Abstract
The p53 protein activates cellular death programs through multiple pathways. Because the high frequency of p53 mutations in human tumors is believed to contribute to resistance to commonly used chemotherapeutic agents, it is important to identify drugs that induce p53-independent cell death and to define the mechanisms of action of such drugs. Here we screened a drug library (the National Cancer Institute mechanistic set; 879 compounds with diverse mechanisms of actions) and identified 175 compounds that induced caspase cleavage of cytokeratin-18 in cultured HCT116 colon cancer cells at <5 μM. Interestingly, whereas most compounds elicited a stronger apoptotic response in cells with functional p53, significant apoptosis was observed also in p53-null cells. A subset of 15 compounds showing weak or no dependence on p53 for induction of apoptosis was examined in detail. Of these compounds, 11 were capable of activating caspase-3 in enucleated cells. Seven such compounds with nonnuclear targets were found to induce lysosomal membrane permeabilization (LMP). Translocation of the lysosomal proteases cathepsin B and cathepsin D into the cytosol was observed after treatment with these drugs, and apoptosis was inhibited by pepstatin A, an inhibitor of cathepsin D. Apoptosis depended on Bax, suggesting that LMP induced a mitochondrial apoptotic pathway. We conclude that a large number of potential anticancer drugs induce p53-independent apoptosis and that LMP is a mediator of many such responses.
Keywords: anticancer drugs, cathepsin D, M30 antibody
Most commonly used anticancer drugs were discovered through screening of synthetic compounds and natural products by using cell-based cytotoxicity assays. A major advantage of such assays is that they predict drug effects within the context of the whole cell. A major cell-based screening endeavor is conducted by the National Cancer Institute (NCI) that has screened >100,000 compounds for growth inhibitory activity on 60 different human tumor cell lines (1). Numerous patterns of cellular responsiveness were observed, and the results have been analyzed by different methods, including hierarchical clustering (2, 3). Prominent examples of drugs detected by the screen that have advanced to the clinic include flavopiridol, UCN-01, and depsipeptide.
The efficiency of chemotherapeutic drugs is determined by different factors, including the genotype of the tumor cell. Many anticancer agents induce DNA damage that triggers a p53-dependent apoptotic response. The p53 tumor suppressor gene is mutated in up to 50% of human tumors (4), which may contribute to resistance to various types of therapies (5). Identifying drugs that induce p53-independent apoptosis is therefore important. The cytotoxicity of taxol has been reported to be independent of p53 function (6). In addition, various investigational drugs have been described to have p53-independent mechanisms of action (for examples, see refs. 7–11). It is not clear whether p53-independent apoptosis is a common phenomenon generated by a large number of drugs or whether p53-independent pathways are exceptional.
Studies from a number of laboratories have shown that lysosomal rupture is an early event in various apoptotic processes, including apoptosis initiated by oxidative stress, serum withdrawal, and Fas ligation (12–15). Interestingly, it has been demonstrated that p53-induced apoptosis involves early lysosomal membrane permeabilization (LMP) (16). It was recently demonstrated that immortalization and/or transformation increases the susceptibility of cells to lysosomal death pathways (17), suggesting that these pathways are potential targets of anticancer drug development.
Measurement of the number of living cells {using 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide (MTT) or similar assays} in drug-treated and control cultures is the most commonly used endpoint in cell-based screening experiments. A decrease in the cell number may be due to inhibition of cell proliferation or due to cell death (induction of apoptosis or necrosis). This intrinsic property of viability assays complicates the interpretation of drug screening experiments. To examine the sensitivity of p53-deficient human tumor cells to drug-induced apoptosis, we used a recently developed assay that measures the accumulation of caspase-cleaved cytokeratin (CK) 18 fragments in drug-treated cell cultures (18). A wide variety of drugs with different mechanisms of action were obtained from the NCI mechanistic set. This collection consists of 879 compounds selected from a larger set of ≈40,000 compounds based on different patterns of growth inhibition induced in 60 tumor cell lines (www.dtp.nci.nih.gov). The results of our study showed that apoptosis was induced by about one-half of the compounds that score positive in the MTT assay in the lower micromolar range and that many of the compounds were capable of inducing apoptosis in p53-deficient cells. Furthermore, the results suggest that LMP is a common mechanism of action of compounds that induce p53-independent apoptosis.
Experimental Procedures
Materials. The mechanistic set was obtained from the Developmental Therapeutics Program of NCI (www.dtp.nci.nih.gov). Other reagents: pepstatin A (Roche Molecular Biochemicals); cisplatin and doxorubicin (Karolinska Hospital Pharmacy); thapsigargin (Molecular Probes); and ciprofloxacin (A/S GEA, Frederiksberg, Denmark).
Cell Culture. HCT116 colon carcinoma (p53 WT and p53-/-; generously provided by Bert Vogelstein, The Johns Hopkins University Medical Institutions, Baltimore) (19) were maintained in McCoy's 5A modified medium/10% FCS at 37°C in 5% CO2.
Assessment of Cell Viability and Apoptosis. The accumulation of caspase-cleaved CK18, CK18-Asp396, was determined by the M30-Apoptosense ELISA (Peviva, Bromma, Sweden) (18). Cells were seeded at 104 per well in 96-well plates 16 h before treatment and treated with drugs as indicated. At the end of the incubation period, Nonidet P-40 was added to the tissue culture medium to 0.5%, and 25 μl of the content of each well (thus including CK18 released to the medium from attached and floating cells and cell fragments) was assayed for caspase-cleaved CK18 as recommended by the manufacturer (Peviva). In some experiments, the release of total CK18 from cells to the culture medium was used as a marker for loss of membrane integrity (20). CK18 was assayed in untreated medium by using the M65-ELISA from Peviva. Western blot experiments showed that p53 WT and p53-null cells expressed the same levels of CK18 (data not shown).
Caspase-3/7 activity was measured by using a Caspase-Glo 3/7 kit from Promega. Nuclear fragmentation was assessed in air-dried fixed, acetone-methanol (2:1) fixed smears stained with ethidium bromide (5 ng/ml) for 5 min. Loss of cell viability was determined by MTT assay (Promega).
Preparation of Cytoplasts. Preparation of cytoplasts by density gradient centrifugation in the presence of cytochalasin B was conducted as described in ref. 21. The purity of the cytoplast preparation was determined by flow cytometry after resuspension in PBS containing digitonin (50 μg/ml) and labeling for 10 min with 5 μg/ml propidium iodide. Cytoplast preparations were treated with drugs as indicated, fixed and stained with antibodies to caspase-3 or caspase-cleaved CK18, and analyzed by flow cytometry. Cytoplasts were identified by lack of propidium iodide staining and were electronically gated.
Western Blot Analysis. Cell extract proteins were resolved by SDS/PAGE and transferred onto a polyvinylidene difluoride membrane for Western blotting. The antibodies that were used are as follows: anti-p53 (1:500; Santa Cruz Biotechnology), anti-mouse pro-caspase-12 (1:40; kindly provided by J. Yuan, Harvard Medical School, Boston), anti-GRP78 (1:1,000; Becton Dickinson), anti-GRP94 (1:1,000; Calbiochem), and anti-tubulin (1:1,000; Sigma-Aldrich). Tubulin was used as an internal standard for loading.
Flow Cytometric Analysis. Activation of caspase-3 was analyzed by using an antibody detecting the cleaved form of the enzyme (Cell Signaling Technology, Beverly, MA). CK18 cleavage was assessed by using the monoclonal antibody M30 (22). Cells and cytoplasts were fixed with 0.25% paraformaldehyde for 5 min, washed twice in PBS, and incubated for 30 min with primary antibody in the presence of digitonin for permeabilization, followed by incubation with a secondary FITC-conjugated antibody (DAKO A/S)for 1 h in the absence of digitonin. Staining for γ-H2AX was performed by using an antibody from Upstate Biotechnology (Lake Placid, NY) (used at a 1:50 dilution).
Immunofluorescence Staining. Cells were treated for 3 h with various compounds as indicated, fixed for 20 min with 4% paraformaldehyde, and permeabilized with 0.2% Nonidet P-40 for 5 min. After blocking with BSA, cells were incubated with the following primary antibodies: anti-p53 (Santa Cruz Biotechnology, 1:30), anti-cath-B (Oncogene Research; 1:20), or anti-cath-D (Oncogene Research; 1:10). Incubation with primary and FITC-conjugated secondary antibodies were both conducted for 1 h.
Acridine Orange (AO) Staining. Cells were exposed to 5 μg/ml of the lysosomotropic metachromatic fluorophore and weak base AO for 15 min (23). Cells were photographed 3 h after exposure to the various compounds.
Results
Assessment of p53-Dependent Apoptotic Responses to Multiple Drugs. The pair of HCT116 cell lines with WT and disrupted p53 genes described by Bunz and coworkers (19) was used for the present study. Cells were exposed to the 879 compounds of the NCI mechanistic set at concentrations of both 1 and 5 μM. Detergent was added to the culture medium after 24 h, and accumulation of caspase-cleaved cytokeratin-18 was determined by using the M30-ELISA assay (18). Compounds that induced CK18 cleavage were selected and retested at the lower effective concentration (1 or 5 μM; see Fig. 1 legend). A total of 175 compounds in the set were able to generate a signal >2-fold background of the assay (Fig. 1). The vast majority of these compounds elicited a stronger apoptotic response in HCT116 p53 WT cells, whereas only ≈20 compounds induced stronger apoptosis in the p53-null cells. However, all of the 175 drugs were capable of inducing an apoptotic response also in the p53-null cells.
Fig. 1.
Graphic representation of apoptosis induction by the compounds in the NCI mechanistic set. One hundred seventy-five compounds that induced caspase cleavage of CK18 in p53 WT or p53-null HCT116 cells at concentrations of 1 and 5 μM were selected in a first screen. In a second step, p53 WT and p53-null HCT116 cells were exposed to the selected 175 compounds in one experiment (for compounds that induced apoptosis at 1 μM, this concentration was used), and the data were plotted. CK18 caspase cleavage is expressed as U/L, the response in p53 WT cells is shown on the x axis, and the response of p53-null cells is shown on the y axis for each compound. Compounds studied in more detail are indicated.
The growth inhibitory effects of the agents in the mechanistic set have been examined for HCT116 (www.dtp.nci.nih.gov). A total of 337 compounds show GI50, or the dose needed to cause 50% growth inhibition, values of ≤5 μM on these cells; apoptosis was induced by 152 (45%) compounds. Twenty-three compounds induced apoptosis at ≤5 μM but nevertheless showed GI50 values of >5 μM. The signals obtained from these compounds in the M30-ELISA were generally low, suggesting that apoptosis was induced only in a fraction of the cells. A similar relationship between proliferation inhibition and apoptosis induction was obtained by using data from a commercially available MTT assay instead of the NCI data (unpublished data). We conclude that apoptosis is an important mechanism of action of compounds that are growth inhibitory in the NCI cell-based screen.
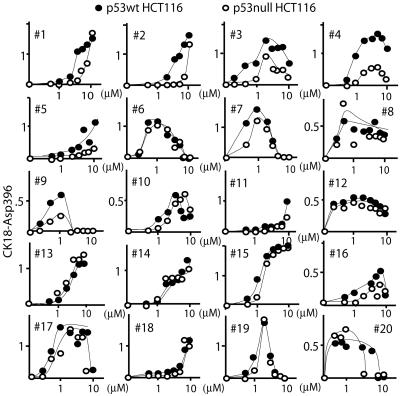
Compounds That Induce p53 Expression Do Not Necessarily Induce p53-Independent Apoptosis. From the initial screen, 20 compounds were selected as follows. Ten compounds induced p53-independent apoptosis at the lower effective concentration (at or close to the dashed line in Fig. 1), and 10 compounds were chosen with different degrees of p53 dependence. Dose–response curves showed that all 20 compounds induced apoptosis in p53-null cells (Fig. 2). Note that some compounds (e.g., 10, NSC13973; and 16, NSC313981) were judged as p53-dependent based on the response at 1 μM (Fig. 1) but induced p53-independent apoptosis at higher concentrations (Fig. 2). Apoptosis by five compounds was classified as “p53-supported” based on the preferential induction of caspase cleavage activity in p53 WT cells (for criteria, see Fig. 2 legend).
Fig. 2.
Dose–response curves of CK18 cleavage of 20 compounds selected from the mechanistic set. HCT116 cells (•, p53wt; ○, p53-null) were incubated with different concentrations of each compound, and the levels of caspase-cleaved CK18 was assayed in medium and cells after 24 h (expressed as U/L × 10-3). Five compounds induced a >2-fold difference in apoptosis in p53 WT and p53-null cells (the apoptotic response being defined as the area under the curve).
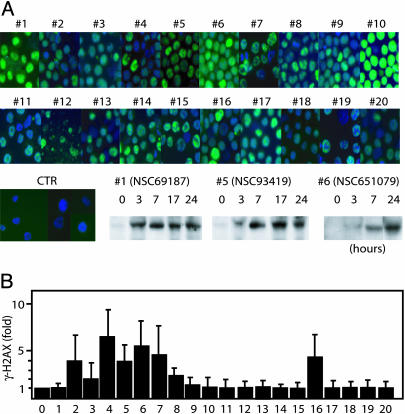
Induction of p53 protein expression was examined by immunofluorescence staining and Western blotting (Fig. 3A). p53 was strongly induced by 5 compounds (1, NSC69187; 5, NSC93419; 6, NSC651079; 10, NSC13973; and 12, NSC24819) and weakly by 12 compounds. No simple relationship was observed between induction of p53 expression and p53-supported apoptosis. This finding is exemplified by NSC651079 (compound 6) that induced strong p53 expression but nevertheless induced identical apoptotic responses in p53 WT and p53-null cells (Fig. 2).
Fig. 3.
Induction of p53 expression and DNA damage. (A) Induction of p53 expression. HCT116 p53 WT cells were treated for 3 h with the indicated compounds, fixed, and stained for p53. Cells were costained with 4′,6-diamidino-2-phenylindole, and overlays of both fluorescent signals are shown. In Bottom Left Center, Bottom Right Center, and Bottom Right, p53 expression was examined by using Western blotting at the indicated times of drug exposure. (B) Induction of DNA damage. HCT116/p53-null cells were treated for 3 h with the indicated compounds, fixed, and stained with an antibody to phosphorylated H2AX. Staining was quantified by flow cytometry and presented as increase compared with control.
To determine whether compounds that induce p53 are DNA damaging, we examined induction of phosphorylation of histone H2AX at serine-139 (γ-H2AX), an event associated with induction of double-strand DNA breaks (24). Eight of the 20 compounds induced γ-H2AX >2-fold above background of the assay by using HCT116/p53-null cells (Fig. 3B and Table 1). These eight compounds all induced p53, and four of eight preferentially induced apoptosis in p53 WT cells.
Table 1. Summary of the biological effects of selected compounds.
| Apoptosis supported by
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | Compound NSC no. | Compound name | p53* | BAX† | Induction of p53 | DNA damage | Cytoplasmic target | LMP‡ | ER stress§ |
| 1 | 69187 | 9-methoxy-ellipticine | + | + | ++ | – | – | – | – |
| 2 | 622627 | + | + | + | + | + | – | – | |
| 3 | 102811 | Formycin A | + | + | + | + | – | – | – |
| 4 | 72961 | 8-azaadenosine | + | + | + | + | – | – | – |
| 5 | 93419 | Steffimycin | + | + | ++ | + | – | – | – |
| 6 | 651079 | – | + | ++ | + | + | + | – | |
| 7 | 687852 | – | + | + | + | + | + | – | |
| 8 | 106408 | Anthramycin meth.-ether | – | + | + | + | – | – | – |
| 9 | 267461 | Nanaomycin | – | + | + | – | + | + | – |
| 10 | 13973 | Acridine yellow | – | + | ++ | – | + | – | – |
| 11 | 166381 | – | + | – | – | – | – | – | |
| 12 | 24819 | β-Peltatin A | – | + | ++ | – | – | – | – |
| 13 | 10010 | – | + | + | – | + | – | – | |
| 14 | 647889 | – | + | + | – | + | + | – | |
| 15 | 285116 | Siomycin A | – | + | + | – | + | + | + |
| 16 | 313981 | Gold Cl-3-ethylphosphine | – | + | + | + | + | + | – |
| 17 | 140377 | Arnebin | – | + | + | – | + | – | – |
| 18 | 157389 | – | + | – | – | + | – | – | |
| 19 | 85236 | Helenalin | – | + | – | – | + | + | – |
| 20 | 24818 | Podophyllotoxin | – | + | + | – | – | – | – |
p53-supported apoptosis was defined as 2-fold higher CK18 cleavage in HCT116 WT cells (area under curve in Fig. 2)
Drugs that showed Bax-supported apoptosis induced <10% caspase activation in HT116 Bax (null) cells compared with HCT116 control cells
See Fig. 5 for details
ER, endoplasmic reticulum. Induction of GRP78 in HCT116 p53 WT cells; cleavage of caspase-12 in mouse CT51 cells
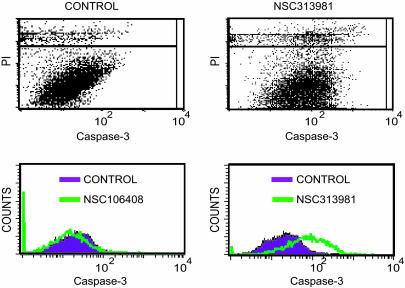
Demonstration of Cytoplasmic Targets. The majority of the selected 20 proapoptotic compounds did not induce DNA damage detected by the γ-H2AX assay. To examine whether these drugs have nonnuclear targets, we enucleated HCT116 p53 WT cells by centrifugation in the presence of cytochalasin B and treated the enucleated cells with drugs. Caspase activation was analyzed after 16 h of treatment by using flow cytometry. Examples of results obtained are shown in Fig. 4. A total of 12 compounds were capable of inducing caspase activity in enucleated cells (Table 1). Three of the compounds that induced DNA damage were also capable of triggering caspase activation in enucleated cells.
Fig. 4.
Induction of caspase-3 activation in enucleated cells. HCT116 p53 WT cells were enucleated by centrifugation in the presence of cytochalasin B. Cytoplasts were incubated with NSC106408 (drug 8) or NSC313981 (drug 16) for 16 h, fixed, and incubated with an antibody to activated caspase-3. After staining with propidium iodide, cytoplasts were gated during fluorescence-activated cell sorter analysis (see Upper Right for NSC313981). Some of the compounds used were fluorescent in the FL1 channel and disturbed the analysis. Caspase cleavage of CK18 was analyzed by using the M30 antibody and a rhodamine-conjugated secondary antibody in these instances. Results from all experiments are shown in Table 1.
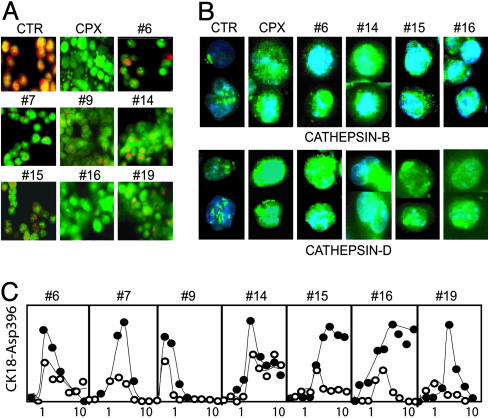
Induction of LMP by Drugs That Activate p53-Independent Apoptosis. Some forms of apoptosis have been found to be associated with a lysosomal pathway (25, 26). Partial permeabilization of lysosome membranes results in relocation of lysosomal proteases to the cytosol where they induce apoptotic signaling (27). The occurrence of LMP was investigated by using vital staining with AO. AO preferentially accumulates in secondary lysosomes, resulting in red fluorescence after excitation with blue light (28) (Fig. 5A Top Left). Relocation of AO from the lysosomes to the cytosol results in green fluorescence (28, 29). The quinolone antibiotic ciprofloxacin is known to accumulate in lysosomes, to induce LMP (30), and was found to induce the expected shift from red to green fluorescence (Fig. 5A). We found that seven compounds induced LMP in HCT116/p53-null cells at concentrations that induced maximal apoptosis (Fig. 5A and Table 1). These seven compounds all induced caspase-3 activation in enucleated cells (Table 1).
Fig. 5.
Induction of LMP. (A) HCT116/p53-null cells were incubated with AO, washed, and treated with the indicated compounds for 3 h. Untreated cells show red fluorescence due to the high concentration of AO in lysosomes. An increase in green fluorescence is observed when AO is released from lysosomes into the cytosol. (B) Release of cathepsin B and cathepsin D from lysosomes. HCT116/p53-null cells were treated with the indicated drugs, fixed, and subjected to immunofluorescence staining by using antibodies to cathepsin B and cathepsin D. (C) Inhibition of apoptosis by pepstatin A. HCT116/p53-null cells were incubated with each compound for 24 h in the presence (○) or absence (•) of pepstatin A (100 μM), and the levels of caspase-cleaved CK18 were assayed in medium and cells.
Staining of HCT116/p53-null cells with antibodies to the cysteine protease cathepsin B and the aspartyl protease cathepsin D resulted in lysosomal staining (Fig. 5B). Treatment with ciprofloxacin and with the LMP-inducing compounds resulted in relocation of cathepsin B to the cytosol. Cathepsin D also showed a cytoplasmic staining after treatment with these drugs. In addition, cathepsin D-positive cytoplasmic aggregates were also observed in drug-treated cells (Fig. 5B). The aspartyl protease inhibitor pepstatin A, which inhibits lysosomal cathepsin D, partially inhibited apoptosis by the seven LMP-inducing compounds (Fig. 5C).
To determine whether agents that induce LMP induce a mitochondrial apoptotic pathway, we used a HCT116 Bax-null cell line (31). We found that none of the 20 compounds studied, including the ones that induce LMP, caused caspase-3 activation in HCT116 cells defective in Bax (Table 1).
Shift from Apoptosis to Necrosis at High Drug Concentrations of LMP-Inducing Drugs. For 10 compounds, CK18 caspase cleavage was found to decline at higher drug concentrations (Fig. 2). Cell viability, measured by the MTT assay, did not, however, increase at higher drug concentrations (data not shown). Five of seven compounds that induce LMP showed this type of dose–response curve. The basis for this phenomenon was examined in detail for drug 9 (NSC267461; nanaomycin). The highest levels of CK18 cleavage were observed at 0.5 μM, whereas no cleavage was observed at >5 μM (Fig. 6A). Similar patterns were observed for caspase-3/7 activity and nuclear fragmentation (Fig. 6 B and C). In contrast, cell viability measurements with the MTT assay showed an increasing toxicity at higher concentration (Fig. 6D). At drug concentrations >4 μM, intracellular CK18 protein was released into the medium (Fig. 6E), and membrane integrity was lost (Fig. 6F). Altogether, these data suggest that NSC267461 triggers an apoptotic response at low concentrations and that apoptosis shifts to necrosis at higher concentrations.
Fig. 6.
NCS267461 induces necrosis at higher concentrations. (A) Dose–response of caspase cleavage of CK18 in HCT116/p53-null cells over 24 h. The total levels of cleaved CK18 were measured in cells and medium after addition of nonionic detergent to the culture medium (A450 readings in ELISA). Negative values (lower levels in drug-treated compared with untreated cells) were observed at high drug concentrations but were displayed as zero for simplicity. (B) Dose–response of caspase-3/7 cleavage in HCT116/p53-null cells over 24 h (arbitrary units). (C) Nuclear fragmentation of HCT116/p53-null cells induced by different concentrations of NCS267461 over 24 h (percent fragmented cells). (D) Loss of cell viability after 48 h incubation with different concentrations of NCS267461 measured by the MTT assay. (E) Higher concentrations of NCS267461 induce release of CK18 into the tissue culture medium. The levels of CK18 were measured in the culture medium by the M65-ELISA assay at 24 h (no detergent added; A450 readings in ELISA). Values of untreated control cells were subtracted. (F) Loss of membrane integrity induced by increasing concentrations of NCS267461. Cells were stained by Trypan blue after 24 h of incubation (percent positive cells).
Discussion
We identified 175 compounds in the NCI mechanistic set that induce apoptosis at ≤5 μM. None of these compounds was totally dependent on p53 for induction of apoptosis (Fig. 1). Five selected compounds were defined as being p53-supported based on preferential induction of apoptosis in p53 WT cells. Four of these p53-supported compounds induced DNA damage. This association was expected because sensors of DNA damage, such as DNA-PK and ataxia telangiectasia mutated kinase and ataxia telangiectasia mutated-Rad3-related kinase, transmit signals that lead to the activation of the p53 pathway (32). However, apoptosis induction by four other compounds that induced DNA damage and p53 expression was only weakly dependent or even independent of p53. An illustrative example is compound 6 (NSC651079) that induced DNA damage and strong p53 expression, but nevertheless generated identical apoptotic responses in p53 WT and p53-null cells. Similarly, compound 7 (NSC687852) induced DNA damage and p53 expression, but only a small difference in dose–response in p53 WT and p53-null cells. These results clearly show that p53-inducing drugs do not necessarily induce p53-dependent apoptosis.
The majority (11 of 15) of the selected compounds that induced p53-independent apoptosis were capable of activating caspase-3 in enucleated cells. Further experiments showed that seven of the compounds with a nonnuclear target induced LMP. The mechanisms of induction of LMP by these compounds are unknown and are likely to be diverse. A number of different mechanisms leading to LMP have been described, including a p53-activated pathway (16), increased cellular generation of reactive oxygen species, and a sphingosine mediated mechanism (reviewed in ref. 26). Lysosomotropic agents such as ciprofloxacin (30) and hydroxychloroquine (33) also induce LMP. LMP is known to cause translocation of lysosomal proteases to the cytosol (26, 34). We here documented release of cathepsin B and cathepsin D from lysosomes after treatment with LMP-inducing drugs. Cathepsin B has been shown to be necessary for tumor cell apoptosis induced by tumor necrosis factor in WEHI-S fibrosarcoma cells (15). Cathepsin D mediates apoptosis in other models (35–37), and injection of cathepsin D induces caspase-dependent apoptosis of fibroblasts (38). Cathepsin D belongs to the comparatively small family of aspartyl proteases, and we examined the effect of the aspartyl protease inhibitor pepstatin A on apoptosis induced by the LMP-inducing compounds. A varying degree of inhibition was observed, suggesting that cathepsin D released from the lysosomes is involved in induction of the apoptotic response. Cathepsin D has been demonstrated to induce activation of Bax (37). Consistent with a role of LMP and cathepsin D translocation upstream from activation of Bax, we found that Bax-deficient HCT116 cells did not undergo apoptosis (Table 1).
Three agents were found to induce both DNA damage and LMP. It is possible that LMP is the primary event caused by these agents and that DNA damage is a secondary event. Alternatively, these compounds induce DNA damage and loss of lysosomal integrity in parallel. Regardless, LMP induction offers an explanation to the phenomenon of p53-independent apoptosis by these drugs. In addition to the drugs that induce both DNA damage and LMP, we found that one compound (NSC285116, siomycin A) was capable of inducing both LMP and endoplasmic reticulum stress (Table 1 and data not shown). Apoptosis is a complex biological phenomenon, probably developed during evolution as a defense strategy against infections (39), to remove damaged cells (40), and to remove cells during tissue remodeling (41). Considering the wide variety of apoptotic-sensing mechanisms and the complex network of apoptotic-signaling pathways in mammalian cells (25), it is not surprising that chemical agents at concentrations of 5–10 μM will trigger more than one apoptotic pathway. Despite reports that cytotoxic drugs often induce diverse biological responses (21, 42, 43), such drugs are generally classified according to a major proposed mechanism of action. Although clearly useful, this type of classification may also be misleading.
Caspase cleavage of CK18 was found to decrease with increasing concentrations in five of seven of the LMP-inducing compounds. Decreased caspase activity was paralleled by decreased cell viability at increasing drug concentrations of all these compounds (data not shown). At high drug concentrations, membrane integrity was lost, suggesting a shift from apoptosis to necrosis. It is a well known fact that many drugs produce apoptosis at low doses and necrosis at higher doses (29, 30), and we have previously shown that induction of strong LMP by a lysosomotropic detergent will induce necrosis (44). It was, however, slightly surprising to find that so many compounds induced necrosis at concentrations at or below 10 μM, and it is possible that induction of necrosis by anticancer drugs is more commonly occurring than generally believed.
Caspase activation was induced by ≈45% of the compounds in the NCI mechanistic set that induce growth arrest at ≤5 μM. The M30-ELISA assay is capable of detecting apoptosis in <5% of the cells in a well of a 96-well plate and is therefore expected to be more sensitive compared with a GI50 determination by MTT. Twenty-three compounds were identified that did not reduce cell growth by 50% but scored positive in the apoptosis assay. However, even after taking the higher sensitivity of the M30-ELISA assay in to account, it is clear that induction of apoptosis is an important mechanism of action of compounds that score positive in the NCI cell-based screen. It is important to point out that our data were obtained by using a 24-h apoptosis assay and that changing the experimental conditions is likely to generate different results. For example, it has been demonstrated that treatment with low concentrations of the DNA-damaging agent adriamycin induces a stronger apoptotic response in p53-deficient cells. This apoptosis appears to be a secondary consequence of a failure to induce a p53/p21 dependent cell cycle checkpoint (45).
The development of anticancer agents with different modes of action is of key importance in overcoming clinical therapy resistance. The lysosomal cell death pathway has been suggested to represent a potential cancer target (46), but anticancer drugs that trigger this pathway are not in clinical use. Considering the high frequency of p53 mutations in human cancer cells, the demonstration that LMP is potentially a common mechanism of action of p53-independent drugs is interesting in terms of future drug development.
Acknowledgments
We thank the Developmental Therapeutics Program of the NCI for supplying the mechanistic set and Dr. Bert Vogelstein for generously sending the HCT116 cell lines. This work was supported by the Swedish Cancer Society (Cancerfonden) and the King Gustaf V Jubilee Foundation.
Author contributions: H.E., M.B., and J.C. performed research; H.E., M.B., M.C.S., and S.L. analyzed data; U.B., M.C.S., and S.L. designed research; and U.B., M.C.S., and S.L. wrote the paper.
Abbreviations: AO, acridine orange; CK, cytokeratin; LMP, lysosomal membrane permeabilization; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; NCI, National Cancer Institute.
References
- 1.Holbeck, S. L. (2004) Eur. J. Cancer 40, 785-793. [DOI] [PubMed] [Google Scholar]
- 2.Weinstein, J. N., Myers, T. G., O'Connor, P. M., Friend, S. H., Fornace, A. J., Jr., Kohn, K. W., Fojo, T., Bates, S. E., Rubinstein, L. V., Anderson, N. L., et al. (1997) Science 275, 343-349. [DOI] [PubMed] [Google Scholar]
- 3.Shi, L. M., Fan, Y., Lee, J. K., Waltham, M., Andrews, D. T., Scherf, U., Paull, K. D. & Weinstein, J. N. (2000) J. Chem. Inf. Comput. Sci. 40, 367-379. [DOI] [PubMed] [Google Scholar]
- 4.Beroud, C. & Soussi, T. (1998) Nucleic Acids Res. 26, 200-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gasco, M. & Crook, T. (2003) Drug Resist. Updat. 6, 323-328. [DOI] [PubMed] [Google Scholar]
- 6.Wahl, A. F., Donaldson, K. L., Fairchild, C., Lee, F. Y., Foster, S. A., Demers, G. W. & Galloway, D. A. (1996) Nat. Med. 2, 72-79. [DOI] [PubMed] [Google Scholar]
- 7.Del Bufalo, D., Biroccio, A., Soddu, S., Laudonio, N., D'Angelo, C., Sacchi, A. & Zupi, G. (1996) J. Clin. Invest. 98, 1165-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi, L. M., Myers, T. G., Fan, Y., O'Connor, P. M., Paull, K. D., Friend, S. H. & Weinstein, J. N. (1998) Mol. Pharmacol. 53, 241-251. [DOI] [PubMed] [Google Scholar]
- 9.Marchini, S., Ciro, M. & Broggini, M. (1999) Apoptosis 4, 39-45. [DOI] [PubMed] [Google Scholar]
- 10.Pepper, C., Thomas, A., Hoy, T., Milligan, D., Bentley, P. & Fegan, C. (2003) Blood 101, 2454-2460. [DOI] [PubMed] [Google Scholar]
- 11.Kuroda, J., Kimura, S., Segawa, H., Sato, K., Matsumoto, S., Nogawa, M., Yuasa, T., Kobayashi, Y., Yoshikawa, T., Ottmann, O. G. & Maekawa, T. (2004) Cancer Sci. 95, 186-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberg, K. & Öllinger, K. (1998) Am. J. Pathol. 152, 1151-1156. [PMC free article] [PubMed] [Google Scholar]
- 13.Brunk, U. T. & Svensson, I. (1999) Redox Rep. 4, 3-11. [DOI] [PubMed] [Google Scholar]
- 14.Guicciardi, M. E., Deussing, J., Miyoshi, H., Bronk, S. F., Svingen, P. A., Peters, C., Kaufmann, S. H. & Gores, G. J. (2000) J. Clin. Invest. 106, 1127-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foghsgaard, L., Wissing, D., Mauch, D., Lademann, U., Bastholm, L., Boes, M., Elling, F., Leist, M. & Jäättelä, M. (2001) J. Cell Biol. 153, 999-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan, X.-M., Li, W., Dalen, H., Lotem, J., Kama, R., Sachs, L. & Brunk, U. T. (2002) Proc. Natl. Acad. Sci. USA 99, 6286-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fehrenbacher, N., Gyrd-Hansen, M., Poulsen, B., Felbor, U., Kallunki, T., Boes, M., Weber, E., Leist, M. & Jäättelä, M. (2004) Cancer Res. 64, 5301-5310. [DOI] [PubMed] [Google Scholar]
- 18.Hägg, M., Biven, K., Ueno, T., Rydlander, L., Björklund, P., Wiman, K. G., Shoshan, M. & Linder, S. (2002) Invest. New Drugs 20, 253-259. [DOI] [PubMed] [Google Scholar]
- 19.Bunz, F., Dutriaux, A., Lengauer, C., Waldman, T., Zhou, S., Brown, J. P., Sedivy, J. M., Kinzler, K. W. & Vogelstein, B. (1998) Science 282, 1497-1501. [DOI] [PubMed] [Google Scholar]
- 20.Kramer, G., Erdal, H., Mertens, H. J., Nap, M., Mauermann, J., Steiner, G., Marberger, M., Biven, K., Shoshan, M. C. & Linder, S. (2004) Cancer Res. 64, 1751-1756. [DOI] [PubMed] [Google Scholar]
- 21.Mandic, A., Hansson, J., Linder, S. & Shoshan, M. C. (2003) J. Biol. Chem. 278, 9100-9106. [DOI] [PubMed] [Google Scholar]
- 22.Leers, M. P., Kolgen, W., Björklund, V., Bergman, T., Tribbick, G., Persson, B., Björklund, P., Ramaekers, F. C., Björklund, B., Nap, M., Jornvall, H. & Schutte, B. (1999) J. Pathol. 187, 567-572. [DOI] [PubMed] [Google Scholar]
- 23.Kågedal, K., Zhao, M., Svensson, I. & Brunk, U. T. (2001) Biochem. J. 359, 335-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. (1998) J. Biol. Chem. 273, 5858-5868. [DOI] [PubMed] [Google Scholar]
- 25.Ferri, K. F. & Kroemer, G. (2001) Nat. Cell Biol. 3, E255-E263. [DOI] [PubMed] [Google Scholar]
- 26.Guicciardi, M. E., Leist, M. & Gores, G. J. (2004) Oncogene 23, 2881-2890. [DOI] [PubMed] [Google Scholar]
- 27.Leist, M. & Jäättelä, M. (2001) Cell Death Differ. 8, 324-326. [DOI] [PubMed] [Google Scholar]
- 28.Robbins, E. & Marcus, P. I. (1963) J. Cell Biol. 18, 237-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olsson, G. M., Rungby, J., Rundquist, I. & Brunk, U. T. (1989) Virchows Arch B Cell Pathol. Incl. Mol. Pathol. 56, 263-269. [DOI] [PubMed] [Google Scholar]
- 30.Boya, P., Andreau, K., Poncet, D., Zamzami, N., Perfettini, J. L., Metivier, D., Ojcius, D. M., Jäättelä, M. & Kroemer, G. (2003) J. Exp. Med. 197, 1323-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, L., Yu, J., Park, B. H., Kinzler, K. W. & Vogelstein, B. (2000) Science 290, 989-992. [DOI] [PubMed] [Google Scholar]
- 32.Lakin, N. D. & Jackson, S. P. (1999) Oncogene 18, 7644-7655. [DOI] [PubMed] [Google Scholar]
- 33.Boya, P., Gonzalez-Polo, R. A., Poncet, D., Andreau, K., Vieira, H. L., Roumier, T., Perfettini, J. L. & Kroemer, G. (2003) Oncogene 22, 3927-3936. [DOI] [PubMed] [Google Scholar]
- 34.Jäättelä, M. (2004) Oncogene 23, 2746-2756. [DOI] [PubMed] [Google Scholar]
- 35.Deiss, L. P., Galinka, H., Berissi, H., Cohen, O. & Kimchi, A. (1996) EMBO J. 15, 3861-3870. [PMC free article] [PubMed] [Google Scholar]
- 36.Kågedal, K., Johansson, U. & Öllinger, K. (2001) FASEB J. 15, 1592-1594. [DOI] [PubMed] [Google Scholar]
- 37.Bidere, N., Lorenzo, H. K., Carmona, S., Laforge, M., Harper, F., Dumont, C. & Senik, A. (2003) J. Biol. Chem. 278, 31401-31411. [DOI] [PubMed] [Google Scholar]
- 38.Roberg, K., Kågedal, K. & Öllinger, K. (2002) Am. J. Pathol. 161, 89-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.James, E. R. & Green, D. R. (2002) Cell Death Differ. 9, 355-357. [DOI] [PubMed] [Google Scholar]
- 40.Wyllie, A. H. (1997) Eur J. Cell Biol. 73, 189-197. [PubMed] [Google Scholar]
- 41.Horvitz, H. R. (1999) Cancer Res. 59, Suppl. 7, S1701-S1706. [Google Scholar]
- 42.Robertson, J. D., Gogvadze, V., Zhivotovsky, B. & Orrenius, S. (2000) J. Biol. Chem. 275, 32438-32443. [DOI] [PubMed] [Google Scholar]
- 43.Kidd, J. F., Pilkington, M. F., Schell, M. J., Fogarty, K. E., Skepper, J. N., Taylor, C. W. & Thorn, P. (2002) J. Biol. Chem. 277, 6504-6510. [DOI] [PubMed] [Google Scholar]
- 44.Li, W., Yuan, X., Nordgren, G., Dalen, H., Dubowchik, G. M., Firestone, R. A. & Brunk, U. T. (2000) FEBS Lett 470, 35-39. [DOI] [PubMed] [Google Scholar]
- 45.Bunz, F., Hwang, P. M., Torrance, C., Waldman, T., Zhang, Y., Dillehay, L., Williams, J., Lengauer, C., Kinzler, K. W. & Vogelstein, B. (1999) J. Clin. Invest. 104, 263-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jäättelä, M. (2002) Ann. Med. 34, 480-488. [DOI] [PubMed] [Google Scholar]