ABSTRACT
Five genes encoding PhaP family proteins and one phaR gene have been identified in the genome of Burkholderia symbiont strain RPE75. PhaP proteins function as the surface proteins of polyhydroxyalkanoate (PHA) granules, and the PhaR protein acts as a negative regulator of PhaP biosynthesis. Recently, we characterized one phaP gene to understand the molecular cross talk between Riptortus insects and Burkholderia gut symbionts. In this study, we constructed four other phaP gene-depleted mutants (ΔphaP1, ΔphaP2, ΔphaP3, and ΔphaP4 mutants), one phaR gene-depleted mutant, and a phaR-complemented mutant (ΔphaR/phaR mutant). To address the biological roles of four phaP family genes and the phaR gene during insect-gut symbiont interaction, these Burkholderia mutants were fed to the second-instar nymphs, and colonization ability and fitness parameters were examined. In vitro, the ΔphaP3 and ΔphaR mutants cannot make a PHA granule normally in a stressful environment. Furthermore, the ΔphaR mutation decreased the colonization ability in the host midgut and negatively affected the host insect's fitness compared with wild-type Burkholderia-infected insects. However, other phaP family gene-depleted mutants colonized well in the midgut of the fifth-instar nymph insects. However, in the case of females, the colonization rate of the ΔphaP3 mutant was decreased and the host's fitness parameters were decreased compared with the wild-type-infected host, suggesting that the environment of the female midgut may be more hostile than that of the male midgut. These results demonstrate that PhaR plays an important role in the biosynthesis of PHA granules and that it is significantly related to the colonization of the Burkholderia gut symbiont in the host insects' midgut.
IMPORTANCE Bacterial polyhydroxyalkanoate (PHA) biosynthesis is a complex process requiring several enzymes. The biological roles of PHA granule synthesis enzymes and the surface proteins of PHA granules during host-gut symbiont interactions are not fully understood. Here, we report the effects on colonization ability in the host midguts and the fitness of host insects after feeding Burkholderia mutant cells (four phaP-depleted mutants and one phaR-depleted mutant) to the host insects. Analyses of both synthesized PHA granule amounts and CFU numbers suggest that the phaR gene is closely related to synthesis of the PHA granule and the colonization of the Burkholderia gut symbiont in the host insect's midgut. Like our previous report, this study also supports the idea that the environment of the host midgut may not be favorable to symbiotic Burkholderia cells and that PHA granules may be required to adapt in the host midgut.
KEYWORDS: symbiosis, polyhydroxyalkanoate, PhaP, PhaR, Burkholderia, Riptortus pedestris
INTRODUCTION
Many insects are known to harbor symbiotic bacteria in their cells, tissues, and guts (1). Most of the obligate symbionts, such as Buchnera spp. in aphids, are vertically transmitted from mother to offspring (2). However, some symbionts, like gut symbionts, are horizontally acquired from the environment in every generation (3). These symbionts provide some benefits to host insects. For example, they have a good influence on the host insects by providing nutrient sources (4) and by defending hosts from attacks by natural enemies or by helping hosts adapt to specific environments (5–7). They can also induce pathogenic or parasitic effects on hosts by causing a decrease in fitness or reproductive aberrations (8). However, the molecular mechanisms of these effects in insect-symbiont systems are not clearly understood. Therefore, many researchers seek to understand these effects by using various insect-symbiont systems (9).
Many insects' symbionts are difficult to culture in vitro because they are highly adapted to the unique environment of their host insects (10, 11). Recently, the bean bug, Riptortus pedestris (Hemiptera: Alydidae), has shown some merit as a good model of insect symbiosis (12–14). This insect has a specialized symbiotic organ in a posterior midgut fourth region named M4. In this region, the Riptortus insect harbors a specific gut symbiont of the betaproteobacterial genus Burkholderia (15, 16). This gut symbiont is acquired orally from the environment by second-instar Riptortus nymphs each generation, and this symbiont is easily cultivated in vitro and can be manipulated genetically (3, 17). Based on these advantages, we can compare the biochemical differences between in vitro-cultured Burkholderia spp. and in vivo-colonized symbiotic Burkholderia cells at the molecular level (13, 16).
Polyhydroxyalkanoate (PHA) is a linear polyester that is produced in many bacteria. PHAs are accumulated as intracellular granules of up to 90% of the dry cell weight and are used as storage compounds of carbon and energy sources in bacterial cells (18, 19). Generally, the biosynthesis of PHA granules is promoted when bacteria face stressful environments, such as nutrition-deficient conditions, especially nitrogen source deficiency (20). Under nutrient-rich conditions, bacteria do not need to synthesize PHA granules because they do not need to store energy or carbon sources. PHA granules are mainly synthesized from acetyl coenzyme A (acetyl-CoA) by three different enzymes, such as the products of phaA (ketothiolase), phaB (acetyl-CoA reductase), and phaC (PHA synthase) (18, 21). The surfaces of PHA granules are surrounded by various proteins, such as PhaP (a surface protein of PHA granules; phasin), PhaR (a negative regulator of PhaP), and PhaZ (PHA depolymerase) (22–24).
In the genome of the Burkholderia symbiont, there are five phaP genes and three genes encoding enzymes related to PHA synthesis named phaA, phaB, and phaC. One phaP family gene (phaP) and three genes encoding enzymes (phaA, phaB, and phaC) were characterized in our previous work (25). These Burkholderia mutants did not synthesize PHA granules normally in vitro and were less colonized in the host midgut than wild-type Burkholderia strains. Among them, especially, fitness parameters of host insects infected with phaC-depleted Burkholderia spp. were significantly decreased compared with those of host infected with wild-type Burkholderia (25). Also, our previous study demonstrated that PhaP proteins are more abundant in M4-colonized symbiotic Burkholderia cells than in cultured Burkholderia cells (25). PhaP is the surface protein of PHA granules that is known to be important in the stabilization of PHA granules (26), but why M4-colonized bacteria accumulate larger amounts of PhaP in their cells remains unknown. To address this question, we constructed a phaP-depleted Burkholderia mutant and fed this ΔphaP mutant to the second-instar nymphs of Riptortus host insects. The insects infected with the ΔphaP mutant showed negative effects in fitness parameters, including a decrease in body length, suggesting that the PhaP protein might be necessary for colonization of gut symbionts in the host midgut. These results also suggest that the environment of the host midgut may not be favorable to symbiotic Burkholderia cells, which indicates that PHA granules may be required to resist external stresses and adapt in the host midgut (24). PhaR is known as a negative regulator of PhaP in several species, with activities of binding to the promoter region of phaP genes and of blocking their transcription (27, 28). In Ralstonia eutropha, PhaP is overexpressed when phaR is deleted from R. eutropha's genome, suggesting that PhaR is a negative regulator of PhaP (23, 29). PhaR is also identified as a negative regulator of PhaP in Paracoccus denitrificans (27). Also, PhaR was reported to control PHA synthesis in Rhizobia species (30–32). Although one phaP gene was characterized in our previous study (25), the functions of four other phaP family genes and one phaR gene have not been determined in the Burkholderia genome. We hypothesized that PhaR, a negative regulator of PhaP synthesis, may also play an important role in Riptortus-Burkholderia gut symbiont interaction. Therefore, in this paper, we explore the biological effects of phaR and four other phaP genes by feeding these gene-depleted Burkholderia mutants to Riptortus host insects.
RESULTS AND DISCUSSION
Bioinformatic characterization of PHA-related genes in the genome of Burkholderia symbiont strain RPE75.
The loci of PHA-related genes are indicated in an illustration in Fig. S2A in the supplemental material. There are three genes encoding enzymes needed for PHA biosynthesis (phaA, phaB, and phaC) and one phaP gene encoding PHA surface protein on chromosome 1. These genes were identified in our previous study (25). Also, there are phaR genes encoding a negative regulator of phaP on chromosome 1, phaP2 and phaP3 genes on chromosome 2, and phaP1 and phaP4 genes on chromosome 3, which are demonstrated in this study. We identified these phaP family genes by searching the genome for the Phasin2 domain, because the phaP gene identified in our previous study has this Phasin2 domain (25). These five phaP genes have 8 to 39% sequence identity, and the phaR gene has no homologous genes in the Burkholderia genome. Also, we analyzed domains of phaP family genes, phaR, and phaC (Fig. S2B). PhaP family proteins have the same domain, called the Phasin2 domain, and PhaR has one helix-turn-helix motif DNA-binding domain with two poly-3-hydroxybutyrate (PHB) accumulation regulatory domains. PhaC has one PHB polymerase domain at the N-terminal region. Additionally, we analyzed the phylogenetic relationships of PHA-related genes of Burkholderia strains and other species (Fig. S3). These PHA-related genes are highly conserved in Burkholderia species. The reason for the redundancy of phaP genes seems to be that the gene product might have redundant functions. In Sinorhizobium meliloti, PHA granules were detected when either one of the phaP genes was mutated but not in the double phaP-depleted mutant (33).
PhaR protein is important for biosynthesis of PHA granules in vitro and in vivo.
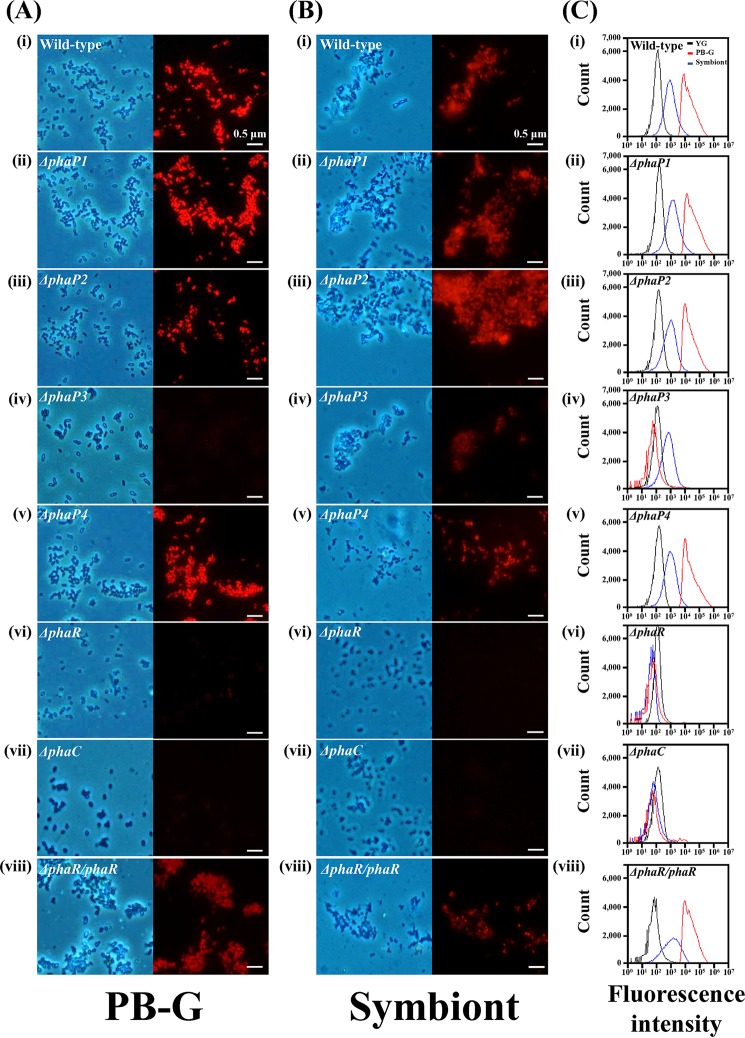
Previously, we characterized one phaP gene among five phaP family genes in the Burkholderia genome (25). In this study, we constructed six Burkholderia mutants: four other phaP-depleted mutants (ΔphaP1, ΔphaP2, ΔphaP3, and ΔphaP4 mutants), one phaR gene-depleted mutant, and a phaR-complemented mutant (ΔphaR/phaR mutant). Because the biological roles of the phaR gene in Burkholderia spp. during symbiosis were unknown, we first examined the biological effects of the PhaR protein after feeding wild-type Burkholderia and ΔphaR mutant Burkholderia strains to the second-instar nymph host insects. When the insects became fifth-instar nymphs, we collected colonized Burkholderia cells from the M4 region. Bacterial proteins from colonized wild-type and colonized ΔphaR Burkholderia mutant cells were analyzed by SDS-PAGE (Fig. 1A). A 19-kDa protein was more highly expressed in colonized ΔphaR mutant cells than in wild-type Burkholderia cells. To identify this 19-kDa protein, we analyzed the N-terminal sequences of that protein by automated Edman degradation. This 19-kDa protein turned out to be the Burkholderia PhaP protein, which is the same protein identified in our previous study (25) (Fig. 1B). These results showed that PhaP is overexpressed by the deletion of the phaR gene in the symbiotic Burkholderia genome, which suggests that PhaR functions as a negative regulator of PhaP biosynthesis in Burkholderia cells. Additionally, we examined the in vitro PHA granule synthesis in PHA-related-gene mutants by fluorescence microscopy. We used phaC-depleted mutant Burkholderia as a negative control because this Burkholderia mutant cannot synthesize PHA granules (25). In vitro-cultured wild-type, phaC-depleted, and phaP family gene-depleted Burkholderia mutants were stained with Nile blue A dye to stain their PHA granules. No Burkholderia cells synthesized PHA granules when they were cultured in yeast-glucose (YG) medium, because there are sufficient nutrient sources (Fig. S4). However, in nutrient-deficient sodium phosphate dibasic-glucose (PB-G) medium, fluorescence derived from PHA granules was highly detected in wild-type Burkholderia cells (Fig. 2Ai). Additionally, fluorescence was evident in ΔphaP1, ΔphaP2, and ΔphaP4 mutant cells, which suggests that these genes are not essential for the synthesis of PHA granules (Fig. 2Aii, iii, and v). However, as with the ΔphaP mutant, which was identified in our previous study (24), and the ΔphaC mutant, which is a negative control (Fig. 2Avii), ΔphaP3 and ΔphaR mutant Burkholderia cells did not synthesize PHA granules (Fig. 2Aiv and vi). These results suggested that the phaP3 and phaR genes are required for PHA granule synthesis in stressful environments, such as PB-G medium. To further confirm whether the phaR gene is crucial for PHA synthesis, we made a ΔphaR mutant complemented with a plasmid containing a normal phaR gene (ΔphaR/phaR mutant). As expected, upon complementation with the phaR gene, the level of PHA biosynthesis was restored to that of wild-type Burkholderia spp. (Fig. 2Aviii). Also, we examined the in vivo PHA granule synthesis of Burkholderia mutants that were colonized in the midgut of fifth-instar nymphs (Fig. 2B). Wild-type and phaP family gene-depleted mutants, including a ΔphaP3 mutant, synthesized PHA granules well in the fifth-instar nymphs' midgut (Fig. 2Bi to v). However, the ΔphaR mutant cannot synthesize PHA granules in vivo, like a negative-control ΔphaC mutant (Fig. 2Bvi and vii). When the phaR gene was inserted into the ΔphaR mutant Burkholderia cells, the ability to synthesize PHA was restored (Fig. 2Bviii). Additionally, flow cytometric analyses were used to reconfirm the PHA synthesis abilities of wild-type and mutant Burkholderia strains in vitro and in vivo. When Burkholderia strains were cultured in YG medium, the wild-type strain and all mutants showed low fluorescence intensities because they did not synthesize PHA granules in their cells (Fig. 2C). However, in PB-G medium, ΔphaP3 mutant Burkholderia strains showed lower fluorescence intensities than other mutants, including the wild-type strain, like the ΔphaP (24), ΔphaR, and ΔphaC mutants (Fig. 2Civ, vi, and vii). This result suggests that PhaR plays a crucial role in the synthesis of PHA granules, and PhaP3 is also needed for synthesis of PHA granules in PB-G medium, which is a very stressful environment. When the ΔphaR Burkholderia mutant was complemented with the phaR plasmid, fluorescence intensity was restored to levels similar to those of wild-type Burkholderia (Fig. 2Cviii). PHA-derived fluorescence intensities of all phaR family gene-depleted mutants that were colonized in the midgut of fifth-instar nymph insects were higher than those of Burkholderia strains cultured in YG medium (Fig. 2Ci to v). However, colonized ΔphaR and ΔphaC mutants did not synthesize PHA granules in the midgut of fifth-instar nymphs (Fig. 2Cvi and vii). Also, the fluorescence intensity of the colonized ΔphaR mutant was restored after complementation with the phaR gene (Fig. 2Cviii). These results confirm that PhaR is necessary for PHA granule synthesis in stressful environments, such as in vitro in PB-G medium and in vivo in the host midgut.
FIG 1.
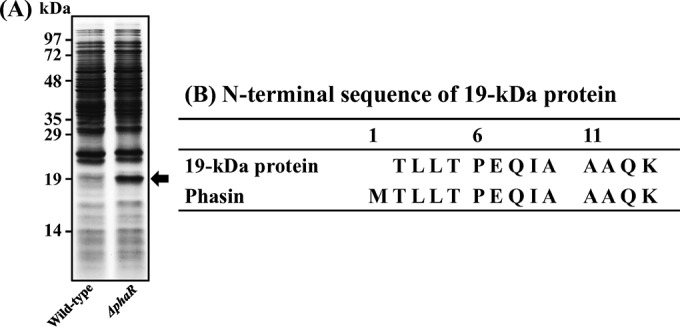
PhaP is overexpressed by deletion of the phaR gene in the genome of symbiont Burkholderia cells. (A) Protein analysis of wild-type and ΔphaR mutant Burkholderia cells colonized in Riptortus midgut. In total, 5 × 107 Burkholderia cells were lysed in 1× Laemmli sample buffer and analyzed on a 12% SDS-PAGE gel. The 19-kDa protein (black arrow) was more abundant in the colonized ΔphaR Burkholderia mutant than in wild-type Burkholderia. (B) N-terminal sequencing of a 19-kDa protein by the automated Edman method. This 19-kDa protein was identified as PhaP, known as a PHA surface protein.
FIG 2.
In vitro PHA production by wild-type and PHA-related gene-depleted mutants of the Burkholderia symbiont. (A) Images of PHA-derived fluorescence under the fluorescence microscope. Phase-contrast images (left) and fluorescent images (right) of Burkholderia cells cultured in PB-G medium are stained with Nile blue A. (i) Wild type; (ii) ΔphaP1 mutant; (iii) ΔphaP2 mutant; (iv) ΔphaP3 mutant; (v) ΔphaP4 mutant; (vi) ΔphaR mutant; (vii) ΔphaC mutant; and (viii) ΔphaR/phaR Burkholderia mutant (scale bars, 0.5 μm). (B) Stained PHA granules of wild-type and mutant Burkholderia cells colonized in the midgut of fifth-instar nymph. (i) Wild type; (ii) ΔphaP1 mutant; (iii) ΔphaP2 mutant; (iv) ΔphaP3 mutant; (v) ΔphaP4 mutant; (vi) ΔphaR mutant; (vii) ΔphaC mutant; and (viii) ΔphaR/phaR Burkholderia mutant. (C) Flow cytometric histograms of PHA-derived fluorescence from Burkholderia cells cultured in YG medium (black), PB-G medium (red), and colonized symbiotic Burkholderia (blue). Burkholderia cells were stained with Nile blue A. (i) Wild type; (ii) ΔphaP1 mutant; (iii) ΔphaP2 mutant; (iv) ΔphaP3 mutant; (v) ΔphaP4 mutant; (vi) ΔphaR mutant; (vii) ΔphaC mutant; and (viii) ΔphaR/phaR Burkholderia strain.
PhaR is important for colonization of symbionts in their host midgut.
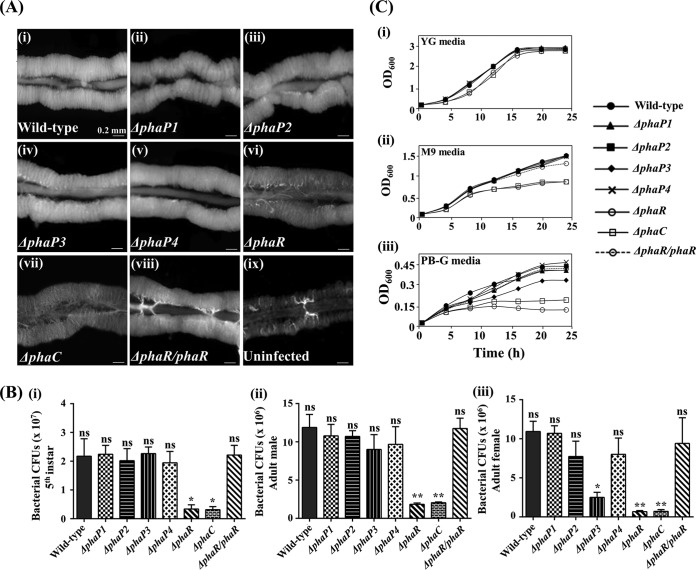
Next, we fed wild-type and mutant Burkholderia cells to the second-instar nymphs to assess whether PhaR and PhaP family proteins are important for the colonization of symbiotic Burkholderia strains. Infected insects were reared to fifth-instar nymphs; their symbiotic organs were dissected, specifically the M4 region, and their morphologies were observed by light microscopy. The M4 crypt infected with the wild-type strain was well developed and fully filled with symbiotic Burkholderia cells (Fig. 3Ai). In a previous study, we identified that M4 crypts infected with the ΔphaP mutant were less conspicuous than those infected with the wild type (24). However, M4 crypts infected with the ΔphaP1, ΔphaP2, ΔphaP3, and ΔphaP4 mutants were well developed and filled with colonized symbionts, unlike the ΔphaP mutant (Fig. 3Aii to v). In contrast, the morphology of crypts infected with the ΔphaR Burkholderia mutant was different from those of other Burkholderia mutants (Fig. 3Avi); the crypts of ΔphaR mutant-infected insects were thinner and more translucent than those of wild-type-infected hosts, which were similar to the crypts infected with ΔphaC mutants and uninfected insects (Fig. 3Avii and ix). When the host insects were infected with ΔphaR/phaR Burkholderia cells, they displayed restored phenotypes with well-developed and well-filled M4 crypts (Fig. 3viii). Additionally, we estimated the number of CFU to determine how many mutant Burkholderia cells were colonized in the host midgut. When we quantified the colonized bacterial numbers, bacterial titers of the third- and fourth-instar nymphs infected with Burkholderia mutants were not significantly different from those of wild-type cells (Fig. S5). However, the fifth-instar nymphs infected with the ΔphaR mutant showed lower symbiont titers in their midguts than did those infected with wild-type cells (Fig. 3Bi). When the host was infected by the ΔphaR/phaR strain, the number of CFU recovered to the level of wild-type cells (Fig. 3Bi). Additionally, we examined symbiont titers in the midgut of adult male and female insects (Fig. 3Bii and iii). In the midgut of adult male insects, the ΔphaR mutant did not colonize normally, as with the ΔphaC mutant (Fig. 3Bii). The colonization rate of the ΔphaP3 mutant was decreased slightly, but the difference was not statistically significant (Fig. 3Bii). However, in the midgut of female insects, ΔphaP3 and ΔphaR mutant Burkholderia strains did not colonize, indicating that the midgut of the female insect is a more hostile environment than that of the male insect (Fig. 3Biii), supporting our hypothesis that the midgut of female insects may be a more stressful environment than that of male insects. Also, we measured PHA levels of Burkholderia strains colonized in the female midgut using fluorescence microscopy and flow cytometry to address the question of why the ΔphaP3 mutant did not colonize normally. The result showed that the ΔphaP3 mutant cannot synthesize PHA granules ordinarily in the female midgut, resulting in a decrease in the colonization rate (Fig. S6). These results support the hypothesis that granule formation in general might be more important for Burkholderia symbiosis with females than with males. To know whether decreased colonization rates are affected by symbiosis-specific conditions or just stressful conditions, such as a nutrient-deficient environment, we measured the growth rates of each Burkholderia mutant by culturing in the YG (nutrient-sufficient), PB-G (severely nutrient-deficient), and M9 (minimal-nutrient) media (Fig. 3C). All Burkholderia strains grew well in YG medium because they do not need a PHA granule (Fig. 3Ci). When Burkholderia mutants were cultured in M9 medium, the ΔphaR and ΔphaC mutants did not grow well (Fig. 3Cii). However, in PB-G medium containing only 1% glucose as a nutrient source, all mutant strains did not grow well because of extremely limited nutrients. Among them, the ΔphaP3, ΔphaR, and ΔphaC mutants did not grow as well as other mutants (Fig. 3Ciii). These results indicate that the lower colonization rates of the ΔphaP3 and ΔphaR Burkholderia mutants are not due to a symbiosis-specific environment. These mutants also showed a growth defect in the nutrient-deficient media, suggesting that they cannot grow well in a stressful environment. These results showed that the ΔphaP3, ΔphaC, and ΔphaR mutants do not grow well in the nutrient-depleted media and likely also in the midgut, due to their reduced ability to produce PHA granules (Fig. 3Avi), indicating that the environment of host midgut is not favorable to the symbiont Burkholderia. All results indicated that PhaR is important for the synthesis of PHA granules and colonization of symbiont Burkholderia in their host midgut, and PhaP3 is also important in maintaining PHA granules when Burkholderia strains faced a more stressful environment.
FIG 3.
Morphologies of host midgut and symbiont densities after feeding with PHA-related gene-depleted mutants of Burkholderia. (A) Morphologies of the M4 region of the fifth-instar nymph midgut. (i) Wild type; (ii) ΔphaP1 mutant; (iii) ΔphaP2 mutant; (iv) ΔphaP3 mutant; (v) ΔphaP4 mutant; (vi) ΔphaR mutant; (vii) ΔphaC mutant; and (viii) ΔphaR/phaR Burkholderia-infected and (ix) uninfected strains (scale bars, 0.2 mm). (B) Bacterial titers of wild-type and mutant Burkholderia symbionts colonized in the M4 region of the fifth-instar nymph and adult insects. (i) Symbiont titers of the fifth-instar nymph, (ii) the adult male, and (iii) the adult female insects. (C) Growth curves of Burkholderia after culture in YG (i), M9 (ii), and PB-G (iii) media. Asterisks indicate a significant difference compared to the wild-type-infected group (ns, nonsignificant; *, P < 0.05; **, P < 0.005 [one-way analysis of variance {ANOVA} with Tukey's correction]).
Host fitness is negatively affected by infection with ΔphaR mutant compared with wild-type Burkholderia-infected insects.
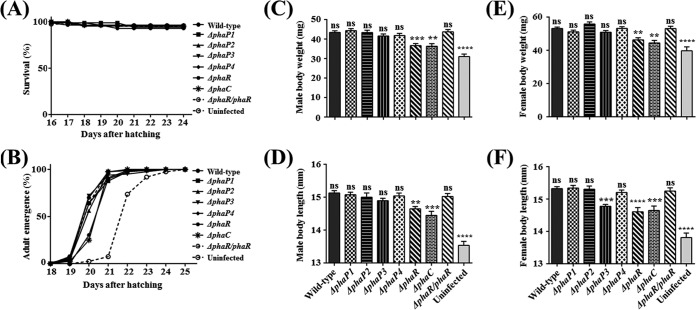
Because the colonization rate was affected by the ΔphaR mutant, we further examined host fitness after feeding with the wild-type and mutant Burkholderia strains. No Burkholderia mutants were toxic to the host insects, because the survival rates were not different among wild-type-fed, mutant-fed, and uninfected insects (Fig. 4A). The times to adulthood were also not different among mutant-fed insects, except for ΔphaR mutant- and ΔphaC mutant-fed insects. Insects fed with ΔphaR mutant cells reached adulthood approximately 1 day later than did wild-type-fed insects, as with a ΔphaC mutant (Fig. 4B). The fitness parameters of hosts infected with the ΔphaP mutant are decreased slightly (24). Likewise, the body weights of male and female insects fed with ΔphaR and ΔphaC mutant cells were decreased compared with those of wild-type-infected insects (Fig. 4C and E). Also, the body lengths of adult males fed with ΔphaR and ΔphaC mutant cells were shorter than those fed with wild-type cells (Fig. 4D). For the adult female's body length, insects fed with ΔphaR, ΔphaC and ΔphaP3 mutant Burkholderia strains had a shorter body length than did insects fed with wild-type Burkholderia strains (Fig. 4F). However, insects fed with other Burkholderia mutants (ΔphaP1, ΔphaP2, and ΔphaP4 mutants) exhibited no remarkable differences compared with those fed with wild-type Burkholderia strains (Fig. 4C to F). Every fitness value of ΔphaR mutant-fed insects was restored to be similar to those of wild-type insects when they were fed with ΔphaR/phaR Burkholderia strains (Fig. 4B to F). Overall, the host bean bugs fed with the ΔphaR Burkholderia mutant tended to exhibit lower fitness than the host fed with the wild-type Burkholderia strain, as with the host fed with the ΔphaC mutant. ΔphaP3 mutant-infected female bean bugs display decreased fitness parameters, because this ΔphaP3 mutant did not colonize well in the midgut of the adult female host (Fig. 3Biii), which may relate to the fact that female bean bugs use more energy for reproduction, so the environment of the female midgut may be more stressful than those of the nymph and male midgut.
FIG 4.
Host insect development and fitness after feeding with wild-type and PHA-related gene-depleted mutants of Burkholderia. (A) Survival rate was estimated each day after hatching (n = 80) by counting dead insects. (B) Adult emergence rate was observed by counting newly molted adult insects from late-fifth-instar nymphs (n = 50). (C to F) Parameters of host insect fitness. Body weight and length were measured after killing insects with acetone and drying completely in a 70°C oven (n = 40). Body weight of adult males (C), body length of adult males (D), body weight of adult females (E), and body length of adult females (F) are shown. Asterisks indicate a significant difference compared to the wild-type-infected group (ns, nonsignificant; **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001 [one-way ANOVA with Tukey's correction]).
Conclusion and perspective.
In this paper, we established the biological importance of the phaR gene, which is known as a negative regulator of PhaP biosynthesis, by using the Riptortus-Burkholderia symbiosis system. We observed that when the phaR gene was deleted from the symbiotic Burkholderia genome, PhaP was overexpressed in Burkholderia cells, confirming that PhaR functions as a negative regulator of PhaP biosynthesis. In addition, we discovered that the ΔphaR Burkholderia mutant cannot synthesize PHA granules normally, not only in vitro but also in vivo. Additionally, ΔphaR mutant cells were unable to colonize normally in their host's midgut, so ΔphaR mutant Burkholderia cells negatively affected host fitness parameters in terms of body weight and length compared with the wild-type Burkholderia strain. These results indicate that the Burkholderia phaR gene is important for maintaining the Riptortus-Burkholderia symbiosis system.
To verify the role of PhaP proteins in our symbiosis model, we generated four other phaP gene-depleted Burkholderia mutants in addition to the phaP gene that was characterized in a previous study (25). We expected that all phaP family gene-depleted mutants would show defective synthesis of PHA granules, but three ΔphaP mutants (ΔphaP1, ΔphaP2, and ΔphaP4 mutants) synthesized PHA granules well, with the exception being the ΔphaP3 mutant in vitro. The ΔphaP3 Burkholderia mutant showed defects in PHA granule synthesis in vitro. However, this mutant also showed no defective phenotypes in vivo, except in the female host insects. The colonization rate of the ΔphaP3 mutant was decreased only in the midgut of a female host, and the fitness parameters of hosts infected with the ΔphaP3 mutant were decreased only in the case of the female insect. We supposed that this phenomenon may be attributed to the unique function of the female's reproduction. Namely, female insects may need more energy for reproduction, which may cause a more stressful environment in the female's midgut, but this possibility requires more research. We supposed that the phaP3 gene has an important function in maintaining PHA granule biosynthesis when the Burkholderia symbiont is faced with a stressful environment, such as PB-G medium and the female midgut.
In summary, PhaR is important for maintaining the colonization of symbiotic Burkholderia strains in their host midgut by keeping the intracellular PHA granules at a normal level. In Burkholderia symbionts, PhaR seems to regulate other genes related with PHA biosynthesis, as well as phaP genes, because PHA levels in the ΔphaR Burkholderia mutant were significantly decreased. There are recent studies supporting this observation. In Rhizobium etli, which is a symbiotic bacterium of plants, the ΔaniA (same as ΔphaR) mutant accumulated only 40% of polyhydroxybutyrate (PHB) compared to the wild-type strain by disappearance of PhaB protein expression (30). Also, in the case of the soil bacterium Bradyrhizobium diazoefficiens, when the phaR gene was knocked out, the expression of phaC2 became uncontrolled, and this led to rapid inhibition of PHB synthesis (32). Thus, PhaR is important to PHA biosynthesis, and it seems reasonable that the PHA synthetic mechanism has been primarily selected for survival at the free-living stage of the Burkholderia symbiont and secondarily coopted for survival under the symbiotic condition within the host insect (25).
The PHA granules produced by microorganisms have been spotlighted as biodegradable plastic substitutes (18, 34). The discovery of a new biological role of the PhaR protein in the insect-bacterium symbiosis system highlights another relationship between symbiosis, microbiology, and biotechnology.
MATERIALS AND METHODS
Insect rearing and Burkholderia infection.
The R. pedestris bean bugs were reared in the insect room of our laboratory at 28°C under a long-day condition of 16 h light and 8 h dark, as described previously (25). Briefly, nymphal insects were reared in clean plastic containers (34 cm by 19.5 cm wide and 27.5 cm high) supplied with soybean seeds and distilled water containing 0.05% ascorbic acid (DWA). The plastic containers were cleaned every day, and the soybean seeds and DWA were changed with fresh ones every 2 days. When the insects emerged as adults, they were transferred to larger plastic containers (35 cm by 35 cm wide and 40 cm high) with soybean seeds and DWA. Additionally, cotton pads were attached to the walls of the plastic containers for egg laying. Eggs were collected every day and transferred to new cages for hatching. When newborn nymphs molted to second-instar nymphs, DWA containing 107 cells/ml cultured Burkholderia was provided for the colonization of Burkholderia in a small petri dish.
Bacteria, plasmids, and media.
A list of the Burkholderia and Escherichia coli strains and plasmids used in this study is shown in Table 1. The Burkholderia strain RPE75 used as a gut symbiont of R. pedestris is a spontaneous rifampin-resistant mutant derived from Burkholderia strain RPE64 (3). The RPE75 cells were cultured at 30°C in YG medium (0.5% yeast extract, 0.4% glucose, and 0.1% NaCl) containing 30 μg/ml rifampin. E. coli cells were cultured at 37°C in LB medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl) containing 30 μg/ml kanamycin. Additionally, colonized Burkholderia spp. from M4 were isolated from each stage of insects, as described previously (16).
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| Burkholderia symbiont | ||
| RPE75 | Burkholderia symbiont (RPE64); Rifr | 46 |
| RPE75 ΔphaP1; Rifr | This study | |
| RPE75 ΔphaP2; Rifr | This study | |
| RPE75 ΔphaP3; Rifr | This study | |
| RPE75 ΔphaP4; Rifr | This study | |
| RPE75 ΔphaR; Rifr | This study | |
| RPE75 ΔphaC; Rifr | 25 | |
| RPE75 ΔphaR/phaR; Rifr | This study | |
| Escherichia coli | ||
| DH5α | F− Φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1 | Invitrogen |
| Plasmids | ||
| pK18mobsacB | pMB1ori allelic exchange vector containing oriT; Kmr | 47 |
| pBBR122 | Broad-host-range vector; Cmr Kmr | 38 |
Rifr, rifampin resistance; Kmr, kanamycin resistance; Cmr, chloramphenicol resistance.
Construction of deletion mutant strains.
The phaC-depleted mutant used in this study is the same one as that used in a previous paper (25). The phaP1, phaP2, phaP3, phaP4, and phaR genes were deleted from the chromosome of Burkholderia RPE75 by the two-step homologous recombination method described previously (35). Briefly, we used the pK18mobsacB plasmid as a suicide vector containing the sacB gene to make mutants (36). The gene product of sacB induced by sucrose is lethal to bacteria. To construct deletion mutants, two fragments (left and right fragments) for every target gene were amplified from the Burkholderia RPE75 chromosome by PCR using the specific primers (LF-LR and RF-RR) listed in Table 2. Then, the left fragment was digested by BamHI and XbaI, and the right fragment was digested by XbaI and HindIII. The pK18mobsacB vector was digested by BamHI and HindIII. After enzyme digestion, two fragments and the vector were ligated and transformed into E. coli DH5α by the heat shock method (37). Transformed cells were selected on LB agar plates containing 30 μg/ml kanamycin. The positive colonies were sorted by colony PCR using the left-forward (LF) primers of the left fragment (listed in Table 2) and the vector primer aphII (5′-ATCCATCTTGTTCAATCATGCG-3′). The positive colonies were incubated in LB medium containing 30 μg/ml kanamycin, and plasmids were extracted from the culture-positive colonies. Then, the plasmid was integrated into the RPE75 chromosome by electroporation (first crossover). Electroporated cells were selected on YG agar plates containing 30 μg/ml rifampin and 30 μg/ml kanamycin. The positive colonies were selected by colony PCR using the upstream 5′ region of RPE75 (up) primers (listed in Table 3) and the vector primer aphII. Next, the positive colonies were incubated in YG medium without any drugs for the second crossover. Cultured cells were serially diluted and spread on YG agar plates containing 30 μg/ml rifampin and 10% sucrose. The positive colonies were identified by colony PCR using the upstream 5′ region of RPE75 (up) primers and the downstream 3′ region of RPE75 (down) primers (listed in Table 3). Deletion mutants were finally selected in this step and used for further experiments. The result of the PCR is shown in Fig. S1.
TABLE 2.
Primers used for cloning PCR
| PCR target region | Primer name | Product size (bp) | Sequence (5′ to 3′) | Restriction site |
|---|---|---|---|---|
| 5′ region of phaP1 | phaP1-LF | 1,133 | ACACCAGGATCCCGCTAGGCTCTGCGAAATCT | BamHI |
| phaP1-LR | CACCTATCTAGAAACTTCAAGCAAGCCACCGA | XbaI | ||
| 3′ region of phaP1 | phaP1-RF | 1,121 | ACACCATCTAGAGGGTTGCTTACACCCTCGTT | XbaI |
| phaP1-RR | CACCTAAAGCTTAATGTCCCGGTAGAGATGCG | HindIII | ||
| 5′ region of phaP2 | phaP2-LF | 1,074 | ACACCAGGATCCTGTTTAGCTGGCTGCTGTTC | BamHI |
| phaP2-LR | CACCTATCTAGATCTCGAAGCTCGACACAGTT | XbaI | ||
| 3′ region of phaP2 | phaP2-RF | 1,001 | ACACCATCTAGAAACTCGGCGTACGAAGCTG | XbaI |
| phaP2-RR | CACCTAAAGCTTGAAGCGTATCGACGGGGAAG | HindIII | ||
| 5′ region of phaP3 | phaP3-LF | 1,015 | ACACCAGGATCCCTGTGCCTGAGGCGAGTACG | BamHI |
| phaP3-LR | CACCTATCTAGAGAAGCCTATGCGCTCGTCA | XbaI | ||
| 3′ region of phaP3 | phaP3-RF | 1,040 | ACACCATCTAGACGAACCCGGGAAGCTGATAC | XbaI |
| phaP3-RR | CACCTAAAGCTTAGCGTCCCGTCCATTGAATC | HindIII | ||
| 5′ region of phaP4 | phaP4-LF | 1,018 | ACACCAGGATCCGAGCCGACTCATTTCCACGA | BamHI |
| phaP4-LR | CACCTATCTAGACGGAATGAAGCAAGCGTTCG | XbaI | ||
| 3′ region of phaP4 | phaP4-RF | 1,025 | ACACCATCTAGACGGTCTTGATGGTCTGCACG | XbaI |
| phaP4-RR | CACCTAAAGCTTGGAAGAGTCGCCTGTCGAG | HindIII | ||
| 5′ region of phaR | phaR-LF | 1,027 | ACACCAGGATCCCGACGAGATCGGTCATGCG | BamHI |
| phaR-LR | CACCTATCTAGAGCAGGCGAAGTCGATGTTCA | XbaI | ||
| 3′ region of phaR | phaR-RF | 1,036 | ACACCATCTAGACGTGATGTACGTGCTCGTCT | XbaI |
| phaR-RR | CACCTAAAGCTTGAGATCGCCGTCCGCATAA | HindIII | ||
| Complementation of phaR | phaR-comF | 1,232 | ACACCAGAATTCGCGAAACCATGCCGATTTTC | EcoRI |
| phaR-comR | CACCTAGAATTCGGACATGGTGAAGTCCATCC | EcoRI |
TABLE 3.
Primers used for diagnostic PCR
| Primer-binding region | Primer name | Sequence (5′ to 3′) |
|---|---|---|
| Upstream of phaP1 | phaP1-up | CCTACGTCAACGTGGATTACTT |
| Downstream of phaP1 | phaP1-down | GGCAGTGACAGAAAATACCAAT |
| Upstream of phaP2 | phaP2-up | GGCAGTGACAGAAAATACCAAT |
| Downstream of phaP2 | phaP2-down | AGTCCGCCTTTTTCTATACGAC |
| Upstream of phaP3 | phaP3-up | AGCGGGTTTGTTATTGTCTTC |
| Downstream of phaP3 | phaP3-down | ACCATTAAGGTCATCATACGGTAG |
| Upstream of phaP4 | phaP4-up | GTAAAGAGCGACCAGAACGTAT |
| Downstream of phaP4 | phaP4-down | AGTGATGAAGCGCTATGAAGAG |
| Upstream of phaR | phaR-up | TTGACGATTGAATCCAAAGAAT |
| Downstream of phaR | phaR-down | ACGCATCATCAATATTTCGTC |
| Upstream of phaC | phaC-up | GCTCATGTTTTCCTGACCGC |
| Downstream of phaC | phaC-down | AACCCCCTGCGAATGCAATA |
Generation of the complemented strain.
To construct the ΔphaR mutant complemented with the phaR plasmid, a broad-host-range vector, pBBR122, was used as described previously (38). To do this, the whole sequence of the phaR gene-harboring promoter region was amplified by PCR using the primers listed in Table 2. The amplified DNA fragment and pBBR122 vector were digested by EcoRI and ligated together. The ligated plasmid was transformed into E. coli DH5α by the heat shock method, and colonies harboring the cloned plasmid were selected on LB agar plates containing 30 μg/ml kanamycin. Then, the cloned plasmid was extracted from E. coli DH5α and electroporated into Burkholderia ΔphaR mutant competent cells. Finally, the Burkholderia ΔphaR/phaR strain was selected on YG agar plates containing 30 μg/ml rifampin and 30 μg/ml kanamycin.
Protein analyses of M4-colonized and cultured Burkholderia cells by SDS-PAGE.
The colonized Burkholderia cells in the M4 region were collected from fifth-instar nymphs. M4 regions of host midguts were collected by dissection in a sterile 1.5-ml centrifuge tube containing 100 μl of PB buffer (10 mM sodium phosphate dibasic [pH 7.0]). The collected organs were homogenized using an electric homogenizer until the symbionts had completely come out from M4. Next, 900 μl of PB buffer was added and filtered through a 5-μm-pore-size filter. The filtered cells were centrifuged (8,000 × g for 10 min) and adjusted to 5 × 107 cells/10 μl. In vitro-cultured Burkholderia cells grown in YG medium containing 30 μg/ml rifampin until mid-log phase were centrifuged (8,000 × g for 10 min) to harvest bacterial cells. Next, bacterial cells were washed with PB buffer and adjusted to 5 × 107 cells/10 μl. These M4-colonized and cultured Burkholderia cells (total 5 × 107 cells) were lysed in 1× Laemmli sample buffer and boiled at 100°C for 5 min. The lysed samples were loaded in a 12% SDS-PAGE gel. After running the gel, protein bands were stained with Coomassie brilliant blue R250 and destained.
N-terminal sequencing.
The N-terminal amino acid sequence of the PhaP protein (19 kDa) was determined by the automated Edman degradation method (39).
Fluorescence staining of PHA granules.
A single colony of Burkholderia sp. was inoculated into YG medium containing 30 μg/ml rifampin and incubated overnight at 30°C. Cultured cells were transferred to fresh YG or PB-G medium (PB buffer containing 1% glucose) and incubated for 12 h with vigorous shaking (25). After incubation, the bacterial cells were washed with PB buffer and adjusted to 5 × 107 cells/ml in PB buffer. Twenty microliters of these cells was dropped onto a glass slide and fixed by heat treatment. Then, 20 μl of 0.05% Nile blue A dissolved in ethanol was applied to fixed cells and incubated at room temperature for 10 min in darkness to stain the PHA granules in the bacteria (40). Subsequently, the glass slide was washed with tap water three times, and 8% acetic acid was added to reduce overstaining. Then, the glass slide was washed again with tap water, air dried, and observed under a fluorescence microscope (Olympus BX50). The PHA granules in bacterial cells were observed in fluorescence mode using a 550-nm dichroic mirror with the exciter filter BP450∼480 and the barrier filter BA515.
PHA measurement by flow cytometry.
Burkholderia cells incubated in YG medium containing 30 μg/ml rifampin were transferred to fresh YG or PB-G medium. These cells were incubated until mid-log phase and centrifuged (8,000 × g for 10 min) to harvest the cells. The cells were washed with distilled water three times and fixed with 70% ethanol at 4°C for 1 h. One hundred microliters of 0.05% Nile blue A dye was added to stain PHA granules and incubated for 10 min at room temperature in darkness. The samples were then washed with distilled water three times, treated with 8% acetic acid for 1 min, and washed with distilled water three more times. Washed cells were suspended in 1 ml of distilled water and analyzed on a flow cytometer machine (model FC500; Beckman Coulter). Fluorescence intensity was measured in channel FL2 (585 ± 42 nm bandpass filter).
CFU assay.
The second-instar nymphs of host insects were fed with in vitro-cultured wild-type and mutant Burkholderia strains. Upon emerging, the M4 region of each nymphal and adult stage was dissected and transferred to a 1.5-ml centrifuge tube containing 200 μl of PB buffer. Organs were completely homogenized with an electric homogenizer and diluted with PB buffer to 1:100 for the third-instar nymph, 1:1,000 for the fourth-instar nymph, and 1:10,000 for the fifth-instar nymph and adult insects. Diluted samples were spread on YG agar plates containing 30 μg/ml rifampin and incubated for 2 days at 30°C. The colonies were counted following incubation.
Growth curves of bacteria.
Growth curves of wild-type and mutant Burkholderia strains were estimated in YG, M9 (0.2% glucose, 0.6% Na2HPO4·2H2O2, 0.3% KH2PO4, 0.1% NH4Cl2, 0.05% NaCl, 0.0003% CaCl2, 1 mM MgSO4), and PB-G media. Burkholderia strains were primarily cultured in YG medium at 30°C for 18 h. Using the primary culture, starting bacterial solutions were prepared by adjusting the optical density at 600 nm (OD600) to 0.05 in each YG, M9, and PB-G medium and incubating at 30°C for 24 h with vigorous shaking. The bacterial cultures were monitored by measuring the turbidity at OD600 every 4 h using a spectrophotometer.
Measurement of fitness parameters.
The survival rate after feeding Burkholderia cells to the second-instar nymphs was estimated every day until 25 days after hatching by counting the dead insects. The adult emergence rate was estimated by counting the newly molted adult insects from the late-fifth-instar nymphs. To measure the body length and weight, adult insects were killed 3 days after molting in acetone for 5 min and dried completely in a 70°C oven for 30 min. Finally, fitness parameters were obtained. Soybean was not supplied to insects 24 h before killing to exclude the weight of the soybean.
Domain and phylogenetic analyses of PHA-related genes.
Domain analyses of PHA-related genes were performed using the InterProScan 5.0 and the BLASTP programs (41–43). Phylogenetic analysis of PHA-related proteins was performed by Clustal X2 and MEGA7 programs (44, 45). The nucleotide and amino acid sequences were obtained from GenBank (https://www.ncbi.nlm.nih.gov/).
Statistical analyses.
The statistical significance of differences in the data was determined using an unpaired t test with the GraphPad Prism software.
Accession number(s).
The sequences reported in this paper have been deposited in the DNA Data Bank of Japan/European Molecular Biology Laboratory/GenBank (accession numbers AB787502 for phaP, LC216413 for phaP1, LC216414 for phaP2, LC216415 for phaP3, LC216416 for phaP4, LC216417 for phaR, and AB787504 for phaC).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Global Research Laboratory grant of the National Research Foundation of Korea (grant 2011-0021535) to B.L.L.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00459-17.
REFERENCES
- 1.Buchner P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience Publishers, New York, NY. [Google Scholar]
- 2.Baumann P, Baumann L, Lai CY, Rouhbakhsh D, Moran NA, Clark MA. 1995. Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu Rev Microbiol 49:55–94. doi: 10.1146/annurev.mi.49.100195.000415. [DOI] [PubMed] [Google Scholar]
- 3.Kikuchi Y, Hosokawa T, Fukatsu T. 2011. Specific developmental window for establishment of an insect-microbe gut symbiosis. Appl Environ Microbiol 77:4075–4081. doi: 10.1128/AEM.00358-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 5.Kim JK, Lee JB, Huh YR, Jang HA, Kim C-H, Yoo JW, Lee BL. 2015. Burkholderia gut symbionts enhance the innate immunity of host Riptortus pedestris. Dev Comp Immunol 53:265–269. [DOI] [PubMed] [Google Scholar]
- 6.Jaenike J, Unckless R, Cockburn SN, Boelio LM, Perlman SJ. 2010. Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329:212–215. doi: 10.1126/science.1188235. [DOI] [PubMed] [Google Scholar]
- 7.Oliver KM, Degnan PH, Burke GR, Moran NA. 2010. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol 55:247–266. doi: 10.1146/annurev-ento-112408-085305. [DOI] [PubMed] [Google Scholar]
- 8.Werren JH, Baldo L, Clark ME. 2008. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 9.Douglas AE. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol 43:17–37. doi: 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- 10.Baumann P, Moran NA. 1997. Non-cultivable microorganisms from symbiotic associations of insects and other hosts. Antonie Van Leeuwenhoek 72:39–48. doi: 10.1023/A:1000239108771. [DOI] [PubMed] [Google Scholar]
- 11.Pontes MH, Dale C. 2006. Culture and manipulation of insect facultative symbionts. Trends Microbiol 14:406–412. doi: 10.1016/j.tim.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Kim JK, Lee BL. 2015. Symbiotic factors in Burkholderia essential for establishing an association with the bean bug, Riptortus pedestris. Arch Insect Biochem Physiol 88:4–17. doi: 10.1002/arch.21218. [DOI] [PubMed] [Google Scholar]
- 13.Kim JK, Lee JB, Am Jang H, Han YS, Fukatsu T, Lee BL. 2016. Understanding regulation of the host-mediated gut symbiont population and the symbiont-mediated host immunity in the Riptortus-Burkholderia symbiosis system. Dev Comp Immunol 64:75–81. doi: 10.1016/j.dci.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Takeshita K, Kikuchi Y. 2016. Riptortus pedestris and Burkholderia symbiont: an ideal model system for insect-microbe symbiotic associations. Res Microbiol 168:175–187. doi: 10.1016/j.resmic.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Kikuchi Y, Meng XY, Fukatsu T. 2005. Gut symbiotic bacteria of the genus Burkholderia in the broad-headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae). Appl Environ Microbiol 71:4035–4043. doi: 10.1128/AEM.71.7.4035-4043.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JK, Son DW, Kim C-H, Cho JH, Marchetti R, Silipo A, Sturiale L, Park HY, Huh YR, Nakayama H, Fukatsu T, Molinaro A, Lee BL. 2015. Insect gut symbiont susceptibility to host antimicrobial peptides caused by alteration of the bacterial cell envelope. J Biol Chem 290:21042–21053. doi: 10.1074/jbc.M115.651158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kikuchi Y, Hosokawa T, Fukatsu T. 2007. Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl Environ Microbiol 73:4308–4316. doi: 10.1128/AEM.00067-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson AJ, Dawes EA. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev 54:450–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SY. 1996. Plastic bacteria? Progress and prospects for polyhydroxyalkanoate production in bacteria. Trends Biotechnol 14:431–438. [Google Scholar]
- 20.Poblete-Castro I, Escapa IF, Jäger C, Puchalka J, Lam CM, Schomburg D, Prieto MA, Martins dos Santos VA. 2012. The metabolic response of P. putida KT2442 producing high levels of polyhydroxyalkanoate under single- and multiple-nutrient-limited growth: highlights from a multi-level omics approach. Microb Cell Fact 11:1–21. doi: 10.1186/1475-2859-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madison LL, Huisman GW. 1999. Metabolic engineering of poly (3-hydroxyalkanoates): from DNA to plastic. Microbiol Mol Biol Rev 63:21–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stubbe J, Tian J. 2003. Polyhydroxyalkanoate (PHA) homeostasis: the role of the PHA synthase. Nat Prod Rep 20:445–457. doi: 10.1039/b209687k. [DOI] [PubMed] [Google Scholar]
- 23.York GM, Stubbe J, Sinskey AJ. 2002. The Ralstonia eutropha PhaR protein couples synthesis of the PhaP phasin to the presence of polyhydroxybutyrate in cells and promotes polyhydroxybutyrate production. J Bacteriol 184:59–66. doi: 10.1128/JB.184.1.59-66.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.York GM, Junker BH, Stubbe J, Sinskey AJ. 2001. Accumulation of the PhaP phasin of Ralstonia eutropha is dependent on production of polyhydroxybutyrate in cells. J Bacteriol 183:4217–4226. doi: 10.1128/JB.183.14.4217-4226.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JK, Won YJ, Nikoh N, Nakayama H, Han SH, Kikuchi Y, Rhee YH, Park HY, Kwon JY, Kurokawa K, Dohmae N, Fukatsu T, Lee BL. 2013. Polyester synthesis genes associated with stress resistance are involved in an insect-bacterium symbiosis. Proc Natl Acad Sci U S A 110:E2381–E2389. doi: 10.1073/pnas.1303228110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida K-i, Takemoto Y, Sotsuka T, Tanaka K, Takenaka S. 2013. PhaP phasins play a principal role in poly-β-hydroxybutyrate accumulation in free-living Bradyrhizobium japonicum. BMC Microbiol 13:1. doi: 10.1186/1471-2180-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maehara A, Doi Y, Nishiyama T, Takagi Y, Ueda S, Nakano H, Yamane T. 2001. PhaR, a protein of unknown function conserved among short-chain-length polyhydroxyalkanoic acids producing bacteria, is a DNA-binding protein and represses Paracoccus denitrificans phaP expression in vitro. FEMS Microbiol Lett 200:9–15. doi: 10.1111/j.1574-6968.2001.tb10685.x. [DOI] [PubMed] [Google Scholar]
- 28.Pötter M, Müller H, Steinbüchel A. 2005. Influence of homologous phasins (PhaP) on PHA accumulation and regulation of their expression by the transcriptional repressor PhaR in Ralstonia eutropha H16. Microbiology 151:825–833. doi: 10.1099/mic.0.27613-0. [DOI] [PubMed] [Google Scholar]
- 29.Pötter M, Madkour MH, Mayer F, Steinbüchel A. 2002. Regulation of phasin expression and polyhydroxyalkanoate (PHA) granule formation in Ralstonia eutropha H16. Microbiology 148:2413–2426. doi: 10.1099/00221287-148-8-2413. [DOI] [PubMed] [Google Scholar]
- 30.Encarnación S, del Carmen Vargas M, Dunn MF, Davalos A, Mendoza G, Mora Y, Mora J. 2002. AniA regulates reserve polymer accumulation and global protein expression in Rhizobium etli. J Bacteriol 184:2287–2295. doi: 10.1128/JB.184.8.2287-2295.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Povolo S, Casella S. 2000. A critical role for aniA in energy-carbon flux and symbiotic nitrogen fixation in Sinorhizobium meliloti. Arch Microbiol 174:42–49. [DOI] [PubMed] [Google Scholar]
- 32.Quelas JI, Mesa S, Mongiardini EJ, Jendrossek D, Lodeiro AR. 2016. Regulation of polyhydroxybutyrate synthesis in the soil bacterium Bradyrhizobium diazoefficiens. Appl Environ Microbiol 82:4299–4308. doi: 10.1128/AEM.00757-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C, Sheng X, Equi RC, Trainer MA, Charles TC, Sobral BW. 2007. Influence of the poly-3-hydroxybutyrate (PHB) granule-associated proteins (PhaP1 and PhaP2) on PHB accumulation and symbiotic nitrogen fixation in Sinorhizobium meliloti Rm1021. J Bacteriol 189:9050–9056. doi: 10.1128/JB.01190-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rehm BH. 2003. Polyester synthases: natural catalysts for plastics. Biochem J 376:15–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JK, Jang HA, Won YJ, Kikuchi Y, Han SH, Kim C-H, Nikoh N, Fukatsu T, Lee BL. 2014. Purine biosynthesis-deficient Burkholderia mutants are incapable of symbiotic accommodation in the stinkbug. ISME J 8:552–563. doi: 10.1038/ismej.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 37.Froger A, Hall JE. 2007. Transformation of plasmid DNA into E. coli using the heat shock method. J Vis Exp 2007(6):253. doi: 10.3791/253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szpirer CY, Faelen M, Couturier M. 2001. Mobilization function of the pBHR1 plasmid, a derivative of the broad-host-range plasmid pBBR1. J Bacteriol 183:2101–2110. doi: 10.1128/JB.183.6.2101-2110.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niall HD. 1973. Automated Edman degradation: the protein sequenator. Methods Enzymol 27:942–1010. [DOI] [PubMed] [Google Scholar]
- 40.Ostle AG, Holt JG. 1982. Nile blue A as a fluorescent stain for poly-beta-hydroxybutyrate. Appl Environ Microbiol 44:238–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. 2005. InterProScan: protein domains identifier. Nucleic Acids Res 33:W116–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 44.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 45.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kikuchi Y, Hosokawa T, Fukatsu T. 2011. An ancient but promiscuous host-symbiont association between Burkholderia gut symbionts and their heteropteran hosts. ISME J 5:446–460. doi: 10.1038/ismej.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schäfer A, Schwarzer A, Kalinowski J, Pühler A. 1994. Cloning and characterization of a DNA region encoding a stress-sensitive restriction system from Corynebacterium glutamicum ATCC 13032 and analysis of its role in intergeneric conjugation with Escherichia coli. J Bacteriol 176:7309–7319. doi: 10.1128/jb.176.23.7309-7319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.