ABSTRACT
The Czech v351 strain of rabbit hemorrhagic disease virus (RHDV1) is used in Australia and New Zealand as a biological control agent for rabbits, which are important and damaging introduced vertebrate pests in these countries. However, nonpathogenic rabbit caliciviruses (RCVs) can provide partial immunological cross-protection against lethal RHDV infection and thus interfere with effective rabbit biocontrol. Antibodies that cross-reacted against RHDV antigens were found in wild rabbits before the release of RHDV1 in New Zealand in 1997, suggesting that nonpathogenic RCVs were already present in New Zealand. The aim of this study was to confirm the presence of nonpathogenic RCV in New Zealand and describe its geographical distribution. RCV and RHDV antibody assays were used to screen serum samples from 350 wild rabbits from 14 locations in New Zealand. The serological survey indicated that both RCV and RHDV are widespread in New Zealand wild rabbits, with antibodies detected in 10 out of 14 and 12 out of 14 populations, respectively. Two closely related RCV strains were identified in the duodenal tissue from a New Zealand wild rabbit (RCV Gore-425A and RCV Gore-425B). Both variants are most closely related to Australian RCV strains, but with 88% nucleotide identity, they are genetically distinct. Phylogenetic analysis revealed that the New Zealand RCV strains fall within the genetic diversity of the Australian RCV isolates, indicating a relatively recent movement of RCVs between Australia and New Zealand.
IMPORTANCE Wild rabbits are important and damaging introduced vertebrate pests in Australia and New Zealand. Although RHDV1 is used as a biological control agent, some nonpathogenic RCVs can provide partial immunological cross-protection against lethal RHDV infection and thus interfere with its effectiveness for rabbit control. The presence of nonpathogenic RCVs in New Zealand wild rabbits has been long hypothesized, but earlier attempts to isolate a New Zealand RCV strain have been unsuccessful. Therefore, it is important to determine if such nonpathogenic viruses exist in New Zealand rabbits, especially considering the proposed introduction of new RHDV strains into New Zealand as biocontrols.
KEYWORDS: ELISA, RHDV, rabbit, rabbit calicivirus, virus discovery
INTRODUCTION
Rabbit hemorrhagic disease virus (RHDV) is a highly pathogenic member of the family Caliciviridae within the genus Lagovirus (1). RHDV was first reported in China in 1984 and causes acute necrotizing hepatitis with a high mortality rate in European rabbits (Oryctolagus cuniculus) (2). Due to the high level of pathogenicity and species specificity of the virus, the Czech v351 strain of RHDV (designated RHDV1) was investigated as a biological control agent in Australia and New Zealand, where wild rabbits are major introduced pests.
In 1995, initial testing to assess the transmission of RHDV1 in wild rabbits was undertaken on Wardang Island, off the coast of South Australia. However, the virus escaped quarantine, likely aided by insect vectors, and was found on mainland Australia (3, 4). RHDV1 quickly disseminated across the country, reducing rabbit numbers greatly, particularly in arid areas (5, 6). Following an unsuccessful application to legally import and release RHDV1 into New Zealand, in August 1997, RHDV1 was illegally released in the South Island of New Zealand (7). This strain was found to be closely related to the RHDV1 strain released in Australia (8).
The introduction of RHDV1 into New Zealand significantly reduced wild rabbit populations, although its long-term effectiveness as a biocontrol agent has been varied. One factor contributing to this variability is the increasing number of rabbits acquiring immunity to RHDV1 early in life (9). RHDV immunity can also be influenced by the presence of nonpathogenic or moderately pathogenic rabbit caliciviruses (RCVs), which can confer various degrees of cross-protection against the pathogenic RHDV (10–13). RCVs have been shown to provide complete protection (RCV in Italy) (10), partial protection (RCV-A1 in Australia) (11, 12, 14), and no protection (06-11 in France) (15, 16) against RHDV.
The first nonpathogenic rabbit calicivirus was isolated in Italy from domestic rabbits when animals in a rabbitry appeared to seroconvert to RHDV without the symptoms and clinical signs of disease (10, 17). This lagovirus caused a nonpathogenic infection of the intestinal tract and was given the name rabbit calicivirus (RCV). Since the discovery of this first nonpathogenic RCV, other nonpathogenic and moderately pathogenic strains have been isolated. A rabbit calicivirus (RCV-A1) was discovered in Australia in 2009 (11), and further viruses have since been isolated from rabbitries and wild populations across Australia (18, 19). The distribution and prevalence of Australian RCV-A1 are associated with cool high-rainfall areas found in the south of the continent (20), similar to conditions that prevail in New Zealand.
The presence of nonpathogenic RCVs in New Zealand wild rabbits has long been hypothesized. Antibodies that cross-reacted with RHDV antigens were found in serum from wild rabbits sampled before 1997, suggesting that nonpathogenic RCVs were present in New Zealand prior to the arrival of RHDV1 (21, 22). However, early attempts to isolate a New Zealand RCV strain were unsuccessful (23). The identification and genetic characterization of RCV-A1 in Australia have allowed for the development of specific serological tools for the detection of RCV antibodies (24, 25), warranting a reinvestigation of putative RCV-like viruses in New Zealand.
RESULTS
Serum antibodies.
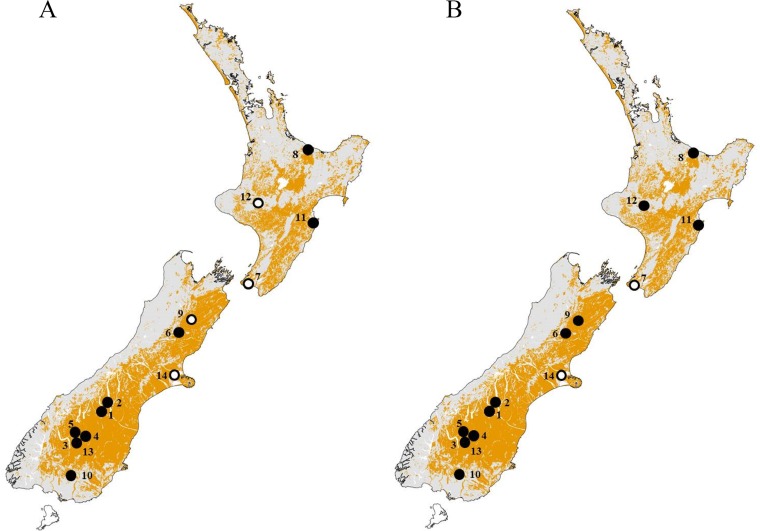
RCV-seropositive animals were detected at 10 out of 14 sample sites tested in both the North and South Islands of New Zealand, with seroprevalences ranging from 16% to 100% (Fig. 1 and Table 1). It should be noted that of the four sample sites that tested negative for RCV antibodies, all sites except Marlborough had a low sample size of ≤10 animals. RHDV antibodies were found in 12 out of the 14 sample sites tested.
FIG 1.
Distribution of RCV-A1 and RHDV in New Zealand at 14 sampling locations. Areas where RHDV occurs in New Zealand are shaded in yellow (based on rabbit prevalence data, modified from Kerr and Ross [44]). Seronegative sample sites are denoted by open circles and seropositive sample sites by closed circles for RCV-blocking ELISA (A) and RHDV cELISA (B). Numbers correspond to sites listed in Table 1. Maps created with ArcGIS version 10.3.
TABLE 1.
Rabbit serum antibodies to RHDV and RCV from 14 sites across New Zealand
| Site no.a | No. of samples | Date collected (mo/yr) | Seropositivity status (%) |
|||||
|---|---|---|---|---|---|---|---|---|
| RCV-A1 seropositive | RHDV seropositive | RHDV and RCV-A1 seropositive | RHDV and RCV-A1 seronegative | RCV-A1 only seropositive | RHDV only seropositive | |||
| 1 | 30 | 11/2012 | 16.7 | 56.7 | 6.7 | 33.3 | 10.0 | 50.0 |
| 2 | 30 | 10/2012 | 53.3 | 83.3 | 43.3 | 6.7 | 10.0 | 40.0 |
| 3 | 31 | 03/2013 | 87.1 | 71.0 | 58.1 | 0.0 | 29.0 | 12.9 |
| 4 | 30 | 03/2013 | 63.3 | 73.3 | 53.3 | 16.7 | 10.0 | 20.0 |
| 5 | 31 | 03/2013 | 35.5 | 51.6 | 19.4 | 32.3 | 16.1 | 32.3 |
| 6 | 29 | 09/2013 | 37.9 | 69.0 | 31.0 | 24.1 | 6.9 | 37.9 |
| 7 | 9 | 12/2012 | 0.0 | 0.0 | 0.0 | 100 | 0.0 | 0.0 |
| 8 | 20 | 05-06/2013 | 100 | 85.0 | 85.0 | 0.0 | 15.0 | 0.0 |
| 9 | 30 | 06/2013 | 0.0 | 60.0 | 0.0 | 40.0 | 0.0 | 60.0 |
| 10 | 30 | 05/2013 | 96.7 | 76.7 | 73.3 | 0.0 | 23.3 | 3.3 |
| 11 | 30 | 06/2013 | 93.3 | 50.0 | 50.0 | 6.7 | 43.3 | 0.0 |
| 12 | 10 | 09/2013 | 0.0 | 90.0 | 0.0 | 10.0 | 0.0 | 90.0 |
| 13 | 30 | 02/2015 | 80.0 | 60.0 | 46.7 | 6.7 | 33.3 | 13.3 |
| 14 | 10 | 04/2013-12/2014 | 0.0 | 0.0 | 0.0 | 100 | 0.0 | 0.0 |
| 350 | 54.3 | 63.4 | 37.7 | 20.0 | 16.6 | 25.7 | ||
Site numbers: 1, Mackenzie Country: Pukaki Flat, Simon's Hills; 2, Mackenzie Country: Tekapo River, Iron Bridge; 3, Otago: Cromwell, Bendigo Station, 2013; 4, Otago: Taras, Ardgour Valley, Cloudy Peak; 5, Otago: Wanaka, Queensberry Hills; 6, North Canterbury: Leslie Hills Station; 7, Wellington: Orongorongo Valley; 8, Bay of Plenty: Te Puke/Pongakawa; 9, Marlborough: Molesworth Station, Isolation Flat; 10, Southland: Gore, Wantwood Station; 11, Hawke's Bay: Cape Kidnappers; 12, Whanganui: Mangapurua Valley; 13, Otago: Cromwell, Bendigo Station, 2015; 14, Canterbury: Selwyn, Motukarara.
According to the enzyme-linked immunosorbent assay (ELISA) results, 20% of all rabbits tested were seronegative for both RCV and RHDV (Table 1). Approximately one-quarter of the rabbits (25.7%) exhibited RHDV antibodies only, 37.7% had both RCV and RHDV antibodies, and 16.6% of animals had RCV antibodies only. In populations where RCV antibodies were present, the proportion of rabbits with RCV antibodies varied with the sex and age of the individual. A higher proportion of females tested RCV seropositive (74.5%) than did males (58.1%).
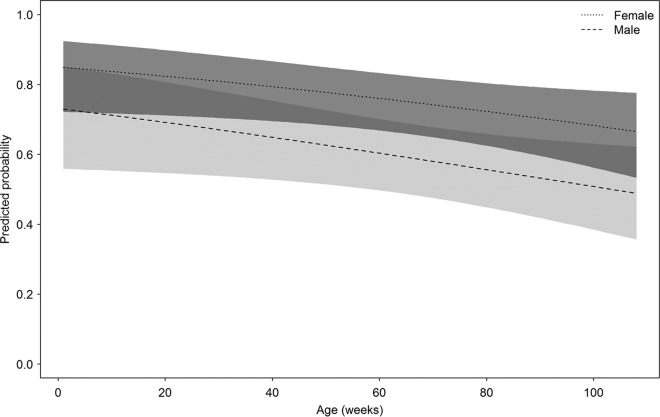
The generalized linear model illustrated that there was evidence for an effect of age (P = 0.036) and sex (P = 0.019) on the seroprevalence of RCV, which was best explained using a multiterm model (i.e., ∼age + sex). The model coefficients were age = −0.01 (standard error [SE] = 0.05) and sex = −0.73 (SE = 0.31). This model had the lowest Akaike's information criterion value (245.5) of all models, and the addition of age as a quadratic term showed no evidence of strengthening the model based on likelihood ratio test (P = 0.70). There was also no evidence of an age-by-sex interaction. Based on the most parsimonious model, the predicted probability of RCV antibody status can be calculated using the age and sex of a rabbit (Fig. 2). There was clearly a difference in the predicted probability between males and females, despite the overlap between 95% confidence intervals. Also, younger rabbits have a higher probability of being RCV seropositive than do older rabbits.
FIG 2.
Predicted probability of a rabbit exhibiting RCV-A1-seropositive antibodies based on the most parsimonious general linear model with age (in weeks) and sex as factors. Shaded areas for both male and females show the 95% confidence intervals around the mean.
Detection of nonpathogenic rabbit calicivirus.
Other nonpathogenic RCVs have been predominantly detected in the small intestine (10, 11, 26), so duodenal tissue was targeted for the detection of a putative nonpathogenic calicivirus. Approximately 100 duodenal RNA samples were screened using universal lagovirus PCR primers. Initially, young rabbits were preferentially selected because other RCV viruses are more commonly isolated from younger animals (10, 11). As there were few very young rabbits in the RCV antibody-positive populations, 143 rabbits up to 35 weeks of age were subsequently screened. A positive PCR product was obtained from a 33-week-old male rabbit. Other samples from the same location were weakly positive (n = 9), but good-quality sequencing product was obtained from only one rabbit (Gore-425). This sample was collected on a property near Gore in Southland and was seropositive for RCV antibodies at a 1:40 dilution using the RCV-blocking ELISA, and it was also seropositive for RHDV antibodies at a 1:40 dilution. Initial sequencing of the 320-nucleotide product revealed 86% nucleotide identity with RHDV Czech v351 isolate (GenBank accession no. KF594473) and 93% nucleotide identity with the Australian RCV-A1 MIC3-3 (GenBank accession no. GU368892.1). The initial sequenced product matched the sequence from RCV Gore-425A.
Inoculum production trial.
To reproduce RCV infection and generate inocula for future studies, two New Zealand White rabbits were infected orally with material from rabbit Gore-425 (inoculum production trial). Dosed animals appeared healthy and were euthanized at 4 days postinfection. RNA was extracted from the duodenum of each rabbit and the presence of RCV confirmed by universal lagovirus PCR and Sanger sequencing. RNA extracts from the duodenum were used for the sequencing of the viral capsid gene (VP60).
VP60 sequencing.
Two variants of RCV were detected, each selectively amplified by a different primer set, indicating a mixed infection with two strains of RCV in rabbit Gore-425. The variant RCV Gore-425A was obtained using primers capsRCV-A1[HindIII]/capsRCV-A1rev[NotI], and the variant RCV Gore-425B was obtained using primers RCV-4/ALR-6. The variants differ by 22 nucleotides across the VP60 gene (1,740 nucleotides), which equates to 98.7% nucleotide identity and results in four amino acid changes. Sequencing analysis revealed that both RCV Gore-425 variants have approximately 79% nucleotide identity to RHDV Czech v351 (accession no. KF594473) and the Italian RCV VP60 genes (accession no. X96868), as well as 71% nucleotide identity to European brown hare syndrome virus (EBHSV; accession no. NC_002615), a related lagovirus that infects European brown hares. RCV Gore-425A and Gore-425B are most closely related to Australian RCV CAT3-4 (accession no. GU368890) isolated from Cattai National Park, NSW, Australia, with 88.4% and 88.6% nucleotide identity, respectively.
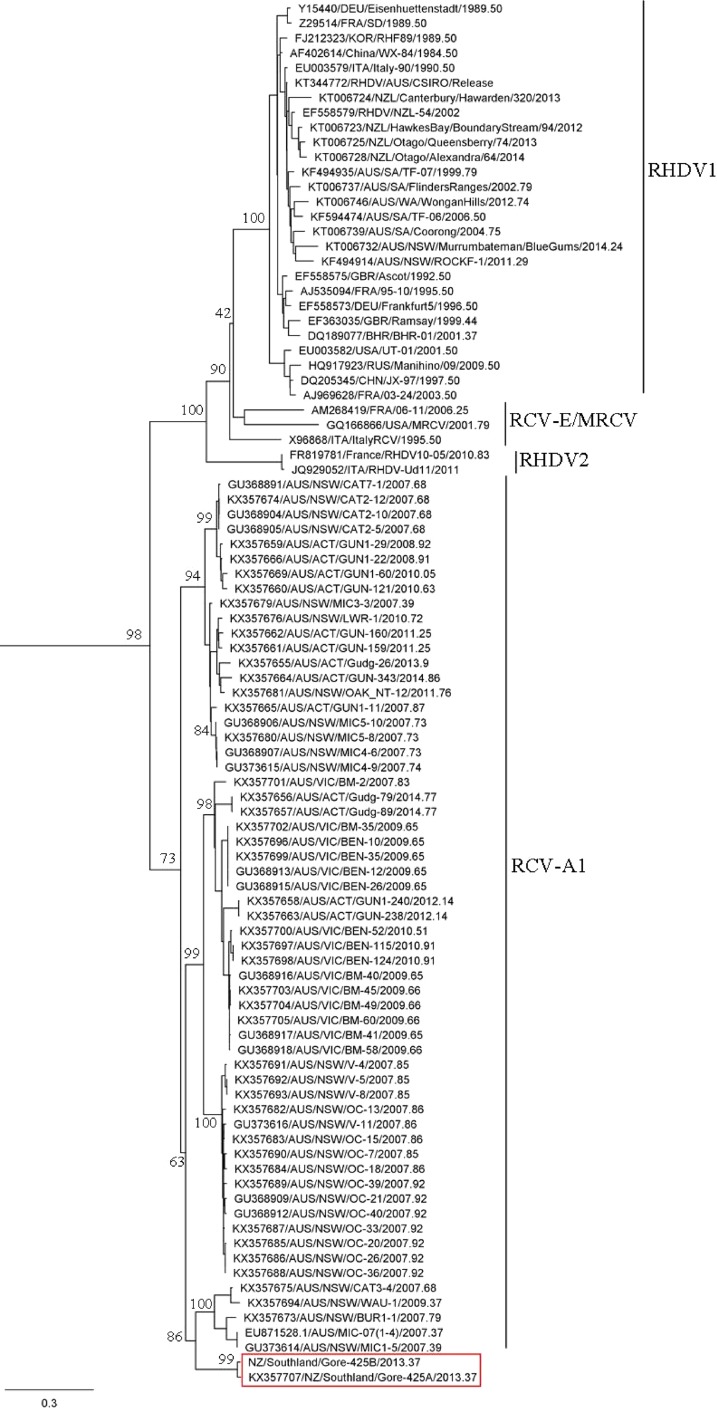
A maximum likelihood phylogenetic tree was assembled using the VP60 capsid gene sequences of an EBHSV strain (utilized as an outgroup sequence, not shown), 29 RHDV isolates, 62 RCV isolates, and the newly sequenced RCV Gore-425A and Gore-425B isolates (Fig. 3), confirming that these viruses sit within the level of genetic diversity of RCV-A1 that was isolated from rabbits in Australia (19).
FIG 3.
Maximum likelihood phylogenetic analysis of Gore-425A and Gore-425B (red box) with 62 published RCV and 29 RHDV VP60 capsid gene sequences (1,740 nucleotides [nt]). The phylogeny was generated using the PhyML program in Geneious and was rooted using EBHSV as the outgroup (not shown). Support for individual nodes was estimated from 1,000 bootstrap replicates, and values are indicated at major nodes. The GenBank accession numbers of published sequences included in the tree are indicated in the taxon name. The scale bar represents the number of nucleotide substitutions per site.
Time scale of New Zealand RCV evolution.
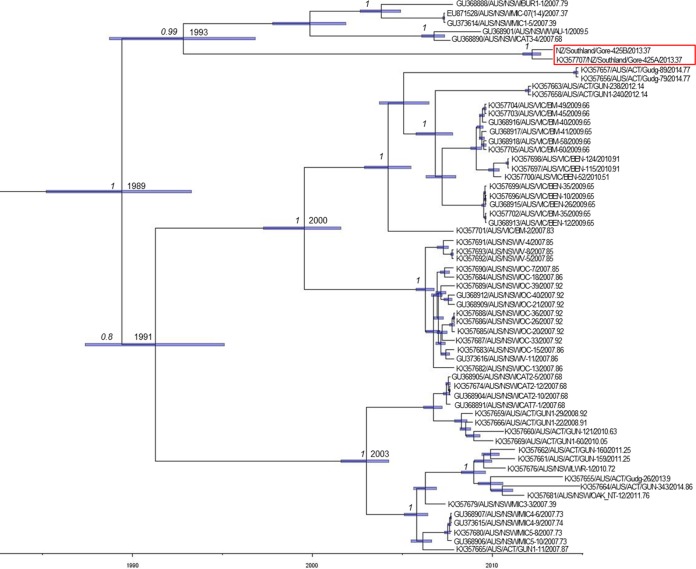
Evolutionary analysis was conducted using the RCV-A1 VP60 gene data set to estimate the nucleotide substitution rate (substitutions [subs]/site/year) and the time to most recent common ancestor (TMRCA) of these viruses using a Bayesian Markov chain Monte Carlo approach. Accordingly, we estimated the mean rate of evolutionary change to be 5.75 × 10−3 subs/site/year, with 95% highest posterior density (HPD) intervals spanning 4.61 × 10−3 to 6.85 × 10−3 subs/site/year. Notably, the two New Zealand RCV isolates were estimated to share a most recent common ancestor with Australian RCV-A1 strains in the early 1990s (mean, 1993; 95% HPD, 1989 to 1997) (Fig. 4). The TMRCA for all RCV-A1 strains, including the two New Zealand variants, was estimated to be in the late 1980s (mean, 1989; 95% HPD, 1985 to 1993) (Fig. 4).
FIG 4.
Bayesian Markov chain Monte Carlo tree of RCV-A1 sequences and New Zealand RCV-A1 isolates. The x axis provides a time scale (years). Mean TMRCA values are shown at each node, and node bars (purple) depict 95% HPD values on node height (age). Posterior probability values, a measure of support, are shown at key nodes when >0.8. The year of sampling is represented in decimal form at the end of the taxa name. The GenBank accession numbers of published sequences included in the tree are indicated at the beginning of the taxon name. The red box represents the isolates used in this study (Gore-425A and Gore-425B).
RCV-blocking ELISA validation.
The RCV-blocking ELISA was originally developed for RCV-A1 Australian virus. To validate the use of RCV-blocking ELISA for the detection of RCV Gore-425 antibodies, serum from three rabbits that had been experimentally infected with RCV Gore-425 (RCV-blocking ELISA validation trial) was tested in both the RCV-blocking ELISA and RHDV competition ELISA (cELISA). All rabbits were seronegative for RCV and RHDV antibodies prior to infection and remained seronegative for RHDV antibodies for all time points. From day 7, rabbits were positive for RCV antibodies at a serum titer cutoff of ≥1:160. No signs of disease were observed.
DISCUSSION
This study has identified an apparently nonpathogenic rabbit calicivirus isolated from a wild rabbit in New Zealand. The existence of a nonpathogenic RCV in New Zealand that may cause a level of cross-protective immunity toward RHDV has long been predicted (22, 27), and the isolation and sequencing of viral RNA from rabbit duodenum have now confirmed its presence, which contributes to the growing number of RCVs already identified. Sequence data indicate that RCV Gore-425A and Gore-425B are most similar to the RCV-A1 cohort and are distinctly different from RHDV and the European RCVs. It is interesting to note that RCV Gore-425A and Gore-425B were isolated from the same rabbit. We can speculate that the laboratory rabbit was dosed with a mixed infection, but there is a possibility that the rabbit was already infected with another nonpathogenic calicivirus, which would explain why two viruses were amplified. The idea of a mixed infection appears to be more likely, and the use of different primer sets selectively amplified each virus.
Based on phylogenetic analysis of the VP60 gene, these viruses cluster with RCV-A1 in a tight monophyletic group, despite being found in different countries. In the current study, evolutionary analysis of the VP60 gene for all published RCV-A1 isolates and the New Zealand RCV variants produced a mean evolutionary rate of 5.57 × 10−3 subs/site/year (95% HPD interval, 4.61 × 10−3 to 6.85 × 10−3 subs/site/year) and hence equivalent to those rates previously estimated in RCV-A1 (19). Interestingly, these rates are higher than those observed for the pathogenic RHDV (mean, 2.77 × 10−3 subs/site/year) (29) and hence may be inflated because of the presence of transient deleterious mutations that are yet to be removed by purifying selection (19). The VP60 data set from this study placed the common ancestor of all RCV-A1s in the late 1980s, whereas dating based on the nonstructural gene data set (19) gave a TMRCA closer to the early 1980s. Because of the likely rate inflation in VP60, it is possible that the true date of divergence of the New Zealand isolates was closer to the lower estimate of the HPD interval, perhaps in the early to mid-1980s. Such a recent date of divergence, and the fact that the New Zealand isolates cluster within the Australian RCV-A1 isolates, strongly suggests that the RCV Gore-425 may have come from Australia rather than having arrived from Europe when rabbits were first introduced to New Zealand in the mid-19th century (30). Due to the limited number of New Zealand isolates available for sequencing, it is unclear if this is the only strain of RCV present or if there are other more divergent lineages circulating in New Zealand wild rabbits. Detection and sequencing of more nonpathogenic RCV isolates in New Zealand will be important for understanding the evolutionary history of this virus in this country.
Antibody data from the RCV-blocking ELISA show that RCV antibodies are widespread across the country and present in both the North and South Islands. In Australia, RCV-A1 antibodies are predominately detected in rabbit populations situated in cooler higher-rainfall areas in the southeast of the continent. The high prevalence of RCV-A1 antibodies in these areas is thought to also be due to other factors relating to the climate, as higher rainfall supports year-round breeding compared to the more arid areas of Australia and provides a constant source of susceptible rabbits to help the transmission of RCV infection. The comparatively widespread distribution of RCV antibodies across New Zealand may in part be explained by the region's generally more temperate climate. According to annual climate summary data collected by the National Institute of Water and Atmospheric Research (NIWA), the average rainfall and temperature for all the sampling sites in New Zealand fell within the rainfall and temperature conditions that correlate with high RCV-A1 prevalence in Australia (20). It therefore appears unlikely that climatic conditions limit the prevalence and distribution of RCV in New Zealand.
The distribution of RCV antibodies versus that of RHDV antibodies in New Zealand rabbit populations could be attributed to the different transmission modes of the two viruses. In total, four sample sites tested negative for RCV antibodies, and two samples sites tested negative for RHDV antibodies. RHDV is likely to be spread over greater distances due to the recognized role of insect transmission, whereas RCV-A1 is believed to spread between rabbits via direct contact or the fecal-oral route (18, 31). The sample sites that tested negative for RCV tended to be geographically isolated sites with relatively low rabbit densities at the time of collection. Populations from Orongorongo Valley, Mangapurua Valley, and Motukarara exhibited low numbers, estimated at <1 rabbit per km spotlight counting, and Isolation Flat recorded moderate numbers, at 2.4 rabbits per km. However, it should be noted that at several of these sites, only small sample numbers were obtained, which lowers the confidence that this population is truly negative. Most of the RCV-seropositive populations had higher rabbit counts: six of seven sites had more than 2.5 rabbits per km spotlight count (range, 1.5 to 108 rabbits/km spotlight count).
This study showed that the RCV-blocking ELISA is able to detect the presence of serum antibodies for nonpathogenic RCVs in New Zealand wild rabbit populations. In light of the sequence analysis, this is not surprising, as the New Zealand strains sit within the diversity of Australian RCV-A1 isolates and are thus genetically closely related. Not only is the RCV-blocking ELISA able to detect RCV antibodies, but the results indicate that RHDV antibodies detected by the cELISA do not cross-react with the RCV-blocking ELISA. For example, the ELISA results from both the Marlborough and Whanganui populations (Table 1) are seronegative for RCV antibodies, but both populations exhibit RHDV antibodies based on the RHDV competition ELISA. Not surprisingly, RCV Gore-425A and Gore-425B have significant amino acid substitutions (11/25) at the RHDV monoclonal antibody (MAb) 1H8 putative binding site, the key reagent in the cELISA (32). This finding supports existing evidence that rabbits that survive RHDV infection have serum antibodies that bind in the RHDV competition ELISA but do not cross-react with antigens in the RCV-blocking ELISA (24).
There is evidence to suggest there are different prevalences of RCV antibodies between male and female rabbits. In populations where RCV was present, females had a higher seroprevalence of RCV antibodies than males, irrespective of age. The reason for this difference is not clear. However, RCV-A1 is believed to be transmitted by the fecal-oral route and may be spread between females sharing burrows and by females to young rabbits while they are in close proximity with each other in the nest (18), thereby increasing the likelihood of exposure to RCV-A1. In contrast, males have larger home ranges and may lead solitary lives when young (30). It is yet to be experimentally determined if infection with the New Zealand RCV isolate provides partial cross-protection against lethal RHDV infection.
Across New Zealand, it is clear that rabbit numbers in certain areas are increasing, while numbers in some regions remain stable (33). In Canterbury, although rabbit numbers show peaks and troughs attributable to breeding pulses and RHDV epidemics, overall, there has been a general increase in rabbit numbers despite the application of secondary control, such as 1080, pindone pellets, and shooting (33). In some sites in Central Otago, rabbit densities have recovered to pre-RHDV levels despite annual RHDV outbreaks (27). The introduction of RHDV1 and fluctuations in rabbit density in some areas may have contributed to regional differences in RCV-A1 prevalence (20). Populations that were seronegative for RCV tended to be geographically isolated populations not immediately adjacent to high-rabbit-density areas. It may be that RCV has never established in these areas or that high rabbit density plays a role in maintaining the prevalence of RCV in a population. If rabbit density is a factor for RCV prevalence, it is possible that RCV could establish itself in currently seronegative populations if rabbit numbers were to increase.
Both RCV-A1 isolates (Gore-425A and Gore-425B) were isolated from an apparently healthy wild rabbit, and in both trials where laboratory rabbits were experimentally infected with RCV Gore-425, the animals showed no obvious signs of disease before they were euthanized at 4 days (inoculum production trial) and 21 days (RCV-blocking ELISA validation trial) postinfection. Additional controlled infection studies are under way to confirm the nonpathogenic nature of the New Zealand isolate and to identify sites of viral replication and virus loads within tissues, as well as the potential routes of viral shedding and transmission.
The confirmation of nonpathogenic RCV-A1 in New Zealand and the high prevalence of RCV antibodies (with over 50% of sampled animals testing RCV seropositive) raise new questions with regard to its impact on the efficacy of RHDV as a biocontrol agent in New Zealand. If RCV Gore-425, like the closely related Australia RCV-A1 strains, provides cross-protection against RHDV challenge (12), knowledge of RHDV and RCV antibody prevalence in rabbit populations will be important for predicting the likely success of RHDV biocontrol operations. Ideally, long-term monitoring of antibody status, along with detailed knowledge of RHDV outbreaks and associated mortality rates, would be required. The virus-virus interactions between RCV and RHDV are complicated and not well understood. Antibody data indicate that infections with RCV and RHDV can occur in the same individual (Table 1) and that the nature of virus interactions and infections is likely to be complex and fluid. To fully elucidate whether the New Zealand RCV isolates (RCV Gore-425A and Gore-425B) are able to provide protection against lethal RHDV exposure, challenge studies will need to be undertaken. This will be particularly important when considering the potential impact the introduction of new RHDV strains into New Zealand will have in the future.
MATERIALS AND METHODS
Sampling.
Wild rabbit samples were collected from 14 survey sites across New Zealand (Fig. 1) between August 2012 and February 2015. Sites were located in eastern (Hawke's Bay), western (Whanganui), and southern (Orongorongo Valley) regions in the North Island and in northern (Marlborough) and southern (Southland) regions in the South Island. Sites were selected with the aim of covering a range of rabbit-prone areas in New Zealand. Additional sites, such as Whanganui and Orongorongo Valley, were included because of their isolation and anecdotal reports suggesting a lack of RHDV exposure to rabbits in those areas. Some sites were also assessed for rabbit density based on rabbits per kilometer spotlight count or the modified McLean scale (34).
Tissue and data were collected from freshly killed rabbits that had been shot by experienced contractors, caught by dogs, or live-trapped and humanely killed. The intent was to survey 30 rabbits from each site, but in some cases, rabbit numbers were low and fewer samples were collected (Table 1). Most rabbits were collected as part of regional and district councils' annual rabbit control programs undertaken to assess age structure, breeding condition, and the RHDV antibody status of regional rabbit populations. Animals were assessed for body condition and/or body weight, sex, and reproductive state. Samples of tissue from the liver and duodenum (2 to 12 cm below the pyloric sphincter) were collected; a subsample of each was stored in RNAlater (Sigma-Aldrich, St. Louis, MO, USA) and the remainder frozen at −80°C. Eyeballs were removed and preserved in 10% buffered formalin for aging of the animals by lens weight (35). Blood samples were collected from the heart using a syringe and an 18-gauge needle and transferred to a 5-ml serum separator tube (SST; BD Vacutainer, Plymouth, UK). Blood was allowed to clot for at least 2 h before the serum samples were centrifuged, decanted, and stored at −20°C.
Serology.
Enzyme-linked immunosorbent assays (ELISAs) were used to detect serum antibodies to RHDV and RCV-A1 and to deduce what populations and individuals may have previously been exposed to RCV infection. RHDV antibody titers were measured semiquantitatively using a competition ELISA (cELISA) (36) at AgResearch (Hopkins Research Centre, Palmerston North, New Zealand). The cELISA kit was obtained from OIE-RHDV-RL, Italy, and was carried out according to the instructions. Rabbit serum was tested at two serial dilutions (1:10 and 1:40), and the optical density at 490 nm (OD490) was read. The OD490 of negative serum at each dilution was defined as 100%, and for the purpose of this study, rabbit serum with more than 50% reduction in absorbance at OD490 for a ≥1:40 serum dilution was considered positive. A specific blocking ELISA originally developed for Australian RCV-A1 was used to measure RCV-A1 antibody titers, as described previously (24). Rabbit serum was tested at serial dilutions of 1:10, 1:40, and 1:160 using high-binding 96-well plates (Greiner Bio-One, Austria). The OD at 492 nm was read, and the results were measured as a percent inhibition compared to the negative serum on the same plate. The OD492 of negative serum at each dilution was defined as 100%, and rabbit serum with more than 25% reduction in absorbance at OD492 for a ≥1:10 serum dilution was considered positive.
Virus detection.
RNA was extracted from approximately 15 mg of homogenized tissue from the first 5 cm of the duodenum using the easy-spin total RNA extraction kit (Intron, South Korea), as per the protocols provided by the supplier. First-strand cDNA synthesis was performed using approximately 2 to 6 μg of RNA, oligo(dT15) primer, and SuperScript IV reverse transcriptase (Invitrogen, MA, USA), according to the supplier's instructions.
In an attempt to isolate a New Zealand RCV strain, rabbit duodenal RNA extracts from populations where RCV antibodies were prevalent, as well as all young rabbits (<26 weeks of age) from other locations, were screened by the universal lagovirus PCR assay (11), as described previously. Viral cDNA was detected in 50-μl PCRs using 20 pmol each oligonucleotide Rab1b and Rab2 (Table 2) and 25 μl of BioMix PCR kit (Bioline, London, UK), according to the supplier's instructions. Cycling conditions were 94°C for 3 min, 45 repeats of 15 s at 95°C, 55°C for 20 s, and 72°C for 20 s, followed by a final extension step of 72°C for 1 min. PCR products were run on 2% E-gel general purpose gel (Invitrogen), and positive PCR products were recovered from agarose gel using the Zymoclean gel DNA recovery kit (Zymo Research), according to the supplier's instructions. Potential positive bands were further analyzed by Sanger sequencing at Ecological Genetics Laboratory, EcoGene (Landcare Research, Auckland, New Zealand). Nucleotide Basic Local Alignment Search Tool (BLAST) analysis was used to compare the similarity of isolated sequences with known sequences in the National Center for Biotechnology Information NCBI database (37). RNA from the liver of an animal known to have died from RHDV was included as a positive lagovirus control.
TABLE 2.
Oligonucleotides used for amplification and sequencing of New Zealand RCV Gore-425 strains
| Primer | Sequence 5′ to 3′ | Orientation | Positionsa | Reference or source |
|---|---|---|---|---|
| For virus detection | ||||
| Rab1b | CAGCDSGCACTGCYACCACAGCATC | + | 5309–5333 | 11 |
| Rab2 | GAAKCKRAACTGCATGCCACCAGCCCA | − | 5608–5634 | 11 |
| For cDNA synthesis and amplification of RCV Gore-425 VP60 capsid | ||||
| Oligo dTV | + | |||
| MICV-10 | CCAAACCTAGCTACCATCTTCAACTTGTAGC | − | 4355–4385 | 11 |
| RCV-4 | TCAARATGACAGACATYGGTTGGGT | + | 4946–4970 | 18 |
| ALR-6 | CTCGCCAGTGGTATTATAAATCTTAACAC | − | 7308–7336 | 11 |
| capsRCV-A1[HindIII] | AAAAGCTTATGGAGGGCAAGGCCCGTGCAACG | + | 5264–5295 | 18 |
| capsRCV-A1rev[NotI] | AAGCGGCCGCGTTGCTCAAAACGCCGGCACCTGC | − | 7034–7076 | This study |
| For real-time PCR | ||||
| RCV RT F | GTTGGYAGGAAYGTRCCCATCATGTTTGC | + | 6637–6665 | 18 |
| RCV NZ R2 | AGACAGGTCAATAAGTGTGGTCA | − | 6896–6918 | This study |
Positions in relation to Gore-425A genome (accession no. KX357707).
Real-time PCR quantification of viral RNA.
A specific real-time PCR assay was set up for the quantification and detection of RCV in tissue samples. Standards for real-time PCR are described elsewhere (11). For real-time quantification of viral RNA in rabbit tissues, the instrument Mx3005P (Agilent Technologies) was used with Bioline SensiFAST SYBR Lo-ROX one-step kit (Bioline, London, UK), according to the instructions from the supplier. Each reaction mixture contained a final concentration of 400 nM the target gene primers RCV RT FW and RCV-NZ Rev2 (Table 2). The cycling conditions used were 10 min at 45°C, 2 min at 95°C, followed by 41 repeats of a three-step cycle with 5 s at 95°C, 20 s at 55°C, and 10 s at 78°C. A final step was added to confirm the specificity and absence of primer dimers of 10 s at 95°C, 5 s at 65°C, and 30 s at 95°C. All samples were tested in triplicate and the results analyzed on MxPro software version 4.10 (Agilent Technologies, CA, USA).
Animal experimental infections.
Two animal experiments were carried out. New Zealand White rabbits were housed individually in cages (Tecniplast, Italy), were offered fresh apple and carrots daily, and had ad libitum access to water and commercial rabbit pellets (Weston's Stockfeed, Canterbury, New Zealand). Serum and whole-blood samples were collected from the marginal ear vein. Animals were sedated (0.2 ml/kg of body weight of 50 mg/ml Zoletil [Virbac, Carros, France]) and euthanized by anesthetic overdose (100 mg/kg of pentobarbitone by intraperitoneal injection or 100 mg/kg of Pentobarbitone injected intracardially).
Inoculum production trial.
Two 16-week-old New Zealand White rabbits were experimentally infected with RCV Gore-425. For the experimental infection, gut tissue derived from the wild rabbit (rabbit no. 425) which tested PCR positive for RCV Gore-425 was used to produce inoculum by homogenizing approximately 1.12 g of duodenal tissue in 2.25 ml of sterile chilled 0.01 M phosphate-buffered saline (PBS). The rabbits were dosed orally with 500 μl of gut homogenate applied dropwise into the oral cavity. The genome copy equivalents of the dosing inoculum were determined using quantitative reverse transcription-PCR (qRT-PCR) and measured 8.3 × 103 genome copies/μl (4.2 × 106 copies/dose). Treated animals were euthanized 4 days postinfection, and duodenum tissue was stored in RNAlater or frozen at −80°C. This trial confirmed that the virus collected from wild rabbit no. 425 was infective, and it provided an inoculum for future studies.
RCV-blocking ELISA validation.
Three 16-week-old New Zealand White rabbits were experimentally infected with the RCV Gore-425 inoculum derived from the rabbits infected in the inoculum production trial, by homogenizing approximately 4.52 g of duodenal tissue in 21 ml of sterile chilled 0.01 M PBS. The rabbits were dosed orally with 500 μl of gut homogenate applied dropwise into the oral cavity. The genome copy equivalents of the inoculum were determined using qRT-PCR and measured 3.16 × 104 genome copies/μl (1.6 × 107 copies/dose). Serum samples were taken at 0, 7, 14, and 21 days postinfection. The rabbits remained in good health throughout the trial and were euthanized 21 days postinfection. Serum was used to measure antibody responses with the RCV-blocking ELISA for the New Zealand RCV.
DNA library preparation and Illumina sequencing.
First-strand synthesis of the duodenal RNA extractions from one of the laboratory rabbits from the inoculum production trial was undertaken using SuperScript III reverse transcriptase (Invitrogen), as per the supplier's instructions. The primer OligoDT18 (0.25 μg) was used, as were Oligo dTV (1.5 μM) and an RCV-A1 specific primer, MICV-10 (0.8 μM) (Table 2). The capsid region of Gore-425 was amplified using two primer sets (Table 2), RCV-4/ALR-6 and capsRCV-A1[HindIII]/capsRCV-A1rev[NotI], using Platinum Taq DNA polymerase high-fidelity kit (Invitrogen), as per the manufacturer's instructions. Cycling conditions for RCV-4/ALR-6 were performed as described previously (19). Cycling conditions for capsRCV-A1[HindIII]/capsRCV-A1rev[NotI] were 94°C for 2 min, 16 cycles of 15 s at 94°C, 68°C (−0.5°C per cycle) for 20 s, and 68°C for 3 min, 24 cycles of 15 s at 94°C, 60°C for 20 s, and 68°C for 3 min, followed by a final extension step of 68°C for 5 min. Products were analyzed using 1% agarose gel electrophoresis and purified using the QIAquick gel extraction kit (Qiagen, Hilden, Germany). Purified amplicons were then pooled to equimolar concentrations (0.14 ng/μl).
DNA libraries were prepared from amplicons (0.7 ng of total DNA) using the Nextera XT DNA sample preparation kit (Illumina). Paired-end sequencing was performed using the Illumina MiSeq platform and a 300-cycle MiSeq reagent kit (version 2), as described previously (38).
Sequence data processing and genome assembly.
Low-quality reads were removed, and reads were trimmed and merged as described previously (19). The final cleaned reads were mapped to MIC-07 RCV-A1 (accession no. EU871528) in Geneious version 8.1.6 (39) to generate a majority consensus sequence for each sample.
Compilation of sequence data sets.
For phylogenetic analysis, the complete VP60 capsid gene sequences of RCV Gore-425A and Gore-425B were aligned with previously published RCV isolates (Australian RCV-A1 and European RCV-E) and RHDV (classic and RHDV2) sequences available in GenBank (http://www.ncbi.nlm.nih.gov/GenBank/). The accession numbers of previously published sequences are included in the taxon name.
For evolutionary analysis, the complete VP60 capsid gene sequences of RCV Gore-425A and Gore-425B were aligned with previously published RCV-A1 isolate sequences (18, 19); the accession numbers are included in the taxon names.
Phylogenetic analysis.
Sequence alignments were generated using Clustal W in Geneious, and the best-fit model of nucleotide substitution was determined to be GTR+Γ using JModelTest (40). A maximum likelihood (ML) phylogenetic tree was inferred using the PhyML program (41) available in Geneious, using a combination of nearest neighbor interchange, subtree pruning, and regrafting branch swapping. Support for individual nodes was estimated using 1,000 bootstrap replicates.
Bayesian Markov chain Monte Carlo analysis.
The time to most recent common ancestor (TMRCA) of the Gore-425 variants was estimated using the Bayesian Markov chain Monte Carlo method, available in the BEAST package version 1.8 (42). The RCV-A1 VP60 gene sequence alignment was utilized, and sampling dates (day/month/year) were assigned to all sequences. Marginal likelihood estimation using path sampling/stepping-stone sampling (as available in BEAST) indicated that the strict clock model and the relaxed uncorrelated lognormal deviation clock model fit these data equally (log marginal likelihood estimation values differed by <3.1), and the 95% highest posterior density (HPD) intervals for each model overlapped for all parameters. Accordingly, the simpler strict clock model was selected. The constant population size coalescent model was shown to be the most appropriate model of population growth for RCV-A1 data (19). Each BEAST analysis was run for 50,000,000 generations until convergence was achieved, and at least two independent runs were performed for each set of priors. A maximum clade credibility tree with mean node heights and Bayesian posterior probability values indicating the degree of support for each node was created using the TreeAnnotator program (as available in the BEAST package) from the posterior set of trees.
Statistical analysis of antibody data.
Statistical analysis assessing the effects of age and sex on RCV serum antibody status used a generalized linear model (binomial distribution) in RStudio (43). All models were completed using the lmtest package in RStudio. We explored the influence of age and sex on RCV serum antibody status using candidate models including age, sex, and age as a quadratic.
Models were ranked using Akaike's information criterion, and likelihood ratio tests from the lmtest package were used to decide the model of best fit. Based on the likelihood ratio test, there was no support for treating RCV status as a random effect. The most parsimonious model was used to plot predicted probabilities of RCV antibodies using age and sex as factors. Predictions were averaged, and 95% confidence intervals were plotted using RStudio (43).
Approval for experiments involving animals.
All experimental methods involving animals were approved by the Landcare Research Animal Ethics Committee (AEC approvals 13/07/02 and 15/02/02).
Accession number(s).
The accession number for RCV Gore-425B is listed in GenBank as KY426993. RCV Gore-425A has been previously published by Mahar et al. (19) under the accession number KX357707.
ACKNOWLEDGMENTS
We thank Grant Morris, Carlos Rouca, Don Robson, Brent Glentworth, Jono Underwood, David Latham, Bill Tree, Alby Anderson, Dion Burgess, and Wayne Godfrey for providing samples; Jane Arrow, Sam Brown, and Mike Wehner for animal care support; and Katherine Trought for technical assistance in the laboratory.
REFERENCES
- 1.Clarke I, Estes M, Green K, Hansman G, Knowles N, Koopmans M, Matson D, Meyers G, Neill J, Radford A, Smith AW, Studdert MJ, Thiel H-J, Vinjé J. 2012. Caliciviridae, p 977–986. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy: classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses. Elsevier, San Diego, CA. [Google Scholar]
- 2.Liu SJ, Xue HP, Pu BQ, Qian NH. 1984. A new viral disease in rabbits. Anim Husb Vet Med 16:253–255. [Google Scholar]
- 3.Cooke BD, Fenner F. 2002. Rabbit haemorrhagic disease and the biological control of wild rabbits, Oryctolagus cuniculus, in Australia and New Zealand. Wildl Res 29:689–706. doi: 10.1071/WR02010. [DOI] [Google Scholar]
- 4.Schwensow NI, Cooke B, Kovaliski J, Sinclair R, Peacock D, Fickel J, Sommer S. 2014. Rabbit haemorrhagic disease: virus persistence and adaptation in Australia. Evol Appl 7:1056–1067. doi: 10.1111/eva.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kovaliski J. 1998. Monitoring the spread of rabbit hemorrhagic disease virus as a new biological agent for control of wild European rabbits in Australia. J Wildl Dis 34:421–428. doi: 10.7589/0090-3558-34.3.421. [DOI] [PubMed] [Google Scholar]
- 6.Mutze G, Cooke B, Alexander P. 1998. The initial impact of rabbit hemorrhagic disease on European rabbit populations in South Australia. J Wildl Dis 34:221–227. doi: 10.7589/0090-3558-34.2.221. [DOI] [PubMed] [Google Scholar]
- 7.Thompson J, Clark G. 1997. Rabbit calicivirus disease now established in New Zealand. Surveillance 24:5–6. [Google Scholar]
- 8.O'Keefe JS, Tempero J, Atkinson PH, Pacciarini L, Fallacara F, Horner GW, Motha J. 1998. Typing of rabbit haemorrhagic disease virus from New Zealand wild rabbits. N Z Vet J 46:42–43. doi: 10.1080/00480169.1998.36053. [DOI] [PubMed] [Google Scholar]
- 9.Parkes JP, Glentworth B, Sullivan G. 2008. Changes in immunity to rabbit haemorrhagic disease virus, and in abundance and rates of increase of wild rabbits in Mackenzie Basin, New Zealand. Wildl Res 35:775–779. doi: 10.1071/WR08008. [DOI] [Google Scholar]
- 10.Capucci L, Fusi P, Lavazza A, Pacciarini ML, Rossi C. 1996. Detection and preliminary characterization of a new rabbit calicivirus related to rabbit hemorrhagic disease virus but nonpathogenic. J Virol 70:8614–8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strive T, Wright JD, Robinson AJ. 2009. Identification and partial characterisation of a new lagovirus in Australian wild rabbits. Virology 384:97–105. doi: 10.1016/j.virol.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Strive T, Wright J, Kovaliski J, Botti G, Capucci L. 2010. The non-pathogenic Australian lagovirus RCV-A1 causes a prolonged infection and elicits partial cross-protection to rabbit haemorrhagic disease virus. Virology 398:125–134. doi: 10.1016/j.virol.2009.11.045. [DOI] [PubMed] [Google Scholar]
- 13.Bergin IL, Wise AG, Bolin SR, Mullaney TP, Kiupel M, Maes RK. 2009. Novel calicivirus identified in rabbits, Michigan, USA. Emerging Infect Dis 15:1955–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strive T, Elsworth PG, Liu J, Wright JD, Kovaliski J, Capucci L. 2013. The non-pathogenic Australian rabbit calicivirus RCV-A1 provides temporal and partial cross protection to lethal rabbit haemorrhagic disease virus infection which is not dependent on antibody titres. Vet Res 44:51. doi: 10.1186/1297-9716-44-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchandeau S, Le Gall-Recule G, Bertagnoli S, Aubineau J, Botti G, Lavazza A. 2005. Serological evidence for a non-protective RHDV-like virus. Vet Res 36:53–62. doi: 10.1051/vetres:2004049. [DOI] [PubMed] [Google Scholar]
- 16.Le Gall-Reculé G, Zwingelstein F, Fages M-P, Bertagnoli S, Gelfi J, Aubineau J, Roobrouck A, Botti G, Lavazza A, Marchandeau S. 2011. Characterisation of a non-pathogenic and non-protective infectious rabbit lagovirus related to RHDV. Virology 410:395–402. doi: 10.1016/j.virol.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Capucci L, Nardin A, Lavazza A. 1997. Seroconversion in an industrial unit of rabbits infected with a non-pathogenic rabbit haemorrhagic disease-like virus. Vet Rec 140:647–650. doi: 10.1136/vr.140.25.647. [DOI] [PubMed] [Google Scholar]
- 18.Jahnke M, Holmes EC, Kerr PJ, Wright JD, Strive T. 2010. Evolution and phylogeography of the nonpathogenic calicivirus RCV-A1 in wild rabbits in Australia. J Virol 84:12397–12404. doi: 10.1128/JVI.00777-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahar JE, Nicholson L, Eden J-S, Duchêne S, Kerr PJ, Duckworth J, Ward VK, Holmes EC, Strive T. 2016. Benign rabbit caliciviruses exhibit similar evolutionary dynamics to their virulent relatives. J Virol 90:9317–9329. doi: 10.1128/JVI.01212-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Fordham DA, Cooke BD, Cox T, Mutze G, Strive T. 2014. Distribution and prevalence of the Australian non-pathogenic rabbit calicivirus is correlated with rainfall and temperature. PLoS One 9:e113976. doi: 10.1371/journal.pone.0113976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lough RS. 2000. Serological survey of feral rabbit populations in New Zealand before the arrival of rabbit haemorrhagic disease. Otago Regional Council, Dunedin, New Zealand. [Google Scholar]
- 22.O'Keefe JS, Tempero JE, Motha MXJ, Hansen MF, Atkinsona PH. 1999. Serology of rabbit haemorrhagic disease virus in wild rabbits before and after release of the virus in New Zealand. Vet Microbiol 66:29–40. doi: 10.1016/S0378-1135(98)00307-1. [DOI] [PubMed] [Google Scholar]
- 23.Hall DCA. 2002. Molecular and serological studies of rabbit haemorrhagic disease virus in Otago. Ph.D. thesis. University of Otago, Dunedin, New Zealand. [Google Scholar]
- 24.Liu J, Kerr PJ, Strive T. 2012. A sensitive and specific blocking ELISA for the detection of rabbit calicivirus RCV-A1 antibodies. Virol J 9:182–186. doi: 10.1186/1743-422X-9-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Kerr PJ, Wright JD, Strive T. 2012. Serological assays to discriminate rabbit haemorrhagic disease virus from Australian non-pathogenic rabbit calicivirus. Vet Microbiol 157:345–354. doi: 10.1016/j.vetmic.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 26.Hoehn M, Kerr PJ, Strive T. 2013. In situ hybridisation assay for localisation of rabbit calicivirus Australia-1 (RCV-A1) in European rabbit (Oryctolagus cuniculus) tissues. J Virol Methods 188:148–152. doi: 10.1016/j.jviromet.2012.11.043. [DOI] [PubMed] [Google Scholar]
- 27.Parkes JP, Norbury GL, Heyward RP, Sullivan G. 2002. Epidemiology of rabbit haemorrhagic disease (RHD) in the South Island, New Zealand. Wildl Res 29:543–555. doi: 10.1071/WR00108. [DOI] [Google Scholar]
- 28.Reference deleted.
- 29.Eden J-S, Read AJ, Duckworth JA, Strive T, Holmes EC. 2015. Resolving the origin of rabbit hemorrhagic disease virus: insights from an investigation of the viral stocks released in Australia. J Virol 89:12217–12220. doi: 10.1128/JVI.01937-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norbury G, Reddiex B. 2005. Order Lagomorpha: European rabbit, p 131–150. In King CM. (ed), The handbook of New Zealand mammals, 2nd ed Oxford University Press, South Melbourne, Victoria, Australia. [Google Scholar]
- 31.McColl KA, Merchant JC, Hardy J, Cooke BD, Robinson A, Westbury HA. 2002. Evidence for insect transmission of rabbit haemorrhagic disease virus. Epidemiol Infect 129:655–663. doi: 10.1017/S0950268802007756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng T, Parkes JP. 2011. Rabbit haemorrhagic disease: advantages of cELISA in assessing immunity in wild rabbits (Oryctolagus cuniculus). Vet Microbiol 153:387–392. doi: 10.1016/j.vetmic.2011.05.049. [DOI] [PubMed] [Google Scholar]
- 33.Lough RS. 2009. The current state of rabbit management in New Zealand. MAF Biosecurity, Wellington, New Zealand. [Google Scholar]
- 34.Kimura T, Mitsui I, Okada Y, Furuya T, Ochiai K, Umemura T, Itakura C. 2001. Distribution of rabbit haemorrhagic disease virus RNA in experimentally infected rabbits. J Comp Pathol 124:134–141. doi: 10.1053/jcpa.2000.0440. [DOI] [PubMed] [Google Scholar]
- 35.Myers K, Gilbert N. 1968. Determination of age of wild rabbits in Australia. J Wildl Manage 32:841–849. doi: 10.2307/3799559. [DOI] [Google Scholar]
- 36.Capucci L, Scicluna MT, Lavazza A. 1991. Diagnosis of viral haemorrhagic disease of rabbits and the European brown hare syndrome. Rev Sci Tech 10:347–370. [DOI] [PubMed] [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 38.Eden J-S, Kovaliski J, Duckworth JA, Swain G, Mahar JE, Strive T, Holmes EC. 2015. Comparative phylodynamics of rabbit hemorrhagic disease virus in Australia and New Zealand. J Virol 89:9548–9558. doi: 10.1128/JVI.01100-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 42.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.RStudio Team 2015. RStudio: integrated development for R. RStudio Inc. Boston, MA. [Google Scholar]
- 44.Kerr IGC, Ross WD. 1990. Rabbit management in Central Otago: classification of land for rabbit proneness. New Zealand Mountain Lands Institute, Lincoln University and MAF Technology, Alexandra, New Zealand. [Google Scholar]