ABSTRACT
meso-Diaminopimelate dehydrogenase (meso-DAPDH) from Symbiobacterium thermophilum (StDAPDH) is the first member of the meso-DAPDH family known to catalyze the asymmetric reductive amination of 2-keto acids to produce d-amino acids. It is important to understand the catalytic mechanisms of StDAPDH and other enzymes in this family. In this study, based on an evolutionary analysis and examination of catalytic activity, the meso-DAPDH enzymes can be divided into two types. Type I showed highly preferable activity toward meso-diaminopimelate (meso-DAP), and type II exhibited obviously reversible amination activity with a broad substrate spectrum. StDAPDH belongs to type II. A quaternary structure analysis revealed that insertions/deletions (indels) and a loss of quaternary structure resulted in divergence among members of the meso-DAPDH family. A structure alignment of StDAPDH with a representative of type I, the meso-DAPDH from Corynebacterium glutamicum (CgDAPDH), indicated that they had the same folding. Based on sequence and conservation analyses, two amino acid residues of StDAPDH, R35 and R71, were found to be highly conserved within type II while also distinct from each other between the subtypes. Site mutagenesis studies identified R71 as a substrate preference-related residue of StDAPDH, which may serve as an indicator of the amination preference of type II. These results deepen the present understanding of the meso-DAPDH family and provide a solid foundation for the discovery and engineering of meso-DAPDH for d-amino acid biosynthesis.
IMPORTANCE The l-form of amino acids is typically more abundant than the d-form. However, the d-form has many important pharmaceutical applications. meso-Diaminopimelate dehydrogenase (meso-DAPDH) from Symbiobacterium thermophilum (StDAPDH) was the first member of meso-DAPDH known to catalyze the amination of 2-keto acids to produce d-amino acids. Accordingly, we analyzed the evolution of meso-DAPDH proteins and found that they form two groups, i.e., type I proteins, which show high preference toward meso-diaminopimelate (meso-DAP), and type II proteins, which show a broad substrate spectrum. We examined the differences in sequence, ternary structure, and quaternary structure to determine the mechanisms underlying the functional differences between the type I and type II lineages. These results will facilitate the identification of additional meso-DAPDHs and may provide guidance to protein engineering studies for d-amino acid biosynthesis.
KEYWORDS: d-amino acid, functional divergence, meso-diaminopimelate dehydrogenase, protein evolution, subfamily
INTRODUCTION
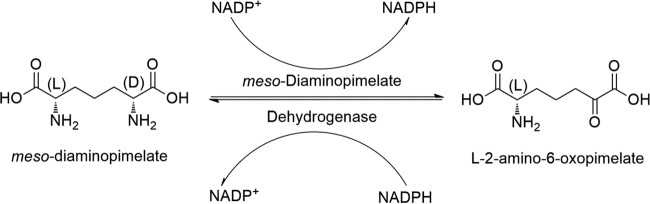
A key enzyme in the l-lysine synthesis pathway is meso-diaminopimelate dehydrogenase (meso-DAPDH; EC 1.4.1.16). It is widely distributed in bacteria and plants, including Corynebacterium glutamicum, Bacillus sphaericus, and Glycine max (1–4). It catalyzes the amination of l-tetrahydrodipicolinate to produce meso-diaminopimelate (meso-DAP) (5). meso-DAPDH is an NADP+-dependent oxidoreductase that catalyzes the reversible deamination of meso-diaminopimelate at the d-configuration center (depicted in Fig. 1). Almost all meso-DAPDHs reported to date show high specificity toward meso-DAP and extremely weak activities toward lanthionine and some 2-keto acids but do not act on other d-amino acids or 2-keto acids (2, 6–8).
FIG 1.
The reversible catalytic reaction via meso-DAPDH.
d-amino acids have broad applications in the pharmaceutical, food, and cosmetics industries. With the increasing requirement for d-amino acids, increasing research has focused on the asymmetric amination of 2-keto acids by d-amino acid dehydrogenase (9). Owing to the high stereospecificity of meso-DAPDH toward the d-center, it is well suited as the starting enzyme of engineering to obtain d-amino acid dehydrogenases for synthetic purposes. Since a 2006 study by Vedha-Peters and coworkers (6), it has been a focus of genetic engineering studies on d-amino acid dehydrogenase. Using meso-DAPDH from C. glutamicum ATCC 13032 (CgDAPDH) as a starting material, 10,000 variants were screened in three rounds of mutagenesis. The variant BC621, which contains Arg196Met, Thr170Ile, His245Asn, Gln151Leu, and Asp155Gly, exhibited high activity and stereospecificity toward cyclohexyl pyruvate. The same mutations have been introduced into other wild-type meso-DAPDHs from Ureibacillus thermosphaericus and B. sphaericus for the synthesis of d-branched-chain amino acids and (R)-5,5,5-trifluoronorvaline, respectively (10, 11).
In 2012, a meso-DAPDH from Symbiobacterium thermophilum IAM14863 (StDAPDH) was cloned, overexpressed in Escherichia coli, and characterized as the first wild-type DAPDH that effectively catalyzed the reductive reaction from 2-keto acids to d-amino acids (12). Since StDAPDH prefers pyruvic acid as its substrate, a subsequent site-directed mutagenesis study was performed to enlarge its substrate-binding pocket for large substrate phenyl pyruvic acid (13). Therefore, a question arises as to whether there are other wild-type members of the meso-DAPDH family that also catalyze reductive amination as StDAPDH does, and if so, how are their molecular, functional, and evolutionary properties different from those meso-DAPDHs with substrate specificity toward meso-DAP. In this study, we performed a phylogenetic analysis using available meso-DAPDH sequences, and their catalytic activities were studied. Comparative sequence and structural analyses were performed to explore the possible molecular mechanisms underlying differences among meso-DAPDHs.
RESULTS AND DISCUSSION
meso-DAPDHs can be classified into two types according to phylogenetic analysis.
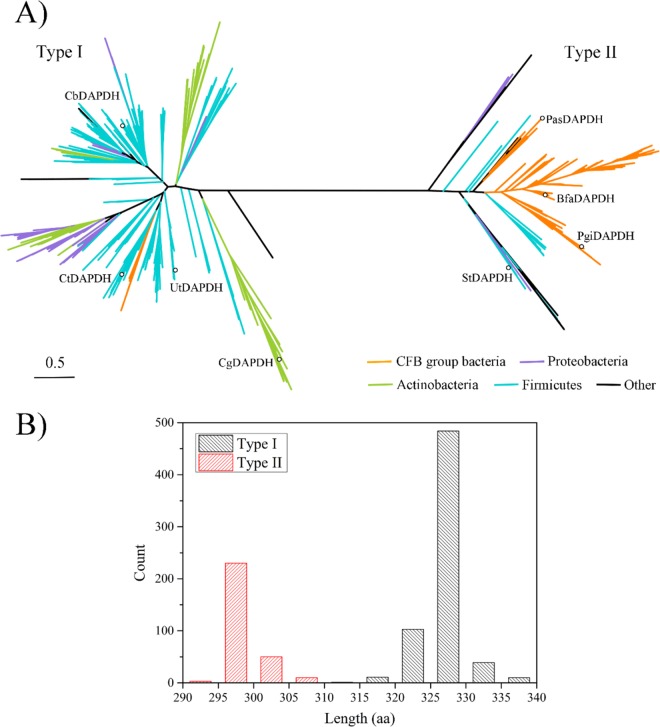
Using the amino acid sequences of CgDAPDH and StDAPDH as query sequences, 941 homologous sequences were identified in the U.S. National Center for Biotechnology Information (NCBI) reference sequence (RefSeq) database using the position-specific iterated basic local alignment search tool (PSI-BLAST). All of these sequences were from bacteria, including Actinobacteria, Firmicutes, Cytophaga-Flexibacter-Bacteroides (CFB) group bacteria, Proteobacteria, and other bacterial taxa. As shown in Fig. 2, a phylogenetic analysis indicated that there were two branches of the meso-DAPDH family; these two clades were named type I and type II. In total, 648 proteins belonged to the type I lineage whereas 293 were type II. The known CgDAPDH (Protein Data Bank [PDB] accession number 1F06) and meso-DAPDHs from U. thermosphaericus (UtDAPDH; PDB accession number 3WYB) (7, 14), Clostridium tetani E88 (CtDAPDH; PDB accession number 3WGQ), and B. sphaericus (BasDAPDH) were located in the type I clade, while the first meso-DAPDH, StDAPDH (PDB accession number 3WBF), with a relatively broader substrate spectrum, was classified as type II. The proteins located in the type I lineage were mainly from Firmicutes, Actinobacteria, and Proteobacteria, whereas type II proteins were mainly from CFB group bacteria, with a few from Firmicutes and Proteobacteria. According to Bergey's Manual of Systematic Bacteriology (15), the bacteria from the CFB group, Firmicutes, and Proteobacteria are distantly related. Therefore, it can be deduced from the tree that type I and type II meso-DAPDHs arose from a recent common ancestor and exhibit divergent evolution.
FIG 2.
Sequence analysis of 941 meso-DAPDHs from bacteria. (A) Phylogenetic tree analysis of 941 meso-DAPDHs. The evolutionary history was inferred using the maximum likelihood method. Estimates of branch support were calculated using approximate likelihood ratio tests (aLRT SH-like). The tree is drawn to scale with branch lengths proportionate to evolutionary distances. Colors indicate different bacterial phyla. The proteins marked with circles were used for the activity assay. (B) Distribution of amino acid sequence lengths of meso-DAPDHs.
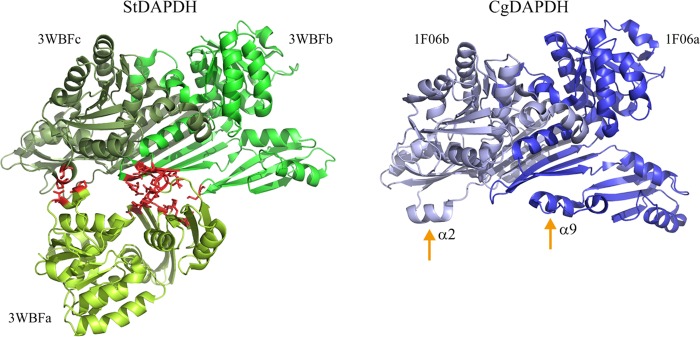
Insertions and deletions (indels) in protein sequences can lead to a series of structural changes (16, 17), and indels and the loss of quaternary structure are commonly associated with the rate of protein evolution (17). Based on the multiple-sequence alignment generated in our previous work (12), there are some obvious indels that distinguish StDAPDH from three other meso-DAPDHs, i.e., CgDAPDH, UtDAPDH, and BasDAPDH. Therefore, we analyzed the lengths of all meso-DAPDHs, including 648 type I and 293 type II meso-DAPDHs. All type I meso-DAPDHs contained more amino acid residues than type II. The average length of type I meso-DAPDHs was approximately 327 amino acids (aa), whereas the average length was 299 aa for type II meso-DAPDHs. The indels were conserved within each type. According to previous crystal structural studies (14, 18, 19), StDAPDH aggregates as a hexamer, whereas CgDAPDH forms a dimer, and indels are located at α2, α9, and some linkers of CgDAPDH but not in the catalytic pocket. Quaternary structures indicated that, as shown in Fig. 3, the b and c subunits of StDAPDH were assembled with protein-protein interfaces that were similar to those of CgDAPDH and then further interacted with subunit a. Therefore, the protein-protein interface of StDAPDH subunits was analyzed. There were 55 pairs of interacting residues between subunit b and c; however, for subunit a, there were only 13 and 2 residues that interacted with subunits b and c, respectively. Notably, the protein-protein interfaces between subunits a and b/c were directly related to the indels at α2 and α9 in CgDAPDH. As such, it is possible that indels in CgDAPDH hinder the interactions between subunits a and b/c and thus result in conformational changes and a loss of the hexameric structure.
FIG 3.
Protein-protein interfaces of two types of meso-DAPDHs. The interface of StDAPDH between subunit a and subunit b/c is indicated in red. α2 and α9 of CgDAPDH are also marked.
Type II meso-DAPDHs showed broader substrate spectra than type I meso-DAPDHs.
As shown in Table 1, CgDAPDH and CtDAPDH, which are type I, have been reported to have a high preference toward meso-DAP (6, 18). Additionally, the gene encoding meso-DAPDH from Clostridium bolteae 90A9 (CbDAPDH; NCBI RefSeq accession number WP_002575100.1), which has a distant phylogenetic relationship with CgDAPDH and CtDAPDH, was chemically synthesized, and the recombinant protein was purified. The activity assay showed that CbDAPDH also had a higher preference toward meso-DAP but not 2-keto acids (Table 1). As we know, StDAPDH, which is type II, catalyzes not only the oxidative deamination of meso-DAP but also the reductive amination of 2-keto acids. This difference in asymmetric reductive amination raised a question regarding the general difference in activity between these two types of meso-DAPDHs. Therefore, according to the evolutionary relationships of different subclades in type II (shown in Fig. 2), several members of the type II lineage were selected for overexpression, purification, and activity assays, including the meso-DAPDHs from Porphyromonas asaccharolytica DSM 20707 (PasDAPDH; GenBank accession number AEE12144.1), Porphyromonas gingivalis W83 (PgiDAPDH; GenBank accession number AAQ65966.1), and Bacteroides faecis MAJ27 (BfaDAPDH; NCBI accession number ZP_09861452.1). As shown in Table 1, all of the four enzymes exhibited clear asymmetric oxidative deamination activity toward meso-DAP. For the reverse reaction, these meso-DAPDHs showed much higher activity than type I members. Among the 2-keto acids, pyruvic acid was the best substrate, and activity toward other 2-keto acids was approximately 1 order of magnitude lower. Although CgDAPDH, CtDAPDH, and CbDAPDH exhibited detectable activity toward some 2-keto acids, the enzymes in the type II lineage were much more active toward these 2-keto acids. Based on these observations, we can draw the following two conclusions: (i) type I meso-DAPDH has high substrate preference toward meso-DAP, which is consistent with the substrate spectra of CgDAPDH, CtDAPDH, and CbDAPDH, and (ii) the broad substrate spectrum of type II enzymes suggests that reductive amination activity toward 2-keto acids is a common feature of the type II lineage and that this functional feature differentiates the two types of enzymes.
TABLE 1.
Catalytic activities of meso-DAPDHs from various taxa
| Substrate | Specific activity (U · mg−1)a |
||||||
|---|---|---|---|---|---|---|---|
| Type I |
Type II |
||||||
| CgDAPDHb | CtDAPDHc | CbDAPDHf | StDAPDHd | PasDAPDHf | PgiDAPDHf | BfaDAPDHf | |
| meso-DAP | 131.6 | 1.04 ± 0.03 | 3.18 ± 0.10 | 11.33 ± 0.46 | 17.22 ± 0.85 | 2.58 ± 0.05 | 13.24 ± 0.55 |
| Pyruvic acid | 0.009 | 0.22 ± 0.01 | 0.08 ± 0.00 | 2.87 ± 0.16 | 2.31 ± 0.10 | 1.27 ± 0.01 | 1.76 ± 0.02 |
| 4-Methyl-2-oxopentanoic acid | 0.004 | 0.02 ± 0.00 | 0.04 ± 0.00 | 0.11 ± 0.03 | 0.15 ± 0.00 | 0.06 ± 0.00 | 0.06 ± 0.00 |
| Sodium 3-methyl-2-oxobutanoate | 0.05 | NRe | 0.04 ± 0.00 | 0.13 ± 0.01 | 0.39 ± 0.11 | 0.28 ± 0.02 | 0.18 ± 0.00 |
| 2-Oxosuccinic acid | NR | NR | 0.02 ± 0.00 | 0.26 ± 0.01 | 0.16 ± 0.01 | 0.13 ± 0.00 | 0.15 ± 0.00 |
One unit (U) was defined as the amount of enzyme necessary to produce or consume 1 μmol NADPH per minute.
This information is taken from reference 6. The typical reductive amination system contained 100 mM sodium carbonate buffer, pH 9.0, 200 mM NH4Cl, 25 mM 2-keto acid, and 0.2 mM NADPH; the oxidative deamination system contained 100 mM sodium carbonate buffer, pH 9.5, 25 mM meso-DAP, and 1 mM NADP+.
This information is taken from reference 32. The typical reductive amination system contained 100 mM sodium carbonate buffer, pH 9.0, 200 mM NH4Cl, 20 mM 2-keto acid, and 0.5 mM NADPH; the oxidative deamination system contained 100 mM sodium carbonate buffer, pH 9.5, 5 mM meso-DAP, and 0.5 mM NADP+.
This information is taken from reference 12. The activity assay system was the same as that used for CtDAPDH.
NR, not reported.
The reductive amination system contained 100 mM sodium carbonate buffer, pH 8.6, 200 mM NH4Cl, 20 mM 2-keto acid, and 0.5 mM NADPH; the oxidative deamination system contained 100 mM sodium carbonate buffer, pH 9.6, 5 mM meso-DAP, and 0.5 mM NADP+. The reactions were performed at 25°C.
Sequence and ternary structure alignments between type I and type II meso-DAPDHs.
According to the analyses described above, including the phylogenetic analysis, the meso-DAPDH family exhibits clear functional diversity. Therefore, we investigated the differences in sequence and structure between type I and type II meso-DAPDH to explore the possible molecular basis for their functional divergence. CgDAPDH and StDAPDH were used as type I and type II representatives, respectively, and sequence and structural alignments were performed.
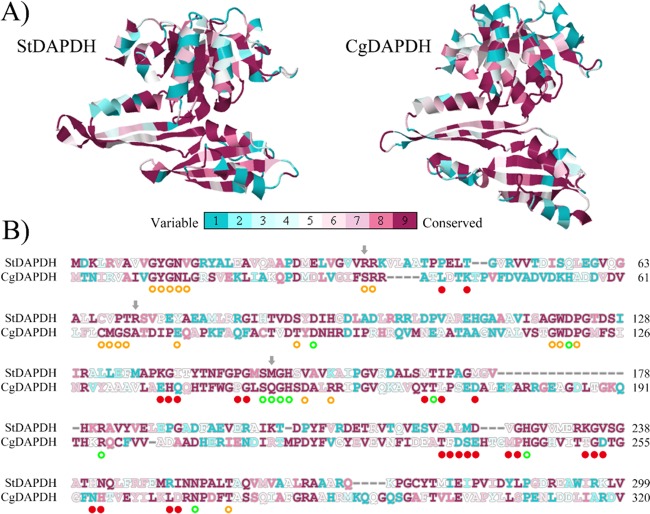
A series of analyses were performed, beginning with analyses of three-dimensional structures. First, based on the ternary structure alignment, the root mean square deviation (RMSD) was only 3.53, and the average template modeling (TM) score was 0.76. This means that although the amino acid sequence identity is only 28% between CgDAPDH and StDAPDH, the crystal structures have the same folding. Subsequently, the conserved positions in CgDAPDH and StDAPDH were compared. Although interaction models for proteins and meso-DAP or NADP+ have been reported (14, 18, 19), there are no obvious definitions of the substrate/NADP+-binding residues for CgDAPDH and StDAPDH. Therefore, MSDsite (28) was used to predict substrate/NADP+-binding residues for the two enzymes. As shown in Fig. 4B, 19 NADP+-binding residues and 10 meso-DAP-binding residues were identified. Conservation analyses of all amino acid residues were performed based on evolutionary conservation scores calculated using ConSurf (20). As shown in Fig. 4A, the substrate-binding pockets of CgDAPDH and StDAPDH were both highly conserved. Among all of the meso-DAP-binding residues of the two enzymes, Met152/Gln150 (Met152 of StDAPDH and its counterpart Gln150 in CgDAPDH) was the only pair of amino acid residues with obvious differences (Fig. 4B). Among NADP+-binding residues of StDAPDH, 73.7% were highly conserved; a comparison of these residues in CgDAPDH and StDAPDH revealed that nearly all of the different NADP+-binding residues belong to the same amino acid type, except Arg35/Ser35. In addition, Arg71/Ala69 was next to NADP+-binding residues (Fig. 4B), and from the perspective of the crystal structure, it was located on the protein surface far from the active site (as shown in Fig. 5). Mutation of Met152 to Gln resulted in a decrease of kcat/Km to 1/30 and 1/14 of the wild type toward meso-DAP and pyruvic acid, respectively, which indicated that M152 is not a critical site for substrate preference (12). Considering the high conservation of residues at positions 35 (R35) and 71 (R71) within their respective subtypes and the substantially different characteristics between R35, R71, and their counterparts, we examined the two Arg residues of StDAPDH to determine their roles in catalytic activity.
FIG 4.
ConSurf analysis of two types of meso-DAPDH. In total, 648 proteins of type I and 293 proteins of type II were used for the evolutionary conservation analysis. (A) Evolutionary conservation of amino acid positions in the structures of meso-DAPDH. (B) Sequence alignment between StDAPDH and CgDAPDH. The predicted substrate-binding residues of StDAPDH are marked with green circles, and NADP+-binding residues are marked with orange circles. Residues involved in multimerization are marked with red dots. On the conservation scale, grade 1 was defined as the most highly variable amino acid positions (turquoise), and grade 9 was defined as the most highly conserved positions (maroon).
FIG 5.
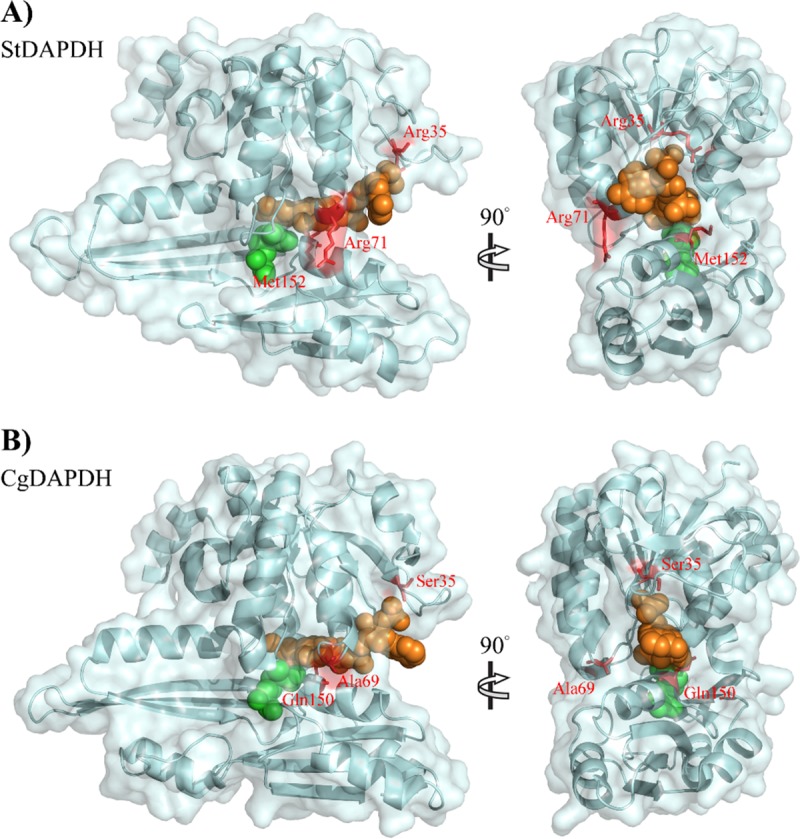
Structural alignment of the two types of meso-DAPDHs. (A) Type II meso-DAPDH, with StDAPDH (PDB accession number 3WBF_a) as a representative; (B) type I meso-DAPDH, with CgDAPDH (PDB accession number 1F06_a) as a representative. meso-DAP is shown in green; NADPH is shown in orange.
Therefore, site-directed mutagenesis was performed on Arg35 and Arg71, with the gene of StDAPDH as the template. The two residues were replaced with the corresponding amino acid residues in CgDAPDH. The mutant and wild-type enzymes were purified, and their kinetic parameters for meso-DAP, NADP+, pyruvic acid, and NADPH were investigated. As shown in Table 2, when Arg35 was swapped to Ser in CgDAPDH, the catalytic efficiencies toward meso-DAP and pyruvic acid were 76% and 92% of the wild-type enzyme, respectively. Amino acid swapping with Ser at position 35 negatively affected amination and deamination activities, indicating that Arg35 of StDAPDH was an activity-related residue but was not related to the reaction preference of StDAPDH. The decreased catalytic efficiency of the deamination reaction was mainly attributed to the lower kcat values. For Arg at position 71, when it was replaced with Ala in CgDAPDH, the kcat values toward pyruvic acid and NADPH were decreased and the Km value toward pyruvic acid was increased; consequently, the catalytic efficiencies of the mutant toward meso-DAP and pyruvic acid were 98% and 35% of the wild-type enzyme, respectively. In other words, amino acid replacement with Ala at position 71 in StDAPDH did not have a substantial effect on the catalytic ability of deamination but had a strong influence on amination. Therefore, R71 was an activity-related residue as well as a substrate preference-related one. Furthermore, the evolutionary conservation analysis indicated that the amino acid residues of substrate- and cofactor-binding residues are highly conversed in each type; thus, we can draw a general conclusion that the amino acid residue of this position may be an indicator of the amination preference of type II meso-DAPDHs. The important role of Arg71 and its counterpart Ala69 in CgDAPDH, which are not in the active site, suggests that there may be other auxiliary residues responsible for the substrate preference of type II enzymes. Future studies should explore the specific positions related to the observed functional divergence using bioinformatics and protein engineering.
TABLE 2.
Kinetic parameter analysis of StDAPDH and R35S and R71A variants
| Reactiona | Substrate | Wild type |
R35S |
R71A |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Km (mM) | kcat (s−1) | kcat/Km (mM−1 · s−1) | Km (mM) | kcat (s−1) | kcat/Km (mM−1 · s−1) | Km (mM) | kcat (s−1) | kcat/Km (mM−1 · s−1) | ||
| Deamination | meso-DAP | 0.41 ± 0.07 | 40.47 ± 2.95 | 98.48 | 0.39 ± 0.06 | 29.44 ± 1.33 | 75.07 | 0.42 ± 0.08 | 40.46 ± 2.88 | 96.54 |
| NADP+ | 0.35 ± 0.04 | 25.47 ± 1.32 | 73.83 | 0.46 ± 0.06 | 17.91 ± 0.91 | 39.02 | 0.39 ± 0.08 | 58.16 ± 6.11 | 148.06 | |
| Amination | Pyruvic acid | 16.31 ± 2.62 | 7.00 ± 0.58 | 0.43 | 7.45 ± 1.72 | 2.96 ± 0.31 | 0.40 | 21.77 ± 3.06 | 3.31 ± 0.18 | 0.15 |
| NADPH | 0.10 ± 0.01 | 6.97 ± 0.21 | 69.29 | 0.35 ± 0.05 | 2.92 ± 0.18 | 8.32 | 0.18 ± 0.02 | 4.17 ± 0.15 | 23.49 | |
The deamination reaction was performed in 100 mM Na2CO3-NaHCO3 buffer (pH 9.6) while the amination reaction was carried out in 100 mM Na2CO3-NaHCO3 buffer (pH 8.6) at 25°C.
MATERIALS AND METHODS
Materials.
The restriction enzyme DpnI was purchased from Fermentas (St. Leon-Rot, Germany). The 2-keto acids and meso-DAP were purchased from Sigma (St. Louis, MO, USA) or TCI (Tokyo, Japan). NADP(H) was obtained from Roche (Basel, Switzerland).
Collection of meso-DAPDH sequences.
The position-specific iterated basic local alignment search tool (PSI-BLAST) was used to search for meso-DAPDHs in the U.S. National Center for Biotechnology Information (NCBI) reference sequence (RefSeq) database. The amino acid sequences of CgDAPDH and StDAPDH (21, 22) were used as query sequences.
Phylogenetic analysis.
A multiple-sequence alignment of the full-length protein sequences of the meso-DAPDHs was obtained using MAFFT (FFT-NS-i, BLOSUM62) (23). A phylogenetic tree was constructed using PhyML with the LG substitution model and four substitution rate categories (24). Estimates of branch support were obtained using approximate likelihood ratio tests (aLRT SH-like) (25). The phylogenetic tree was visualized using iTOL (26).
Site-directed mutagenesis, overexpression, and purification of meso-DAPDHs and variants.
The genes encoding meso-DAPDHs were synthesized chemically and inserted into the pET32a(+) vector by Shanghai Xuguan Biotechnological Development Co., Ltd. (Shanghai, China) or Genewiz (Suzhou, China). Variants of StDAPDH were created following the protocols reported previously (12) using PCR. Recombinant vectors were introduced into Escherichia coli Rosetta (DE3)pLysS. Gene expression was induced by treatment with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h at 37°C. After centrifugation, cells were collected and disrupted by sonication (Sonics, Newtown, CT, USA). For purification, the His tag protein purification magnetic bead kit (Beaver Bioscience, Suzhou, China) was used. The supernatant was mixed with magnetic beads and then incubated for affinity adsorption. Stepwise elution was performed to obtain purified proteins. The protein concentration was determined using the Bradford kit (BioTeke, Beijing, China).
Activity assay and kinetic parameter determination.
Enzymatic activity and kinetic parameters were determined following the previously reported protocols (12) at 25°C for both deamination and amination. The differences were as follows. The pH of the reaction for amination was 8.8, whereas that for deamination was 9.6. When the kinetic parameters of NADPH/NADP+ were determined, the concentration of NADPH/NADP+ varied from 0.1 to 1.5 mM, whereas the concentrations of pyruvic acid and NH4Cl for amination were 20 mM and 200 mM, respectively, and the concentration of meso-DAP for deamination was 1 mM. The absorbance at 340 nm was monitored using a Varioskan Flash (Thermo, Waltham, MA, USA). The molar absorption coefficient of NADPH was 6.22 mM−1 · cm−1. All tests were performed with three replicates. The kinetic values were determined by fitting the Michaelis-Menten curve to the data using the nonlinear regression method.
Structural analysis.
The meso-DAPDH structures were obtained from the PDB database (27). The substrate-binding residues of meso-DAPDH were predicted using MSDsite (28). The protein-protein interfaces were predicted using PDBsum (29). Pairwise structural differences in meso-DAPDHs were determined using TM-align (30). The TM score, which ranges from 0 to 1, was used as a measure of the structural similarity of two proteins (31).
Evolutionary conservation analysis.
The evolutionary conservation of the meso-DAPDHs was estimated using the ConSurf algorithm (20). The LG substitution matrix and the empirical Bayesian paradigm were used to estimate the accuracy of the conservation scores. The continuous conservation scores were divided into a discrete scale of nine grades for visualization, in which grade 1 indicates the most variable positions (grade 1) and grade 9 represents the most highly conserved positions.
ACKNOWLEDGMENTS
We thank Dunming Zhu, Jinhui Feng, and Hong Liu for helpful discussions and constructive suggestions.
This work was financially supported by the National Natural Science Foundation of China (grant 21402109) and the China Postdoctoral Science Foundation (grant 2016M592191).
REFERENCES
- 1.Misono H, Togawa H, Yamamoto T, Soda K. 1979. Meso-alpha, epsilon-diaminopimelate d-dehydrogenase: distribution and the reaction product. J Bacteriol 137:22–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Misono H, Ogasawara M, Nagasaki S. 1986. Characterization of meso-diaminopimelate dehydrogenase from Corynebacterium glutamicum and its distribution in bacteria. Agric Biol Chem 50:2729–2734. doi: 10.1080/00021369.1986.10867837. [DOI] [Google Scholar]
- 3.Wenko LK, Treick RW, Wilson KG. 1985. Isolation and characterization of a gene encoding meso-diaminopimelate dehydrogenase from Glycine max. Plant Mol Biol 4:197–204. doi: 10.1007/BF02418236. [DOI] [PubMed] [Google Scholar]
- 4.Sakamoto S, Seki M, Nagata S, Misono H. 2001. Cloning, sequencing, and expression of the meso-diaminopimelate dehydrogenase gene from Bacillus sphaericus. J Mol Catal B: Enzym 12:85–92. doi: 10.1016/S1381-1177(00)00207-1. [DOI] [Google Scholar]
- 5.White P. 1983. The essential role of diaminopimelate dehydrogenase in the biosynthesis of lysine by Bacillus sphaericus. J Gen Microbiol 129:739–749. [Google Scholar]
- 6.Vedha-Peters K, Gunawardana M, Rozzell JD, Novick SJ. 2006. Creation of a broad-range and highly stereoselective d-amino acid dehydrogenase for the one-step synthesis of d-amino acids. J Am Chem Soc 128:10923–10929. doi: 10.1021/ja0603960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akita H, Fujino Y, Doi K, Ohshima T. 2011. Highly stable meso-diaminopimelate dehydrogenase from an Ureibacillus thermosphaericus strain A1 isolated from a Japanese compost: purification, characterization and sequencing. AMB Express 1:43–50. doi: 10.1186/2191-0855-1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Misono H, Soda K. 1980. Properties of meso-alpha, epsilon-diaminopimelate d-dehydrogenase from Bacillus sphaericus. J Biol Chem 255:10599–10605. [PubMed] [Google Scholar]
- 9.Gao X, Ma Q, Zhu H. 2015. Distribution, industrial applications, and enzymatic synthesis of d-amino acids. Appl Microbiol Biotechnol 99:3341–3349. doi: 10.1007/s00253-015-6507-3. [DOI] [PubMed] [Google Scholar]
- 10.Akita H, Suzuki H, Doi K, Ohshima T. 2014. Efficient synthesis of d-branched-chain amino acids and their labeled compounds with stable isotopes using d-amino acid dehydrogenase. Appl Microbiol Biotechnol 98:1135–1143. doi: 10.1007/s00253-013-4902-1. [DOI] [PubMed] [Google Scholar]
- 11.Hanson RL, Johnston RM, Goldberg SL, Parker WL, Goswami A. 2013. Enzymatic preparation of an R-amino acid intermediate for a γ-secretase inhibitor. Org Process Res Dev 17:693–700. doi: 10.1021/op400013e. [DOI] [Google Scholar]
- 12.Gao X, Chen X, Liu W, Feng J, Wu Q, Hua L, Zhu D. 2012. A novel meso-diaminopimelate dehydrogenase from Symbiobacterium thermophilum: overexpression, characterization, and potential for d-amino acid synthesis. Appl Environ Microbiol 78:8595–8600. doi: 10.1128/AEM.02234-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao X, Huang F, Feng J, Chen X, Zhang H, Wang Z, Wu Q, Zhu D. 2013. Engineering the meso-diaminopimelate dehydrogenase from Symbiobacterium thermophilum by site saturation mutagenesis for d-phenylalanine synthesis. Appl Environ Microbiol 79:5078–5081. doi: 10.1128/AEM.01049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akita H, Seto T, Ohshima T, Sakuraba H. 2015. Structural insight into the thermostable NADP+-dependent meso-diaminopimelate dehydrogenase from Ureibacillus thermosphaericus. Acta Crystallogr D Biol Crystallogr 71:1136–1146. doi: 10.1107/S1399004715003673. [DOI] [PubMed] [Google Scholar]
- 15.Kersters K, Vancanneyt M. 2005. Bergey's manual of systematic bacteriology. Springer Verlag, Berlin, Germany. [Google Scholar]
- 16.Zhang Z, Huang J, Wang Z, Wang L, Gao P. 2011. Impact of indels on the flanking regions in structural domains. Mol Biol Evol 28:291–301. doi: 10.1093/molbev/msq196. [DOI] [PubMed] [Google Scholar]
- 17.Odokonyero D, Sakai A, Patskovsky Y, Malashkevich VN, Fedorov AA, Bonanno JB, Fedorov EV, Toro R, Agarwal R, Wang C, Ozerova ND, Yew WS, Sauder JM, Swaminathan S, Burley SK, Almo SC, Glasner ME. 2014. Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family. Proc Natl Acad Sci U S A 111:8535–8540. doi: 10.1073/pnas.1318703111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cirilli M, Scapin G, Sutherland A, Vederas JC, Blanchard JS. 2000. The three-dimensional structure of the ternary complex of Corynebacterium glutamicum diaminopimelate dehydrogenase-NADPH-l-2-amino-6-methylene-pimelate. Protein Sci 9:2034–2037. doi: 10.1110/ps.9.10.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W, Li Z, Huang C-H, Guo R-T, Zhao L, Zhang D, Chen X, Wu Q, Zhu D. 2014. Structural and mutational studies on the unusual substrate specificity of meso-diaminopimelate dehydrogenase from Symbiobacterium thermophilum. Chembiochem 15:217–222. doi: 10.1002/cbic.201300691. [DOI] [PubMed] [Google Scholar]
- 20.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. 2010. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O'Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, et al. . 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 25.Anisimova M, Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 26.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rose PW, Prlic A, Bi C, Bluhm WF, Christie CH, Dutta S, Green RK, Goodsell DS, Westbrook JD, Woo J, Young J, Zardecki C, Berman HM, Bourne PE, Burley SK. 2015. The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res 43:D345–D356. doi: 10.1093/nar/gku1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golovin A, Dimitropoulos D, Oldfield T, Rachedi A, Henrick K. 2005. MSDsite: a database search and retrieval system for the analysis and viewing of bound ligands and active sites. Proteins 58:190–199. doi: 10.1002/prot.20288. [DOI] [PubMed] [Google Scholar]
- 29.de Beer TA, Berka K, Thornton JM, Laskowski RA. 2014. PDBsum additions. Nucleic Acids Res 42:D292–D296. doi: 10.1093/nar/gkt940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Skolnick J. 2005. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res 33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Skolnick J. 2004. Scoring function for automated assessment of protein structure template quality. Proteins 57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 32.Liu W, Guo R-T, Chen X, Li Z, Gao X, Huang C-H, Wu Q, Feng J, Zhu D. 2015. Structural analysis reveals the substrate-binding mechanism for the expanded substrate specificity of mutant meso-diaminopimelate dehydrogenase. Chembiochem 16:924–929. doi: 10.1002/cbic.201402632. [DOI] [PubMed] [Google Scholar]