Abstract
The eukaryotic six-subunit origin recognition complex (ORC) governs the initiation site of DNA replication and formation of the prereplication complex. In this report we describe the isolation of the wild-type Homo sapiens (Hs)ORC and variants containing a Walker A motif mutation in the Orc1, Orc4, or Orc5 subunit using the baculovirus-expression system. Coexpression of all six HsORC subunits yielded a stable complex containing HsOrc subunits 1–5 (HsORC1-5) with virtually no Orc6 protein (Orc6p). We examined the ATPase, DNA-binding, and replication activities of these complexes. Similar to other eukaryotic ORCs, wild-type HsORC1-5 possesses ATPase activity that is stimulated only 2-fold by single-stranded DNA. HsORC1-5 with a mutated Walker A motif in Orc1p contains no ATPase activity, whereas a similar mutation of either the Orc4 or Orc5 subunit did not affect this activity. The DNA-binding activity of HsORC1-5, using lamin B2 DNA as substrate, is stimulated by ATP 3- to 5-fold. Mutations in the Walker A motif of Orc1p, Orc4p, or Orc5p reduced the binding efficiency of HsORC1-5 modestly (2- to 5-fold). Xenopus laevis ORC-depleted extracts supplemented with HsORC1-5 supported prereplication complex formation and X. laevis sperm DNA replication, whereas the complex with a mutation in the Walker A motif of the Orc1, Orc4, or Orc5 subunit did not. These studies indicate that the ATP-binding motifs of Orc1, Orc4, and Orc5 are all essential for the replication activity associated with HsORC.
Keywords: ATP, replication
In eukaryotes, DNA replication is initiated by the origin recognition complex (ORC), which binds DNA at replication origins and promotes the assembly of the prereplication complex (pre-RC) (1, 2). The role of ORC in the initiation of DNA replication was first revealed in Saccharomyces cerevisiae (Sc), in which well defined sequences of ≈150 bp serve as replication origins (3, 4). Based on its origin binding affinity, ScORC was purified from budding yeast and shown to interact with origins in an ATP-dependent reaction. The ScORC contained six unique proteins (Orc1p–Orc6p), the genes of which are all essential for viability and DNA replication (5). Subsequently, homologues of ORC have been identified in all eukaryotes, and ORCs have been purified from Schizosaccharomyces pombe (Sp), Drosophila melanogaster (Dm), Xenopus laevis (Xl), and Homo sapiens (Hs) cells. Similar to ScORC, both genetic and biochemical studies revealed that ORCs from these organisms are required for the initiation of DNA replication, suggesting that the mechanism of initiation is conserved (5).
Although there is striking conservation of ORC and various initiation proteins, the sequences recognized by the various eukaryotic ORCs differ (6, 7). The origin regions from S. pombe have been genetically defined and do not resemble those from S. cerevisiae in either size or sequence (8, 9). DNA binding by SpORC is ATP-independent and depends solely on the SpOrc4p subunit, which uniquely contains nine repeats of an AT-hook domain at its N terminus that targets the binding of either the SpORC or the SpOrc4p subunit alone to AT-rich DNA (10–12). In vivo, the unique AT-hook domain of SpOrc4p was shown to be essential (11).
Studies in metazoans suggest that DNA initiation may be controlled by both local sequences and those markedly distant from putative origins (13). The definition of origins in higher eukaryotes has been limited by the lack of genetic assays. In vivo studies, using a variety of approaches, have been used to map bidirectional origins of replication in mammalian cells (7), some of which, including the lamin B2 origin, were shown to interact with ORC in vivo (14–16). However, detection of ORC binding to specific sequences within these regions remains unclear. Biochemical experiments with DmORC and HsORC indicated that they both bind to AT-rich DNA with no striking sequence preference, and this interaction is stimulated by ATP (5, 17, 18). However, DmORC was shown to bind to negatively superhelical DNA more selectively than linear DNA (19).
All eukaryotic ORCs contain three subunits (Orc1, Orc4, and Orc5) that belong to the AAA+ family of ATPases (5), which undergoes conformational changes or induces changes in interacting partners after binding of ATP (20). Among the characterized eukaryotic ORCs, the three AAA+ subunits contain a conserved Walker A motif (except for ScOrc4, which contains a YKT sequence) and a fairly well conserved Walker B motif, although all Orc5 subunits possess a questionable Walker B motif (5). All eukaryotic ORCs examined to date possess ATPase activity. Studies with both ScORC and DmORC indicate that ATP binding to Orc1p is essential for their ATPase activity, ability to bind DNA, and support replication (17, 21, 22). In contrast, mutations in the Walker A motif of Orc4p or Orc5p did not compromise these activities.
The properties of the eukaryotic ORCs as well as homology among the Orc subunits differ. Both ScORC and DmORC formed a stable, stoichiometric six-subunit complex, whereas HsORC, isolated either from cells or after expression by using the baculovirus insect cell system, contained low levels of Orc6p (18, 23). The isolated XlORC is comprised of only the Orc1–Orc5 subunits, and it is surprising that no Xl Orc6p homologue has been identified to date (24). ScORC devoid of Orc6p (ScORCΔ6) binds DNA as efficiently as the holocomplex, whereas DmORCΔ6 does not bind DNA (17, 25). Studies in Drosophila embryo extracts detected a pool of free Orc6p devoid of other Orc subunits (17, 26). This material was found localized to the cell membrane along the cleavage furrow, suggesting that Orc subunits may have additional roles distinct from replication.
In this report, the properties of baculovirus-expressed wild-type HsORC and HsORC containing a mutated Walker A motif in Orc1p, Orc4p, or Orc5p were examined. Analogous to previous reports (18, 23), infection of cells with viruses expressing all six HsOrc subunits leads to the isolation of a complex containing near-stoichiometric levels of Orc1–Orc5 subunits with low levels of Orc6p (which we call HsORC1-6). The properties of HsORC preparations formed by expression of Orc1–Orc5 subunits (HsORC1-5) were identical to those of HsORC1-6. Like the DmORC and ScORC, the integrity of the HsOrc1 Walker A motif is essential for ATP hydrolysis, whereas mutations of this motif in Orc4p and Orc5p did not affect the ATPase activity of the complex. As reported by Vashee et al. (18), the DNA-binding activity of HsORC1-5 (or HsORC1-6) is partially stimulated by ATP, and we find that this activity is reduced marginally by mutations in the Walker A motifs. We show that HsORC1-5 supports Xenopus sperm DNA replication by Xenopus cell-free extracts as reported (18), whereas complexes containing a mutant Walker A motif in Orc1p, Orc4p, or Orc5p do not. Thus, the ATP-binding motifs of Orc1, Orc4, and Orc5 subunits are required for the biological activities associated with HsORC.
Materials and Methods
Reagents. Anti-FLAG M2 Ab-agarose and FLAG peptide were from Sigma. Rabbit polyclonal antibodies were generated by Cocalico Biologicals (Reamstown, PA), and mouse anti-HsOrc1 antibodies were obtained from NeoMarkers (Fremont, CA). Subcloning efficiency DH5α-competent cells were obtained from Invitrogen. The Xenopus Orc1 and Orc2 antibodies were a gift from Ronald Laskey (Hutchison MRC Research Centre, Cambridge, U.K.). Labeled lamin B2 DNA, used for the DNA-binding assays described below, was prepared as follows. The genomic sequence of the lamin B2 origin region was amplified by PCR using HeLa DNA as template and primers specific to the lamin B2 genomic region (sequences are available on request). The DNA was digested with PstI and labeled with [α-32P]dATP and [α-32P]dCTP with Klenow polymerase. The recovered product contained the sequence from nucleotides 3832 to 4409 of the HUMLAMBBB locus (GenBank accession no. M94363).
Cloning of Various HsORC Genes into Baculovirus Vectors. The cDNA sequences of human Orc1 (accession no. NM_004153), Orc2 (accession no. NM_006190), Orc3 (accession no. AF125507), Orc4 (accession no. AF132596), Orc5 (accession no. HSU92538), and Orc6 (accession no. AF139658) were obtained from the GenBank database. Primers were used to amplify their ORFs from a Marathon-Ready human cDNA library, and these PCR products then were cloned into the pCR2.1-TOPO vector (Invitrogen). Individual clones were sequenced and subcloned into pET vectors (Novagen). Sequence-specific primers were designed to amplify each cDNA such that the start and stop codons were flanked with BssHII and NotI restriction sites, respectively. PCRs were conducted by using either bacterial clones or human ZAPII cDNA library (for Orc6) as template. The PCR products were digested and cloned into the BssHII and NotI restriction sites of pFastBacI vector. To construct Orc1, Orc3, and Orc6 clones with a FLAG epitope at the N terminus, a linker containing the FLAG-coding sequence was inserted into the BamHI and BssHII sites of the pFastBacI vectors containing these cDNAs. The nucleotide sequence of PCR products and linker region were confirmed by DNA sequencing. All recombinant viruses were produced according to manufacturer protocols (BAC-TO-BAC baculovirus-expression systems, Invitrogen). Primer sequences are available on request.
Preparation of Antibodies Against the Orc6 Protein. For the preparation of polyclonal antibodies against Orc6p, Orc6 cDNA was cloned into the BamHI and XhoI sites of pET28-a (Novagen). PCR amplification was used to create the restriction sites in the cDNA by using specific primers (sequences are available on request) and a human ZAPII cDNA library as template. The protein was expressed and purified from DH5α cells by Ni2+-agarose chromatography with denaturing conditions. After SDS/10% PAGE, protein bands were excised from the gel and used for immunization of rabbits.
Generation of GKT Mutant Orc Subunits. Site-directed mutagenesis of lysine to alanine in the GKT motifs of Orc1, Orc4, and Orc5 was carried out by PCR amplification of cDNAs with mutagenic primers (sequences are available on request). PCR products were digested with DpnI to linearize the DNA and used to transform DH5α-competent cells. Recombinant viruses were produced according to manufacturer protocols (BAC-TO-BAC baculovirus-expression systems).
Expression of ORC Proteins in Sf9 Cells. To express recombinant Orc proteins, Sf9 cells (≈2.0 × 106 cells per ml), cultured in Grace's medium supplemented with 10% FBS, were infected with HsOrc-expressing baculoviruses at a multiplicity of infection of 0.5–3 and incubated at 27°C for 48 h. To facilitate purification of complexes, a baculovirus expressing the Orc3 cDNA with a FLAG epitope at its N terminus was used. For purification of the HsOrc1p and HsOrc6p, baculoviruses expressing each cDNA with a FLAG epitope at its N′ terminus were used.
Purification of Recombinant HsORCs. Sf9 cells (≈2.0 × 106 cells per ml, 500 ml) were infected with recombinant viruses and incubated at 27°C for 48 h to generate the HsORC1-6, HsORC1-5, and HsORC2-5 complexes. Cells were harvested, washed with ice-cold PBS, and Dounce-homogenized with 5 pellet volumes of buffer H (20 mM Hepes-NaOH, pH 7.5/5 mM magnesium acetate/1 mM ATP/1 mM DTT/1 mM EDTA/1 mM EGTA/0.02% Nonidet P-40/1 mM PMSF/5 μg/ml each of leupeptin, pepstatin A, and aprotinin) containing 0.3 M sodium glutamate and 0.5% Nonidet P-40 (final concentration). The pelleted fraction, collected by centrifugation at 28,000 × g at 4°C for 20 min, was resuspended in 2.5 pellet volumes of buffer H containing 0.5 M NaCl. After Dounce homogenization, the pellet was collected by centrifugation at 28,000 × g at 4°C for 20 min, and the supernatant was recentrifuged at 34,000 × g at 4°C for 10 min. The supernatant was incubated with anti-FLAG M2 agarose overnight at 4°C with rocking using 1 ml of 50% slurry of beads per 140 mg of protein (Bradford assay). The beads were washed four times, each for 10 min, with 10 ml of buffer H containing 0.5 M NaCl. Bound protein was eluted by incubation with 1 bead volume of 1 mg/ml FLAG peptide in buffer H containing 0.5 M NaCl at 4°C for 2 h with rocking, and the eluted proteins were analyzed by SDS/PAGE and Coomassie blue or silver staining. The yield of various HsORC preparations by using this procedure was 0.1–0.2 μg per 108 cells. For additional purification, 0.2 ml of FLAG-peptide-eluted material was loaded onto a 5-ml 15–35% glycerol gradient containing buffer A (25 mM Tris·HCl, pH 7.5/0.4 M NaCl/1 mM EDTA/1 mM DTT/0.01% Nonidet P-40 plus protease inhibitors). Gradients were centrifuged at 48,000 rpm in a Sorvall ultracentrifuge for 16 h at 4°C, and fractions (≈180 μl) were collected from the bottom. The distribution of proteins across the gradient was determined by SDS/PAGE followed by Coomassie blue or silver staining.
ATPase Assay. ATP hydrolysis was measured in reactions (50 μl) containing 25 mM Hepes-KOH (pH 7.5), 5 mM magnesium acetate, 1 mM DTT, 0.1 mg/ml BSA, 100 μM [α-32P]ATP (4,000–5,000 cpm/pmol), and 0.25 μM poly(dT)300, where indicated. After incubation at 37°C for 1 h, 0.5-μl aliquots were spotted onto a polyethyleneimine-cellulose thin-layer plate and chromatographed by using 1 M formic acid/0.5 M LiCl. The extent of ATP hydrolysis was quantitated by phosphorimager (Fuji) analysis.
Nitrocellulose Filter Binding Assay. Nitrocellulose filter binding assays were carried out in reaction mixtures (15 μl) containing 25 mM Hepes-NaOH (pH 7.5), 5 mM magnesium acetate, 1 mM DTT, 0.1 mg/ml BSA, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 10 fmol of [α-32P]lamin B2 DNA (4,800 cpm/fmol), and enzyme fractions with or without 1 mM ATP, as indicated. After incubation at 4°C for 30 min, mixtures were filtered through alkaline-washed nitrocellulose HA filters (Millipore, 0.45 μm), washed with buffer containing 25 mM Hepes-NaOH (pH 7.5), 5 mM magnesium acetate, 1 mM DTT, 1 mM EDTA, 1 mM EGTA, ±1 mM ATP, and the level of 32P adsorbed to the filter was determined.
Xenopus Cell-Free Extract and Chromatin Preparations. Cytostatic factor-arrested extracts were freshly prepared as described (27). Cytostatic factor extracts were released into interphase with 0.4 mM CaCl2 and incubated at 22°C for 15 min. Immunodepletion of XlORC from extracts was carried out by using polyclonal α-XlOrc1 and α-XlOrc2 coupled to protein A-Sepharose 6MB. Mock depletions were performed with preimmune serum coupled to protein A beads. Two rounds of depletions were carried out by rotation at 4°C for 30 min and monitored by Western blotting with α-XlOrc1 and α-XlOrc2. Demembranated Xenopus sperm nuclei were prepared as described (27) and frozen in aliquots in liquid nitrogen.
Replication Assays. Replication assays (12 μl) contained 10 μl of interphase extract with FLAG-peptide-eluted HsORC preparations or buffer H, as indicated. Demembranated Xenopus sperm nuclei were added at a final concentration of 1,000 nuclei per μl, and DNA synthesis was monitored by [α-P32]dATP incorporation after 90 min at 22°C and by agarose gel electrophoresis and autoradiography.
Chromatin-Binding Assays. Reaction mixtures (50 μl) containing 45 μl of interphase extract with either FLAG-peptide-eluted HsORC proteins or buffer H and demembranated Xenopus sperm nuclei (final concentration of 10,000 nuclei per μl) were incubated at 22°C for 15 min and stopped with 0.8 ml of chromatin-isolation buffer (50 mM Hepes, pH 7.8/2.5 mM MgCl2/100 mM KCl) supplemented with 0.125% Triton X-100. Chromatin was isolated by spinning reaction mixtures at 6,000 × g for 30 min at 4°C through a 30% sucrose cushion in chromatin-isolation buffer. Pellets were resuspended in Laemmli loading buffer and analyzed by SDS/PAGE and Western blotting.
Results
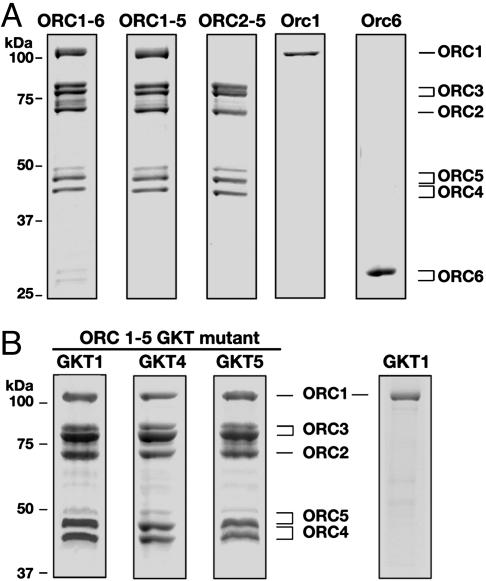
Reconstitution and Isolation of HsORCs and Subunits. Purification of HsORC and Orc subunits was facilitated by insertion of a FLAG tag at the N termini of Orc1p, Orc3p, and Orc6p. After infection of cells with viruses expressing the FLAG-tagged Orc3p and other untagged Orc subunits, the HsORC1-6, HsORC1-5, and HsORC2-5 preparations were purified by M2-agarose-affinity chromatography. SDS/PAGE and Coomassie staining of the HsORC1-6 and HsORC1-5 revealed the presence of variable levels of Orc subunits with mobilities consistent with the molecular weights of the human subunits (Fig. 1A) (23, 28). The HsORC1-6 preparation, however, contained levels of Orc6p that were barely detected by Coomassie staining, but the presence of Orc6p was confirmed by Western blotting (data not presented). Treatment of preparations with λ phosphatase did not significantly alter the migration of the subunits from those shown in Fig. 1A (data not presented).
Fig. 1.
Isolation of wild-type and GKT mutant HsORCs. (A) Subunits present in HsORC and individual subunits were isolated as described in Materials and Methods. FLAG-peptide-eluted proteins were analyzed by SDS/PAGE, and individual subunits were detected by Coomassie blue staining. The level of protein loaded was 2 μg for each complex and 0.5 μg each of Orc1p and Orc6p. (B) Analysis of HsORC1-5 and HsOrc1 containing mutant GKT motifs. Complexes were isolated as described in Materials and Methods. The level of protein loaded was 5 μg of each complex and 0.5 μg of Orc1 GKT.
All eukaryotic ORCs contain three subunits (Orc1, Orc4, and Orc5) that are members of the AAA+ family of ATPases. To characterize the functional role of the Walker A motifs of the HsOrc1, HsOrc4, and HsOrc5 subunits in HsORC, complexes containing these mutations were generated. SDS/PAGE analysis of the different GKT mutants of HsORC1-5 preparations after M2-affinity chromatography are shown in Fig. 1B. The ratio of subunits present in the various mutant HsORC1-5 preparations was similar to those of the wild-type complex. The corresponding GKT mutants of the HsORC1-6 were also prepared. Both mutant and wild-type complexes contained similar low levels of HsOrc6p. In addition to the HsORC preparations described above, FLAG-tagged HsOrc6p and HsOrc1p and the HsOrc1p GKT mutant were isolated. These preparations contained primarily a single protein band of high purity (Fig. 1).
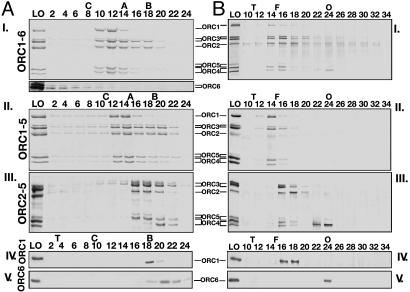
The FLAG-peptide-eluted HsORC preparations described above were purified further by glycerol-gradient sedimentation and sizing-column separation (Fig. 2). Sedimentation of the wild-type ORC1-6 and ORC1-5 revealed the presence of a five-subunit complex that cosedimented to a position between the catalase (220 kDa) and aldolase (150 kDa) markers (Fig. 2 AI and AII). A substantial level of subunits that sedimented more slowly was detected. Surprisingly, the low level of HsOrc6p present in the immunopurified HsORC1-6 did not cosediment with the other Orc subunits and appeared aggregated (lower part of Fig. 2AI). The ratios of subunits present in both the HsORC1-6 and HsORC1-5 glycerol-gradient fractions (fractions 10–12 and 12–14, respectively) were almost identical and more stoichiometric than the material loaded onto the gradients (FLAG-peptide-eluted material; see Fig. 1A). The ratio of the subunits Orc1p:Orc2p:Orc3p:Orc4p + Orc5p in the HsORC1-5 preparation determined by densitometric scanning (with Orc2 set as 1) was 1:1:1.31:2.03, similar to that observed with the HsORC1-6. The level of Orc4 and Orc5 are reported as a sum, because Western blotting detected overlapping bands of each subunit. Sizing-column separation of these complexes also indicated the presence of a five-subunit complex that eluted in the ferritin region (440 kDa) and smaller subcomplexes that eluted later (Fig. 2 BI and BII). Both gel-filtration and glycerol-gradient sedimentation of the HsORC2-5 indicated that this complex was less stable than HsORC1-5 (Fig. 2AIII and BIII). Variable dissociation of Orc4p from the complex was noted. HsOrc1p and HsOrc6p yielded single protein peaks after glycerol-gradient centrifugation and filtration (Fig. 2AIV, AV, BIV, and BV). Based on their sedimentation coefficients and Stokes radii, both HsORC1-6 and HsORC1-5 preparations possessed a molecular mass of 270 kDa, calculated by using the Siegel–Monty equation (29), somewhat lower than the molecular mass based on their amino acid composition (346 kDa, HsORC1-5), suggesting that the complexes were monomeric in structure. The calculated frictional coefficient of HsORC1-5 was 1.50, suggesting that the complex is elongated in shape. Similar calculations for Orc1 and Orc6 indicated that they, too, were monomeric.
Fig. 2.
Glycerol-gradient sedimentation and gel filtration analyses of the HsORCs. FLAG-peptide-eluted proteins were sedimented through 15–35% glycerol gradient (A) or chromatographed over a Superdex 200 gel-filtration column (B) and then analyzed by SDS/PAGE and Coomassie blue or silver staining, respectively. The distribution of Orc6 presented below the ORC1-6 gel (AI) was a silver-stained gel of the same fractions and only shows the distribution of Orc6p. The LO (load on) lanes represent the material loaded onto gradients or sizing columns. For gradient analysis (A), the levels of protein loaded (LO) were 2 μg (ORC1-6), 3.5 μg (ORC1-5), 6 μg (ORC2-5), and 1 μg (Orc1p and Orc6p); for gel-filtration analysis, the amount was ≈100 ng per subunit (B). The volume of each glycerol-gradient fraction (≈180 μl total) gel-loaded was 5 μl each (ORC1-6 and ORC1-5), 20 μl (ORC2-5), and 10 μl each (Orc1p and Orc6p), whereas 15 μl of the gel-filtration fractions (50 μl total) was used. Protein markers: C, catalase (232 kDa); A, aldolase (158 kDa); B, albumin (67 kDa); T, thyroglobin (669 kDa); F, ferritin (440 kDa); and O, ovalbumin (43 kDa).
The properties of the different HsORC1-5 GKT mutant preparations were examined. The ratio of subunits present in these complexes, their sedimentation in glycerol gradients, and elution from the sizing columns were indistinguishable from the wild-type complex (data not presented).
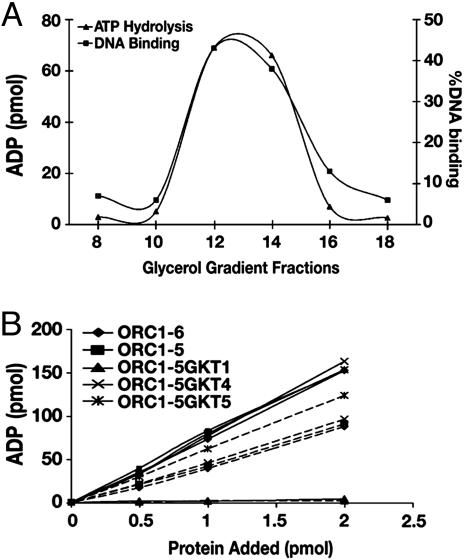
ATP Hydrolysis and DNA-Binding Properties of Wild-Type and Mutant HsORCs. We characterized the ATPase activity of the various HsORC preparations derived from the peak fractions after glycerol-gradient sedimentation. As shown in Fig. 3A, ATPase activity cosedimented with HsORC1-5. The rate of ATP hydrolysis, catalyzed by HsORC1-6 and HsORC1-5, was stimulated maximally 2-fold by various synthetic single-stranded DNAs [poly(dT), poly(dA), and poly(dC)] and inhibited 3- to 5-fold by double-stranded DNAs [poly(dA-dT)•(dA-dT) and poly(dA-dC)•(dG-dT)] (data not presented). In the presence or absence of poly(dT)300, the rate of ATP hydrolysis was linear up to 1.5 h with a Vmax of 1.3 or 0.65 pmol of ATP hydrolyzed per pmol of protein per min, respectively, with either HsORC1-6 or HsORC1-5 (Fig. 3B). ATPase activity was not detected with the HsORC2-5, suggesting that the Orc1 subunit is critical for the ATPase activity of the HsORC (data not presented). To further characterize the subunits contributing to its ATPase activity, HsORCs containing mutation in the Walker A motif of Orc1p, Orc4p, or Orc5p were assayed. The complex with a mutated Orc1 subunit (HsORC1-5 GKT1) was devoid of ATPase (Fig. 3B), whereas the Orc4 and Orc5 mutants were as active as the wild-type complex in the presence of poly(dT)300. In the absence of poly(dT)300, the ORC1-5 GKT5 mutant was reproducibly marginally more active than the other complexes. Thus, like the ScORC and DmORC, the Walker A motif of the Orc1p of HsORC is essential for ATP hydrolysis.
Fig. 3.
Cosedimentation of activities associated with HsORC1-5 and ATPase activity of wild-type and mutant HsORCs. (A) Cosedimention of ATPase and DNA-binding activities with ORC subunits. Glycerol-gradient centrifugation of HsORC1-5 was carried out as described for Fig. 2. An aliquot (2 μl) of the gradient fractions indicated was assayed for ATPase activity as described in Materials and Methods in the presence of poly(dT)300. DNA-binding assays were carried out as described in Materials and Methods with an aliquot (0.1 μl) of the indicated fraction. The distribution of the complex across the gradient was analyzed as described for Fig. 2. (B) ATPase activity associated with wild-type and mutant HsORC1-5. Reaction mixtures prepared and analyzed as described in Materials and Methods were incubated for 1 h at 37°C in the presence (—) or absence (- - -) of poly(dT)300, and ADP formed is shown.
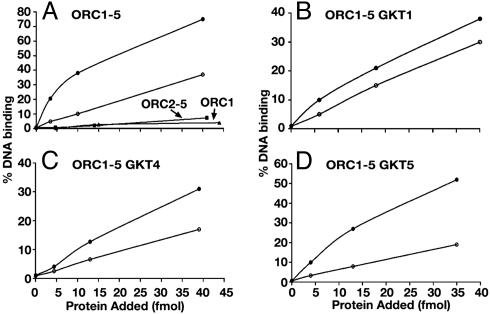
The DNA-binding activity of the wild-type HsORC and HsORC1-5 GKT mutants were determined by using a nitrocellulose filter binding assay with lamin B2 DNA as substrate. No competitor DNA was used in these experiments. Both the DNA-binding activity and the HsORC1-5 cosedimented during glycerol-gradient centrifugation, indicating that this activity was intrinsic to the complex (Fig. 3A). In the presence of increasing levels of protein, wild-type HsORC1-5 and HsORC1-6 (data not presented) bound DNA identically and were stimulated ≈3- to 5-fold by ATP (Fig. 4A). At low protein levels (5 fmol), these complexes bound 2.5 fmol of DNA in the presence of ATP. The HsORC1-5 GKT mutants bound DNA less efficiently than the wild-type complex. In the presence of ATP and at a low protein level (5 fmol), the HsORC1-5 GKT1, GKT4, and GKT5 mutants bound DNA ≈70%, 80%, and 50% less efficiently than the wild-type complex, respectively. The DNA-binding activity observed with the HsORC1-5 GKT1, GKT4, and GKT5 preparations was stimulated by ATP 1.3-, 1.5-, and 2.5-fold, respectively.
Fig. 4.
DNA-binding activity of wild-type and GKT mutant HsORC preparations. Various ORC preparations purified by glycerol-gradient sedimentation were incubated with 10 fmol of [32P]lamin B2 DNA in the presence (•) or absence (○) of ATP as described in Materials and Methods. Preparations of HsORC2-5 and Orc1p were also assayed in the presence of ATP (as shown in A).
The HsORC2-5, devoid of Orc1p, did not bind DNA, nor did the HsOrc1 subunit alone (Fig. 4A). These findings suggest that the HsOrc1p plays a role in the DNA-binding activity of the HsORC1-5 even in the absence of ATP.
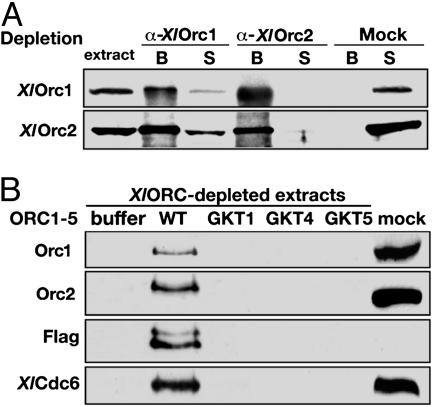
Analysis of the Replication Activity of Various HsORC Preparations by Xenopus Cell-Free Extracts. A fundamental property of ORC is its ability to bind chromosomes and support the recruitment of Cdc6p and the minichromosome maintenance (MCM) complex to form the pre-RC. This function is critical in replication. We examined the binding of various HsORC preparations to chromatin and the subsequent loading of XlCdc6p in Xenopus cell-free extracts. Endogenous XlORC was quantitatively depleted from the extract by using a combination of antibodies against XlOrc1p and XlOrc2p (Fig. 5A;also see Materials and Methods). After addition of recombinant HsORCs to ORC-depleted extracts, the formation of chromatin-bound protein was assessed by SDS/PAGE followed by Western blotting (Fig. 5B). Both HsOrc1p and HsOrc2p were detected with XlOrc1p and XlOrc2p antibodies, because they crossreacted with the human proteins, whereas HsOrc3p, which was FLAG-tagged, was detected with anti-FLAG monoclonal antibody. As shown (Fig. 5B, lane 2), depleted extracts supplemented with wild-type HsORC1-5 supported the association of HsOrc1, HsOrc2, and HsOrc3 subunits, XlCdc6, and the XlMCM complex (data not shown). This result established that recombinant HsORC can substitute for XlORC in the extract and support two of the critical roles of ORC (chromatin assembly and pre-RC recruitment), similar to previous results (18). In contrast, none of the HsORC1-5 GKT mutant preparations bound to chromatin or recruited XlCdc6 (Fig. 5B), which indicates that intact Walker A domains are required for all three Orc1, Orc4, and Orc5 subunits to support productive chromatin association of the HsORC.
Fig. 5.
Assembly of HsORC complex onto chromatin requires the wild-type Walker A motifs. (A) XlORC-depleted and mock-depleted extracts (1 μl) were analyzed on SDS/PAGE, followed by Western blot with anti-XlOrc1p (Upper) and anti-XlOrc2p (Lower) antibodies. The lanes labeled “B” refer to proteins associated with antibody-coupled protein A-Sepharose beads, and lanes labeled “S” refer to proteins present in depleted supernatant extracts. (B) Chromatin-binding assays were performed as described in Materials and Methods. FLAG-peptideeluted recombinant complexes, wild-type HsORC1-5 (WT), and GKT mutant complexes (GKT1, GKT4, and GKT5), 1 μg of each, were incubated with XlORC-depleted extracts supplemented with sperm nuclei. No protein was added to ORC-depleted extracts (buffer) or to untreated extracts (mock). Chromosomal DNA was purified from each reaction, and chromatin-bound proteins were detected by SDS/PAGE and Western blotting with anti-Xenopus Orc1 and Orc2 antibodies, anti-FLAG antibody, and anti-Xenopus Cdc6 antibody. The anti-FLAG antibody was used to detect the human FLAG-tagged Orc3p.
Next we examined whether the different HsORC and HsORC GKT mutants supported DNA replication in cell-free extracts derived from Xenopus eggs. Previous studies demonstrated that the DNA-replication activity of X. laevis extracts can be selectively inhibited by the immunodepletion of XlORC and that supplementation of the depleted extracts with either HsORC or DsORC preparations restored DNA-replication activity (18, 30, 31).
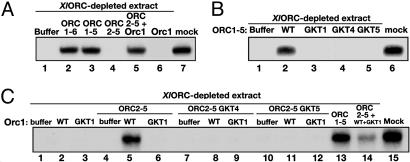
Supplementation of XlORC-depleted extracts (Fig. 6A) with either wild-type HsORC1-6 or HsORC1-5 supported X. laevis sperm DNA replication to levels observed with mock-depleted extracts (Fig. 6A, compare lanes 2 and 3 to lane 6). The addition of HsOrc6p with either of these complexes did not affect the level of replication (data not presented). Neither HsORC2-5 nor HsOrc1p alone supported DNA replication by depleted extracts (Fig. 6A, lanes 4 and 6), whereas their combination restored DNA-replication activity to a level near that of mock-depleted extract. In agreement with the chromatin-binding data, this result showed that both HsORC1-6 and HsORC1-5 supported all known replication functions in Xenopus extracts (Fig. 6A, lane 5). Furthermore, these findings suggest that HsOrc1p can interact with the HsORC2-5. In contrast to wild-type HsORC1-5, the three HsORC1-5 preparations, containing a mutated Walker A motif of the Orc1, Orc4, or Orc5 subunit, did not support DNA replication (Fig. 6B). These findings correlate with the chromatin-binding data (Fig. 5B). Furthermore, only wild-type HsORC2-5 supplemented with wild-type Orc1p rescued DNA replication in ORC-depleted extracts (Fig. 6C). It is interesting to note that when equal amounts of wild-type and mutated GKT HsOrc1 subunits were added to the depleted extract with the wild-type HsORC2-5, the level of replication was lower than that observed with the wild-type HsOrc1 + HsORC2-5 (Fig. 6C, compare lanes 5 and 14). It is likely that the two different HsOrc1 subunits competed for the input HsORC2-5 and that the HsORC1-5 reconstituted with the mutant Orc1p is replication-incompetent. This proposal further suggests that the inability of mutated GKT-Orc1 to support replication when combined with HsORC2-5 is not caused by its failure to assemble with HsORC2-5.
Fig. 6.
HsOrc6p is not required for recombinant complex to support DNA replication, whereas Walker A motifs are essential. DNA-replication assays were performed as described in Materials and Methods. Genomic DNA replication was monitored by incorporation of [α-32P]dATP followed by agarose gel electrophoresis. (A) FLAG-peptide-eluted wild-type ORC1-6 (lane 2), ORC1-5 (lane 3), ORC2-5 (lane 4), and ORC2-5 + Orc1p (lane 5) were added to XlORC-depleted extracts at 80 ng per Orc subunit. (B) FLAG-peptide-eluted wild-type HsORC1-5 (lane 2) and GKT mutant complexes (lanes 3–5) were added at 400 ng each to XlORC-depleted extracts. (C) Wild-type or mutant Orc1p (80 ng each) was added either alone (lanes 2 and 3) or together with FLAG-peptide-eluted wild-type (lanes 5 and 6) or GKT mutant HsORC2-5 (320 ng) (lanes 8, 9, 11, and 12) in XlORC-depleted extracts. Lane 13, ORC1-5 (400 ng); lane 14, ORC2-5 (320 ng) and Orc1p (80 ng) wild type and GKT mutant (80 ng); lane 15, mock-depleted extract.
Taken together, our findings suggest that the HsORC1-5, devoid of the HsOrc6 subunit, and the HsORC1-6, containing substoichiometric levels of Orc6p, support replication in the Xenopus cell-free system. These observations indicate that Orc1p is essential for the association of HsORC with chromatin, formation of the pre-RC, and replication. These findings are similar to the crucial role played by this subunit in the function of the ScORC and DmORC. However, the results described in the Fig. 6B legend differ from those reported for the DmORC containing either a mutated Walker A motif in Orc4p or Orc5p. Both mutant DmORC variants formed stable chromatin complexes and supported replication in the Drosophila cell-free system (17). One difference between the HsORC and DmORC is that the latter contains a stoichiometric level of the Orc6 subunit, whereas the former lacks this subunit. However, we find that HsORC1-5 is as efficient as HsORC1-6 (with a low Orc6p content) for chromatin loading and replication, arguing that HsOrc6p is dispensable in our experimental system. In addition, our data support an important role for intact Walker A motifs in Orc4 and Orc5, because the inability of the HsORC1-5 GKT4 and HsORC1-5 GKT5 to support replication correlated with their inability to bind chromatin and reduced in vitro DNA-binding activity.
Discussion
In this study, the properties of the wild-type HsORC and mutants with altered Walker A motifs in Orc1, Orc4, or Orc5 subunits were examined. We show that HsORC, purified from Sf9 cells infected with viruses expressing all six Orc subunits, contained near-stoichiometric amounts of the Orc1p–Orc5p polypeptides but only low levels of HsOrc6p. These results are similar to those reported by Vashee et al. (18, 23). HsORC containing low levels of Orc6p and HsORC1-5, lacking Orc6p, bound to DNA, hydrolyzed ATP, and supported pre-RC formation and DNA replication in X. laevis extracts identically. However, HsORC2-5, devoid of Orc1p, possessed none of these activities.
All eukaryotic ORCs contain ATPase and duplex DNA-binding activities. The properties of the ATPase activity associated with HsORC1-5 are similar to those observed with both DmORC and ScORC. All three ORCs require a functional Orc1p Walker A motif to support ATP hydrolysis, whereas mutations of this motif in Orc4p and Orc5p do not affect this activity. The ATPase activities of DmORC, ScORC, and HsORC are stimulated by single-stranded DNA ≈2-fold and inhibited by duplex DNAs (17, 21, 22). In contrast, the DNA-binding properties of these ORCs differ. Only the ScORC interacts specifically with an origin sequence, and this interaction shows an absolute dependency on ATP. The DNA-binding activities of DmORC and HsORC1-5 are similar, because they bind to AT-rich duplex DNAs with no apparent sequence specificity. The DNA-binding activity of DmORC is stimulated 10- to 20-fold by ATP, whereas DNA binding by HsORC is stimulated only 3- to 5-fold (17). DmORC and ScORC require a functional ATP-binding site only in Orc1 for DNA binding. Mutation of the Walker A motif in HsOrc1p, HsOrc4p, or HsOrc5p reduced the DNA-binding activity of the HsORC1-5 between 2- and 5-fold, and the stimulation of DNA binding by ATP observed with HsORC1-5 was reduced only slightly by the GKT mutations.
A critical property of ORC is its ability to support DNA replication. HsORC1-5 addition to X. laevis extracts depleted of XlORC resulted in the formation of the pre-RC and DNA replication (Figs. 5 and 6). Similar observations have been reported for HsORC (18) and DmORC (31). In contrast to wild-type HsORC1-5, none of the GKT Walker A motif mutants supported replication or associated with chromatin when incubated with the X. laevis extracts. The lack of activity observed with the HsORC1-5 GKT1 is similar to that reported for DmORCs and ScORCs containing a mutant Orc1p. However, our findings that HsORC1-5 must contain intact P loops in both Orc4p and Orc5p to support replication in the Xenopus cell-free system differ from those made with ScORCs and DmsORCs. HsORC1-5 containing mutations in Orc4 or Orc5 displayed full ATPase activity in vitro but did not support pre-RC assembly and replication in cell-free extracts, which suggests that nucleotide binding by Orc4 and Orc5 is essential for chromatin binding and subsequent pre-RC assembly and replication.
We have no explanation for these differences, especially between the DmORC and HsORC, which contain almost identical Walker A and B motifs in their Orc4 and Orc5 subunits. However, other differences between DmORC and HsORC have been noted, particularly concerning the function of the Orc6p. As described above, Orc6p is associated stably with DmORC but not with HsORC. In contrast to HsORC1-5, DmORCΔ6 did not associate with chromatin or promote replication in Drosophila cell-free extracts (17). Our results, described in the legends to Figs. 5 and 6, however, do not rule out the possibility that X. laevis extracts depleted of XlORC contain a pool of free XlOrc6p that interacts with the HsORC1-5 to form the six-subunit complex that is replication-competent.
It is likely that ATP binding to the AAA+ subunits in ORC induces conformational changes in the complex. Studies with ScORC indicate that the Walker A motif of Orc1p and Orc5p bind ATP. Orc4p, because of its modified Walker A motif, does not bind ATP directly but can be crosslinked to ATP, provided Orc1p contains an ATP-binding site, suggesting that the formation of an ATP–Orc1p complex influences the structure of the other Orc subunits. The function of ATP interaction with Orc5p is unclear, but mutation of its Walker A motif reduced ORC functions in vivo (32). Marked conformational changes of ScORC occur after binding single-stranded DNA (22). Although this alteration was ATP-independent, it indicates that the complex shows some structural plasticity.
The role of the GKT motifs of Orc4p and Orc5p in the function of HsORC is unknown. Additional studies on the binding of ATP to the HsOrc1, HsOrc4, and HsOrc5 subunits of HsORC and complexes containing mutated Walker A motifs may help evaluate their role. How these subunits participate in the interaction of HsORC with other critical proteins, however, will require a more systematic analysis of the formation of the mammalian pre-RC.
Acknowledgments
We thank Dr. Vladimir Bermudez for his critical help during these studies. This work was supported by National Institutes of Health Grants R01 CA92245 (to J.G.) and GM 34559 (to J.H.).
Author contributions: J.G.-C., C.Y.Y., J.G., and J.H. designed research; J.G.-C., C.Y.Y., J.G., and J.H. performed research; J.G.-C., C.Y.Y., J.G., and J.H. contributed new reagents/analytic tools; J.G.-C., C.Y.Y., J.G., and J.H. analyzed data; and J.G.-C., C.Y.Y., J.G., and J.H. wrote the paper.
Abbreviations: ORC, origin recognition complex; pre-RC, prereplication complex; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe; Dm, Drosophila melanogaster; Xl, Xenopus laevis; Hs; Homo sapiens.
References
- 1.Diffley, J. F. (2001) Curr. Biol. 11, R367-R370. [DOI] [PubMed] [Google Scholar]
- 2.Kelly, T. J. & Brown, G. W. (2000) Annu. Rev. Biochem. 69, 829-880. [DOI] [PubMed] [Google Scholar]
- 3.Bell, S. P. & Stillman, B. (1992) Nature 357, 128-134. [DOI] [PubMed] [Google Scholar]
- 4.Rao, H. & Stillman, B. (1995) Proc. Natl. Acad. Sci. USA 92, 2224-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell, S. P. (2002) Genes Dev. 16, 659-672. [DOI] [PubMed] [Google Scholar]
- 6.Dutta, A. & Bell, S. P. (1997) Annu. Rev. Cell Dev. Biol. 13, 293-332. [DOI] [PubMed] [Google Scholar]
- 7.Todorovic, V., Falaschi, A. & Giacca, M. (1999) Front Biosci. 4, D859-D868. [DOI] [PubMed] [Google Scholar]
- 8.Clyne, R. K. & Kelly, T. J. (1997) Methods 13, 221-233. [DOI] [PubMed] [Google Scholar]
- 9.Antunes, D. F., Kim, S. M., Huberman, J. A. & de Morais, M. A. (2003) Plasmid 50, 113-119. [DOI] [PubMed] [Google Scholar]
- 10.Lee, J. K., Moon, K. Y., Jiang, Y. & Hurwitz, J. (2001) Proc. Natl. Acad. Sci. USA 98, 13589-13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chuang, R. Y., Chretien, L., Dai, J. & Kelly, T. J. (2002) J. Biol. Chem. 277, 16920-16927. [DOI] [PubMed] [Google Scholar]
- 12.Kong, D. & DePamphilis, M. L. (2001) Mol. Cell. Biol. 21, 8095-8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalejta, R. F., Li, X., Mesner, L. D., Dijkwel, P. A., Lin, H. B. & Hamlin, J. L. (1998) Mol. Cell 2, 797-806. [DOI] [PubMed] [Google Scholar]
- 14.Bielinsky, A. K., Blitzblau, H., Beall, E. L., Ezrokhi, M., Smith, H. S., Botchan, M. R. & Gerbi, S. A. (2001) Curr. Biol. 11, 1427-1431. [DOI] [PubMed] [Google Scholar]
- 15.Ladenburger, E. M., Keller, C. & Knippers, R. (2002) Mol. Cell. Biol. 22, 1036-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdurashidova, G., Danailov, M. B., Ochem, A., Triolo, G., Djeliova, V., Radulescu, S., Vindigni, A., Riva, S. & Falaschi, A. (2003) EMBO J. 22, 4294-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chesnokov, I., Remus, D. & Botchan, M. (2001) Proc. Natl. Acad. Sci. USA 98, 11997-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vashee, S., Cvetic, C., Lu, W., Simancek, P., Kelly, T. J. & Walter, J. C. (2003) Genes Dev. 17, 1894-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Remus, D., Beall, E. L. & Botchan, M. R. (2004) EMBO J. 23, 897-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, D. G. & Bell, S. P. (2000) Curr. Opin. Cell Biol. 12, 280-285. [DOI] [PubMed] [Google Scholar]
- 21.Klemm, R. D., Austin, R. J. & Bell, S. P. (1997) Cell 88, 493-502. [DOI] [PubMed] [Google Scholar]
- 22.Lee, D. G., Makhov, A. M., Klemm, R. D., Griffith, J. D. & Bell, S. P. (2000) EMBO J. 19, 4774-4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vashee, S., Simancek, P., Challberg, M. D. & Kelly, T. J. (2001) J. Biol. Chem. 276, 26666-26673. [DOI] [PubMed] [Google Scholar]
- 24.Gillespie, P. J., Li, A. & Blow, J. J. (2001) BMC Biochem. 2, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, D. G. & Bell, S. P. (1997) Mol. Cell. Biol. 17, 7159-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chesnokov, I. N., Chesnokova, O. N. & Botchan, M. (2003) Proc. Natl. Acad. Sci. USA 100, 9150-9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray, A. W. (1991) Methods Cell Biol. 36, 581-605. [PubMed] [Google Scholar]
- 28.Dhar, S. K., Delmolino, L. & Dutta, A. (2001) J. Biol. Chem. 276, 29067-29071. [DOI] [PubMed] [Google Scholar]
- 29.Siegel, L. M. & Monty, K. J. (1966) Biochim. Biophys. Acta 112, 346-362. [DOI] [PubMed] [Google Scholar]
- 30.Carpenter, P. B., Mueller, P. R. & Dunphy, W. G. (1996) Nature 379, 357-360. [DOI] [PubMed] [Google Scholar]
- 31.Chesnokov, I., Gossen, M., Remus, D. & Botchan, M. (1999) Genes Dev. 13, 1289-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loo, S., Fox, C. A., Rine, J., Kobayashi, R., Stillman, B. & Bell, S. (1995) Mol. Biol. Cell 6, 741-756. [DOI] [PMC free article] [PubMed] [Google Scholar]