Abstract
Group I and II introns self-splice in vitro, but require proteins for efficient splicing in vivo, to stabilize the catalytically active RNA structure. Recent studies showed that the splicing of some Neurospora crassa mitochondrial group I introns additionally requires a DEAD-box protein, CYT-19, which acts as an RNA chaperone to resolve nonnative structures formed during RNA folding. Here we show that, in Saccharomyces cerevisiae mitochondria, a related DEAD-box protein, Mss116p, is required for the efficient splicing of all group I and II introns, some RNA end-processing reactions, and translation of a subset of mRNAs, and that all these defects can be partially or completely suppressed by the expression of CYT-19. Results for the aI2 group II intron indicate that Mss116p is needed after binding the intron-encoded maturase, likely for the disruption of stable but inactive RNA structures. Our results suggest that both group I and II introns are prone to kinetic traps in RNA folding in vivo and that the splicing of both types of introns may require DEAD-box proteins that function as RNA chaperones.
Keywords: bakers' yeast, mitochondria, splicing factor
DExH/D-box proteins are a large, ubiquitous protein family, whose members use the energy of ATP hydrolysis to mediate RNA structural rearrangements in a variety of cellular processes (1, 2). These proteins have a core region containing nine conserved motifs flanked by unique N- and/or C-terminal extensions, which in some cases target the proteins to their sites of action by specific RNA or protein interactions. The proteins are named for the amino acid sequence of motif II, which in different subfamilies is DEAD, DEAH, or some variant thereof. Experiments with model substrates show that DExH/D-box proteins can act as RNA helicases (3) or can disrupt ribonucleoprotein (RNP) complexes independently of their helicase activity (4). However, how these proteins function on their natural substrates has remained largely unknown.
Group I and II introns self-splice in vitro but require proteins for efficient splicing in vivo to help fold the intron RNA into the catalytically active structure (5). In the fungus Neurospora crassa, the splicing of a subset of mitochondrial (mt) group I introns depends on two proteins encoded by nuclear genes, the mt tyrosyl-tRNA synthetase (CYT-18 protein), which stabilizes the catalytically active RNA structure, and the DEAD-box protein CYT-19 (6, 7). Recently, CYT-19 was shown to function as an ATP-dependent RNA chaperone to destabilize nonnative structures that constitute kinetic traps in the CYT-18-assisted RNA folding pathway (7). A mutation in the cyt-19 gene did not affect the splicing of non-CYT-18-dependent group I introns or a group II intron, but did inhibit some 5′ and 3′ end processing reactions and, possibly, mt translation (7, 8).
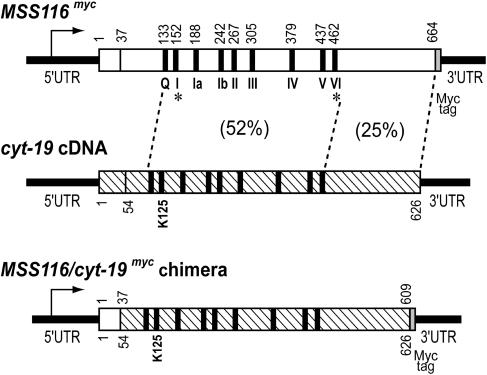
The Saccharomyces cerevisiae nuclear genome encodes three DExH/D-box proteins (Suv3p, Mrh4p, and Mss116p) that function in mitochondria (9–11). Of these, Mss116p is the most closely related to CYT-19. The two proteins have 32% identity and 52% similarity in the region containing the conserved ATPase motifs, but less similarity (25%) in the C-terminal region and no significant similarity in the short N-terminal region (Fig. 1).
Fig. 1.
Diagrams of the MSS116myc, cyt-19, and chimeric MSS116/CYT-19 myc genes. MSS116myc encodes the 664-aa MSS116 ORF (open box) with a 36-aa mt targeting sequence and a C-terminal myc tag. The cyt-19 cDNA (hatched box) encodes the 626-aa cyt-19 ORF with a putative 53-aa mt targeting sequence. Both Mss116p and CYT-19 contain a conserved core with nine motifs characteristic of the DEAD-box subfamily of DExH/D-box proteins (black bars, labeled for MSS116 myc). Percent similarity is shown for indicated regions of MSS116p and CYT-19. The chimeric MSS116/cyt-19 myc gene consists of the promoter and mt targeting sequence of MSS116 fused to cyt-19 codons 54–626 with a C-terminal myc tag, and the MSS116 3′ UTR. Asterisks indicate the locations of MSS116 mutations analyzed in this work.
MSS116 was identified in a screen for nuclear mutants that are unable to grow by respiration on nonfermentable carbon sources (e.g., glycerol) when their mtDNAs contain introns, but not when their mtDNAs lack them (12). The effect of MSS16 point mutations on the growth phenotype of yeast strains containing different combinations of mtDNA introns suggested that Mss116p functions in splicing just three or four of the nine group I introns (aI5α, aI5β, bI2, and/or bI3) and only two of the four group II introns (aI1 and bI1, but not aI2 and aI5γ) (10). [Note that introns in the cytochrome oxidase subunit I (COX1) gene are aI1, -2, -3, -4, -5α, -5β, and -5γ; introns in the cytochrome b (COB) gene are bI1, -2, -3, -4, and -5; and the single intron in the 21S rRNA gene is ω.) A possible additional function of Mss116p was suggested by the finding that disruption of MSS116 in a strain lacking mtDNA introns (I0) blocked the glycerol growth of cells at 30°C (10). mss116Δ strains containing some COB and COX1 introns appeared defective in translating COB and COX1 mRNAs (10). However, it could not be distinguished whether the MSS116 disruption inhibits translation of those mRNAs directly or indirectly by affecting RNA splicing. Similarly, because the splicing of some yeast mtDNA introns depends on an intron-encoded protein (IEP) with maturase activity, it could not be excluded that MSS116 disruption inhibits their splicing indirectly by inhibiting mt protein synthesis. The splicing of the group II intron bI1 does not require a mitochondrially synthesized protein, and the possibility that Mss116p functions directly in its splicing was supported by the finding that a mt extract from cells overexpressing Mss116p promoted ATP-dependent splicing of a bI1-containing in vitro transcript (13). However, the role of Mss116p in splicing group I and II introns and the nature of its additional function(s) have remained unclear.
Here we show that (i) Mss116p is, in fact, required for the efficient splicing of all nine group I introns and all four group II introns of S. cerevisiae mtDNA, some RNA end-processing reactions, and translation of a subset of mt mRNAs; (ii) all of the phenotypic defects in mss116Δ strains can be partially or completely suppressed by the expression of the N. crassa RNA chaperone, CYT-19; and (iii) Mss116p functions in splicing the aI2 group II intron at a step after maturase binding, likely the disruption of stable intermediate or nonnative RNA structures that are kinetic traps in RNA folding. Our results indicate that, in addition to maturases and other splicing factors, the efficient splicing of yeast mt group I and II introns in vivo requires a DEAD-box protein with RNA chaperone function.
Materials and Methods
Yeast Strains. Unless noted, yeast strains have the nuclear background of strain 161-U7 (MATa ade1 lys1 ura3). The MSS116/kanr disruption cassette was generated by PCR of plasmid pFA6-kanMX2 (14) with primers containing 50 nt at the beginning and end of the MSS116 ORF. The gel-purified PCR product was transformed into a 161-U7 strain containing a respiratory-competent (ρ+) mtDNA with 12 introns, and disruptants were selected by G418 resistance. To obtain mss116Δ strains with different mtDNA introns, the initial disruptant was grown in ethidium bromide to generate a ρ0 petite derivative, lacking mtDNA. The ρ0 strain was then mated with derivatives of MCC109 (MATα ade2-101 ura3-52 kar1-1) carrying ρ+ mtDNAs with zero, one, or two introns, the construction of which has described (15).
To construct MSS116/cyt-19 knock-in (cyt-19-ki) strains, a chimeric MSS116/cyt-19 gene (see below) was transformed into an mss116Δ I0 strain, and Gly+ colonies were selected at 18°C. Derivatives carrying different mtDNA introns were generated as above. PCR-based site-directed mutagenesis was used to construct mss116 alleles in which AKTGTGKT in motif I was changed to VKTATANA, and HRIGRTAR in motif VI was changed to AAAGATAA. The mutant alleles were placed in the mss116Δ strains on the CEN plasmids pHRH161 and pHRH186, respectively (15).
Construction of MSS116myc and MSS116/cyt-19myc Chimera. The MSS116myc allele was constructed by fusing a 2.7-kb segment containing the promoter and MSS116 ORF to a 0.34-kb segment containing a C-terminal myc tag followed by the MSS116 3′ UTR. Both segments were generated by PCR of plasmid pHRH108 containing the MSS116 gene of 161-U7, cloned as a 3-kb HindIII fragment in pRS416 (15). The PCR for the 5′ segment used primers T3 (5′-AATTAACCCTCACTAAAGGG) and 5′-CAAATCTTCTTCAGAAATCAATTTTTGTTCATATATGTTGCTGTTTCTACTGGAGT), and the PCR for the 3′ segment used primers T7 (5′-TAATACGACTCACTATAGGG) and 5′-GAACAAAAATTGATTTCTGAAGAAGATTTGTAGAAAAGATAAAAAAGGAGGACCAAGAG. The segments were fused by PCR with primers T3 and T7, and the product was cloned into the HindIII site of pRS416, yielding pHRH110.
The MSS116/cyt-19myc chimera consists of three fused segments: a 2.5-kb segment containing the 5′ UTR and codons 1–36 of MSS116, amplified by PCR of pHRH108 with primers 5′-CTGGGATAGTGGAAGCTTGCAAGGCA and 5′-GGCAGTGGCTTCGGCCGATCTTCTTGAAACAGCCCA; codons 54–626 of cyt-19 cDNA, amplified by PCR of pTWC19 (7) with primers 5′-TGGGCTGTTTCAAGAAGATCGGCCGAAGCCACTGCC and 5′-CCAGCTCTTACTAGTGAAGCTGGCCTGCTGGCGACGGGT; and a 0.34-kb segment containing the myc tag and 3′UTR of MSS116, amplified by PCR of pHRH110 with primers 5′-AACAGCAACACTAGTGAACAAAAATTGATTTCTGAAGAA and 5′-GTAAATTCTCCGCGGCTTTCTTCCATCCGTTTAATTAAG. The first two segments were combined by PCR splicing by overlap extension (16), generating a 2.5-kb product that was digested with HindIII and SpeI and cloned between the corresponding sites of pRS416 to yield pHRH400, and the third segment was cloned into the SacII site of pHRH400. The MSS116/cyt-19myc chimera contains two inadvertent silent changes, A51G and T159C.
Western Blotting and Pulse Labeling. Proteins were isolated from cells grown in YPR medium containing 2% raffinose (17), and aliquots containing ≈10 μg of protein were run in 1% SDS/7.5% polyacrylamide gels (18). Gels were transferred with a semidry transfer unit to a poly(vinylidene difluoride) (PVDF) membrane (Amersham Pharmacia) and probed with antibodies as described (18). Anti-myc antibody (9E10) and anti-HA antibody (16B12) (Covance, Princeton, NJ) were used at 1:5,000 dilution. Anti-porin antibody (Molecular Probes) was used at 1:1,000 dilution. Mss116p levels were measured by using a guinea pig polyclonal antibody against full-length recombinant Mss116p expressed in E. coli.
In vivo labeling of mt translation products was as described (19) with the following modifications: cells were grown to A600 = 6 at 30°C in YNB medium containing 1% casamino acids and 2% raffinose, supplemented with appropriate nutritional requirements. The cells were then refreshed and labeled at the specified temperatures in YPR medium. Proteins from crude mt fractions were separated in 1% SDS/10–16% polyacrylamide gradient gels, which were dried and scanned with a PhosphorImager.
Biochemical Methods. For Northern hybridizations, RNA was isolated from cells grown at 30°C to A600 = 1–1.5 in YPR medium and separated on 1% formaldehyde–Mops agarose gels. The gels were blotted to nylon membranes (Nytran, Schleicher and Schuell) and hybridized with 32P-end-labeled DNA oligonucleotide probes (15). The hybridized blots were washed, air-dried, and scanned with a PhosphorImager. Splicing levels are expressed as the fraction of fully spliced mRNA relative to total gene transcripts based on quantitation with imagequant.
HG1 primer-dependent reverse transcriptase assays were carried out with mtRNP particles as described (20), except that 0.25 A260 RNP particles were incubated at 37°C for 30 min. The 32P-labeled cDNAs were extracted with phenol/chloroform/isoamyl alcohol (25:24:1), ethanol precipitated, heated to 95°C for 3 min, and hybridized to Southern blots of EcoRI-digested mtDNAs (20).
Results
Mss116p Is Required for Efficient Splicing of all S. cerevisiae mt Group I and II Introns. At the outset, we confirmed that disruption of the MSS116 gene in the yeast strain 161-U7 blocks respiration-dependent growth on glycerol medium at 28–37°C (Gly- phenotype) when the strain's mtDNA contains a standard complement of eight group I and four group II introns, but has a Gly+ phenotype when the strain contains I0 mtDNA lacking introns. This intron-dependent Gly- phenotype indicates that Mss116p is required for RNA splicing in mitochondria.
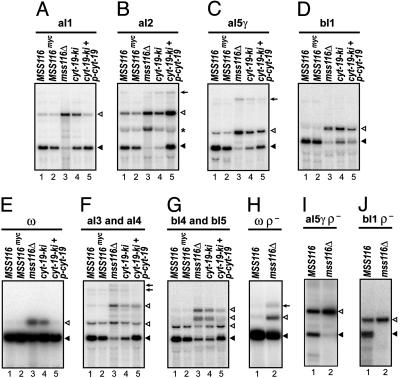
To identify those introns whose splicing requires Mss116p, we carried out Northern hybridizations of mt RNAs isolated from sets of MSS116 and mss116Δ strains grown at 30°C whose mt genes contain only one (aI1, aI2, aI5γ, bI1, or ω) or two (aI3 and aI4 or bI4 and bI5) introns (Fig. 2). The blots show that the disruption of MSS116 inhibits the splicing of all four group II introns (aI1, aI2, aI5γ, and bI1) by 88–97% (Fig. 2 A–D, lanes 1 and 3) and inhibits the splicing of five group I introns (ω, aI3, aI4, bI4, and bI5) by 20–60% (Fig. 2 E–G, lanes 1 and 3). Northern blots for the four remaining group I introns (aI5α, aI5β, bI2, and bI3) also showed partial splicing inhibition (data not shown). In the case of the aI2 group II intron, mss116Δ accumulates not only unspliced precursor RNA, but also the intermediate containing the intron and downstream exons (asterisk in Fig. 2B), indicating that both steps of splicing are inhibited. Strains expressing wild-type levels of Mss116p with mutations in the conserved ATPase motifs were phenocopies of mss116Δ (data not shown; ref. 15).
Fig. 2.
Splicing phenotypes of yeast strains containing different MSS116 and cyt-19 alleles. (A–G) Northern hybridizations of mt RNAs from 30°C-grown strains containing the following mtDNA introns: aI1 (A), aI2 (B), aI5γ (C), bI1 (D), ω (E), aI3 and aI4 (F), and bI4 and bI5 (G). The blots were hybridized with 32P-labeled probes complementary to COX1 exon 6 (A–C and F), COB exon 6 (D and G), and 21S rRNA exon 2 (E). (H–J) Northern hybridizations of mt RNAs from MSS116 and mss116Δ strains with ρ- petite mtDNAs containing a 21S rRNA gene with the ω intron (H), a COX1 gene with aI5γ (I), and a COB gene with bI1 (J). Spliced mRNAs and unspliced precursor RNAs are indicated by filled and open triangles, respectively. Multiple precursor RNAs containing different combinations of introns are present in F and G. Arrows in B, C, F, and H indicate precursor RNAs with 3′ extensions. The asterisk in B indicates an aI2 splicing intermediate containing the intron and downstream exons.
Because the group I introns aI4, bI2, bI3, and bI4 and the group II introns aI1 and aI2 require an IEP with maturase activity for splicing, it is possible that their defective splicing in mss116Δ results from impaired synthesis of the maturase (see Introduction and below). The remaining introns do not encode a maturase, and these include the group II introns aI5γ and bI1, two of the introns whose splicing is most severely inhibited in mss116Δ. To exclude that defective splicing of these introns is caused by impaired mt translation, we constructed matching wild-type MSS116 and mss116Δ strains carrying ρ- petite mtDNAs that retain a COX1 gene with aI5γ, a COB gene with bI1, or a 21S rRNA gene with ω, but lack other mtDNA regions that encode most components of the mt translation apparatus.
The Northern blots show that despite the lack of mt protein synthesis, the splicing of all three introns continues in ρ- strains with a wild-type MSS116 allele, but is inhibited in ρ- strains with the mss116Δ allele (Fig. 2 H–J, lanes 1 and 2). These findings exclude the possibility that the inhibition of splicing in mss116Δ is caused by impaired mt translation. Similar results were obtained for the group I introns aI3 and bI5 (data not shown). We note that in strains with functional Mss116p, the group II introns aI5γ and bI1 splice less efficiently in the ρ- derivatives than in the ρ+ strains (compare lane 1 of Fig. 2 I and J with lanes 1 and 2 of Fig. 2 C and D). That inhibition may reflect that a nuclear-encoded splicing factor for these introns is down-regulated in the respiration-deficient strain (21).
Together, the above results demonstrate that Mss116p is required for the efficient splicing of all group I and II introns in S. cerevisiae mitochondria. A number of these introns are known to require an additional, intron-specific splicing factor to stabilize the active RNA structure (a maturase for aI1, aI2, aI4, bI2, bI3, and bI4 and nuclear-encoded factors Cbp2p for bI5, Nam2p for bI4 and aI4, Mss18p for aI5β, and Mrs1p for bI3) (5, 22). Thus, the splicing of most, if not all, yeast mt introns depends on one or more intron-specific splicing factors and a broadly acting DEAD-box protein, Mss116p.
Mss116p Is also Required for Efficient Translation in Yeast Mitochondria. Although the mss116Δ I0 strain is Gly+ at 28–37°C, it is cold-sensitive (cs), becoming Gly- at 18–24°C. This behavior differs from that of the previously characterized mss116Δ I0 strain, which was Gly- even at 30°C (10), likely reflecting different nuclear backgrounds. The cs Gly- phenotype of the mss116Δ I0 strain indicates that Mss116p has one or more additional functions unrelated to splicing.
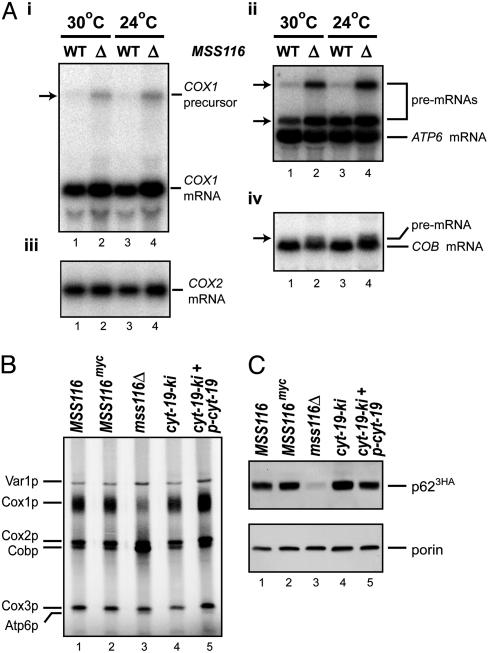
Fig. 3A shows Northern blots of mt RNAs from I0 wild-type and mss116Δ strains grown on raffinose medium at 24°C and 30°C, which are restrictive and permissive temperatures for respiration-dependent growth of mss116Δ I0, respectively. The blots show similar levels of most mt mRNAs, suggesting that the cs defect is posttranscriptional. S. cerevisiae mt RNAs are transcribed as polygenic precursors that are processed via endonucleolytic cleavages. The mss116Δ I0 strain shows some accumulation of larger COX1, ATP6, and COB precursors at both 24°C and 30°C (arrows in Fig. 3A i, ii, and iv), indicating partial defects in 5′ and 3′ end processing (see also ref. 15). It also has somewhat elevated levels of COX1 mRNA (Fig. 3Ai).
Fig. 3.
Mt RNA processing and protein synthesis in yeast strains containing different MSS116 and cyt-19 alleles. (A) Northern hybridization of mt RNAs from wild-type MSS116 and mss116Δ strains with intronless (I0) mtDNA grown on raffinose at 30°C or 24°C. The blot was hybridized sequentially with 32P-labeled probes complementary to COX1 exon 6 (i), ATP6 (ii), COX2 (iii), and COB exon 6 (iv). Arrow in i and upper arrow in ii indicate a polygenic precursor RNA containing COX1-ATP8-ATP6-RF3. Lower arrow in ii indicates an ATP6 precursor RNA with a 5′ extension, and arrow in iv indicates a COB precursor RNA with a 5′ extension. (B) Pulse labeling of mt translation products in strains with I0 mtDNA grown at 24°C. Cells were labeled with 35S-SO4 in the presence of cycloheximide, and mt proteins were analyzed in a 1% SDS/10–16% polyacrylamide gradient gel, which was scanned with a PhosphorImager. Mt translation products are identified to the left. (C) Western blots showing levels of the aI2 IEP (p623HA) in different strains grown at 30°C. Whole-cell proteins (≈20 μg) were analyzed in a 1% SDS/7.5% polyacrylamide gel. After blotting, the membrane was divided at the position of the 40-kDa marker, and the upper and lower halves were probed with anti-hemagglutinin and anti-porin monoclonal antibodies, respectively. Porin is a mt outer membrane protein used to confirm equal loading.
To investigate whether Mss116p is needed for mt translation, the I0 MSS116 and mss116Δ strains were grown at 24°C and mt translation products were pulse-labeled with 35S-SO4 in the presence of cycloheximide, an inhibitor of cytosolic protein synthesis (Fig. 3B). Mt proteins were then analyzed by SDS/PAGE, and the gel was scanned with a PhosphorImager. The results show that all of the major mt translation products are synthesized in mss116Δ (Fig. 3B, lane 3), but the labeling of Cox1p is decreased substantially relative to that in the MSS116 control (Fig. 3B, lane 1). At lower temperature (20°C), Cox1p synthesis was further inhibited, and some inhibition of Cox3p synthesis was also evident (data not shown and ref. 15). On the other hand, the labeling of Cobp relative to the other proteins appears elevated in mss116Δ at both temperatures, presumably reflecting that Cobp synthesis is least dependent on Mss116p (Fig. 3B, lane 3; in contrast to ref. 10).
The steady-state levels of the aI2 IEP (p62) were measured directly by immunoblotting proteins from strains grown at 30°C in which aI2 encoding 3HA-tagged p62 (p623HA) was the only mt intron (Fig. 3C) (18). The blots, which were balanced for the outer membrane protein porin, show that the level of p623HA is strongly decreased in mss116Δ (Fig. 3C, lanes 1 and 3). Because the mss116Δ strain accumulates unspliced precursor RNA from which p62 is translated (Fig. 2B, lane 3), the degree of inhibition of p62 synthesis must be very high.
Expression of the N. crassa CYT-19 Protein in Yeast. To investigate whether the N. crassa CYT-19 protein could rescue the defects in mss116Δ strains, we constructed yeast strains expressing a chimeric MSS116/cyt-19myc gene (Fig. 1). This chimera consists of the promoter, 5′ UTR, and mt import sequence of MSS116 fused in-frame to the coding sequence of mature CYT-19 protein with a C-terminal myc tag and the MSS116 3′ UTR. The site of leader processing was determined by N-terminal sequencing of mature Mss116p (15). One derivative of the wild-type yeast strain contains a single copy of the chimeric cyt-19myc construct “knocked-in” at the chromosomal location of MSS116 (cyt-19-ki), and a second contains the same knock-in plus an extra copy of cyt-19myc on a CEN plasmid (cyt-19-ki + p-cyt-19). For comparison, we constructed a control strain encoding Mss116p with the same C-terminal myc tag.
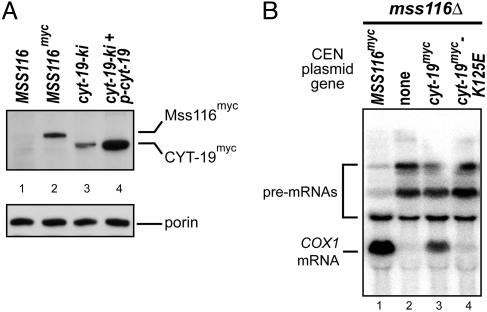
Immunoblots of mt proteins probed with an anti-myc antibody (Fig. 4A) show that the level of CYT-19myc in the cyt-19-ki strain is ≈60% that of Mss116pmyc in the control strain (Fig. 4A compare lane 2 with lane 3), whereas the level of CYT-19myc in cyt-19-ki + p-cyt-19, with an extra copy of cyt-19, is ≈2-fold higher than that of Mss116pmyc (Fig. 4A, compare lane 2 with lane 4; values based on densitometry against serially diluted protein standards; data not shown). In both strains, all of the CYT-19myc detected in the blot is the size expected for cleavage of the mt import sequence, and subcellular fractionation confirmed that the CYT-19myc is present in highly purified mitochondria (data not shown).
Fig. 4.
Expression of CYT-19myc in S. cerevisiae and inability of the CYT-19 K125E mutant to support RNA splicing. (A) CYT-19myc expression. A Western blot of whole-cell protein (≈20 μg) from the indicated strains grown at 30°C was divided at the position of 40-kDa marker, and the top and bottom halves were probed with anti-myc and anti-porin antibodies, respectively. (B) The CYT-19 K125E does not support splicing of COX1 introns. Northern hybridization was carried out with RNAs from strains grown at 30°C, which have a COX1 gene containing aI3, aI4, and aI5γ and carry CEN plasmids with the nuclear genes indicated in the figure. The blot was hybridized with a 32P-labeled probe complementary to COX1 exon 6.
CYT-19 Ameliorates the Translation and Splicing Defects in mss116Δ. Initial growth experiments showed that both cyt-19-ki and cyt-19-ki + p-cyt-19 rescue the cs growth phenotype of the mss116Δ I0 strain, suggesting that CYT-19 can substitute for Mss116p to support mt translation. This inference was confirmed by pulse-labeling with 35S-SO4 in the presence of cycloheximide, which showed that the mt translation defects of the mss116Δ I0 strain are rescued by both levels of CYT-19 expression (Fig. 3B, lanes 4 and 5). Immunoblots show that synthesis of the aI2 maturase (p623HA) is also rescued in the strains expressing CYT-19 (Fig. 3C, lanes 4 and 5).
To assess the effect of CYT-19 on RNA splicing, cyt-19-ki and cyt-19-ki + p-cyt-19 derivatives were constructed containing the same mtDNAs with just one or two introns used in Fig. 2, and mtRNAs were analyzed by Northern hybridization. As noted earlier, the splicing of each of the four group II introns is inhibited 88–97% in mss116Δ (Fig. 2 A–D, lanes 3). The single copy of cyt-19 increased aI1 splicing 7-fold to ≈60% of the wild-type MSS116 level, with smaller increases for the other group II introns (aI2, 4-fold to 20% wild type; aI5γ, 3.9-fold to 37% wild type; and bI1, 1.5-fold to 23% wild type; Fig. 2 A–D, lanes 4). The higher level of CYT-19 expression rescued aI1 splicing to the wild-type level, and rescued aI2 and aI5γ splicing to >50% wild type, but bI1 splicing was restored to only ≈35% wild type (Fig. 2 A–D, lanes 5). CYT-19 also ameliorated the group I intron splicing defects in a dose-dependent fashion (Fig. 2 E–G, lanes 3–5).
To test whether the ability of CYT-19 to rescue the phenotypic defects in mss116Δ requires its ATPase activity, we constructed a chimeric gene in which K125 in motif I was changed to E. This mutation was shown previously to block CYT-19's ATPase and RNA chaperone activities in vitro (7). The K125E mutant protein expressed from a CEN plasmid accumulates in the mss116Δ strain to the same level as wild-type CYT-19 (data not shown; ref. 15), but failed to suppress the splicing defects of a strain with three COX1 introns (aI3, aI4, and aI5γ; Fig. 4B). The K125E allele also failed to suppress the cs Gly- phenotype of the mss116Δ I0 strain (data not shown). Thus, the ability of CYT-19 to ameliorate the splicing and translation defects of mss116Δ requires the ATPase activity of the protein.
Mss116p Functions in Splicing aI2 After Maturase Binding. The splicing of the group II intron aI2 depends on the intron-encoded p62 maturase, which by analogy to the Lactobacillus lactis Ll.LtrB-encoded maturase (23, 24) likely functions by stabilizing the catalytically active structure of the intron RNA. We were intrigued by the finding that expression of one copy of CYT-19 in mss116Δ grown at 30°C restores wild-type levels of p62 (Fig. 3C, lane 4), but only slightly rescues aI2 splicing (Fig. 2B, lane 4). This inefficient splicing could reflect either that Mss116p is required to act on the pre-mRNA to enable it to bind p62 or for an RNA folding step after binding p62.
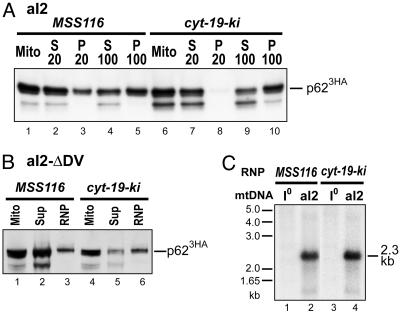
To distinguish these possibilities, we tested whether p62 in cyt-19-ki is associated with aI2 in RNP particles. We showed previously that the recovery of p62 in RNPs after mt lysis in high salt (500 mM KCl) depends on its tight binding to aI2 RNA (18). Fig. 5A shows fractionation experiments carried out under similar conditions with the MSS116 and cyt-19-ki strains. In both strains, p623HA detected by immunoblotting remains mostly in the 20,000 × g supernatant (S20) after mt lysis and then partitions ≈50:50 between the 100,000 × g supernatant (S100) and RNP pellet. We also found that similar proportions of p623HA were recovered in RNPs from MSS116 and cyt-19-ki strains in which aI2 splicing was blocked completely by deletion of intron domain V, eliminating complications from intron excision (Fig. 5B). By contrast, previous experiments showed that p623HA remains entirely in the S100 supernatant when its binding to aI2 is impaired by deletion of the high-affinity binding site in DIVa (see figure 6 of ref. 18).
Fig. 5.
The aI2-encoded protein p62 is associated with aI2 RNA in RNP particles from cyt-19-ki. (A and B) Fractionation of p62. In A, flotation gradient-purified mitochondria from 30°C-grown MSS116 and cyt-19-ki strains, with aI23HA as the only mtDNA intron, were lysed with 1% Nonidet P-40 in a high-salt (500 mM KCl) buffer, and the lysates were centrifuged at 20,000 × g and 100,000 × g (18, 20). In B, flotation gradient-purified mitochondria from MSS116 and cyt-19-ki strains, with aI23HAΔDV as the only mtDNA intron, were lysed as above and centrifuged through a 1.85 M sucrose cushion containing 500 mM KCl buffer (18, 20). In both panels, aliquots of supernatant (S) and pellet (P) fractions were analyzed for p623HA by Western blotting. The band under p623HA in some lanes is a sporadically occurring degradation product. (C) Southern blot of 32P-labeled cDNA synthesized in RNPs. RNP preparations from the MSS116 and cyt-19-ki strains with wild-type aI23HA were used in reverse transcription reactions with 20-mer DNA primer HG1, which is complementary to a 3′-exon sequence 10 nt downstream of aI2. The resulting 32P-labeled cDNAs were hybridized to Southern blots of EcoRI-digested I0 (lanes 1 and 3) and aI2-containing (lanes 2 and 4) mtDNA.
In addition to maturase activity, p62 has reverse transcriptase activity that functions in intron mobility (5), and we have shown that p62 bound tightly to aI2-containing precursor RNAs in RNPs could use a 20-mer DNA primer (HG1) complementary to a 3′ exon sequence to synthesize a cDNA copy of the intron (20). To confirm that the p623HA in the RNP pellet is associated with aI2 in unspliced precursor RNA, we carried out reverse transcription reactions in which RNPs were incubated with 32P-dCTP and other unlabeled dNTPs in the presence of the HG1 primer. The resulting 32P-labeled cDNAs were then hybridized to a Southern blot of EcoRI-digested yeast mtDNAs. Fig. 5C shows that the labeled cDNAs synthesized in both MSS116 and cyt-19-ki hybridized specifically to a 2.3-kb EcoRI fragment that contains aI2 sequences, with only background hybridization to I0 mtDNA, which lacks aI2 (Fig. 5C). Together, these findings indicate that p623HA in cyt-19-ki RNPs is tightly bound to aI2 in unspliced precursor RNA and consequently, that the defective splicing of aI2 in cyt-19-ki reflects a requirement for Mss116p at a subsequent RNA folding step.
Discussion
Our results show that the DEAD-box protein Mss116p is required for the efficient splicing of all S. cerevisiae mt group I and II introns in vivo and that all of the splicing defects in an mss116Δ strain can be partially or completely suppressed by the related N. crassa DEAD-box protein CYT-19, which has demonstrated RNA chaperone activity. In yeast, both Mss116p and CYT-19 act broadly on a variety of different group I and II introns, most if not all of which additionally require a maturase and/or nuclear-encoded splicing factor to stabilize the active RNA structure (see Introduction). Results for the yeast aI2 intron suggest that Mss116p functions at a step after maturase binding, presumably the disruption of stable intermediate or nonnative structures. Furthermore, the splicing defects in mss116Δ are diminished when cells are grown at higher temperature (37°C), as expected if the disruption of stable but inactive RNA structures is rate-limiting (15). Together, our results indicate that the splicing of yeast mt group I and II introns requires both specific proteins that stabilize the active RNA structure and a DEAD-box protein that functions broadly as an ATP-dependent RNA chaperone to alleviate kinetic traps in RNA folding. Our findings provide evidence that the folding of group II introns in vivo is subject to kinetic traps.
In N. crassa, CYT-19 functions together with the CYT-18 protein to promote the splicing of a subset of group I introns (7). CYT-18 binds tightly and specifically to the intron RNA to stabilize the active RNA structure, whereas CYT-19 disrupts nonnative structures, enabling iterative refolding to the final active structure. Moreover, the phenotype of the cyt-19-1 mutant suggests that CYT-19 functions specifically in the splicing of CYT-18-dependent group I introns (7, 8). This apparent specificity could reflect that CYT-19 is targeted to CYT-18-dependent group I introns, possibly by specific RNA structural features or interaction with CYT-18, or that CYT-18 exacerbates kinetic traps, making CYT-18-requiring introns more dependent on RNA chaperone function (7). The finding that the cyt-19-1 mutant is not defective in splicing a group II intron in N. crassa mitochondria may reflect that this intron folds without kinetic traps or uses an RNA chaperone other than CYT-19.
By contrast to the apparent specificity of CYT-19 in N. crassa mitochondria, we find that Mss116p contributes to the splicing of all S. cerevisiae mt group I and II introns. This wider range of action could reflect the lack of a specific targeting mechanism and/or a more general requirement for RNA chaperone function for efficient splicing of yeast mt introns. Similarly, the wider range of action of CYT-19 in yeast mitochondria suggests that it is not targeted and/or more broadly required in that milieu than it is in N. crassa mitochondria. Lack of targeting of CYT-19 in yeast could reflect the absence of specific targeting interactions or simply that CYT-19 is expressed to higher levels in yeast from the chimeric construct than it is in N. crassa. We also note that, even when expressed at 2-fold higher levels than Mss116p, CYT-19 does not fully suppress defective splicing of some yeast mt introns (e.g., bI1) in an mss116Δ strain, implying that the expressed CYT-19 either has lower specific activity than Mss116p or interacts somewhat differently with the intron RNAs.
In addition to splicing, Mss116p is required for some RNA end-processing reactions and for translation of COX1 and COX3 mRNAs at lower growth temperatures. Mss116p may be targeted specifically to these mRNAs, or they may be particularly prone to formation of stable secondary structures that must be disrupted to allow access of translation factors or ribosomes. The phenotype of the cyt-19-1 mutant suggests that CYT-19 also has functions in 5′ and 3′ end processing and possibly mt translation in N. crassa mitochondria (8). The finding that CYT-19 rescues those defects in a yeast mss116Δ strain (Figs. 2 C and F and 3 B and C) indicates either that these functions do not require targeting to a specific site of action or that the targeting occurs by means of interactions that are conserved between yeast and N. crassa.
Our results have implications for the mechanism of folding of group II intron RNAs. In vitro studies of a streamlined ribozyme derived from aI5γ suggested that it folds in a single step without kinetic traps (25). By contrast, our results indicate that the folding of wild-type aI5γ and three other yeast mt group II introns present in their natural precursor RNAs is in fact limited by kinetic traps in vivo, necessitating the recruitment of an RNA chaperone for efficient splicing.
Finally, we note that S. cerevisiae and N. crassa mitochondria contain multiple DExH/D-box proteins that do not substitute for the loss-of-function of Mss116p or CYT-19 in mutants. It is possible that these other DExH/D-box proteins are sequestered in specific complexes or are present at concentrations that are too low to act broadly as RNA chaperones. A recent study estimates 10,300 Mss116p molecules per yeast cell, compared to 1,440 Suv3p and 736 Mrh4p molecules (26). However, an alternate possibility suggested by finding that Mss116p and CYT-19 contain closely related core regions is that only specific types of DExH/D-box proteins can function as RNA chaperones. Such proteins may not be required to unwind long helical regions and thus may be poorly processive RNA helicases, a common characteristic of proteins with the DEAD motif (27). However, they may need to carry out multiple cycles of binding, disruption, and rebinding, necessitating relatively high off-rates after ATP hydrolysis. Dedicated RNA chaperones may be particularly critical for the folding and rearrangement of large RNA assemblies. Indeed, multiple DEAD-box proteins are known to function in ribosome assembly and in the splicing of nuclear pre-mRNA introns, which are thought to be evolutionarily related to group II introns (1, 2, 5).
Acknowledgments
We thank Geen-Dong Chang (National Taiwan University, Taipei) for preparing guinea pig antiserum against Mss116p, and Marlene Belfort (Wadsworth Center, Albany, NY) and R. J. Lin (City of Hope, Duarte, CA) for comments on the manuscript. This work was supported by National Institutes of Health Grant GM31480 and Welch Grant I-1211 (to P.S.P.) and National Institutes of Health Grant GM37951 (to A.M.L.).
Author contributions: H.-R.H., C.E.R., A.M.L., and P.S.P. designed research; H.-R.H., C.E.R., and Y.J. performed research; S.M. contributed new reagents/analytic tools; H.-R.H., S.M., Y.J., A.M.L., and P.S.P. analyzed data; H.-R.H., S.M., A.M.L., and P.S.P. wrote the paper.
Abbreviations: mt, mitochondrial; IEP, intron-encoded protein; RNP, ribonucleoprotein; cs, cold sensitive.
References
- 1.Silverman, E., Edwalds-Gilbert, G. & Lin, R. J. (2003) Gene 312, 1-16. [DOI] [PubMed] [Google Scholar]
- 2.Rocak, S. & Linder, P. (2004) Nat. Rev. Mol. Cell. Biol. 5, 232-241. [DOI] [PubMed] [Google Scholar]
- 3.Jankowsky, E., Gross, C. H., Shuman, S. & Pyle, A. M. (2000) Nature 403, 447-451. [DOI] [PubMed] [Google Scholar]
- 4.Fairman, M. E., Maroney, P. A., Wang, W., Bowers, H. A., Gollnick, P., Nilsen, T. W. & Jankowsky, E. (2004) Science 304, 730-734. [DOI] [PubMed] [Google Scholar]
- 5.Lambowitz, A. M., Caprara, M. G., Zimmerly, S. & Perlman, P. S. (1999) in The RNA World, eds. Gesteland, R. F., Cech, T. R. & Atkins, J. F. (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed., pp. 451-485.
- 6.Akins, R. A. & Lambowitz, A. M. (1987) Cell 50, 331-345. [DOI] [PubMed] [Google Scholar]
- 7.Mohr, S., Stryker, J. M. & Lambowitz, A. M. (2002) Cell 109, 769-779. [DOI] [PubMed] [Google Scholar]
- 8.Henderson, M. F. (1981) M.A. thesis (Saint Louis University, St. Louis).
- 9.Margossian, S. P., Li, H., Zassenhaus, H. P. & Butow, R. A. (1996) Cell 84, 199-209. [DOI] [PubMed] [Google Scholar]
- 10.Séraphin, B., Simon, M., Boulet, A. & Faye, G. (1989) Nature 337, 84-87. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt, U., Lehmann, K. & Stahl, U. (2002) FEMS Yeast Res. 2, 267-276. [DOI] [PubMed] [Google Scholar]
- 12.Séraphin, B., Boulet, A., Simon, M. & Faye, G. (1987) Proc. Natl. Acad. Sci. USA 84, 6810-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niemer, I., Schmelzer, C. & Börner, G. V. (1995) Nucleic Acids Res. 23, 2966-2972. [PMC free article] [PubMed] [Google Scholar]
- 14.Wach, A., Brachat, A., Pohlmann, R. & Philippsen, P. (1994) Yeast 10, 1793-1808. [DOI] [PubMed] [Google Scholar]
- 15.Huang, H.-R. (2004) Ph.D. dissertation (University of Texas Southwestern Medical Center, Dallas).
- 16.Horton, R. M., Cai, Z. L., Ho, S. N. & Pease, L. R. (1990) BioTechniques 8, 528-535.2357375 [Google Scholar]
- 17.Sherman, F., Fink, G. R. & Hicks, J. B. (1986) Laboratory Course: Manual for Methods in Yeast Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 18.Huang, H.-R., Chao, M. Y., Armstrong, B., Wang, Y., Lambowitz, A. M. & Perlman, P. S. (2003) Mol. Cell. Biol. 23, 8809-8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Douglas, M., Finkelstein, D. & Butow, R. A. (1979) Methods Enzymol. 56, 58-66. [DOI] [PubMed] [Google Scholar]
- 20.Zimmerly, S., Moran, J. V., Perlman, P. S. & Lambowitz, A. M. (1999) J. Mol. Biol. 289, 473-490. [DOI] [PubMed] [Google Scholar]
- 21.Butow, R. A. & Avadhani, N. G. (2004) Mol. Cell 14, 1-15. [DOI] [PubMed] [Google Scholar]
- 22.Bassi, G. S., de Oliveira, D. M., White, M. F. & Weeks, K. M. (2002) Proc. Natl. Acad. Sci. USA 99, 128-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuura, M., Noah, J. W. & Lambowitz, A. M. (2001) EMBO J. 20, 7259-7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noah, J. W. & Lambowitz, A. M. (2003) Biochemistry 42, 12466-12480. [DOI] [PubMed] [Google Scholar]
- 25.Su, L. J., Brenowitz, M. & Pyle, A. M. (2003) J. Mol. Biol. 334, 639-652. [DOI] [PubMed] [Google Scholar]
- 26.Ghaemmaghami, S., Huh, W. K., Bower, K., Howson, R. W., Belle, A., Dephoure, N., O'Shea, E. K. & Weissman, J. S. (2003) Nature 425, 737-741. [DOI] [PubMed] [Google Scholar]
- 27.Bizebard, T., Ferlenghi, I., Iost, I. & Dreyfus, M. (2004) Biochemistry 43, 7857-7866. [DOI] [PubMed] [Google Scholar]