determined by the ferrozine technique.
Further enrichment of wetland sediment with external NOM, in the form of HS derived from Pahokee peat (Florida Everglades, 2.5 g · liter−1), provided complementary electron-accepting capacity, which significantly elicited AOM up to ∼1.88 μmol 13C-methane oxidized · cm−3 · day−1 and extended the exponential phase to 20 days (Fig. 1). In this experimental treatment, electron balances revealed methanotrophic activity responsible for ∼100 nmol 13CH4 oxidized · cm−3 · day−1 linked to microbial reduction of NOM (including both intrinsic and externally added as Pahokee peat HS). As hypothesized before, the consumption of intrinsically produced methane was confirmed by experimental controls enriched with HS from Pahokee peat and incubated in the absence of 13C-methane, which showed significant consumption of 12CH4 (Fig. S2). This was also confirmed by an increase in quantified 12CO2 production, which was reflected in the 2- to 4-fold-lower enrichment of 13CO2 in HS-enriched incubations compared to unamended controls (see 13FCO2 values in Fig. 1). Reports (23, 24) indicate that methanotrophic microorganisms prefer to oxidize 12CH4 compared to 13CH4, which may partly explain our findings.
The role of sulfate reduction on AOM when wetland sediment was enriched with HS was not possible to assess (Table S2) due to the large degree of endogenous sulfate reduction elicited by the degradation of the labile fraction of externally added NOM (Fig. 2), which also triggered methanogenesis in these microcosms. Since no significant differences in iron reduction were detected between microcosms with or without added 13CH4, the only microbial process clearly identified as driving AOM in Pahokee peat-enriched sediments was the microbial reduction of HS (Table S2).
Spectroscopic evidence on the presence and reduction of redox functional groups in NOM.
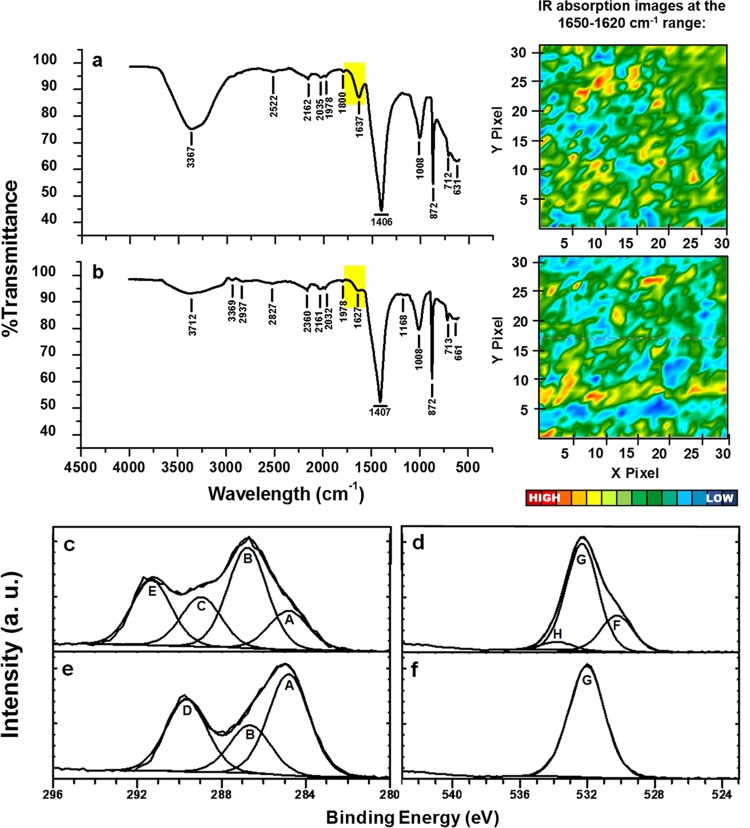
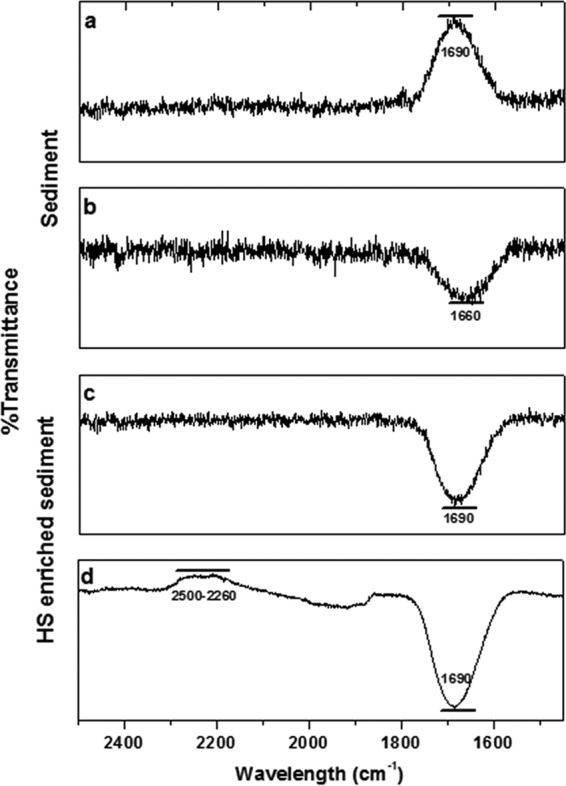
Initial exploration of the solid-phase NOM present in wetland sediment by microattenuated total reflection-Fourier transform infrared (micro-ATR-FTIR) spectra revealed the presence of electron-accepting moieties in both unamended and HS-enriched wetland sediments. By mapping of acquisition points at 1,650 to 1,620 · cm−1, the presence and heterogeneous distribution of quinone functional groups were evidenced in sediments, confirming the presence of nonsoluble electron-accepting moieties classically attributed to humic-like materials (Fig. 3a and b). To further confirm this, we looked for double-bonded carbon and oxygen (C=O) by use of X-ray photoelectron spectra (XPS); this technique supported the existence of quinone-like functional groups in unamended sediment and furthermore provided evidence of the reduction of these moieties by showing the disappearance of the C=O signal from C1s and O1s high-resolution spectra in a comparison of signals from sediment analyzed before and after incubation with 13CH4 in the absence of external HS (Fig. 3c to f). Another missing signal after the AOM process was that which corresponds to metallic oxides, evidenced by an analysis of the O1s high-resolution spectra (Fig. 3d and f), which may imply the reduction of intrinsic iron oxides that supported ∼0.5% of methanotrophy, according to electron balances (Table S2). Further analysis of the liquid phase of pristine sediment microcosms also revealed the reduction of quinone-like moieties during the course of AOM (Fig. 4). Initial samples exhibited a well-defined and strong peak at 1,690 · cm−1 associated with quinone moieties, while reduced samples, at the end of the incubation period, showed an increase in the signal related to phenolic groups (1,660 · cm−1). Additional signals of phenolic groups were detected after incubation with 13CH4 and Pahokee peat by spectral signals detected around 2,260 to 2,500 · cm−1 (25).
FIG 3.
Spectroscopic evidence of the presence of quinone moieties and their reduction in wetland sediment samples. (a and b) Micro-ATR-FTIR representative spectra taken from imaged areas generated after processing quinone functional groups (1,650 to 1,620 · cm−1) of sediment samples before incubation in the absence (a) and in the presence (b) of external NOM in the form of Pahokee peat HS. (c and e) XPS high-resolution profiles of C1s. (d and f) O1s signal. (c to f) Sediment samples prior to incubation (c and d) and sediment samples after incubation (e and f) with 13C-methane. Regions and components were corrected at 284.8 eV for the C—C adventitious carbon A, B and G components belong to C—O bond (∼286.6 and ∼532 eV, respectively), C and H correspond to C=O functional group (∼288.9 and ∼533.3 eV, respectively), D belongs to –COOH (∼289.6 eV), E is typical of the presence of carbonate (∼291 eV), and F suggests the occurrence of a metallic oxide (∼530 eV). a.u., absorbance units.
FIG 4.
High-performance UV-visible-near infrared spectra obtained from liquid samples before and after incubation with 13CH4. (a and c) Spectra obtained before incubation with 13CH4. (b and d) Spectra obtained after incubation with 13CH4.
Microbial communities performing AOM.
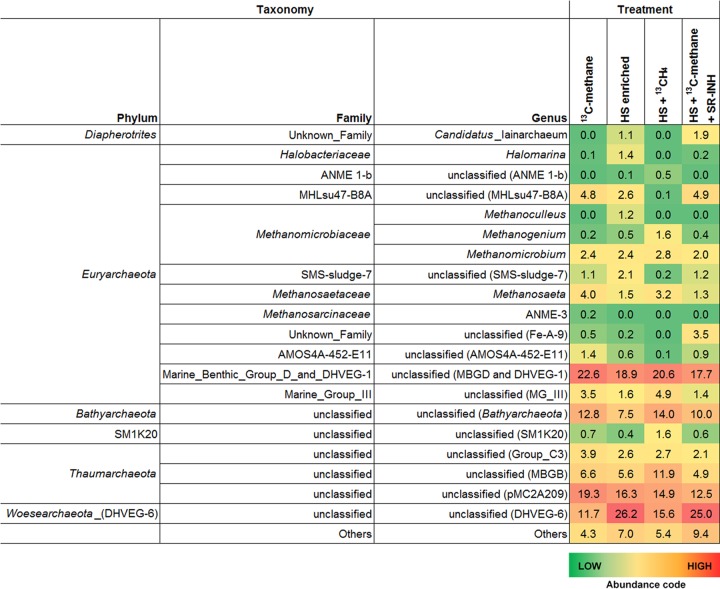
According to 16S rRNA gene sequences from wetland sediment samples performing AOM, anaerobic methanotrophic archaea (ANME), which are traditionally linked to anaerobic methanotrophy under sulfate-reducing (2, 26), Fe(III)-reducing (6, 8), and artificial electron acceptor-reducing conditions (27), were barely detected in our experiments, with ANME-1b and ANME-3 representing less than 0.5% and 0.2%, respectively, from the archaeal community in all experimental treatments (Fig. 5). The only abundant Euryarchaeota members detected were affiliated with an unclassified genus of the marine benthic group D (MBG-D) family (deep hydrothermal vent euryarchaeotal group 1 [DHVEG-1]), which accounted for 18 to 23% of the archaeal biota in all treatments. Outside the Euryarchaeota phylum, members from the newly named Bathyarchaeota lineage (formerly known as Miscellaneous Crenarchaeotic Group) were another cluster of microorganisms that remained in high percentages (from 8 to 14%) in all treatments. Two genera from the Thaumarchaeota phylum, one belonging to the pMC2A209 class, and the other from marine benthic group B (MBG-B), were also consistently present in all sediment samples showing AOM, with the MBG-B genus increasing its proportion up to 12% when sulfate reduction was inhibited (Fig. 5). From the bacterial counterpart, the most abundant bacteria in two of the treatments were from the genus Oceanimonas from the Aeromonadaceae family (Gammaproteobacteria), whose presence was diminished when sulfate reduction was inhibited and when 13CH4 was absent (Fig. S3), suggesting that this microorganism might have been involved in sulfate-dependent AOM. Other evident changes in the bacterial community included the increase in Clostridia and Bacilli members when external NOM was supplied (Fig. S3), which agrees with their capacity to reduce HS (28).
FIG 5.
Archaeal composition in wetland sediment samples performing AOM. Shown are the most abundant archaeal genera detected, based on 16S rRNA amplicon gene libraries, on selected experimental treatments shown in Fig. 1 at the end of the incubation period (30 days).
AOM linked to AQDS reduction.
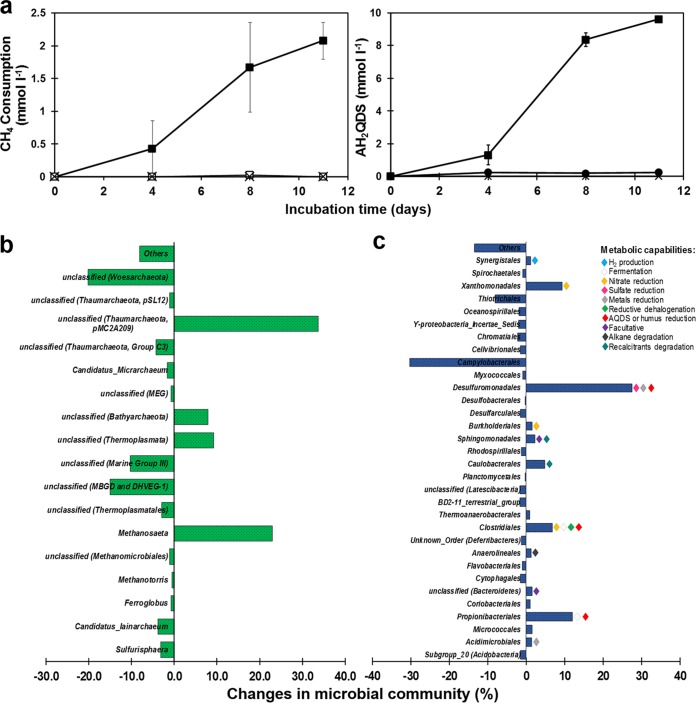
In order to confirm the capacity of the sediment biota to channel 13C-methane-derived electrons to quinone groups, the humic analogue anthraquinone-2,6-disulfonate (AQDS) was added as an electron acceptor to the artificial basal medium for sediment enrichments. AQDS reduction and methane consumption were observed since the first enrichment cycle, although no clear relationship between net methane consumption and anthrahydroquinone-2,6-disulfonate (AH2QDS) production was observed due to high concentrations of intrinsic electron donors and acceptors (data not shown). Nevertheless, during the third incubation cycle, net AOM was observed within 11 days, which corresponded to a final ratio of oxidized methane to reduced AQDS of 1:4.7, corrected for endogenous controls, which is very close to the stoichiometric 1:4 ratio, according to the following equation (Fig. 6a):
FIG 6.
AOM with AQDS as electron acceptor by an enrichment derived from wetland sediment. (a) Kinetics of methane consumption linked to AQDS reduction (to AH2QDS) observed during the last 11 days of the entire enrichment process lasting 151 days: filled squares (■) represent microcosms with CH4 as an electron donor and AQDS as an electron acceptor (complete experiments, n = 3), open squares (□) represent controls without an electron acceptor provided (without AQDS control, n = 3), filled circles (●) represent CH4-free microcosms (endogenous controls, n = 3), and crosses (×) represent heat-killed controls (sterile controls, n = 2). Error bars represent the standard error among replicates. (b and c) Microbial community changes at the end of the enrichment (151 days of incubation) at the phylum level based on Illumina sequencing of 16S rRNA V3 to V4 region. Fresh sediment composition was used as a reference.
Analysis of 16S rRNA gene sequences from enriched sediment sampled at the end of the third cycle of AQDS-dependent AOM activity (Fig. S1) displayed a significant decrease in the diversity of the microbial community, evidenced by a decrease in Shannon index, from 5.52 in freshly sampled sediment to 3.56 after enrichment with CH4 and AQDS. Significant increments and decreases in specific groups of archaea and bacteria did occur in this enrichment (Fig. 6b and c). From the archaeal fraction, the pMC2A209 class from the Thaumarchaeota and the Methanosaeta genus were archaeal clusters that significantly increased their presence in the AQDS enrichment (34% and 23%, respectively). Also in the AQDS enrichment, the Bathyarchaeota phylum previously detected in wetland sediments, both in the presence and in the absence of external NOM, significantly increased its proportion in the archaeal community (by around 10% with respect to the original composition), suggesting potential metabolic arrangements that thrive under AQDS-dependent AOM conditions (Fig. 6b). Humus-reducing bacteria that proliferated throughout the 5 months of enrichment included genera from the Desulfuromonadales (29, 30), Clostridiales (14, 28), and Propionibacteriales (31) orders, which increased by 27%, 7%, and 12%, respectively, with respect to the original composition (Fig. 6c).
DISCUSSION
NOM as terminal electron acceptor fueling AOM in wetland sediment.
Although the complex composition of the studied wetland sediment challenged efforts to elucidate the microbial processes responsible for the high methanotrophic activities quantified, the present study provides multiple lines of evidence demonstrating that electron-accepting functional groups present in its NOM fueled AOM by serving as a terminal electron acceptor. Indeed, while sulfate reduction was the predominant process, accounting for up to 42.5% of AOM activities, microbial reduction of NOM concomitantly occurred. Furthermore, enrichment of wetland sediment with external NOM, as Pahokee peat HS, significantly promoted AOM, with ∼100 nmol 13CH4 oxidized · cm−3 · day−1 attributed to this microbial process. Spectroscopic evidence also demonstrated that quinone moieties, which are main redox functional groups in HS (19), were heterogeneously distributed in the studied wetland sediment and that their reduction occurred during the course of AOM. Moreover, an enrichment derived from wetland sediments performing AOM linked to NOM reduction stoichiometrically oxidized methane coupled to AQDS. Sediment incubations performed in the presence of the sulfate reduction inhibitor molybdate further confirmed the role of HS in AOM. Certainly, even though sulfate-reducing activities significantly decreased in the presence of molybdate, AOM activities remained high, while microbial reduction of NOM was doubled under these conditions. These interesting findings suggest that methanotrophic microorganisms performing sulfate-dependent AOM might have directed electrons derived from AOM toward NOM when sulfate reduction became blocked, as has been suggested based on experiments performed under artificial conditions (27).
Microbial communities in wetland sediments performing AOM.
Archaeal clusters consistently found in wetland sediment incubations performing AOM included members from the MBG-D family, which have already been proposed as players in metal-dependent AOM (6); thus, their presence agrees with evidence indicating that AOM is linked to iron reduction observed in some experimental controls (Table S2). Additionally, these microorganisms were not found in the AQDS enrichment, probably due to the depletion of intrinsic ferric iron throughout the incubation cycles. Archaea constantly present among fresh sediment incubation and AQDS enrichment were those from the pMC2A209 class and the Bathyarchaeota phylum. To our knowledge, the pMC2A209 class of archaea has not been related to AOM, but its close partners from the MBG-B class have been consistently found in environments in which AOM occurs (32–35). In fact, Thaumarchaeota members, including the MBG-B, have been found in consortia performing AOM in the absence of ANME clades (36). Interestingly, the pMC2A209 cluster seemed to duplicate its proportion up to 12% when sulfate reduction was inhibited (by molybdate), which might suggest that the impediment of sulfate reduction enhanced its activity promoting AOM coupled to NOM reduction. With respect to the Bathyarchaeota phylum, increasing evidence suggests that this lineage might be involved in the methane cycle. Recently, it has been demonstrated that this cluster possesses the necessary genetic elements to express the enzymatic machinery required for methane production, and potentially methane consumption (37). Additionally, Saxton and colleagues have found abundant Bathyarchaeota representation in a fulvic acid-rich deep sediment that oxidizes methane uncoupled from sulfate reduction (22). Unexpectedly, a very low percentage within the archaeal population was identified as members from the ANME type archaea, even though it would be expected to find ANME-2 members, since it is the only ANME subgroup with a proven capability to derive electrons extracellularly toward humus and its analogues under artificial conditions (27). Our microcosms, both in fresh sediment as well as in the AQDS long-term enrichment, showed a barely detectable number of copies of ANME-1b and ANME-3 sequences retrieved by the methodology employed, suggesting a low presence of ANME microorganisms in the ecosystem studied.
Regarding the bacterial composition, while Clostridia, Bacilli, and Gammaproteobacteria were significantly represented within the fresh sediment performing AOM (Fig. S3), the AQDS enrichment (Fig. 6) exhibited the most significant increase in Deltaproteobacteria of the Desulfuromonadales order, which includes several humus-reducing microorganisms (14). Since a wide diversity of microorganisms have been proven to reduce humus analogues or HS, we do not rule out that diverse bacterial clusters could have participated in partnership with detected archaea to jointly perform AOM coupled to NOM reduction. Nevertheless, humus-reducing bacteria possess metabolic versatility and capability to reduce miscellaneous electron acceptors, which makes it difficult to come to conclusions about their participation in our experiments. Further investigation must be done to unravel the potential involvement of humus-reducing bacteria in AOM.
Ecological significance.
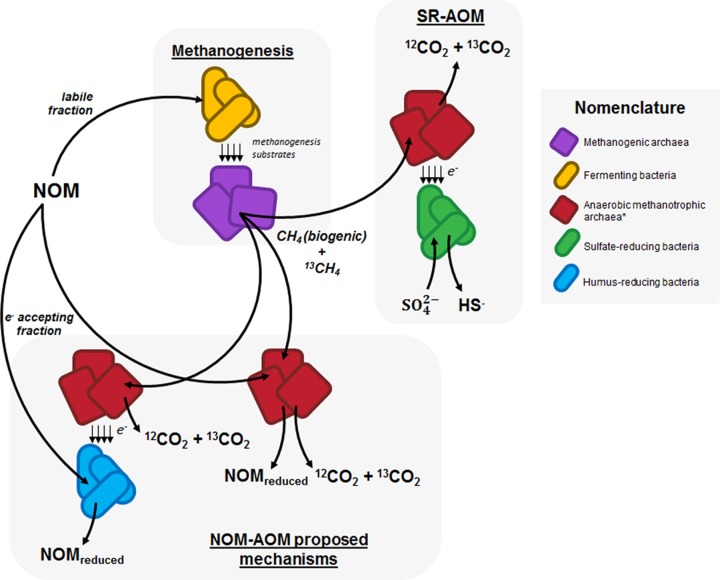
Here, we report AOM coupled to microbial reduction of NOM, which constitutes a missing link within the carbon cycle. HS frequently contribute up to 80% of soil NOM and up to 50% of dissolved NOM in aquatic environments. While the labile fraction of NOM promotes methanogenesis in anaerobic environments, the slowly decomposing humic portion may serve as an important barricade to prevent methane emissions in organotrophic ecosystems by serving as a terminal electron acceptor driving AOM (Fig. 7). As an example, considering the maximum AOM driven by the microbial reduction of NOM measured in humus-enriched sediments and the global area of coastal wetlands (38, 39), we approximate that this microbial process consumes up to 114 Tg of CH4 · year−1. Considering the global wetland area (10), we anticipate suppression of more than 1,300 Tg of CH4 · year−1 (see supplemental material for details). Accordingly, NOM-driven AOM may be more prominent in organotrophic sites with poor sulfate content, such as peatlands, swamps, and organotrophic lakes. This premise is supported by the suppression of methanogenesis by HS observed in different ecosystems (16, 17) and by the widespread AOM activity reported across many peatland types (40–42). The potential role of HS is further emphasized, because their electron-accepting capacity is fully recycled in recurrently anoxic environments. Thus, the suppression of methanogenesis by HS may be much greater than previously considered (it had been estimated to be on the order of 190,000 mol CH4 · km−2 · year−1 [43]).
FIG 7.
Schematic representation of methane generation and consumption by wetland sediment biota. While a fraction of NOM may serve as an electron acceptor to support AOM (NOM-AOM) and decouple sulfate reduction-dependent AOM (SR-AOM), depending on its chemical properties, a labile fraction of NOM could also be degraded following the methanogenesis pathway by a fermenting and methanogenic fraction of the consortia. Equilibrium between these three phenomena must be tightly dependent on thermodynamic conditions, concentration of chemical species, and composition of microbial community. *, anaerobic methanotrophic archaea are considered in a broader perspective than ANME clades from the Euryarchaeota phylum.
MATERIALS AND METHODS
Sediment sampling and characterization.
Sediment cores were collected from the tropical marsh Sisal, located in the Yucatán Peninsula, southeastern Mexico (21°09′26″N, 90°03′09″W) in January 2016. Sediment cores with a depth of 15 cm were collected under a water column of approximately 70 cm. Water samples were also collected from the area of sediment sampling points to be used as liquid medium in anaerobic incubations. All sediment and water samples were sealed in hermetic flasks and were maintained in ice until arrival at the laboratory. Upon arrival, all sampled materials were stored at 4°C in a dark room until analysis and incubation. Sediment cores were opened and homogenized within an anaerobic chamber (atmosphere composed of 95%/5% [vol/vol] N2/H2) before characterization and incubation. No amendments (addition of chemicals, washing, or exposure to air) were allowed on the sediment and water samples in order to reflect the actual conditions prevailing in situ as closely as possible. The characterization of water and sediment samples is described in Table S1.
Sediment incubations.
Water samples collected from sediment sampling points were thoroughly mixed before amendment with HS (2.5 g · liter−1) by magnetic stirring. Pahokee peat (Florida Everglades) HS, purchased from the International Humic Substances Society, was employed as external NOM in sediment incubations. Humus-enriched water was flushed with N2 to blow away any dissolved oxygen. Portions of 15 ml were then distributed in 25-ml serological flasks. Sediment containers were opened inside an anaerobic chamber. Portions of 2.5 ml of previously homogenized wet sediment were then inoculated into each serological bottle. After sealing all bottles with rubber stoppers and aluminum rings inside the anaerobic chamber, they were flushed with N2. Once anaerobic conditions were established, 5 ml of 13C-methane was injected into each vial to reach a 13CH4 partial pressure of 0.67 atm in a headspace of 7.5 ml. Controls incubated in the absence of external HS were also prepared according to an identical protocol. Killed controls included chloroform at a concentration of 10% (vol/vol) to annihilate any microbial activity. Additional incubations were executed in the presence of the sulfate reduction inhibitor molybdate (25 mmol liter−1) in the presence and absence of external NOM. All incubation bottles were statically placed in a dark room at 28°C (the temperature prevailing at the Sisal wetland at the sampling time). The pH remained at 7.5 ± 0.05 throughout all incubations.
Enrichment incubations with AQDS.
Incubations were commenced by inoculating 120-ml serological bottles with 10 g of volatile suspended solids (VSS) per liter of Sisal sediment. Prior to inoculation, portions of 60 ml of artificial medium were distributed into the incubation bottles and flushed for 15 min with a mixture of N2 and CO2 (80%/20% [vol/vol]) for stripping any dissolved oxygen from the medium. AQDS (>98.0% purity; TCI America Chemicals) was added at a concentration of 10 mmol liter−1 as a terminal electron acceptor, along with the following basal medium components (in grams per liter): NaHCO3 (5), NH4Cl (0.3), K2HPO4 (0.2), MgCl2·6H2O (0.03), and CaCl2 (0.1). Trace elements were included in the medium by adding 1 ml · liter−1 of a solution with the following composition (in milligrams per liter): FeCl2·4H2O (2,000), H2BO3 (50), ZnCl2 (50), CuCl2·6H2O (90), MnCl2·4H2O (500), AlCl3·6H2O (90), CoCl2·6H2O (2,000), NiCl·6H2O (920), Na2SeO·5H2O (162), (NH4)6Mo7O24 (500), EDTA (1,000), Na2WO4·H2O (100), and 1 ml · liter−1 HCl at 36%. The final pH of the medium was 7.2, and no changes were observed throughout the incubation time. Once inoculation took place, microcosms were sealed with rubber stoppers and aluminum rings and then flushed with the same N2-CO2 mixture. After anoxic conditions were established, 1 ml of sodium sulfide stock solution was injected into each vial to reach a sulfide concentration of 0.1 g · liter−1 in order to consume any traces of dissolved oxygen. Methane was provided into the microcosms by injecting 30 ml of CH4 (99.9% purity; Praxair), reaching a partial pressure of methane of 0.54 atm. Subsequent incubations were performed after AQDS was reduced (i.e., converted to AH2QDS), coupled to anaerobic oxidation of methane (AOM). A new set of bottles containing basal medium with AQDS (10 mmol liter−1) were inoculated within an anaerobic chamber by transferring 10 ml of slurry (sediment and medium) taken from previous incubations (Fig. S1). The following incubations were completed under the same experimental conditions.
Analytical techniques. (i) Isotopic carbon dioxide and methane measurements.
Ions 16 (12CH4), 17 (13CH4), 44 (12CO2), and 45 (13CO2) were detected and quantified in an Agilent Technologies 7890A gas chromatograph (GC) coupled to an Agilent Technologies 5975C mass spectrometer (detector); ionization was achieved by electronic impact and a quadrupole analyzer. For the analysis, an Agilent Technologies HP-PLOT/Q capillary column with a stationary phase of poly(styrene-divinylbenzene) (30 m by 0.320 mm by 20 μm) was employed as stationary phase using helium as a carrier gas. The chromatographic method was as follows: the starting temperature was 70°C, which was held for 3 min, and then a ramp with an increase of 20°C per min was implemented until 250°C was reached and maintained for 1 min. The total time of the run was 13 min. The temperature of the injection port was 250°C. The injection volume was 20 μl, and there was only one replicate of injection per bottle. The gas injected into the GC was taken directly from the headspace of the incubations and immediately injected into the GC port. Methane calibration curves were made by injection of different methane (99.9% purity) volumes into serological bottles under the same experimental conditions (atmosphere composition, pressure, temperature, and liquid volume). 12CO2 and 13CO2 curves were made using different dried sodium bicarbonate (99% purity; Sigma-Aldrich) and sodium 13C-labeled carbonate (99 atom %13C; Sigma-Aldrich) concentrations, respectively, in serological bottles which contained the same volume of wetland sediment and water used in incubations. Standards were incubated at room temperature for 12 h until equilibrium with the gaseous phase was reached. The linear regression analysis of obtained measurements had a correlation coefficient higher than 0.97. 13CO2 production rates were based on the maximum slope observed on linear regressions considering at least three sampling points.
(ii) Methane quantification in AQDS enrichment.
Net methane consumption was assessed in terms of methane concentration measurements in the headspace of microcosms. These measurements were carried out by injecting 100 μl of gas samples from the headspace of incubation bottles into a gas chromatograph (Agilent Technologies 6890M) equipped with a thermal conductivity detector and a HayeSep D column (Alltech, Deerfield, IL, USA) with the dimensions 3.048 m by 3.185 m by 2.16 mm. Helium was employed as a carrier gas at a flux of 12 ml · min−1. The temperatures of the injection port, oven, and detector were 250, 60, and 250°C, respectively. Calibration curves were made for each reaction volume used by injecting different methane concentrations into serological bottles under the same experimental conditions at which microcosms were studied (atmosphere composition, pressure, temperature, and liquid volume).
(iii) Determination of electron-accepting functional groups in solid phase by XPS.
Sediment samples (solid fraction of microcosms) were dried under a constant nitrogen flow after incubation with methane. Once sediments became dried, bottles were open inside an anaerobic chamber with an atmosphere composed of 95%/5% (vol/vol) N2/H2 and were triturated on an agate mortar. Samples were then kept under anaerobic conditions until analysis in a PHI VersaProbe II X-ray photoelectron spectroscopy analyzer (Physical Electronics, ULVAC-PHI). Two representative spectra were recorded per scanned sample.
(iv) Determination of electron-accepting functional groups in solid phase by micro-ATR-FTIR imaging.
Micro-ATR-FTIR images were collected from each sample with a continuous-scan spectrometer, the Agilent 660 FTIR interfaced to a 620 infrared microscope with a 32 by 32 focal plane array (FPA) detector and Ge ATR objective for micro-ATR. Each pixel obtains a full IR spectrum or a total of 1,024 spectra. Background spectra were collected from a clean ATR crystal (i.e., without sample). The Ge crystal of the ATR microscope was lowered onto the surface of each sample for a contact area of approximately 100 by 100 μm. Spectra were collected by coaddition of 256 scans over a spectral range of 4,000 to 900 · cm−1, at a spectral resolution of 4 · cm−1. In all images, a color scale bar is set within the software to reflect the relative concentration range, from low to high. Agilent Resolutions Pro was used for data acquisition and analysis.
(v) Determination of electron-accepting functional groups in liquid phase by high-resolution UV-Vis–near-infrared spectroscopy.
After each incubation cycle, liquid samples (1.5 ml) were taken in an anaerobic chamber with a disposable syringe and put into a quartz cell, which was sealed with plastic film in order to maintain anoxic conditions during spectrometric analysis. Spectra were obtained in a Varian Cary 5000 UV-Vis (diffuse reflectance) spectrophotometer equipped with an integrating sphere.
(vi) Nitrite and nitrate determinations.
Nitrite and nitrate concentrations were measured according to spectrometric techniques established by Standard Methods (44). Nitrate measurement is taken under acidic conditions at a wavelength of 275 nm, and the value obtained is corrected for dissolved organic matter, which has its maximum absorbance at 220 nm. Nitrite forms a purple complex through a reaction with sulfanilamide and N-(1-naphthyl) ethylene diamine, which presents its maximum absorbance at a wavelength of 543 nm. Samples were taken with a disposable syringe directly from the microcosms, injected into sealed quartz cuvettes or glass tubes (depending on the required lecture wavelength), and immediately taken to the spectrophotometer to avoid any reaction of the sample with atmospheric oxygen.
(vii) Sulfate and sulfide determinations.
Samples were extracted from microcosms and immediately filtered through 0.22-μm-pore-size nitrocellulose membranes. Filtered samples were then diluted (1:10) with deionized water and processed in an Agilent capillary electrophoresis system (Agilent Technologies), according to the methodology proposed by Soga and Ross (45). Dissolved sulfide was measured by the spectrometric method proposed by Cord-Ruwisch (46). Briefly, 100 μl of sample was taken and immediately vortexed with 4 ml of an acidic CuSO4 solution. Absorbance at 480 nm was immediately registered in a UV-Vis spectrophotometer (Thermo Spectronic) to avoid sulfide oxidation before measurements.
(viii) Humic substance reduction and ferrous iron measurements.
Quantification of the reduction of electron-accepting functional groups in HS was performed according to Lovley et al. (18). Slurry samples (∼500 μl) were taken from microcosms with a disposable syringe while bottles were being manually shaken inside an anaerobic chamber. A portion of each sample (200 μl) was mixed with an equal volume of an acidic solution (0.5 mol liter−1 HCl) and allowed to stand for 30 min, while the same volume of sample was reacted with ferric citrate (20 mmol liter−1) for 3 h. After reaction with ferric citrate, samples were mildly resuspended in a vortex, and 200 μl was left repose with the same volume of HCl solution for 30 min. Afterwards, each sample was centrifuged for 10 min at 10,000 × g in a Spectrafuge 16M centrifuge, and 200 μl of supernatant was then recovered and reacted with a 0.2 g · liter−1 solution of 2,4,6-Tris(2-pyridyl)-1,3,5-triazine (ferrozine reagent). Ferrous iron produced due to the chemical reduction of ferric citrate by reduced functional groups in HS forms a purple complex along with ferrozine reagent, which has its maximum absorbance at 562 nm. The ferrozine solution was buffered with HEPES (50 mmol liter−1). Once centrifuged samples were mixed with ferrozine solution, they were left reacting for 10 min before their measurement in a Thermo Scientific Genesis 10 UV spectrometer located inside an anaerobic chamber. All solutions employed in this determination were bubbled with N2 for 30 min to ensure the absence of dissolved oxygen.
(ix) Total carbon, TOC, and total inorganic carbon measurements.
Water samples were filtered through 0.22-μm-pore-size nitrocellulose membranes and diluted with deionized water, while sediment samples were dried until constant weight. Both liquid and solid samples were analyzed in a Shimadzu TOCVCS/TNM-1 total organic carbon (TOC) analyzer equipped with a solids sampling port (SSM-5000A). The solid-sample processing time was 6 min at 900°C using O2 (500 ml · min−1, 99.9% purity) as a carrier gas; all samples were analyzed in triplicate.
(x) Total, volatile, and fixed solids.
Total, fixed, and volatile solids were measured in triplicate according to the Standard Methods procedure (44).
(xi) Elemental composition.
The elemental composition of the sediments was assessed by analyzing acid extracts from 2 g of dry sediment. In the case of iron and manganese measurements in microcosms, supernatant samples were taken with disposable syringes, filtered, and acidified prior to analysis. Samples were then analyzed by inductively coupled plasma-optical emission spectrometry (ICP-OES) in an equipment Varian 730-ES. The operational conditions were: 1 kW potency, 1.5 liters · min−1 auxiliary flow, 0.75 liters · min−1 net flow, 30-s sample-taking delay, and 3 measured replicates by sample. Argon was employed as a carrier gas.
(xii) DNA extraction, PCR amplification, and sequencing.
One microcosm for each selected treatment was randomly chosen at the end of the incubation period (30 days for experiments presented in Fig. 1 and 151 days for experiments depicted in Fig. 6). Before DNA extraction, liquid medium was decanted and extracted from the serological bottles. The total sediment was homogenized afterwards, and a subsample of 0.5 g was taken to proceed with DNA extraction. The remaining sediment and the other microcosms were used for material characterization. The total DNA was extracted from sediment samples using the PowerSoil DNA extraction kit (Mo Bio Laboratories, Carlsbad, CA, USA), according to the protocol described by the manufacturer. DNA isolated from each sample was amplified using primers 341F and 785R, targeting the V3 and V4 regions of the 16S rRNA gene fused with Illumina adapter overhang nucleotide sequences (47). The PCRs were performed in 50-μl reaction mixtures using Phusion Taq polymerase (Thermo Scientific, USA) under the following conditions: denaturation at 98°C for 60 s, followed by 5 cycles of amplification at 98°C for 60 s, 50°C for 30 s, and 72°C for 30 s, followed by 25 cycles of amplification at 98°C for 60 s, 55°C for 30 s, and 72°C for 30 s, and a final extension of 72°C for 5 min. Two independent PCRs were performed for each sample. The products were indexed using Illumina's 16S metagenomic sequencing library preparation protocol and Nextera XT index kit version 2 (Illumina, San Diego, CA). Libraries were deep sequenced with the Illumina MiSeq sequencer.
(xiii) Bioinformatics analysis.
An analysis of 16S rRNA gene libraries was carried out using the mothur open source software package (version 1.34.4) (48). The high-quality sequence data were analyzed for potential chimeric reads using the UCHIME algorithm. Sequences containing homopolymer runs of 9 or more bases, those with more than one mismatch to the sequencing primer, and those with a Q-value average below 25 were eliminated. Group membership was determined prior to the trimming of the barcode and primer sequence. Sequences were aligned against the SILVA 123 16S/18S rRNA gene template using the Nearest Alignment Space Termination (NAST) algorithm and trimmed for the optimal alignment region. A pairwise distance matrix was calculated across the nonredundant sequence set, and reads were clustered into operational taxonomic units (OTUs) at 3% distance using the farthest neighbor method. The sequences and OTUs were categorized taxonomically using mothur's Bayesian classifier and the SILVA 123 reference set. The sequences obtained have been submitted to the NCBI GenBank database.
Accession number(s).
The accession numbers of the sequences in this work were deposited in the GenBank Sequence Read Archive under BioProject number SRP094593.
Supplementary Material
ACKNOWLEDGMENTS
We thank Derek R. Lovley, Jim A. Field, Alfons Stams, Frederic Thalasso, Cesar Nieto, and Sonia Arriaga for discussions. We are also grateful to Matthew Tippett for proofreading the manuscript. We thank Dulce Partida, Ma. Carmen Rocha-Medina, Guadalupe Ortega-Salazar, Mariela Bravo-Sánchez, Roberto Camposeco, Elizabeth Cortez and Guillermo Vidriales, for technical support, as well as Lluvia Korynthia López-Aguiar and Yessica Parera-Valadez for their help during sediment collection expeditions. We thank Vicente Rodríguez for access to the Cary 5000 UV-Vis spectrophotometer. We are also grateful for the use of infrastructure of the National laboratories LINAN and LANBAMA at IPICYT, as well as USMB at UNAM.
This work was financially supported by grants from the Council of Science and Technology of Mexico (Program Frontiers in Science, grant 1289) and the Marcos Moshinsky Foundation to F.J.C.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00645-17.
REFERENCES
- 1.Reeburgh WS. 2007. Oceanic methane biogeochemistry. Chem Rev 107:486–513. doi: 10.1021/cr050362v. [DOI] [PubMed] [Google Scholar]
- 2.Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, Amann R, Jørgensen BB, Witte U, Pfannkuche O. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- 3.Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AK, Ettwig KF, Rijpstra WI, Schouten S, Damste JS, Op den Camp HJ, Jetten MS, Strous M. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918–921. doi: 10.1038/nature04617. [DOI] [PubMed] [Google Scholar]
- 4.Deutzmann JS, Stief P, Brandes J, Schink B. 2014. Anaerobic methane oxidation coupled to denitrification is the dominant methane sink in a deep lake. Proc Natl Acad Sci U S A 111:18273–18278. doi: 10.1073/pnas.1411617111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJCT, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJM, Janssen-Megens EM, Francoijs K-J, Stunnenberg H, Weissenbach J, Jetten MSM, Strous M. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 6.Beal EJ, House CH, Orphan VJ. 2009. Manganese- and iron-dependent marine methane oxidation. Science 325:184–187. doi: 10.1126/science.1169984. [DOI] [PubMed] [Google Scholar]
- 7.Egger M, Rasigraf O, Sapart CJ, Jilbert T, Jetten MSM, Röckmann T, Van Der Veen C, Bânda N, Kartal B, Ettwig KF, Slomp CP. 2015. Iron-mediated anaerobic oxidation of methane in brackish coastal sediments. Environ Sci Technol 49:277–283. doi: 10.1021/es503663z. [DOI] [PubMed] [Google Scholar]
- 8.Ettwig KF, Zhu B, Speth D, Keltjens JT, Jetten MSM, Kartal B. 2016. Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc Natl Acad Sci U S A 113:12792–12796. doi: 10.1073/pnas.1609534113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anthony KM, MacIntyre S. 2016. Biogeochemistry: nocturnal escape route for marsh gas. Nature 535:363–365. doi: 10.1038/535363a. [DOI] [PubMed] [Google Scholar]
- 10.Bridgham SD, Cadillo-Quiroz H, Keller JK, Zhuang Q. 2013. Methane emissions from wetlands: biogeochemical, microbial, and modeling perspectives from local to global scales. Glob Chang Biol 19:1325–1346. doi: 10.1111/gcb.12131. [DOI] [PubMed] [Google Scholar]
- 11.Megonigal JP, Hines ME, Visscher PT. 2004. Anaerobic metabolism: linkages to trace gases and aerobic processes, p 317–424. In Schlesinger WH. (ed), Biogeochemistry. Elsevier-Pergamon, Oxford, United Kingdom. [Google Scholar]
- 12.Segarra KEA, Schubotz F, Samarkin V, Yoshinaga MY, Hinrichs K-U, Joye SB. 2015. High rates of anaerobic methane oxidation in freshwater wetlands reduce potential atmospheric methane emissions. Nat Commun 6:7477. doi: 10.1038/ncomms8477. [DOI] [PubMed] [Google Scholar]
- 13.Segarra KEA, Comerford C, Slaughter J, Joye SB. 2013. Impact of electron acceptor availability on the anaerobic oxidation of methane in coastal freshwater and brackish wetland sediments. Geochim Cosmochim Acta 115:15–30. doi: 10.1016/j.gca.2013.03.029. [DOI] [Google Scholar]
- 14.Martinez CM, Alvarez LH, Celis LB, Cervantes FJ. 2013. Humus-reducing microorganisms and their valuable contribution in environmental processes. Appl Microbiol Biotechnol 97:10293–10308. doi: 10.1007/s00253-013-5350-7. [DOI] [PubMed] [Google Scholar]
- 15.Lehmann J, Kleber M. 2015. The contentious nature of soil organic matter. Nature 528:60–68. [DOI] [PubMed] [Google Scholar]
- 16.Blodau C, Deppe M. 2012. Humic acid addition lowers methane release in peats of the Mer Bleue bog, Canada. Soil Biol Biochem 52:96–98. doi: 10.1016/j.soilbio.2012.04.023. [DOI] [Google Scholar]
- 17.Miller KE, Lai CT, Friedman ES, Angenent LT, Lipson DA. 2015. Methane suppression by iron and humic acids in soils of the Arctic Coastal Plain. Soil Biol Biochem 83:176–183. doi: 10.1016/j.soilbio.2015.01.022. [DOI] [Google Scholar]
- 18.Lovley DR, Coates JD, Blunt-Harris EL, Phillips EJP, Woodward JC. 1996. Humic substances as electron acceptors for microbial respiration. Nature 17:428–437. [Google Scholar]
- 19.Scott DT, Mcknight DM, Blunt-Harris EL, Kolesar SE, Lovley DR. 1998. Quinone moieties act as electron acceptors in the reduction of humic substances by humics-reducing microorganisms. Environ Sci Technol 32:2984–2989. doi: 10.1021/es980272q. [DOI] [Google Scholar]
- 20.Straub KL, Benz M, Schink B. 2000. Iron metabolism in anoxic environments at near neutral pH. FEMS Microbiol Ecol 34:181–186. doi: 10.1111/j.1574-6941.2001.tb00768.x. [DOI] [PubMed] [Google Scholar]
- 21.Timmers PH, Suarez-Zuluaga DA, van Rossem M, Diender M, Stams AJ, Plugge CM. 2015. Anaerobic oxidation of methane associated with sulfate reduction in a natural freshwater gas source. ISME J 10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saxton MA, Samarkin VA, Schutte CA, Bowles MW, Madigan MT, Cadieux SB, Pratt LM, Joye SB. 2016. Biogeochemical and 16S rRNA gene sequence evidence supports a novel mode of anaerobic methanotrophy in permanently ice-covered Lake Fryxell, Antarctica. Limnol Oceanogr 61:S119–S130. doi: 10.1002/lno.10320. [DOI] [Google Scholar]
- 23.Alperin MJ, Reeburgh WS, Whiticar MJ. 1988. Carbon and hydrogen isotope fractionation resulting from anaerobic methane oxidation. Biogeochem Cycles 2:279–288. doi: 10.1029/GB002i003p00279. [DOI] [Google Scholar]
- 24.Martens CS, Albert DB, Alperin MJ. 1999. Stable isotope tracing of anaerobic methane oxidation in the gassy sediments of Eckernförde Bay German Baltic Sea. Am J Sci 299:589–610. doi: 10.2475/ajs.299.7-9.589. [DOI] [Google Scholar]
- 25.Socrates G. 2004. Infrared and Raman characteristic group frequencies. John Wiley and Sons, Ltd., Chichester, United Kingdom. [Google Scholar]
- 26.Orphan VJ, House CH, Hinrichs K-U, McKeegan KD, DeLong EF. 2002. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc Natl Acad Sci U S A 99:7663–7668. doi: 10.1073/pnas.072210299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheller S, Yu H, Chadwick GL, McGlynn SE, Orphan VJ. 2016. Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction. Science 351:703–707. doi: 10.1126/science.aad7154. [DOI] [PubMed] [Google Scholar]
- 28.Xi B, Zhao X, He X, Huang C, Tan W, Gao R, Zhang H, Li D. 2016. Successions and diversity of humic-reducing microorganisms and their association with physical-chemical parameters during composting. Bioresour Technol 219:204–211. doi: 10.1016/j.biortech.2016.07.120. [DOI] [PubMed] [Google Scholar]
- 29.Coates JD, Cole KA, Chakraborty R, Connor SMO, Achenbach LA. 2002. Diversity and ubiquity of bacteria capable of utilizing humic substances as electron donors for anaerobic respiration. Appl Environ Microbiol 68:2445–2452. doi: 10.1128/AEM.68.5.2445-2452.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lovley DR, Fraga JL, Blunt-Harris EL, Hayes LA, Phillips EJP, Coates JD. 1998. Humic substances as a mediator for microbially catalyzed metal reduction. Acta Hydrochim Hydrobiol 26:152–157. doi:. [DOI] [Google Scholar]
- 31.Benz M, Schink B, Brune A. 1998. Humic acid reduction by Propionibacterium freudenreichii and other fermenting bacteria. Appl Environ Microbiol 64:4507–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glass JB, Yu H, Steele JA, Dawson KS, Sun S, Chourey K, Pan C, Hettich RL, Orphan VJ. 2014. Geochemical, metagenomic and metaproteomic insights into trace metal utilization by methane-oxidizing microbial consortia in sulphidic marine sediments. Environ Microbiol 16:1592–1611. doi: 10.1111/1462-2920.12314. [DOI] [PubMed] [Google Scholar]
- 33.Knittel K, Lösekann T, Boetius A, Kort R, Amann R. 2005. Diversity and distribution of methanotrophic archaea at cold seeps. Appl Environ Microbiol 71:467–479. doi: 10.1128/AEM.71.1.467-479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schubert CJ, Vazquez F, Losekann-Behrens T, Knittel K, Tonolla M, Boetius A. 2011. Evidence for anaerobic oxidation of methane in sediments of a freshwater system (Lago di Cadagno). FEMS Microbiol Ecol 76:26–38. doi: 10.1111/j.1574-6941.2010.01036.x. [DOI] [PubMed] [Google Scholar]
- 35.Yoshinaga MY, Lazar CS, Elvert M, Lin YS, Zhu C, Heuer VB, Teske A, Hinrichs KU. 2015. Possible roles of uncultured archaea in carbon cycling in methane-seep sediments. Geochim Cosmochim Acta 164:35–52. doi: 10.1016/j.gca.2015.05.003. [DOI] [Google Scholar]
- 36.Biddle JF, Lipp JS, Lever MA, Lloyd KG, Sørensen KB, Anderson R, Fredricks HF, Elvert M, Kelly TJ, Schrag DP, Sogin ML, Brenchley JE, Teske A, House CH, Hinrichs K-U. 2006. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc Natl Acad Sci U S A 103:3846–3851. doi: 10.1073/pnas.0600035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans PN, Parks DH, Chadwick GL, Robbins SJ, Orphan VJ, Golding SD, Tyson GW. 2015. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350:434–438. doi: 10.1126/science.aac7745. [DOI] [PubMed] [Google Scholar]
- 38.Keller JK, Medvedeff CA. 2016. Soil organic matter, p 165–188. In Vepraskas MJ, Craft CB (ed), Wetland soils: genesis, hydrology, landscapes, and classification. Taylor and Francis Group, CRC Press,, Boca Raton, FL. [Google Scholar]
- 39.Pendleton L, Donato DC, Murray BC, Crooks S, Jenkins WA, Sifleet S, Craft C, Fourqurean JW, Kauffman JB, Marbà N, Megonigal P, Pidgeon E, Herr D, Gordon D, Baldera A. 2012. Estimating global “blue carbon” emissions from conversion and degradation of vegetated coastal ecosystems. PLoS One 7:e43542. doi: 10.1371/journal.pone.0043542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gupta V, Smemo KA, Yavitt JB, Basiliko N. 2012. Active methanotrophs in two contrasting North American peatland ecosystems revealed using DNA-SIP. Microb Ecol 63:438–445. doi: 10.1007/s00248-011-9902-z. [DOI] [PubMed] [Google Scholar]
- 41.Smemo KA, Yavitt JB. 2007. Evidence for anaerobic CH4 oxidation in freshwater peatlands. Geomicrobiol J 24:583–597. doi: 10.1080/01490450701672083. [DOI] [Google Scholar]
- 42.Smemo KA, Yavitt JB. 2011. Anaerobic oxidation of methane: an underappreciated aspect of methane cycling in peatland ecosystems? Biogeosciences 8:779–793. doi: 10.5194/bg-8-779-2011. [DOI] [Google Scholar]
- 43.Klüpfel L, Piepenbrock A, Kappler A, Sander M. 2014. Humic substances as fully regenerable electron acceptors in recurrently anoxic environments. Nat Geosci 7:195–200. doi: 10.1038/ngeo2084. [DOI] [Google Scholar]
- 44.Eaton AD, Clesceri LS, Greenberg AE, Franson MAH. 1988. Standard methods for the examination of water and wastewater. American Public Health Association, Washington, DC. [Google Scholar]
- 45.Soga T, Ross GA. 1999. Simultaneous determination of inorganic anions, organic acids and metal cations by capillary electrophoresis. J Chromatogr A 834:65–71. doi: 10.1016/S0021-9673(98)00692-X. [DOI] [Google Scholar]
- 46.Cord-Ruwisch R. 1985. A quick method for the determination of dissolved and precipitated sulfides in cultures of sulfate-reducing bacteria. J Microbiol Methods 4:33–36. doi: 10.1016/0167-7012(85)90005-3. [DOI] [Google Scholar]
- 47.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:1–11. doi: 10.1093/nar/gks1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.