ABSTRACT
Significant effort has gone into assessing the fate and removal of viruses, bacteria, and protozoan parasites during wastewater treatment to provide data addressing potential health risks associated with reuse options. Comparatively less is known about the fate of parasitic worm species ova in these complex systems. It is largely assumed that these helminths settle, are removed with the sludge, and consequently represent a relatively low risk for wastewater reuse applications. However, helminths are a highly diverse group of organisms that display a wide range of physical properties that complicate the application of a single treatment for helminth reduction during wastewater treatment. Moreover, their diverse biological and physical properties make some ova highly resistant to both disinfection (i.e., with chlorine or UV treatment) and physical removal (settling) through the wastewater treatment train, indicating that there may be reason to broaden the scope of our investigations into whether parasitic worm eggs can be identified in treated wastewater. The ubiquitous human parasitic nematode Enterobius vermicularis (pinworm) produces small, buoyant ova. Utilizing a novel diagnostic quantitative PCR (qPCR), this study monitored E. vermicularis presence at two full-scale wastewater treatment plants over the course of 8 months and demonstrated incomplete physical removal of E. vermicularis ova through tertiary treatment, with removal efficiencies approximating only 0.5 and 1.6 log10 at the two wastewater treatment plants based on qPCR. These findings demonstrate the need for more-diverse surrogates of helminthic ova to fully assess treatment performance with respect to reclaimed wastewaters.
IMPORTANCE Helminths, despite being a diverse and environmentally resistant class of pathogens, are often underestimated and ignored when treatment performance at modern wastewater treatment plants is considered. A one-size-fits-all surrogate for removal of helminth ova may be inappropriate to adequately assess risk and ensure public safety when treated and partially treated wastewaters are encountered. This study argues for the use of human pinworm as a conservative indicator of the presence of helminth ova due to its small size, buoyancy, prevalence in humans, and environmental resistance.
KEYWORDS: Enterobius, helminth, monitoring, ova, qPCR, wastewater
INTRODUCTION
Water serves as a medium for the transmission of many species of parasitic protozoa and helminths (1, 2). Contamination of water sources by parasites (and other causative agents of gastrointestinal illness) often occurs when sewage containing human waste is improperly treated (2, 3). Drinking, irrigation, recreation, and numerous industrial, commercial, and residential applications for water can lead to accidental ingestion, inhalation, or exposure to infectious stages of parasites (4, 5). Many species of nematodes and cestodes indirectly use water as a means of transmission, and infiltration of sewage into waterways and direct reuse of treated or partially treated sewage are common mechanisms by which parasites exploit water to complete their life cycle (4, 6–9). Reuse options of concern may include land application of biosolids and wastewater and additionally the use of treated or partially treated wastewater for toilet flushing, irrigation, or clothes washing (7–11).
Within the context of treated wastewater for reuse purposes, significant effort has gone into characterizing the risk posed to human health from viral, bacterial, and protozoan parasites that may persist through the treatment process and remain viable/infectious (12, 13). However, we understand very little about how treatment of wastewater affects the viability and infectivity of the vast majority of helminths, making it difficult to accurately assess the risk posed by these parasites in water sources contaminated with human waste (14, 15). Surveys to determine the presence and diversity of helminth ova in wastewater and biosolid material have been conducted, predominantly by using microscopic examination (16, 17). These studies represent a logical starting point for assessing the risk of helminth persistence (10, 18). A study by Schwartzbrod and Banas (19), conducted in France, reported that 78% of activated sludge samples analyzed by microscopy contained either parasitic nematode or cestode ova. Genera that were identified included Trichuris, Capillaria, Hymenolepis, Ascaris, and Toxocara, helminth parasites relevant to human or animal health (19). A similar study conducted in Tunisia reported Ascaris spp., Enterobius vermicularis, Hymenolepis nana, and Taenia spp. in raw influent and final wastewater effluent from treatment plants utilizing various secondary and tertiary treatment processes, including activated sludge and waste stabilization ponds, processes that closely parallel wastewater treatment approaches of many facilities in developed countries (17).
The presence of helminthic ova, particularly Ascaris lumbricoides (the traditional reference parasite) along with its surrogate Ascaris suum, has been assayed in biosolids and wastewater effluent by a variety of methods, including microscopic examination and various PCR and quantitative PCR (qPCR) methods (20–23). Because Ascaris is a ubiquitous parasite (infecting ∼1.4 billion people worldwide) and has a direct life cycle that facilitates transmission by water, food, and person-to-person contact, it remains a logical reference pathogen for worm presence in sewage sludge (24, 25). However, parasitic worms are incredibly diverse, and the physical egg properties of the various species of helminths that are considered relevant in a wastewater context differ greatly, likely leading to differential removals during wastewater treatment (18), challenging the assertion that Ascaris spp. represent the best reference helminths for evaluating wastewater treatment removal of parasitic worm ova. Thus, it is important to understand how the diversity of helminth eggs and their various physical properties affect their removal during the wastewater treatment process.
Removing parasitic ova from wastewater is important in water reuse to mitigate risks associated with these pathogens. Although the behavior of bacteria, viruses, and protozoan parasites have traditionally been the focus of disinfection processes during wastewater treatment, numerous studies have demonstrated that helminthic ova actually represent the pathogen class most resistant to both chemical (i.e., chlorine) and UV disinfection processes. Brownell and Nelson (26) demonstrated that a low-pressure (i.e., 255-nm) UV dose of 500 mJ/cm2 resulted in less than a 0.5-log10 inactivation of A. suum ova. Remarkably, a dose as high as 4,000 mJ/cm2 yielded only an ∼2-log10 inactivation of A. suum ova. The UV dose required to achieve a 1-log10 inactivation of A. suum is four times higher than the dose required to inactivate adenoviruses (26). Moreover, free chlorine is virtually ineffective at reducing Ascaris species viability (27). The challenges related to inactivation of Ascaris species ova demonstrate the importance of physically removing these parasites from the wastewater matrix prior to its reuse.
Enterobius vermicularis is a species of parasitic nematode of the family Oxyuridae that infects humans (28). It has a direct life cycle and is transmitted when a gravid female worm exits the anus and deposits ova on the perianal skin of an infected individual, usually while the host is sleeping. The ova become infectious after a few hours, and infected individuals (especially children) scratch the area and subsequently ingest the ova, facilitating hatching and maturation of adult worms in the small intestine (29). E. vermicularis has a high global prevalence and is not limited to tropical regions, making it more relevant to developed nations than many traditionally studied helminths (29, 30). The ova have a lower specific gravity than ova of A. lumbricoides, Trichuris vulpis, Trichuris trichiura, and Ancylostoma caninum in zinc sulfate and are also smaller than the ova of many other helminths (48 to 60 μm by 20 to 35 μm wide) (30, 31). They are also extremely “sticky,” possessing the ability to bind to various biotic or abiotic compounds that may alter the buoyant density and, thus, removal performance of the ova during water treatment. These properties make E. vermicularis one of the most likely among the helminths to persist throughout the wastewater treatment process. Moreover, studies have demonstrated that E. vermicularis ova can be aerosolized; their potential for disease transmission may be increased if they are not effectively inactivated by traditional wastewater treatment processes before the reuse of wastewater (32). For these reasons, E. vermicularis may be a useful surrogate for assessing the presence of helminth ova in treated or partially treated wastewater.
Our understanding of E. vermicularis occurrence through sewage treatment and in water is limited. To date, most parasitological diagnoses both in clinics and for sewage have relied upon microscopic identification of ova or worms (33). While microscopic diagnosis is advantageous in clinical circumstances, it can be challenging in a wastewater matrix due to the presence of free-living nematodes that can confound accurate identification (33, 34). Therefore, the objective of our study was 2-fold: (i) to improve the identification and detection of this helminth parasite in complex samples by developing a novel quantitative PCR assay and (ii) to estimate E. vermicularis physical removal throughout the wastewater treatment process in two full-scale tertiary treatment systems to consider its usefulness as a potential surrogate for the presence of helminths in wastewater.
RESULTS
Enterobius vermicularis 5S rDNA qPCR test development. (i) In silico analysis of the 5S rRNA gene.
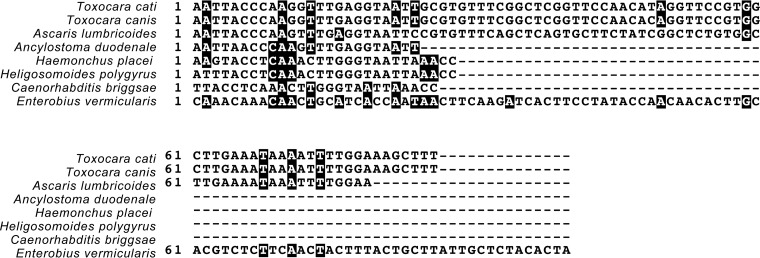
The 5S rRNA gene target was selected by comparing the 5S ribosomal gene and spacer region gene sequence available in the NCBI BLASTn database for E. vermicularis first to the entire NCBI nucleotide collection. This was performed to ensure limited or no cross-reactivity to any other organisms in the wastewater. The results of this BLAST search showed that the 5S ribosomal gene and spacer region were similar to the 5S ribosomal gene of E. vermicularis and other parasitic worms within the phylum Nematoda. However, only the SL-1 transspling region within the spacer region was conserved among other nematodes. Again, using BLAST, the 5S ribosomal DNA (rDNA) region was compared to only those of organisms within the phylum Nematoda expected in wastewater, as illustrated in Fig. 1.
FIG 1.
Alignment of the 5S rRNA gene sequences of E. vermicularis to other helminths. The 5S amplicon region of E. vermicularis was aligned with a number of relevant helminths, which may be present in wastewater. Black shading indicates a >50% sequence similarity to the E. vermicularis 5S rRNA sequence.
(ii) E. vermicularis assay sensitivity and specificity.
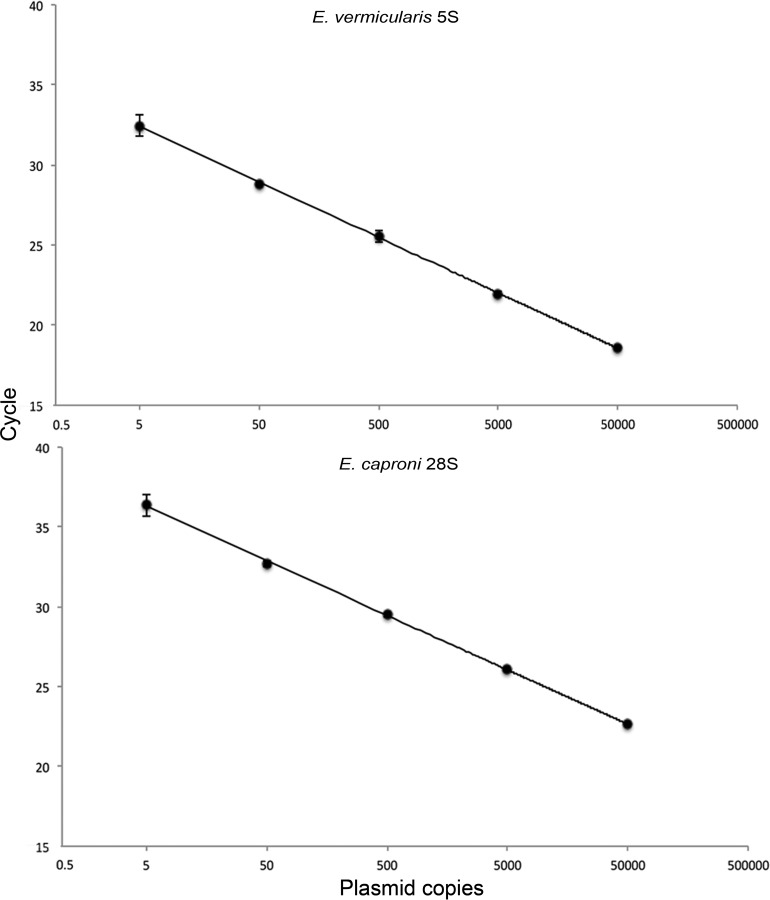
The 95% limit of detection (LOD95) of the E. vermicularis qPCR detection assay was 1.5 gene copies per reaction (upper limit, 2.9; lower limit, 0.8). The average efficiency across 10 standard curves was 92.8% (population standard deviation [σx̄] = 6.93), and the average slope of the standard curve was −3.47 (σx̄ = 0.198). All reactions had correlation coefficients (R2) of 0.99 (Fig. 2, top).
FIG 2.
Standard curves of the E. vermicularis and E. caproni qPCR assays. Plasmid standards were run at 50,000, 5,000, 500, 50, and 5 copies in triplicate. The standard curve is a representative of 10 curves performed independently. Error bars represent standard deviations of the means.
The specificity of the assay for E. vermicularis was tested within the wastewater matrix by performing 15 DNA extractions (concentrated wastewater after grit removal) and attempting to amplify a region of E. vermicularis 5S rDNA using endpoint PCR with primers 5S_fwd and 5S_rev (Table 1). These amplicons were cloned, sequenced, and analyzed using BLASTn, and all had 100% nucleotide identity to E. vermicularis 5S rDNA (Fig. 3).
TABLE 1.
Primer, probe, and inhibition control synthetic gene sequences
| Primer, probe, or control gene name | Sequence (5′–3′)a |
|---|---|
| E. vermicularis 5S_fwd | CAAACAACTGCATCACCAATAAC |
| E. vermicularis 5S_rev | AGTGTAGAGCAATAAGCAGTAAAG |
| E. vermicularis 5S probe | FAM-TACCAACAACACTTGCACGTCTCTTCA-IBFQ |
| E. caproni 28S_fwd | TAGGCAATGTGGTGTTCAGG |
| E. caproni 28S_rev | CATAGAGGGTGAAAGGCCC |
| E. caproni 28S probe | FAM-AGGACTTAGGGTGGAGCAGTATCCC-IBFQ |
| Inhib Con_fwd | GGTTACGTTACCGATTGTGTTAT |
| Inhib Con_rev | GCACGGAGAAGTATTCTTGATATAG |
| Inhibition control probe | JOE-TCGCATACTATGACCGAGACCCTGT-IBFQ |
| Inhibition control sequence | CCAATGCAATGGCTAACACAATATTATATTTTAAAGAATGAATATCATAAGACCATGCTAGCGTATGATAATGGATCTTGGACATAAGATATAGTTCTTATGAAGAGGCAAAAAAACACGATGATGGACTTATCATTATTTCTATATTCTTACAAGTATTGGATCATAA |
FAM, 6-carboxyfluorescein; IBFQ, Iowa black fluorescent quencher; JOE, 6′-carboxy-4′,5′-dichloro-2′,7′-dimethoxyfluorescein, succinimidyl ester.
FIG 3.
Sequences of the E. vermicularis 5S rRNA gene clones, amplified from raw wastewater aligned to the E. vermicularis 5S rRNA sequence (GenBank accession number AB221533.1).
(iii) DNA extraction controls.
Echinostoma caproni ova were used as a DNA extraction control for helminthic ova. E. caproni was selected because we did not expect to find naturally occurring E. caproni ova or larval parasites in wastewater, due to the life history of the parasite. Furthermore, E. caproni is a digenean trematode and thus is phylogenetically distinct from E. vermicularis, making it simple to design a specific assay that does not cross-react with E. vermicularis. Nonetheless, to test these assertions, 15 nonspiked post-grit removal wastewater samples from May 2015 to September 2015 were tested for the presence of naturally occurring E. caproni using the qPCR assay; no naturally occurring E. caproni was detected. The E. caproni primers and probe were also tested against the E. vermicularis 5S rDNA plasmid and vice versa. No cross-reactivity was observed between the two assays with any of the samples examined (see Fig. S1 in the supplemental material).
DNA losses were observed during the lysis and extraction process. Recoveries of spiked E. caproni ova ranged from only 13.8% to 0.6% (Table 2), demonstrating the robustness of helminthic ova to physical DNA extraction methods. We evaluated the effects of freeze-thawing and proteinase K lysis methods to improve recoveries; however, we saw no improvement despite these harsh conditions (Table 3). However, the data demonstrate that there was no difference in lysis efficacy between the influent post-grit removal matrix and the UV-treated effluent matrix. Inhibition was detected in only one sample, that taken on 15 June 2015 from the Bonnybrook treatment plant; the DNA extraction for this sample was repeated, and the subsequent extraction was uninhibited.
TABLE 2.
Average E. caproni ovum recoveriesa
| Date of sampling (mo/day/yr) or additional samples after matrix spikes | Plant | Treatment stage | Avg recovery (%) | Maximum recovery (%) | Minimum recovery (%) |
|---|---|---|---|---|---|
| 10/19/15 | Pine Creek | Post-grit removal | 2.4 | 3.9 | 1.7 |
| Post-UV | 1.0 | 1.6 | 0.7 | ||
| 10/26/15 | Bonnybrook | Post-grit removal | 0.9 | 1.5 | 0.7 |
| Post-UV | 1.1 | 1.8 | 0.8 | ||
| 11/9/15 | Pine Creek | Post-grit removal | 1.5 | 2.4 | 1.1 |
| Post-UV | 0.8 | 1.4 | 0.6 | ||
| 11/9/15 | Bonnybrook | Post-grit removal | 2.2 | 3.5 | 1.6 |
| Post-UV | 3.2 | 5.1 | 2.3 | ||
| 12/7/15 | Pine Creek | Post-grit removal | 0.7 | 1.2 | 0.6 |
| Post-UV | 0.9 | 1.5 | 0.7 | ||
| 12/14/15 | Bonnybrook | Post-grit removal | 0.6 | 1.0 | 0.4 |
| Post-UV | 1.6 | 2.5 | 1.2 | ||
| Additional samples | Pine Creek | Post-grit removal | 9.1 | 14 | 6.6 |
| Post-UV | 6.4 | 10 | 4.7 | ||
| Pine Creek | Post-grit removal | 5.1 | 8.2 | 3.7 | |
| Post-UV | 13 | 22 | 10 | ||
| Pine Creek | Post-grit removal | 8.6 | 13 | 6.2 | |
| Post-UV | 14 | 22 | 10 | ||
| Pine Creek | Post-grit removal | 6.5 | 10 | 4.7 | |
| Post-UV | 9.7 | 15 | 7.0 |
Average recovery is based on a 10-ovum E. caproni spike into the wastewater performed in triplicate. Maximum and minimum recoveries are calculated based on the maximum and minimum values observed for the copy numbers of 10 E. caproni ova in 4 replicates. These values are 1.1 × 106 and 4.7 × 106, respectively. Additional spikes (in triplicate) were performed to better capture variation in the extraction method.
TABLE 3.
Average recoveries of an E. caproni ovum matrix spiked into raw and treated wastewatera
| Method | Matrix | Avg recovery (σx̄) |
|---|---|---|
| A | Post-grit removal | 3.1 (3.2) |
| Post-UV treatment | 3.1 (1.2) | |
| B | Post-grit removal | 3.5 (2.5) |
| Post-UV treatment | 4.1 (5.1) | |
| C | Post-grit removal | 2.4 (0.48) |
| Post-UV treatment | 3.8 (0.05) | |
| D | Post-grit removal | 9.1 (12) |
| Post-UV treatment | 4.0 (0.96) |
Three replicate extractions were performed for each treatment. Four lysis methods were compared. DNA was subsequently purified using the column purification steps in the Mo Bio PowerFecal kit. Samples (250 μl) were subjected to either 10 freeze-thaw cycles (dry ice-ethanol to 98°C) and a 1-h proteinase K digestion (method A) or 5 freeze-thaw cycles and a 1-h proteinase K digestion (method B). In methods C and D, samples were subjected to 10 and 5 freeze-thaw cycles, respectively.
(iv) E. caproni assay sensitivity and determination of copy number per ovum.
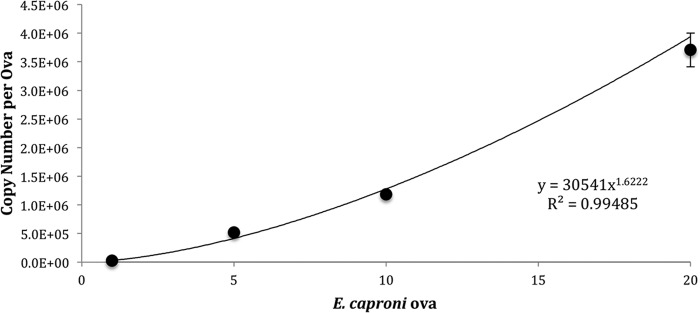
The LOD95 of the E. caproni qPCR detection assay was 2.1 copies/reaction (upper limit, 5.1; lower limit, 0.9). The average efficiency across 10 standard curves was 98.6% (σx̄ = 3.025), and the average slope of the standard curve was −3.37 (σx̄ = 0.11). All reactions had R2 of 0.99 (Fig. 2, bottom). The 28S copy number per egg was determined by creating standard curves consisting of 1, 5, 10, and 20 ova (Fig. 4). Only embryonated ova were selected for use in the standard curve. Embryonated ova were selected because these stages contain developed trematode miracidia and thus should contain roughly the same cell and copy numbers per ovum. Nonetheless, some variation in copy number between trials was observed. The average copy number for a 10-egg embryonated standard was 7.6 × 105 (σx̄ = 1.9 × 105); the maximum and minimum values were determined based on the normally distributed 95% confidence interval surrounding this mean. The maximum and minimum values were 1.0 ×106 and 4.7 × 105, respectively.
FIG 4.
Determination of the E. caproni copy number per egg. Standards (1, 5, 10, and 20 eggs) were run using the E. caproni qPCR assay.
Monitoring of wastewater effluent for E. vermicularis.
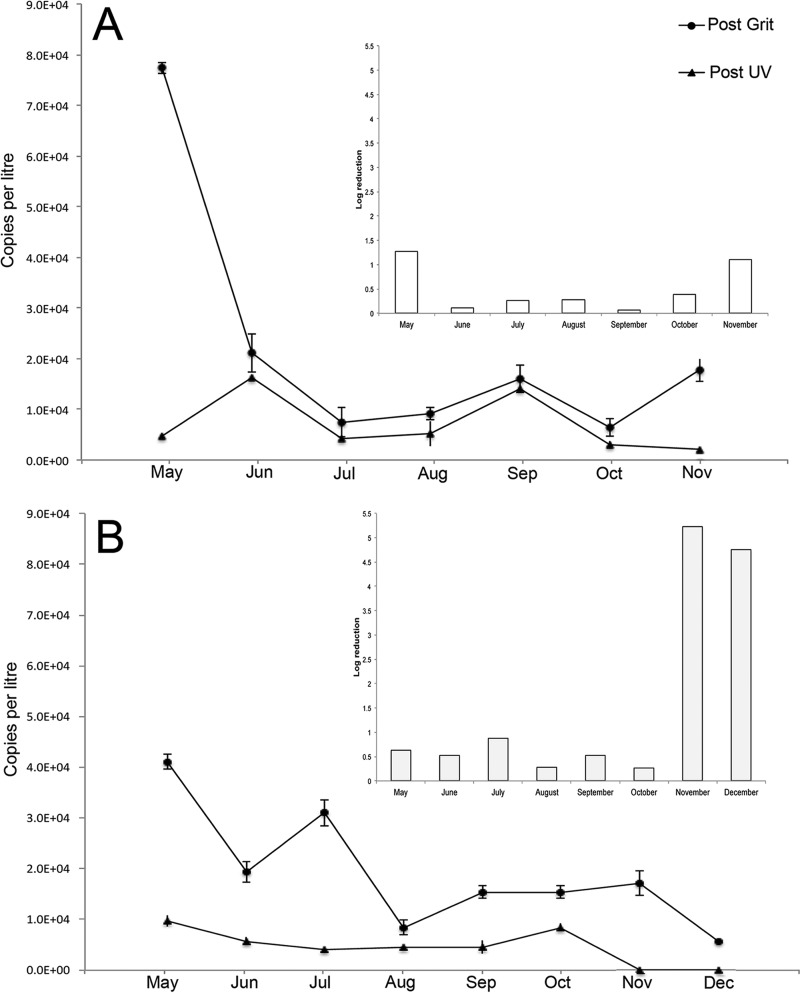
E. vermicularis 5S rDNA was detectable in wastewater influent from May 2015 until November 2015 from both plants (Fig. 5). At Bonnybrook, the adjusted 5S rDNA genome copy numbers (normalized against E. caproni recoveries in spiked samples) in the influent wastewater ranged from as many as 41,000 copies/liter in May 2015 to 5,600 copies/liter in December 2015 (Fig. 5). Copy numbers in treated wastewater effluent, after UV disinfection, ranged from 9,700 copies/liter to as few as 2,500 copies/liter. The Pine Creek plant samples had between 77,000 copies/liter in May 2015 and 17,000 copies/liter in November 2015 in influent wastewater. The amount of E. vermicularis DNA present in the effluent wastewater was similar to that in the effluent wastewater of the Bonnybrook plant, ranging from as many as 4,000 copies/liter in June 2015 to as few as 1,300 copies/liter in November 2015. Four separate DNA extractions per sample were conducted to confirm the absence of E. vermicularis DNA in the wastewater (Fig. 5). There was a significant difference in removal of the 5S gene copies at different locations at each of the treatment plants (Bonnybrook, α = 0.05, P = 0.0071, df = 6; Pine Creek, α = 0.05, P = 0.046, df = 5). However, there was no difference in removal between the two treatment plants (α = 0.05, P = 0.082, df = 5) (Fig. 5). E. vermicularis ova were consistently found in treated wastewater after grit removal at the Bonnybrook plant. In November and December at Pine Creek, no E. vermicularis DNA was detectable in the post-UV samples. At Pine Creek, E. vermicularis DNA was detected in both raw and treated wastewater from May until November. In December, E. vermicularis DNA was present in neither the post-grit removal nor post-UV samples.
FIG 5.
Quantity of E. vermicularis 5S rRNA in wastewater at Pine Creek (A) and Bonnybrook (B) treatment plants. Quantity is expressed in rDNA copies/liter for Post Grit (circles) and Post UV (triangles). The copy number has been adjusted relative to the E. caproni matrix spike. Log removal is shown in the nested pane. A denominator of 0.1 was used to calculate the log removal for November and December due to the absence of 5S rRNA during these months at Bonnybrook.
DISCUSSION
The objective of our study was to devise a quantitative diagnostic qPCR assay for E. vermicularis, as we hypothesize that it may be a useful surrogate in assessing the presence of helminth ova in wastewater. We then applied this assay to determine the physical removal of ova throughout two full-scale wastewater treatment plants. We began by designing a highly sensitive diagnostic qPCR assay specific to E. vermicularis and confirmed the ability of this qPCR assay to detect E. vermicularis in the wastewater environment by DNA sequencing of the amplified product. Key controls were developed to ensure the absence of PCR inhibition and to determine DNA extraction efficiency using ova from the parasitic flatworm Echinostoma caproni, which was not detected in either plant's wastewater. To our knowledge, our methodology for assessing DNA extraction efficiency for any parasitic ova is unique. The DNA extraction findings demonstrate a clear need for future studies to address difficulties lysing these robust helminth ova; especially as detection technologies for both clinical and environmental parasitology move into molecular methods, measures must be implemented to control and improve lysis efficacy. Additionally, we were able to successfully capture E. vermicularis ova in a relatively small volume of wastewater (2.5 liters). However, methodologies to cope with sampling complex matrices for rare ova should be investigated further. Gyawali et al. (35) have worked extensively on developing and validating rapid concentration methods for sampling hookworm ova, and these approaches should be taken up by other groups to improve the methodologies for the detection of ova in complex matrices.
At both plants, the overall concentration of E. vermicularis 5S rDNA decreased in raw wastewater from May 2015 to December 2015. This might be a due to physical factors associated with the wastewater matrix or due to temporal variations in parasite infection in the community. In Alberta, temperature differences in the wastewater may range from 8 to −15°C from winter to summer, and this might influence how ova bind to suspended solids and/or are distributed within the wastewater matrix. Seasonal infection is also common in many parasitic infections (36), and seasonality in enterobiasis occurrence has been noted in epidemiological studies conducted in India and Iraq, but we are unaware of any investigations in North America (29, 37). Further research should be conducted to understand the changes in E. vermicularis prevalences over a longer time frame to understand these fluctuations.
Overall, we estimate a 0.5- to 1.6-log10 removal at the two treatment works. It is important to note that our data do not account for hydraulically paired samples; the hydraulic retention time is approximately 16 h at each plant. Thus, our samples do not measure precisely the same body of water entering and exiting the plant (which is further complicated by recirculation of wastewater during secondary treatment), and the log10 removals should be considered overall estimates. While levels of removal of any pathogen from the wastewater matrix will vary depending on a particular process (waste stabilization ponds, reservoirs, coagulation/flocculation), most studies have reported between a 1- and 2-log10 removal of helminth (Ascaris) ova (38, 39), which is similar to the results reported in this study. E. vermicularis ova persisting through primary and secondary wastewater treatments has been demonstrated in Tunisia (18) but to our knowledge has not previously been demonstrated in North America.
Our study was unable to quantitate the number of ova directly, and we acknowledge this as a limitation of the data. E. vermicularis is multicellular, and ribosomal genes are multicopy in each cell. There are no estimates of 5S rDNA copy number or cell number in E. vermicularis L3 larval stages. Early work on L3 larval stages of other parasitic helminths estimates cell numbers to be around 500 (40). Estimates of the number of ova based on DNA copy number may also be confounded by haplodiploidy in parasitic oxyurids, a sex determination system by which males arise from haploid, unfertilized ova and females arise from diploid, fertilized ova (28, 41). Additionally, although the total number of cells may reach 500 in L3 parasitic nematodes, larval development in unsporulated ova is gradual, and thus ribosomal copy numbers may vary among ova based on their stage of development (40). These considerable knowledge gaps in the literature should be prioritized as molecular methods for the detection of parasitic ova become common.
Helminth ova are incredibly resistant to UV disinfection and chlorine disinfection (18). Two studies have demonstrated E. vermicularis' resistance to monochromatic, low-pressure UV irradiation (38, 39). The first study investigated survival at wavelengths between 228 and 295 nm, using egg hatching as a proxy for viability, and found that E. vermicularis is most sensitive to wavelengths below 240 nm but that parasite killing is poor at higher wavelengths (38). However, determining infectivity in helminth species requires an animal model. Larval worms contained within ova are typically large enough to survive UV disinfection and establish within a host; however, often they are unable to produce ova and reproduce inside the host. This study was unable to assess infectivity because E. vermicularis is a human-specific parasite, and only by using a chimpanzee infection model can infectivity be ascertained. However, it is well established that helminths are highly resistant to most disinfection techniques, and thus, removal of these pathogens from any water that may come into primary or secondary contact with humans is essential to ensure minimal risk. For instance, Ascaris requires a high UV fluence to achieve a >1-log10 pathogen inactivation (26); studies investigating disinfection efficacies of new technologies, like hydrogen peroxide coupled with a Fenton reaction-type nanocatalyst, have shown a greater inactivation of Ascaris organisms than traditional Fenton processes (42). Microwave irradiation is also effective at achieving a 1-log10 or greater inactivation of Ascaris ova (43), and a >1-log10 inactivation of cestode Taenia taeniaeformis ova and nematode Strongyloides ratti ova has also been demonstrated using microwaves (44). Although chemical treatment and/or UV disinfection may be relatively ineffective at inactivating helminthic ova in wastewater, physical removal processes, such as microfiltration, are likely excellent treatment options. Microfiltration (or fine ultrafiltration) is extremely efficient at removing protozoan (oo)cysts (Cryptosporidium spp., Giardia spp.) and therefore would be useful for controlling helminthic ova, especially in reuse options where treated wastewaters may be used to irrigate food crops (45–48). Deposition of eggs on the perianal skin also means that E. vermicularis eggs may become associated with clothing and bed linens, where these ova may be washed into gray water through clothes washing. This suggest that there may be not only a reuse risk associated with black water but also a reuse risk associated with gray water (49, 50). Due to the ability of E. vermicularis ova to be aerosolized, this might pose a risk for transmission and should be investigated (32).
Our study demonstrates that E. vermicularis is capable of escaping sedimentation and persisting through a tertiary wastewater treatment system, which may make it an ideal surrogate helminth for smaller, more buoyant ova. Removal efficacy is likely dependent on differences in the physical egg properties, which lead certain species to perform differently in the treatment environment (54, 55). Although molecular-based methods are useful for evaluating physical removal of pathogens in wastewater, they cannot necessarily be used to interpret UV inactivation efficiency, and consequently, we do not know whether E. vermicularis ova remain infectious after treatment. Our study demonstrates that E. vermicularis ova appear to be relatively persistent during tertiary sewage treatment, compelling us to more broadly consider helminthic ova as potential health risks for water reuse and how we might manage these risks (i.e., membrane filtration) under certain reuse scenarios. We contend that the biological properties of E. vermicularis make this organism an ideal surrogate helminth for evaluating treatment removal and public health risks associated with ova in reused wastewater but emphasize that a one-surrogate-fits-all solution for helminth ova is likely unrealistic; future research should prioritize investigating ova with different physical properties in these complex matrices.
MATERIALS AND METHODS
qPCR assay design. (i) Synthesized plasmid and PCR control preparations.
All control plasmids containing helminth (Echinostoma caproni and Enterobius vermicularis) and inhibition control gene targets (i.e., synthetic genes) were synthesized by GenScript (USA) and inserted into the puc57 vector, which contains an ampicillin resistance gene, and the presence of the insert was confirmed by Sanger sequencing. Plasmid preparations were rehydrated in nuclease-free water and transformed into TOP10 Escherichia coli cells, and the bacteria were plated onto LB agar containing 100 μg/ml carbenicillin. Plasmid purification was performed using the GeneJET plasmid miniprep kit (Thermo Scientific).
(ii) Gene target selection and primer and probe design.
When designing the E. vermicularis qPCR assay, the 5S rRNA gene and spacer region (GenBank accession number AB221533.1) were selected. The primers and probe were designed to amplify a 100-bp region overlapping the first seven nucleotides of the SL-1 transsplicing leader sequence. Diagnostic method validation also included the development of a comparable assay against E. caproni, a helminthic trematode parasite commonly found in small mammals. This parasite assay acted as a control for DNA extraction recovery and PCR detection in a spiked wastewater matrix. The 28S rRNA gene was selected for the E. caproni assay.
A PCR inhibition control was also developed and included a synthetic gene with no known homology to any gene in the NCBI GenBank nucleotide database (Table 1). The primers and TaqMan probe for the E. vermicularis 5S rRNA gene, E. caproni 28S rRNA gene, and the inhibition control were designed using the real-time qPCR assay tool from IDT (USA) and are provided in Table 1.
(iii) qPCR workflow and reaction parameters.
All qPCRs were performed using TaqMan Fast advanced master mix (Life Technologies) and were carried out according to the manufacturer's specifications. Briefly, qPCR mixtures contained 200 nM forward and reverse primers (Table 1), 200 nM probe (Table 1), 1× master mix, and 5 μl of sample DNA to a final volume of 20 μl. Reactions were run on the ABI 7500 Fast real-time PCR system. Thermocycling parameters were as follows: there was a uracil-N-glycosylase (UNG) hold for 2 min at 55°C, a hold step for 20 s at 95°C, initial denaturation at 94°C for 3 s, and annealing and extension at 60°C for 30 s for 40 cycles. Direct quantification of wastewater samples was achieved by utilizing standard-curve settings on the machine's software. Each DNA sample, including standards and controls, was run in triplicate in the qPCR assay. Inhibition controls and extraction controls were performed on each wastewater sample analyzed.
We adhered to PCR workflows throughout the study. All master mix components were prepared in a clean room, and the assembly of PCRs was performed in a separate room using a QIAgility robot (Qiagen). Thermocycling (qPCR and endpoint PCR) and all genetic downstream analyses (agarose gels, plasmid purifications) were carried out in a separate postamplification room.
(iv) Preparation of standard curves and determining the limits of detection.
All qPCRs were quantitated against standard curves consisting of plasmid DNA from E. vermicularis and E. caproni gene targets. Purified plasmid DNA was quantified using the Qubit fluorometer (Thermo). Stocks of 500,000 copies per 5 μl of plasmid DNA were prepared and frozen until the time of use. These stocks were serially diluted to final concentrations of 50,000, 5,000, 500, 50, 5, and 0.5 copies of plasmid per reaction. Each individual standard was run in triplicate and analyzed using the ABI 7500 Fast system software, automatically generating values for PCR efficiency, slope, and the correlation coefficient. The detection limit with 95% confidence (LOD95) of the E. vermicularis, E. caproni, and inhibition control assays was calculated using the probability of detection-limit of detection (POD-LOD) calculation program based on 10 replicate standard curves (51).
(v) Echinostoma caproni ovum control stocks.
The E. caproni parasites were maintained in the laboratory as previously described (52). The harvested ova were stored in Locke's solution and counted using a dissecting microscope into 5 or 10 egg stock suspensions containing approximately 35 μl of Locke's solution. Counted egg stocks were used to create genomic DNA preparations by adding the requisite number of ova to each tube and decanting the supernatant in 10 μl of Locke's solution. DNA was extracted from stocks containing 1, 5, 10, and 20 ova using the DNeasy blood and tissue kit (Qiagen), and the protocol recommended by Webster was used with one exception: eggs were not preserved in RNAlater (Thermo) (53). Copy number per ova was determined using the E. caproni qPCR assay. By plotting the copy numbers versus the number of ova on a standard curve and determining the equation of this line, we were able to quantitate extraction efficiency in wastewater samples.
(vi) Inhibition control.
As indicated above, inhibition controls were run for each sample. The LOD95 for the inhibition control was determined to be 24.1 copies/reaction (upper limit, 55; lower limit, 11). As such, 500 copies of plasmid were spiked into 5 μl of each DNA extract. Inhibition was defined as a three-cycle threshold (CT) shift in the spiked control compared to the CT number of the standard preparation (35).
(vii) Wastewater sampling.
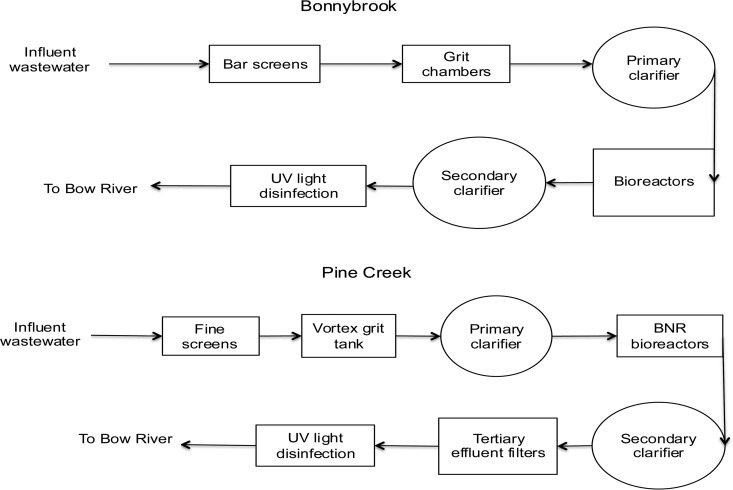
Monthly monitoring of wastewater at two municipal treatment plants (Bonnybrook and Pine Creek facilities) in Calgary, Alberta, Canada, began in May 2015. Influent wastewater at the Bonnybrook facility undergoes bar screening and passes through a grit chamber before entering primary sedimentation. For the purposes of this paper, post-grit removal samples refer to the sampling location after grit removal but before the wastewater entered the primary sedimentation tanks. The wastewater then undergoes an aerobically activated sludge process and secondary treatment clarification and is finally disinfected with UV (low pressure; low intensity) before being discharged to the river. The second sampling point in the study was located after UV treatment, herein referred to as the post-UV sample (Fig. 6). The Pine Creek wastewater treatment plant has a similar design; however, it is a more modern plant and utilizes medium-pressure (polychromatic) UV disinfection. Water enters the treatment plant and undergoes grit removal through fine grit screens and passes through a vortex grit tank. Secondary treatment at Pine Creek involves subjection of water to a biological nutrient reduction (BNR) process; it then undergoes secondary clarification and is subjected to a cloth (∼200-μm) tertiary effluent filter, before undergoing UV (medium-pressure; high-intensity) disinfection and discharge. A post-UV sample was also collected after disinfection at this plant (Fig. 6). The samples were collected once per month per site, with collection alternating between Bonnybrook and Pine Creek and with samples taken from water entering the plant between 9 a.m. and 11 a.m. (approaching peak flows). Samples were shipped on ice to the University of Alberta and were processed the day after collection. Fifty-milliliter wastewater samples were centrifuged at 2,000 × g for 30 min to concentrate and pellet the samples. After the entire volume of wastewater was centrifuged, the supernatant was decanted down to a final volume of 5 ml.
FIG 6.
Flow diagrams of the wastewater treatment process at Bonnybrook and Pine Creek. Water samples were collected after grit processing and after UV disinfection.
(viii) E. caproni extraction control validation.
The wastewater samples that were used to assess background E. caproni concentration in the wastewater were extracted by utilizing the Mo Bio PowerFecal DNA extraction kit according the procedure outlined above for the E. vermicularis samples.
In validating the DNA extraction method, two pretreatment steps were evaluated to improve recoveries. The 250-μl wastewater samples were subjected to one of four pretreatment steps before being purified through the Mo Bio PowerFecal columns. Samples were subjected to either 10 freeze-thaw cycles (dry ice-ethanol, 98°C) and a 1-h proteinase K digestion, 5 freeze-thaw cycles and a 1-h proteinase K digestion, or 10 or 5 freeze-thaw cycles.
(ix) Wastewater DNA extractions.
DNA extractions from wastewater samples were performed using the Mo Bio PowerFecal DNA isolation kit according to the manufacturer's instructions, with two modifications: 250 μl of concentrated wastewater was added to the kit, and ∼35 μl containing 10 E. caproni ova was spiked into the water sample before the protocol was begun. A DNA extraction blank consisting of PCR-grade water was also extracted. Each wastewater sample was independently extracted at least three times to assess variability in DNA extraction efficiency (based on E. caproni spikes). In the rare case of PCR inhibition, a determination based on the results obtained from spiked inhibition control plasmids (as defined by a 3-CT shift) (35), an additional DNA extraction was performed. When we analyzed the qPCR data, any replicate with a copy number below the LOD95 was excluded from subsequent analysis.
(x) E. vermicularis qPCR amplicon sequencing.
To confirm that E. vermicularis was the only amplified target in the qPCR assay, the 5S rDNA qPCR primers were used in an endpoint PCR using the following cycling parameters: initial denaturation at 30 s at 98°C, followed by 30 cycles of denaturation for 5 s at 98°C, annealing for 10 s at 60°C, and an extension for 15 s at 72°C, with a final extension for 5 min at 72°C. The products of this PCR were run out on a 1% agarose gel, and any bands identified were extracted using the GeneJET gel purification kit (Thermo) and cloned using the CloneJET PCR cloning kit (Thermo). After transformation into Top10 E. coli, single colonies were picked and their plasmid DNA was purified, and these plasmid constructs were sequenced using the pJET1.2 primers.
DNA sequence analysis.
All sequence analysis was performed using CLCbio Genomics Workbench software (Qiagen).
Statistics.
A two-tailed paired t test was used to compare the gene copy number reductions at each treatment plant and between the two plants. Log10 removal was calculated using the following formula: log10 removal = [log10 (post-grit removal copy number)/(post-UV treatment copy number)].
Supplementary Material
ACKNOWLEDGMENTS
We thank Graham Banting for his mentorship and comments on the manuscript, as well as the City of Calgary staff from Wastewater Treatment and Water Quality Services for sample collection.
This work was fully funded by Alberta Innovates and the City of Calgary, to whom we are most grateful for their support.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00547-17.
REFERENCES
- 1.Slifko TR, Smith HV, Rose JB. 2000. Emerging parasite zoonoses associated with water and food. Int J Parasitol 30:1379–1393. doi: 10.1016/S0020-7519(00)00128-4. [DOI] [PubMed] [Google Scholar]
- 2.Gaspard P, Wiart J, Schwartzbrod J. 1997. Parasitological contamination of urban sludge used for agricultural purposes. Waste Manag Res 15:429–436. doi: 10.1177/0734242X9701500409. [DOI] [Google Scholar]
- 3.O'Donnell CJ, Meyer KB, Jones JV, Benton T, Kaneshiro ES, Nichols JS, Schaefer FW III. 1984. Survival of parasite eggs upon storage in sludge. Appl Environ Microbiol 48:618–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dickin SK, Schuster-Wallace CJ, Qadir M, Pizzacalla K. 2016. A review of health risks and pathways for exposure to wastewater use in agriculture. Environ Health Perspect 124:900–909. doi: 10.1289/ehp.1509995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bevilacqua PD, Bastos RKX, Mara DD. 2014. An evaluation of microbial health risks to livestock fed with wastewater-irrigated forage crops. Zoonoses Public Health 61:242–249. doi: 10.1111/zph.12063. [DOI] [PubMed] [Google Scholar]
- 6.Shuval HI. 1991. Effects of wastewater irrigation of pastures on the health of farm animals and humans. Rev Sci Tech 10:847–866. [DOI] [PubMed] [Google Scholar]
- 7.Erdogřul Ö, Şener H. 2005. The contamination of various fruit and vegetable [sic] with Enterobius vermicularis, Ascaris eggs, Entamoeba histolyca [sic] cysts and Giardia cysts. Food Control 16:557–560. doi: 10.1016/j.foodcont.2004.06.016. [DOI] [Google Scholar]
- 8.Gupta N, Khan DK, Santra SC. 2009. Prevalence of intestinal helminth eggs on vegetables grown in wastewater-irrigated areas of Titagarh, West Bengal, India. Food Control 20:942–945. doi: 10.1016/j.foodcont.2009.02.003. [DOI] [Google Scholar]
- 9.Kozan E, Gonenc B, Sarimehmetoglu O, Aycicek H. 2005. Prevalence of helminth eggs on raw vegetables used for salads. Food Control 16:239–242. doi: 10.1016/j.foodcont.2004.02.005. [DOI] [Google Scholar]
- 10.Sidhu JPS, Toze SG. 2009. Human pathogens and their indicators in biosolids: a literature review. Environ Int 35:187–201. doi: 10.1016/j.envint.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 11.Navarro I, Jiménez B, Lucario S, Cifuentes E. 2009. Application of helminth ova infection dose curve to estimate the risks associated with biosolid application on soil. J Water Health 7:31–44. doi: 10.2166/wh.2009.113. [DOI] [PubMed] [Google Scholar]
- 12.Payment P, Locas A. 2011. Pathogens in water: value and limits of correlation with microbial indicators. Ground Water 49:4–11. doi: 10.1111/j.1745-6584.2010.00710.x. [DOI] [PubMed] [Google Scholar]
- 13.Harwood VJ, Levine AD, Scott TM, Chivukula V, Lukasik J, Farrah SR, Rose JB. 2005. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl Environ Microbiol 71:3163–3170. doi: 10.1128/AEM.71.6.3163-3170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Victorica J, Galván M. 2003. Preliminary testing of a rapid coupled methodology for quantitation/viability determination of helminth eggs in raw and treated wastewater. Water Res 37:1278–1287. doi: 10.1016/S0043-1354(02)00477-3. [DOI] [PubMed] [Google Scholar]
- 15.deRegnier DP, Cole L, Schupp DG, Erlandsen SL. 1989. Viability of Giardia cysts suspended in lake, river, and tap water. Appl Environ Microbiol 55:1223–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaspard PG, Schwartzbrod J. 2003. Parasite contamination (helminth eggs) in sludge treatment plants: definition of a sampling strategy. Int J Hyg Environ Health 206:117–122. doi: 10.1078/1438-4639-00197. [DOI] [PubMed] [Google Scholar]
- 17.Ben Ayed L, Schijven J, Alouini Z, Jemli M, Sabbahi S. 2009. Presence of parasitic protozoa and helminth in sewage and efficiency of sewage treatment in Tunisia. Parasitol Res 105:393–406. doi: 10.1007/s00436-009-1396-y. [DOI] [PubMed] [Google Scholar]
- 18.Jiménez B, Maya C, Galván M. 2007. Helminth ova control in wastewater and sludge for advanced and conventional sanitation. Water Sci Technol 56:43–51. doi: 10.2166/wst.2007.555. [DOI] [PubMed] [Google Scholar]
- 19.Schwartzbrod J, Banas S. 2003. Parasite contamination of liquid sludge from urban wastewater treatment plants. Water Sci Technol 47:163–166. [PubMed] [Google Scholar]
- 20.Bowman DD, Little MD, Reimers RS. 2003. Precision and accuracy of an assay for detecting Ascaris eggs in various biosolid matrices. Water Res 37:2063–2072. doi: 10.1016/S0043-1354(02)00597-3. [DOI] [PubMed] [Google Scholar]
- 21.Pecson BM, Barrios JA, Johnson DR, Nelson KL. 2006. A real-time PCR method for quantifying viable Ascaris eggs using the first internally transcribed spacer region of ribosomal DNA. Appl Environ Microbiol 72:7864–7872. doi: 10.1128/AEM.01983-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raynal M, Villegas EN, Nelson KL. 2012. Enumeration of viable and non-viable larvated Ascaris eggs with quantitative PCR. J Water Health 10:594–604. doi: 10.2166/wh.2012.101. [DOI] [PubMed] [Google Scholar]
- 23.Johnson PW, Dixon R, Ross AD. 1998. An in-vitro test for assessing the viability of Ascaris suum eggs exposed to various sewage treatment processes. Int J Parasitol 28:627–633. doi: 10.1016/S0020-7519(97)00210-5. [DOI] [PubMed] [Google Scholar]
- 24.Scott ME. 2008. Ascaris lumbricoides: a review of its epidemiology and relationship to other infections. Ann Nestlé (Engl ed) 66:7–22. doi: 10.1159/000113305. [DOI] [Google Scholar]
- 25.McCarty TR, Turkeltaub JA, Hotez PJ. 2014. Global progress towards eliminating gastrointestinal helminth infections. Curr Opin Gastroenterol 30:18–24. doi: 10.1097/MOG.0000000000000025. [DOI] [PubMed] [Google Scholar]
- 26.Brownell SA, Nelson KL. 2006. Inactivation of single-celled Ascaris suum eggs by low-pressure UV radiation. Appl Environ Microbiol 72:2178–2184. doi: 10.1128/AEM.72.3.2178-2184.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bandala ER, González L, Sanchez-Salas JL, Castillo JH. 2012. Inactivation of Ascaris eggs in water using sequential solar driven photo-Fenton and free chlorine. J Water Health 10:20–30. doi: 10.2166/wh.2011.034. [DOI] [PubMed] [Google Scholar]
- 28.Adamson ML. 1989. Evolutionary biology of the Oxyurida (Nematoda): biofacies of a haplodiploid taxon. Adv Parasitol 28:175–228. doi: 10.1016/S0065-308X(08)60333-4. [DOI] [PubMed] [Google Scholar]
- 29.Vermund SH, Wilson CM. 2000. Pinworm (Enterobius vermicularis). Semin Pediatr Infect Dis 11:252–256. doi: 10.1053/spid.2000.9639. [DOI] [Google Scholar]
- 30.Zeibig E. 2014. Clinical parasitology: a practical approach, 2nd ed Elsevier Saunders, St. Louis, MO. [Google Scholar]
- 31.Sawitz W. 1942. The buoyancy of certain nematode eggs. J Parasitol 28:95–102. doi: 10.2307/3272719. [DOI] [Google Scholar]
- 32.Reardon MO, Reardon L. 1939. Studies on oxyuriasis. XX. The distribution of the ova of Enterobius vermicularis in household dust. J Parasitol 25:173–177. [Google Scholar]
- 33.Collender PA, Kirby AE, Addiss DG, Freeman MC, Remais JV. 2015. Methods for quantification of soil-transmitted helminths in environmental media: current techniques and recent advances. Trends Parasitol 31:625–639. doi: 10.1016/j.pt.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang SL, Kabler PW. 1962. Free-living nematodes in aerobic treatment plant effluent. J Water Pollut Control Fed 34:1256–1261. [Google Scholar]
- 35.Gyawali P, Sidhu JPS, Ahmed W, Jagals P, Toze S. 2015. Comparison of concentration methods for rapid detection of hookworm ova in wastewater matrices using quantitative PCR. Exp Parasitol 159:160–167. doi: 10.1016/j.exppara.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 36.Altizer S, Dobson A, Hosseini P, Hudson P, Pascual M, Rohani P. 2006. Seasonality and the dynamics of infectious diseases. Ecol Lett 9:467–484. doi: 10.1111/j.1461-0248.2005.00879.x. [DOI] [PubMed] [Google Scholar]
- 37.Haswell-Elkins MR, Elkins DB, Manjula K, Michael E, Anderson RM. 1987. The distribution and abundance of Enterobius vermicularis in a south Indian fishing community. Parasitology 95:339–354. doi: 10.1017/S0031182000057784. [DOI] [PubMed] [Google Scholar]
- 38.Jones MF, Jacobs L, Hollaender A. 1940. The effects of monochromatic ultraviolet radiation on eggs of the nematode, Enterobius vermicularis. II: sublethal effects. J Parasitol 26:435. doi: 10.2307/3272249. [DOI] [Google Scholar]
- 39.Hollaender A, Jones MF, Jacobs L. 1940. The effects of monochromatic ultraviolet radiation on eggs of the nematode, Enterobius vermicularis I: quantitative response. J Parasitol 26:421–432. doi: 10.2307/3272485. [DOI] [Google Scholar]
- 40.Drag M, Höglund J, Nejsum P, Thamsborg SM, Enemark HL. 2016. The level of embryonation influences detection of Ostertagia ostertagi eggs by semi-quantitative PCR. Parasit Vectors 9:368. doi: 10.1186/s13071-016-1657-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakano T, Murata K, Ikeda Y, Hasegawa H. 2003. Growth of Enterobius vermicularis in a chimpanzee after anthelmintic treatment. J Parasitol 89:439–443. doi: 10.1645/0022-3395(2003)089[0439:GOEVIA]2.0.CO.2. [DOI] [PubMed] [Google Scholar]
- 42.Morales AA, Ramírez-Zamora RM, Schouwenaars R, Pfeiffer H. 2013. Inactivation of Ascaris eggs in water using hydrogen peroxide and a Fenton type nanocatalyst (FeOx/C) synthesized by a novel hybrid production process. J Water Health 11:419–429. doi: 10.2166/wh.2013.203. [DOI] [PubMed] [Google Scholar]
- 43.Mun S, Cho S-H, Kim T-S, Oh B-T, Yoon J. 2009. Inactivation of Ascaris eggs in soil by microwave treatment compared to UV and ozone treatment. Chemosphere 77:285–290. doi: 10.1016/j.chemosphere.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 44.Conder GA, Williams JF. 1983. The microwave oven: a novel means of decontaminating parasitological specimens and glassware. J Parasitol 69:181–185. doi: 10.2307/3281295. [DOI] [PubMed] [Google Scholar]
- 45.Kuhn RC, Oshima KH. 2002. Hollow-fiber ultrafiltration of Cryptosporidium parvum oocysts from a wide variety of 10-L surface water samples. Can J Microbiol 48:542–549. doi: 10.1139/w02-049. [DOI] [PubMed] [Google Scholar]
- 46.Lindquist HDA, Harris S, Lucas S, Hartzel M, Riner D, Rochele P, DeLeon R. 2007. Using ultrafiltration to concentrate and detect Bacillus anthracis, Bacillus atrophaeus subspecies globigii, and Cryptosporidium parvum in 100-liter water samples. J Microbiol Methods 70:484–492. doi: 10.1016/j.mimet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 47.Lipp P, Baldauf G, Schick R, Elsenhans K, Stabel HH. 1998. Integration of ultrafiltration to conventional drinking water treatment for a better particle removal—efficiency and costs? Desalination 119:133–142. doi: 10.1016/S0011-9164(98)00133-7. [DOI] [Google Scholar]
- 48.Drozd C, Schwartzbrod J. 1997. Removal of Cryptosporidium from river water by crossflow microfiltration—a pilot-scale study. Water Sci Technol 35:391–395. doi: 10.1016/S0273-1223(97)00291-6. [DOI] [Google Scholar]
- 49.Jahne MA, Schoen ME, Garland JL, Ashbolt NJ. 9 November 2016. Simulation of enteric pathogen concentrations in locally-collected greywater and wastewater for microbial risk assessments. Microb Risk Anal doi: 10.1016/j.mran.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schoen ME, Ashbolt NJ, Jahne MA, Garland J. 2 March 2017. Risk-based enteric pathogen reduction targets for non-potable and direct potable use of roof runoff, stormwater, greywater, and wastewater. Microb Risk Anal doi: 10.1016/j.mran.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilrich C, Wilrich P-T. 2009. Estimation of the POD function and the LOD of a qualitative microbiological measurement method. J AOAC Int 92:1763–1772. [PubMed] [Google Scholar]
- 52.Adema CM, Hanington PC, Lun C-M, Rosenberg GH, Aragon AD, Stout BA, Lennard Richard ML, Gross PS, Loker ES. 2010. Differential transcriptomic responses of Biomphalaria glabrata (Gastropoda, Mollusca) to bacteria and metazoan parasites, Schistosoma mansoni and Echinostoma paraensei (Digenea, Platyhelminthes). Mol Immunol 47:849–860. doi: 10.1016/j.molimm.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Webster BL. 2009. Isolation and preservation of schistosome eggs and larvae in RNAlater® facilitates genetic profiling of individuals. Parasit Vectors 2:50. doi: 10.1186/1756-3305-2-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmidt-Rhaesa A. 2014. Handbook of zoology: Gastrotricha, Cycloneuralia and Gnathifera. Nematoda, vol 2 De Gruyter, Berlin, Germany. [Google Scholar]
- 55.Sengupta ME, Thamsborg SM, Andersen TJ, Olsen A, Dalsgaard A. 2011. Sedimentation of helminth eggs in water. Water Res 45:4651–4660. doi: 10.1016/j.watres.2011.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.