Abstract
Human immunodeficiency virus type 2 (HIV-2) is generally considered capable of using a broad range of coreceptors. Since HIV-2 variants from individuals with nonprogressive infection were not studied previously, the possibility that broad coreceptor usage is a property of variants associated with progressive infection could not be excluded. To test this, we determined the coreceptor usage of 43 HIV-2 variants isolated from six long-term-infected individuals with undetectable plasma viremia. Using GHOST indicator cells, we showed for the first time that the only coreceptors efficiently used by low-pathogenic HIV-2 variants are CCR5, GPR15 (BOB), and CXCR6 (BONZO). Surprisingly, control HIV-2 variants (n = 45) isolated from seven viremic individuals also mainly used these three coreceptors, whereas use of CCR1, CCR2b, or CCR3 was rare. Nearly a quarter of all HIV-2 variants tested could infect the parental GHOST cells, which could be partially explained by CXCR4 usage. Use of CXCR4 was observed only for HIV-2 variants from viremic individuals. Thirty-eight variants from aviremic and viremic HIV-2-infected individuals were additionally tested in U87 cells. All except one were capable of infecting the parental U87 cells, often with high efficiency. When virus production in parental cells was regarded as background in the coreceptor-transduced cell lines, the results in U87 cells were largely in agreement with the findings in GHOST cells. HIV-2 isolates from aviremic individuals commonly use as coreceptors CCR5, GPR15, and CXCR6, as well as an unidentified receptor expressed by U87 cells. Broad coreceptor usage, therefore, does not appear to be associated with pathogenicity of HIV-2.
Human immunodeficiency viruses (HIV) use CD4 in combination with a chemokine receptor to enter target cells. The first chemokine receptors recognized to function as coreceptors for HIV were CCR5 and CXCR4 (19, 24, 27). Since then, the capacities of numerous members of the chemokine receptor family, e.g., CCR1 through CCR8, CXCR6 (BONZO), GPR15 (BOB), GPR1, APJ, CX3CR1 (V28), CXCR5, and RDC1, to support HIV infection in vitro have been studied (12, 13, 15, 22, 26, 31, 34, 46, 49).
To study which chemokine receptors HIV variants can use for cell entry, cell lines that are nonpermissive for HIV and that have been engineered to express CD4 and a chemokine receptor of interest are generally used. Derivatives of a human glioma cell line (U87) and a human osteosarcoma cell line (HOS or GHOST) have predominantly been used for this purpose. Based on the capacity to productively infect these indicator cell lines, it has been shown that most primary HIV type 1 (HIV-1) variants are restricted to the use of CCR5, although variants exist that can also use CXCR4 and/or CCR3 (4, 8, 16, 21, 45, 55, 59). Only a minority of HIV-1 variants have been shown to be capable of infecting cells expressing other coreceptors, like CCR2b (4, 16), CCR8 (21, 61), CXCR6 (BONZO), or GPR15 (BOB) (8, 21, 59, 61). In contrast, many HIV-2 variants could infect a whole range of indicator cells expressing different coreceptors, indicating a more promiscuous nature of HIV-2 with respect to coreceptor usage (29, 37, 38, 42, 51).
In general, HIV-1 infection is established by CCR5-using variants (R5 variants), whereas CXCR4-using variants (X4 variants) evolve during the course of infection in about half of HIV-1-infected individuals (4, 16, 52). Differential expression of chemokine receptors within CD4-positive cells creates distinct target cell populations and contributes to the different tropisms of HIV variants. Thus, R5 and X4 HIV-1 variants have been shown to infect partially distinct T-lymphocyte subpopulations in vivo (6, 55). The evolution of variants with the capacity to use extra or alternative coreceptors broadens or alters the target cell population and may thereby influence the course of infection and pathogenesis. For HIV-1, the appearance of X4 variants and the acquired capacity to infect thymocytes and naïve T cells coincides with accelerated CD4+-T-cell loss and disease progression (2, 6, 33).
The observation that HIV-2 is generally capable of using a large number of different chemokine receptors as coreceptors suggests that HIV-2 has a broader tropism than HIV-1. Indeed, a broader in vitro tropism for human and animal cell lines has been observed (14, 36). The observed broad coreceptor usage and tropism seem to conflict with the lower pathogenicity of HIV-2 (3, 17, 28, 44). Many HIV-2-infected individuals remain AIDS free for prolonged periods and have low or no detectable levels of viral RNA in their plasma (35). However, the vast majority of HIV-2 variants studied so far have been isolated from HIV-2-infected individuals with clinical symptoms, and the coreceptor usage of variants from individuals with a known nonprogressive course of HIV-2 infection was not determined. Therefore, the possibility that broad coreceptor usage is a feature associated with more pathogenic HIV-2 variants could not be excluded. This bias in selection of viruses can be attributed to the difficulty of isolating virus from asymptomatic HIV-2-infected individuals with high CD4 counts (1, 18, 50), a problem which is likely related to the low viral loads in these individuals (3, 17, 44).
Using an optimized HIV coculture protocol, we previously isolated virus variants from six long-term-nonprogressing HIV-2-infected individuals who had undetectable plasma RNA loads (<500 RNA copies ml−1) for 4 to 10 years (5). In the present study, we established the coreceptor usage of these HIV-2 variants using GHOST and U87 indicator cell lines, and we show for the first time that the majority of HIV-2 variants of aviremic HIV-2-infected individuals efficiently use CCR5, GPR15 (BOB), CXCR6 (BONZO), and an unidentified coreceptor expressed by U87 cells. The coreceptor usage of control variants isolated from individuals with progressive infection was also largely restricted to these chemokine receptors, although some variants used CXCR4 instead. Apparently, the use of CCR5, GPR15, and CXCR6 is a common property of HIV-2 and is not associated with the level of disease progression and pathogenicity.
MATERIALS AND METHODS
Subjects.
Biological virus clones were isolated from peripheral blood mononuclear cells (PBMC) of six HIV-2-infected individuals from the Rotterdam cohort (54) who had undetectable plasma loads (<500 copies ml−1) at multiple time points during a follow-up ranging from 4 to 10.5 years (Table 1). None of the individuals received antiretroviral therapy. In the last 1.5 years of follow-up, an ultrasensitive reverse transcriptase (RT) PCR revealed that the individuals had even less than 50 copies ml−1 at at least one time-point analyzed. CD4+-T-cell numbers were stable and ranged from 550 to 900 μl−1 at the moment of analysis. As a control, biological clones were isolated from seven individuals with low to high plasma RNA loads (1,300 to 330,000 copies ml−1). One of these individuals was asymptomatic, with slightly reduced but stable CD4 counts; the other six had a progressive course of infection, as evidenced by clinical symptoms and/or CD4 counts of <200 μl−1 (Table 1).
TABLE 1.
Characteristics of HIV-2-infected individuals
| Patient | Nationalitya | HIV-2 subtype | Follow-up (mo)b | CDC stagec | CD4/μlc | RNA/mlc | Total fu (mo/yr)d | No. of clones analyzed |
|---|---|---|---|---|---|---|---|---|
| Aviremic HIV-2 infection | ||||||||
| RH2-3 | CVI | A | 96.2 | A1 | 770 | <500 | 126.8/10.6 | 10 |
| RH2-4 | NL | A | 85.5 | A1 | 860 | <500 | 112.3/9.4 | 3 |
| RH2-8 | GH | B | 80.3 | A1 | 900 | <500 | 106.9/8.9 | 1 |
| RH2-13 | CVI | A | 52.0 | A1 | 900 | <500 | 82.4/6.9 | 10 |
| RH2-14 | CVI | A | 50.7 | A1 | 550 | <500 | 75.1/6.3 | 9 |
| RH2-22 | CVI | B | 30.0 | A1 | 670 | <500 | 46.6/3.9 | 10 |
| Viremic HIV-2 infection | ||||||||
| RH2-15a | GB | A | 20.8 | A2 | 410 | 1,300 | 70.8/5.9 | 2 |
| RH2-15b | GB | A | 42.9 | A2 | 400 | 14,000 | 70.8/5.9 | 3 |
| RH2-1 | CVI | A | 56.5 | C1 | 240 | NT | NA | 6 |
| RH2-5 | CVI | A | 38.0 | C3 | 120 | 110,000 | NA | 6 |
| RH2-7 | CVI | A | 32.5 | A3 | 10 | + | NA | 6 |
| RH2-21 | CVI | A | 1.4 | C3 | 60 | 59,000 | NA | 2 |
| RH2-24 | CVI | A | 3.8 | A3 | 70 | 23,000 | NA | 10 |
| RH2-26 | NL | B | 0.9 | C3 | 10 | 330,000 | NA | 10 |
CVI, Cape Verde Islands; GH, Ghana; GB, Guinea Bissau; NL, Netherlands.
Time between first visit to the clinic and the sample of virus isolation.
CDC stage, CD4 numbers and RNA copies at the time point of virus isolation, except RH2-4, RH2-21, and RH2-24, in which cases the previous time point (viremic individuals) or the next time point (aviremic individual) was taken. The time points of virus isolation and CD4/RNA determination were always maximally 6 months apart. NT, not tested; +, positive.
Follow-up (fu) until the last visit to the clinic by August 2003 for the asymptomatic individuals. NA, not applicable.
Isolation of viruses.
Viruses were isolated as described previously (5). Briefly, large quantities of PBMC isolated from fresh blood (aviremic individuals) or cryopreserved PBMC (viremic individuals) were depleted of CD8+ T cells, and a total of 4 · 107 to 1.8 · 108 (aviremic individuals) and 5 · 104 to 2 · 106 (viremic individuals) cells were cocultured in several dilutions with freshly phytohemagglutinin-stimulated CD8-depleted healthy donor peripheral blood lymphocytes in 24- or 96-well plates and maintained for 28 to 40 days.
Propagation of virus stocks.
For each participant, virus stocks were grown from a selection of 10 positive wells (i.e., 10 biological clones) or from all positive wells if <10 were positive (Table 1). Wells were selected to contain a representative mix with respect to the first day of detection of virus production. Further selection from wells that were virus positive on the same day was random. Cultures with the lowest patient cell concentrations were used to maximize the probability that virus variants were clonal. Clonality was confirmed retrospectively by sequence analysis of the V1-to-V3 region of the envelope gene from cultured viruses (not shown). The same sequence analysis also confirmed that the virus variants were HIV-2 subtypes A and B (data not shown). For propagation, half of the cells in virus-positive wells were mixed with 3 · 106 freshly phytohemagglutinin-stimulated (1 μg/ml; Murex; Biotrading Benelux BV, Mijdrecht, The Netherlands) and CD8-depleted (CD8 beads; Dynal Biotech GmbH, Hamburg, Germany) healthy donor peripheral blood lymphocytes in 3 ml of IMDM (Biowhittaker-Cambrex Bio Science, Verviers, Belgium) supplemented with 10% Fetalclone II (HyClone; Perbio Science Nederland BV, Etten-Leur, The Netherlands), 20 U of recombinant interleukin-2 (Proleukin; Chiron Benelux BV, Amsterdam, The Netherlands) and 4 μg of Polybrene (Pb; Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany)/ml in T25 flasks. After 7 days, the cultures were refreshed by the addition of 5 · 106 fresh target cells in 5 ml of medium. On day 14, the cell supernatants were harvested, aliquoted, and stored at −70°C. The virus contents in virus stocks were determined with an enzyme-linked immunosorbent assay-based RT activity detection kit (Lenti-RT Activity kit; Caviditech, Uppsala, Sweden). For infection experiments, cells were exposed to a virus inoculum containing RT activity equivalent to 1 ng of RT ml−1 unless otherwise indicated.
Control viruses.
For control purposes, stocks of four HIV-1 biological clones with known coreceptor usage were grown: ACH208 *4C8 (X4), ACH39 *H4 (R3X4), ACH337 9.2F4 (R5), and ACH583 9.2A2 (R5). These viruses had previously been isolated from individuals participating in the Amsterdam Cohort Studies of HIV infection and AIDS and had been phenotyped at the CLB-Sanquin laboratory in Amsterdam (45, 55). The simian immunodeficiency virus variant SIVrcm had previously been isolated from a naturally infected red-capped mangabey and was provided by P. Marx (10). This virus has been shown to be capable of using human CCR2b, CXCR6, and CX3CR1 as coreceptors (10, 60) and was included to confirm the functionality of the cell lines expressing these receptors. HIV-2 viruses MIR and prCBL20, provided by Á. McKnight (37), and PH2-1 E6 and RH2-5 E4, previously isolated by C. Guillon in our laboratory (29), were included in U87 experiments.
GHOST cells.
GHOST cell lines are human osteosarcoma cells engineered to stably express CD4 (parental cell line) and one chemokine receptor. In addition, GHOST cells contain a green fluorescent protein (GFP) gene under the control of a HIV-2 long terminal repeat. Infection with HIV and subsequent production of the Tat protein induces the expression of GFP. In the present study, cells that express CCR1, CCR2b, CCR3, CCR5, CXCR4, GPR15, or CXCR6 were used. Cells expressing CX3CR1 were used for a subpopulation of viruses. The cells were cultured in RPMI (Biowhittaker) containing 10% fetal bovine serum (FBS) (Biowhittaker), hygromycin (100 μg/ml), Geneticin (500 μg/ml), and puromycin (1 μg/ml). No puromycin was added to the control parental cell line. The cells were passaged two times a week. Batches of cells were stored at different passages. For experiments, only third- and fourth-passage cells were used. Expression of CD4, CCR1, CCR3, CCR5, CXCR4, and CXCR6 on fourth-passage cells was verified by fluorescence-activated cell sorter (FACS) analysis. Expression of CCR2b and GPR15 was confirmed by RT-PCR (not shown). The functionality of CCR2b and CX3CR1 on the respective cell lines was confirmed by the susceptibility of these cells to SIVrcm (10, 60).
U87 cells.
U87 cell lines are human glioma cells engineered to stably express CD4 (parental cell line) and one chemokine receptor. Cells expressing CCR1, CCR2b, CCR3, CCR5, CXCR4, or CXCR6 were used in the present study. The U87-CXCR6 cell line was previously constructed by van Rij et al. (55). CD4 and coreceptor expression levels were determined by FACS analysis, and only batches of cells with known expression levels were used for all experiments (U87-CCR1, 72% CCR1+; U87-CCR3, 91% CCR3+; U87-CCR5, 98% CCR5+; U87-CXCR4, 96% CXCR4+; U87-CXCR6, 99% CXCR6+). CCR2b mRNA expression was confirmed by RT-PCR (not shown). All cells except the parental cell line (see below) contained 92 to 99% CD4+ cells.
Three different batches of the parental cell line that varied in culture history and CD4 expression levels were used. The first batch (A) consisted of cells that had been obtained from the National Institutes of Health AIDS Research and Reference Reagent Program, propagated at CLB-Sanquin in Amsterdam, and then cryopreserved. These cells were thawed again in our laboratory and used for pilot experiments until, after a culture period of 2 to 3 months, FACS analysis revealed that only 15 to 25% of the cells expressed CD4. Then, the cell line was enriched for CD4+ cells by positive selection using magnetic beads coated with anti-CD4 monoclonal antibodies (Dynal Biotech GmbH). The resulting cell line had 90% CD4+ cells and was used as a control cell line for analysis of all tested HIV-2 variants. The second batch (B) was recently obtained from the EVA/MRC Centralised Facility for AIDS Reagents (catalog no. ARP069c; date, May 2002) and propagated for just 1 week before use. The third batch (C) was provided by Áine McKnight (date, October 1998) and was from the same lineage as the cells she had previously used in her studies (37). Batches B and C contained 69 and 85% CD4+ cells, respectively, at the moment of infection. U87 cells were cultured in RPMI (Biowhittaker) containing puromycin (1 μg/ml) and Geneticin (200 μg/ml) and, after the observation of loss of CD4 by the parental cell line, were passaged two times a week, thus refreshing the antibiotics every third or fourth day.
Monoclonal antibodies and primers.
GHOST and U87 cells were stained with phycoerythrin-conjugated α-CCR1, α-CCR3, and α-CXCR6 (clones 53504.111, 61828.111, and 56811, respectively; R&D Systems Europe Ltd., Abingdon, United Kingdom), R-phycoerythrin-conjugated α-CCR5 and α-CXCR4 (clones 2D7 and 12G5, respectively; BD Biosciences-Pharmingen, San Diego, Calif.), and α-CD4 (clone MT310; DAKO Diagnostics BV, Glostrup, Denmark) according to standard flow cytometric procedures.
For determination of expression of GPR15 (in GHOST cells) and CCR2b (in GHOST and U87 cells), total mRNA was isolated from cell lines by using an Oligotex mRNA minikit (QIAGEN GmbH, Hilden, Germany). cDNA was synthesized using random primers, and GPR15 and CCR2b sequences were amplified using CCR2b-specific primers (5′ TGC TGT CCA CAT CTC GTT CTC GG 3′ [sense] and 5′ CCC TAT GCC TCT TCT TCT CGT TTC G 3′ [antisense]) and GPR15-specific primers (5′ TGC AGT GTC CTC CTG CTC ACT TGC 3′ [sense] and 5′ TGT GCT TTC CTG ATT GCT GGT AAT GG 3′ [antisense]).
Infection of GHOST cells.
In total, 88 HIV-2 variants were tested in nine separate experiments, each experiment including an X4-using HIV-1 variant as a positive control and mock-infected cells as a negative control. For every two experiments, a second-passage batch of each of the eight GHOST cell lines was thawed and put into culture. The first experiments were performed on cells that had been allowed to recover from thawing for 4 to 5 days, and the second experiments were performed 3 days thereafter. One day prior to infection, cells were seeded in 24-well plates at 35,000 per well in selection medium. Just prior to infection, the culture medium was removed from the wells, collected in tubes (one for each cell line), and set aside. Cell-free virus (virus stock containing 0.5 ng of RT in 500 μl of RPMI-10% FBS-4 μg of Pb/ml) was added to the cells, and the plates were centrifuged for 15 min at 2,700 × g to enhance the infection efficiency (spinoculation) (30, 43). In preliminary experiments, spinoculation and addition of Polybrene had been shown to increase the infection efficiency 6- to 10-fold (data not shown). After being spun, the plates were placed in a 37°C-5% CO2 incubator for a 4-h incubation. The virus was removed, and 900 μl of the collected 1-day-old selection medium was reapplied to the wells. The cells were incubated for another 3 days at 37°C-5% CO2 and harvested. For that purpose, the culture medium was completely removed, and the cells were incubated with 90 μl of trypsin (GIBCO-Invitrogen). The trypsin was inactivated by the addition of 300 μl of phosphate-buffered saline-20% FBS, and the cells were kept at 4°C until they were analyzed using a flow cytometer (Fig. 1A). As a measure for productive infection, we used the ratio-to-cell-negative (RTCN) that was previously defined by Vödrös et al. (57). This value incorporates both the percentages of GFP-positive cells and the fluorescence levels (i.e., mean fluorescence intensity [MFI]). More specifically, the RTCN equals the ratio of the [percent GFP+ cells × MFI(GFP-positive population)] of infected cells to the [percent GFP+ cells × MFI(GFP-positive population)] of uninfected cells. An RTCN value of 5 was taken as the negative cutoff.
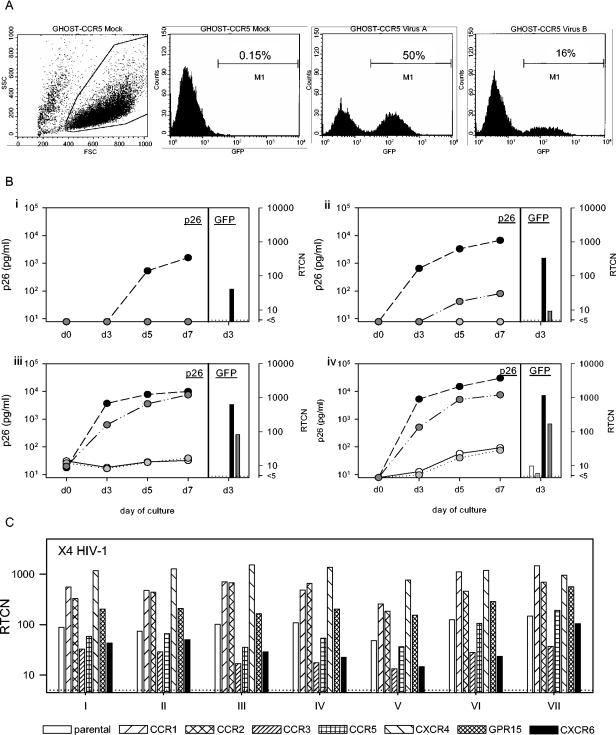
FIG. 1.
Evaluation of GHOST cells. (A) GFP-expression was analyzed by flow cytometry. Three days postinfection, the cells were harvested and live cells were gated based on their forward and side scatter. Instrument settings were adjusted to reduce the mean background signals (caused by constitutive expression of GFP) to <101, and the negative cutoff was set to 0.15% using mock-infected cells. The values obtained were used to calculate the RTCN. Shown are two examples of GHOST-CCR5 cells infected with two different HIV-2 variants. (B) To compare GFP expression (RTCN) with p26 production, GHOST-CCR5 (solid circles), GHOST-CXCR6 (darkly shaded circles), GHOST-CCR2b (lightly shaded circles), and GHOST-CCR3 (open circles) cells were infected with 11 HIV-2 variants, each virus-cell combination in two parallel cultures. Supernatants were harvested and stored on days 0 and 3 (culture A) and days 5 and 7 (culture B) and analyzed for p26 content using a SIV core antigen enzyme-linked immunosorbent assay (Beckman-Coulter Netherlands BV, Mijdrecht, The Netherlands). Culture A was terminated on day 3, when the cells were harvested for analysis of GFP expression. The RTCN and p26 concentrations are shown for four representative viruses (i, RH2-1 A8; ii, RH2-14 1D1; iii, RH2-3 4C5; iv, RH2-4 4B2). (C) The GHOST assay is highly reproducible. Shown are the results of infection with a control X4 HIV-1 variant (ACH208) that was included in multiple independent experiments.
Experiments were repeated with two-times-larger inocula when the infection efficiency of CCR5-using variants in CCR5-expressing cells was low (<10% GFP+) or with equal inocula when the results were inconclusive despite a high infection efficiency of CCR5+ cells.
Infection of U87 cells.
One day prior to infection, cells were seeded in 96-well plates at 5,000 per well in wells previously coated with 0.1% gelatin to enhance cell attachment. Just prior to infection, the culture medium was removed and cell-free virus plus 0.2 ng of RT/200 μl of RPMI-10% FBS-4 μg of Pb/ml was added. Cells and virus were incubated overnight at 37°C-5% CO2. After overnight incubation, the virus was removed and the cells were washed three times with RPMI-10% FBS. After the final wash, 200 μl of selection medium plus Pb was added. The cultures were maintained for 14 days; then, the culture supernatants were harvested, and RT activity was determined using a Lenti-RT Activity kit (Caviditech).
Statistical analyses.
Correlations (p26 concentrations versus RTCN, percentages of CXCR4+ cells versus RTCN, and RT concentrations between batches of parental U87 cells) were determined by calculating Spearman's rho (rs). The susceptibilities of batches of parental U87 cells to HIV-2 were compared using the Kendal W test. All statistical analyses were performed with SPSS version 11 for Windows.
RESULTS
GHOST cells and analysis of GFP expression.
The levels of receptor and coreceptor expression of the cell batches used in the experiments are shown in Table 2. Each cell line expressed CD4 and the coreceptor of given specificity. However, each cell line also contained cells expressing CXCR4, likely due to endogenous gene expression. The proportions of CXCR4-expressing cells in cell lines other than GHOST-CXCR4 were generally low, but they were considerable in the GHOST-CCR1 and GHOST-CCR2b cell lines (11 and 27%, respectively).
TABLE 2.
CD4 and coreceptor expression on GHOST cellsa
| Cell line | CD4+ (%) | CoR+ (%) | CXCR4+ (%) |
|---|---|---|---|
| Parental | 94 | NAb | 3.8 |
| CCR1 | 100 | 96 | 11 |
| CCR2b | 97 | NTc | 27 |
| CCR3 | 98 | 76 | 2.4 |
| CCR5 | 97 | 73 | 2.7 |
| CXCR4 | 94 | 85 | 85 |
| GPR15 | 95 | NT | 2.3 |
| CXCR6 | 98 | 49 | 0.8 |
Percentages of positive cells determined in fourth-passage cultures.
NA, not applicable.
NT, not tested in FACS; mRNA expression was detected by RT-PCR.
Productive infection was determined by analysis of GFP expression using a flow cytometer (Fig. 1A). As a measure of GFP production, the proportion of fluorescent cells and the level of fluorescence were incorporated into one value, the RTCN (57). To validate the assay, we tested whether RTCN values correlated with viral p26 production in supernatants of HIV-2-infected GHOST-CCR2b, -CCR3, -CCR5, and -CXCR6 cells. For all viruses tested, the conclusions regarding coreceptor usage were the same whether GFP expression or p26 production was used as the readout (Fig. 1B). Moreover, the correlation between RTCN values 3 days postinfection and levels of p26 accumulated in culture supernatants on the same day was highly significant (rs = 0.96; P < 0.0001) (data not shown). In some cases, the RTCN was positive while the p26 level measured on day 3 was negative; however, in all these cases, p26 was detected by day 7 (Fig. 1B, virus i, CCR5, and virus ii, CXCR6). Using RTCN, cultures producing only very small amounts of virus (<100 pg of p26/ml) could be identified, demonstrating the high sensitivity of the assay (Fig. 1B, virus iv, CCR2b and CCR3, and virus ii, CXCR6). Moreover, the assay was highly reproducible, as demonstrated by the similar infection patterns of the eight cell lines in independent experiments (Fig. 1C).
Analysis of coreceptor use of HIV-2 variants in GHOST cells.
The coreceptor usage of 43 HIV-2 biological clones obtained from six long-term aviremic HIV-2-infected individuals and 45 biological HIV-2 clones from seven viremic individuals (Table 1) was studied with GHOST cell lines expressing CCR1, CCR2b, CCR3, CCR5, CXCR4, CXCR6, or GPR15 (Table 3). For 62 (70%) of 88 HIV-2 variants tested, infection of the GHOST-CCR5 cells resulted in RTCN values of >500. Consequently, entry via other coreceptors would still be detectable when it was 100 times less efficient than entry via CCR5. Only five variants infected the GHOST-CCR5 cell line with extremely low efficiencies (RTCN < 100). Four of these infected other cell lines with higher efficiencies, indicating that CCR5 was not the major coreceptor for these variants. One variant remained negative on all cell lines and was not included in further analyses.
TABLE 3.
Infection of GHOST cellsa
| Virus | RTCN value
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Parental | CCR1 | CCR2b | CCR3 | CCR5 | CXCR4 | GPR15 | CXCR6 | |
| Aviremic individuals | ||||||||
| RH2-3 2C5 | − | − | − | − | 438 | − | 273 | 29 |
| RH2-3 3A6c | − | − | − | − | 1,142 | − | 950 | 165 |
| RH2-3 3B3 | − | − | − | − | 1,285 | − | 1,154 | 310 |
| RH2-3 3C3 | − | − | − | − | 745 | − | 1,767 | 267 |
| RH2-3 4C5 | − | − | − | − | 704 | − | 725 | 161 |
| RH2-3 5B1 | − | − | − | − | 984 | − | 267 | 59 |
| RH2-3 6C6 | − | − | − | − | 507 | − | 284 | 153 |
| RH2-3 8A5 | − | − | − | − | 1,136 | − | 2,124 | 86 |
| RH2-3 8A3c | − | − | − | − | 1,201 | − | 541 | 98 |
| RH2-3 3A3b | 10 | 10 | 7 | 6 | 1,451 | 8 | 2,629 | 301 |
| RH2-13 1D4 | − | − | − | − | 1,043 | − | 1,378 | 108 |
| RH2-13 1D6 | − | − | − | − | 886 | − | 1,394 | 123 |
| RH2-13 3B4 | − | − | − | − | 972 | − | 1,144 | 39 |
| RH2-13 5C1 | − | − | − | − | 1,209 | − | 1,774 | 117 |
| RH2-13 5C3 | − | − | − | − | 825 | − | 446 | 67 |
| RH2-13 6A3 | − | − | − | − | 895 | − | 382 | 41 |
| RH2-13 6B5b | − | − | − | − | 1,677 | − | 698 | 20 |
| RH2-13 7C3 | − | − | − | − | 1,287 | − | 150 | 18 |
| RH2-13 6A2b | 11 | 14 | 9 | 7 | 1,847 | 6 | 3,812 | 188 |
| RH2-13 6B1c | 6 | 8 | 8 | 7 | 1,052 | 6 | 2,004 | 533 |
| RH2-14 1A6 | − | − | − | − | 375 | − | 124 | 32 |
| RH2-14 1B1 | − | − | − | − | 331 | − | 179 | 38 |
| RH2-14 1C2 | − | − | − | − | 537 | − | 194 | 38 |
| RH2-14 1C3 | − | − | − | − | 407 | − | 239 | 40 |
| RH2-14 1D1 | − | − | − | − | 431 | − | 55 | 13 |
| RH2-14 2A2 | − | − | − | − | 623 | − | 184 | 54 |
| RH2-14 2A4 | − | − | − | − | 414 | − | 275 | 66 |
| RH2-14 2A5 | − | − | − | − | 593 | − | 155 | 53 |
| RH2-14 2D5 | − | − | − | − | 292 | − | 142 | 12 |
| RH2-22 1B4 | − | − | − | − | 491 | − | 97 | 32 |
| RH2-22 1B5b | − | − | − | − | 778 | − | 84 | 18 |
| RH2-22 1C3 | − | − | − | − | 656 | − | 90 | 39 |
| RH2-22 2B2 | − | − | − | − | 514 | − | 46 | 22 |
| RH2-22 2C4 | − | − | − | − | 740 | − | 233 | 65 |
| RH2-22 3A1b | − | − | − | − | 908 | − | 41 | 13 |
| RH2-22 2A4c | 5 | 6 | − | − | 724 | − | 159 | 163 |
| RH2-22 1B1c | 46 | 99 | 37 | 47 | 1,611 | 43 | 1,342 | 383 |
| RH2-22 1B2c | 17 | 27 | 16 | 18 | 1,191 | 13 | 688 | 142 |
| RH2-22 2C2c | 24 | 36 | 22 | 21 | 957 | 17 | 313 | 132 |
| RH2-8 3D5c | 10 | 8 | 8 | 6 | 658 | 6 | 28 | 17 |
| RH2-4 4D1b | − | − | − | − | 1,277 | − | 389 | 61 |
| RH2-4 3B1b | 6 | 5 | − | 5 | 1,147 | − | 168 | 533 |
| RH2-4 4B2b | 12 | 23 | 8 | 15 | 1,298 | 11 | 792 | 298 |
| Viremic individuals | ||||||||
| Asymptomatic | ||||||||
| RH2-15 B9b | − | − | − | − | 953 | − | 170 | 35 |
| RH2-15 E5 | − | − | − | − | 367 | − | 7 | − |
| RH2-15 H11 | − | − | − | − | 964 | − | 73 | 19 |
| RH2-15 A9b | − | − | − | − | − | − | − | − |
| RH2-15 E10b | − | − | − | 37 | 102 | − | 101 | 85 |
| Progressors | ||||||||
| RH2-1 A8b | − | − | − | − | 132 | − | 27 | − |
| RH2-1 A9b | − | − | − | − | 270 | − | 13 | − |
| RH2-1 C7b | − | − | − | − | 129 | − | − | − |
| RH2-1 D8b | − | − | − | − | 268 | − | 14 | − |
| RH2-1 D9b | − | − | − | − | 239 | − | 8 | − |
| RH2-1 G7b | − | − | − | − | 552 | − | 34 | − |
| RH2-5 1F10 | − | − | − | − | 982 | − | 946 | 221 |
| RH2-5 2C8 | − | − | − | − | 1,409 | − | 1,866 | 263 |
| RH2-5 2D11b | − | − | − | − | 368 | − | 249 | 27 |
| RH2-5 2E6b | − | − | − | − | 996 | − | 1,142 | 45 |
| RH2-5 2A12b | − | 10 | − | − | 317 | − | 324 | 87 |
| RH2-5 2F10b | 11 | 6 | 5 | 9 | 2,579 | − | 3,440 | 810 |
| RH2-7 D6 | − | − | − | − | 1,194 | − | 673 | 431 |
| RH2-7 E1 | − | − | − | − | 652 | − | 261 | 112 |
| RH2-7 E4b | − | − | − | − | 423 | − | 211 | 55 |
| RH2-7 F3 | − | − | − | − | 938 | − | 685 | 35 |
| RH2-7 F4b | − | − | − | − | 580 | − | 283 | 126 |
| RH2-7 B3c | 11 | 17 | 9 | 23 | 641 | 11 | 1,192 | 145 |
| RH2-21 2B2c | 62 | 283 | 152 | 23 | 48 | 511 | 34 | 9 |
| RH2-21 2F9c | 81 | 218 | 159 | 16 | 33 | 446 | 28 | 7 |
| RH2-24 1E2 | − | − | − | − | 2,394 | − | 515 | 52 |
| RH2-24 2D7 | − | − | − | − | 2,098 | − | 1,288 | 265 |
| RH2-24 2E10 | − | − | − | − | 1,238 | − | 243 | 21 |
| RH2-24 2E9c | − | − | − | − | 942 | − | 718 | 94 |
| RH2-24 2F11 | − | − | − | − | 1,158 | − | 122 | 17 |
| RH2-24 2G9 | − | − | − | − | 1,374 | − | 543 | 79 |
| RH2-24 2D8c | 183 | 1,022 | 796 | 49 | 163 | 1,504 | 170 | 28 |
| RH2-24 2E8c | 65 | 173 | 127 | 14 | 32 | 543 | 35 | 6 |
| RH2-24 2G7c | 158 | 624 | 788 | 37 | 152 | 1,746 | 144 | 21 |
| RH2-24 2H11c | 17 | 36 | 22 | − | 6 | 96 | − | − |
| RH2-26 1C1 | − | − | − | − | 1,652 | − | − | − |
| RH2-26 1D3 | − | − | − | − | 1,083 | − | − | − |
| RH2-26 2A1 | − | − | − | − | 1,710 | − | − | − |
| RH2-26 2A10 | − | − | − | − | 1,083 | − | − | − |
| RH2-26 2A12 | − | − | − | − | 634 | − | − | − |
| RH2-26 2B4 | − | − | − | − | 2,430 | − | − | − |
| RH2-26 2D3 | − | − | − | − | 2,919 | − | − | − |
| RH2-26 2E2 | − | − | − | − | 2,919 | − | − | − |
| RH2-26 2D9c | 198 | 1,071 | 918 | 40 | 124 | 1,767 | 131 | 18 |
| RH2-26 2E9c | 152 | 620 | 680 | 33 | 103 | 1,274 | 88 | 33 |
| Controls (CoR)d | ||||||||
| ACH208 (X4) | 94 | 708 | 473 | 23 | 85 | 1,140 | 266 | 43 |
| ACH39 (X4R3) | 80 | 429 | 299 | 204 | 53 | 624 | 100 | 33 |
| SIVrcm (R2X6) | − | NT | 710 | NT | − | NT | NT | 628 |
Shown are RTCN values, values <5 are considered negative (−).
Two times the standard amount of virus was used (see Materials and Methods).
Tested in two individual experiments, of which the average is shown.
HIV-1 and SIV variants with known coreceptor usage (indicated by parentheses) were included as controls. HIV-1 was tested multiple times; shown are the averages of nine (ACH208) and four (ACH39) experiments. NT, not tested.
Most HIV-2 variants isolated from the nonprogressing, aviremic individuals efficiently infected the three cell lines expressing CCR5, GPR15, and CXCR6 (n = 33; 76%). These three cell lines, or a subset of them, were also efficiently infected by most variants from viremic individuals (n = 32; 73%). Some HIV-2 variants infected all or a large number of the GHOST cell lines, and this always coincided with permissiveness of the parental cell line. HIV-2 variants with tropism for the parental GHOST cells were detected in 10 individuals, 5 in each study group, and constituted 23% of all variants analyzed for both patient groups. Only rarely, if at all, was infection of the cell lines transduced with CCR1, CCR2b, CCR3, or CXCR4 observed when the parental cell line was not infected.
In a subsequent experiment, a subpopulation of the HIV-2 clones (n = 33) was additionally tested in GHOST-CX3CR1 cells (Table 4). About half of the variants tested (51%) could infect this cell line, albeit with relatively low efficiency compared to SIVrcm.
TABLE 4.
Infection of GHOST-CX3CR1 cellsa
| Virus | RTCN value
|
||
|---|---|---|---|
| Parental | CCR5 | CX3CR1 | |
| Aviremic individuals | |||
| RH2-3 3B3 | − | 1,036 | 31 |
| RH2-3 3C3 | − | 973 | 64 |
| RH2-3 6C6 | − | 540 | − |
| RH2-3 8A5 | − | 1,144 | 65 |
| RH2-13 1D4 | − | 1,359 | 16 |
| RH2-13 5C3 | − | 1,326 | 10 |
| RH2-13 6A3 | − | 900 | 8 |
| RH2-14 1A6 | − | 562 | − |
| RH2-14 1B1 | − | 519 | − |
| RH2-14 2A2 | − | 660 | − |
| RH2-14 2A4 | − | 835 | − |
| RH2-22 1B4 | − | 385 | − |
| RH2-22 1C3 | − | 748 | 10 |
| RH2-22 1B2 | 24 | 1,689 | 55 |
| RH2-22 2B2 | − | 1303 | − |
| RH2-4 4D1 | − | 351 | 10 |
| Viremic individuals | |||
| RH2-15 B9 | − | 642 | 10 |
| RH2-15 E5 | − | 561 | − |
| RH2-15 H11 | − | 1,114 | − |
| RH2-1 A8 | − | 125 | − |
| RH2-1 D8 | − | 254 | − |
| RH2-1 G7 | − | 241 | − |
| RH2-5 1F10 | − | 370 | 10 |
| RH2-5 2C8 | − | 728 | 18 |
| RH2-5 2D11 | − | 239 | − |
| RH2-7 B3 | 25 | 619 | 63 |
| RH2-7 F3 | − | 1,056 | 10 |
| RH2-7 F4 | − | 625 | 11 |
| RH2-24 1E2 | − | 1,303 | 15 |
| RH2-24 2D7 | − | 827 | 17 |
| RH2-24 2E10 | − | 565 | − |
| RH2-26 1C1 | − | 1,201 | − |
| RH2-26 2B4 | − | 1,529 | − |
| Controls (CoR) | |||
| ACH337 (R5) | − | 938 | − |
| SIVrcm (X3R1) | − | − | 1,079 |
Infection of GHOST-CX3CR1 cells was tested in a separate assay. Parental GHOST cells and GHOST-CCR5 cells were included as control cell lines. SIVrcm was previously shown to be capable of using CX3CR1 (60) and was included as a positive control; an R5 HIV-1 variant was included as a negative control. RTCN values of <5 are considered negative (−).
Infection of parental GHOST cells can be partially explained by CXCR4 use.
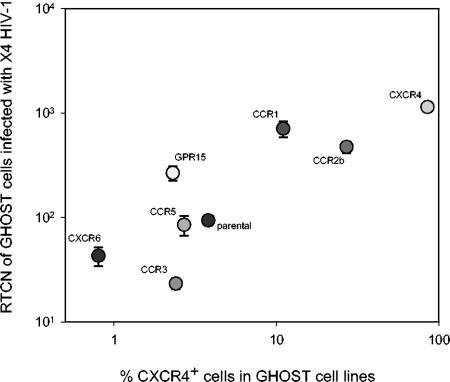
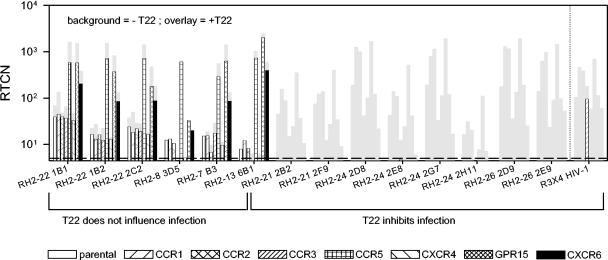
The capacity to infect the whole range of GHOST cells was a property shared with the X4-restricted HIV-1 control virus (Fig. 1C), and the most obvious explanation was that infection is established via endogenous CXCR4. In support of this, levels of virus production (expressed as the RTCN) correlated with the proportion of CXCR4-expressing cells in the cell lines exposed to X4 HIV-1 (Fig. 2). To confirm whether infection of GHOST cells by HIV-2 variants capable of infecting the parental cells was indeed established via endogenous CXCR4 and to determine additional coreceptor use of these variants, infections were repeated in the presence and absence of T22, a CXCR4 antagonist (40, 41) (Fig. 3). In eight cases, T22 completely inhibited virus replication in all cell lines, indicating that these variants were X4 restricted, and infected each cell line via CXCR4 only. In contrast, an R3X4 control HIV-1 variant still infected GHOST-CCR3 cells in the presence of T22, revealing entry via CCR3 and demonstrating the functionality of this coreceptor in GHOST-CCR3 cells. In six other cases, T22 had no effect on infection, suggesting that entry was not established via CXCR4. In line with this, these viruses infected the GHOST-CXCR4 cell line with relatively low efficiency (Table 3). Moreover, the T22-insensitive variants were not capable of infecting CCR5−/− PBMC, in contrast to the T22-sensitive variants, which could infect CCR5−/− PBMC as efficiently as CCR5+/+ PBMC (not shown). Six variants that were capable of infecting parental cells were not tested for T22 sensitivity, but all could infect GHOST-CXCR4 cells only with low efficiency and were incapable of infecting CCR5−/− PBMC (not shown), strongly suggesting that these viruses cannot use CXCR4. The susceptibility of parental GHOST cells to HIV-2 variants independent of CXCR4 may indicate the presence of a second, still unidentified HIV-2 coreceptor on GHOST cells.
FIG. 2.
Correlation between RTCN and percentages of CXCR4+ cells. Eight GHOST cell lines were exposed to an HIV-1 X4-restricted variant, and the RTCN, reflecting virus production, was determined. Each symbol represents a cell line transduced with the indicated chemokine receptor. Parental cells are not transduced with a chemokine receptor. The values depicted are the mean RTCN from nine independent experiments. The vertical bars represent the standard errors.
FIG. 3.
T22 sensitivities of HIV-2 variants with tropism for parental GHOST cells. Fourteen HIV-2 variants capable of infecting parental GHOST cells were tested for their sensitivity to T22 (Bachem, Bubendorf, Switzerland). A R3X4 HIV-1 variant was included as a control. The cells were preincubated in 250 μl of medium containing 6 or 0 μM T22 for 2 h, after which 350 μl of virus inoculum was added, resulting in 2.5 or 0 μM T22 and 1 ng of RT ml−1. After infection, the virus was removed, and selection medium containing 3 μM T22 was added. Each bar represents a cell line. The background bars (gray) represent values obtained in the absence of T22. The overlaid bars represent values obtained in the presence of T22 and are therefore not visible when inhibition was complete.
In contrast to T22-sensitive variants, coreceptor use by the T22-insensitive GHOST-tropic variants could not be accurately determined. However, when virus production in the parental cells is considered as background in the coreceptor-transduced cells, these variants appear to use CCR5, GPR15, and CXCR6 (Table 5). Three out of 12 of these variants infected GHOST-CCR1 cells above background levels, which may indicate weak CCR1 usage.
TABLE 5.
Coreceptor usage of variants with GHOST.CD4 tropism
| Virus | Typea | RTCN value
|
||||||
|---|---|---|---|---|---|---|---|---|
| CCR1 | CCR2b | CCR3 | CCR5 | CXCR4 | GPR15 | CXCR6 | ||
| Aviremic individuals | ||||||||
| RH2-3 3A3 | I | − | − | − | 1,436 | − | 2,614 | 286 |
| RH2-13 6A2 | I | − | − | − | 1,830 | − | 3,795 | 171 |
| RH2-13 6B1 | I | − | − | − | 1,043 | − | 1,995 | 524 |
| RH2-22 2A4 | I | − | − | − | 716 | − | 151 | 155 |
| RH2-22 1B1 | I | 30 | − | − | 1,542 | − | 1,273 | 314 |
| RH2-22 1B2 | I | 1 | − | − | 1,165 | − | 714 | 116 |
| RH2-22 2C2 | I | − | − | − | 921 | − | 277 | 96 |
| RH2-8 3D5 | I | − | − | − | 643 | − | 13 | 2 |
| RH2-4 3B1 | I | − | − | − | 1,138 | − | 159 | 524 |
| RH2-4 4B2 | I | 5 | − | − | 1,280 | − | 774 | 280 |
| Viremic individuals | ||||||||
| RH2-5 2F10 | I | − | − | − | 2,562 | − | 3,423 | 793 |
| RH2-7 B3 | I | − | − | − | 624 | − | 1,175 | 128 |
| RH2-21 2B2 | II | − | − | − | − | 352 | − | − |
| RH2-21 2F9 | II | − | − | − | − | 396 | − | − |
| RH2-24 2D8 | II | − | − | − | − | 1,650 | − | − |
| RH2-24 2E8 | II | − | − | − | − | 510 | − | − |
| RH2-24 2G7 | II | − | − | − | − | 1,891 | − | − |
| RH2-24 2H11 | II | − | − | − | − | 110 | − | − |
| RH2-26 2D9 | II | − | − | − | − | 1,915 | − | − |
| RH2-26 2E9 | II | − | − | − | − | 1,422 | − | − |
| Control virus | ||||||||
| ACH39 X4R3 | II | − | 175 | − | 680 | − | − | |
Coreceptor usage was deduced for T22 insensitive variants (I) by subtracting 1.5 times the value of parental cells from that of coreceptor-transduced cells (values from Table 3), and for T22 sensitive variants (II) by translating the effect of T22 (values from Fig. 3). RTCN values <1.5 times that in the parental cells (I) or <5 in the presence of T22 (II) are considered negative (−).
HIV-2 phenotyping based on GHOST infection and sensitivity to T22.
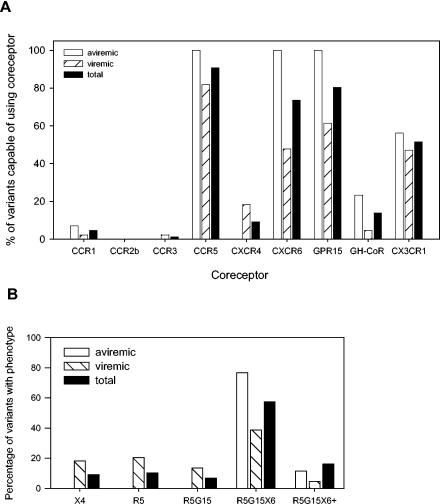
Based on the capacity of the HIV-2 variants to infect the different GHOST cells and their sensitivity to T22, HIV-2 coreceptor use could be determined for all variants except one. The significance of the eight chemokine receptors in functioning as HIV-2 coreceptors is reflected by the frequency of HIV-2 variants capable of using these chemokine receptors as coreceptors (Fig. 4A). CCR5, GPR15, and CXCR6 were used with high efficiency by 91, 81, and 74% of all viruses, respectively, and appeared to be the major HIV-2 coreceptors. About half of the variants tested (51%) also used CX3CR1, but only with low efficiency. Smaller fractions of HIV-2 variants could use CXCR4 efficiently (9%) or use an unidentified GHOST coreceptor inefficiently (14%). CXCR4 use was observed only for variants isolated from viremic individuals. Use of CXCR6, GPR15, and the unidentified GHOST coreceptor was more often observed in the aviremic than in the viremic study group. CCR1, CCR2b, and CCR3 appeared not to be significant HIV-2 coreceptors.
FIG. 4.
HIV-2 coreceptors and phenotypes determined using GHOST cells. (A) The frequency with which chemokine receptors are used as HIV-2 coreceptors is indicated by the percentages of HIV-2 variants capable of using the different chemokine receptors, which are specified on the x axis. Use of a potential unidentified coreceptor expressed by GHOST cells is indicated as GH-CoR. Depicted are the percentages of coreceptor-using variants among all successfully characterized HIV-2 variants (total) and of variants from aviremic and viremic individuals separately. (B) Using GHOST cells in combination with T22, phenotypes were determined; occurrences of the different phenotypes are depicted as a proportion of total HIV-2 variants and of variants from aviremic and viremic individuals separately. The abbreviations X4, R5, G15, and X6 represent the different chemokine receptors used by the variant phenotypes.
The majority of HIV-2 variants could use CCR5, GPR15, and CXCR6 (Fig. 4B). This R5G15X6 phenotype was the most common phenotype in individuals with and without plasma viremia. In addition to these three coreceptors, some variants could use the unidentified GHOST coreceptor (n = 12) and/or CCR1 (n = 4) inefficiently or use CCR3 (n = 1) efficiently; together, these variants are designated R5G15X6+ variants. This type of HIV-2 variant, capable of using four or more coreceptors, was detected in 8 out of 13 infected individuals. Together, the R5G15X6 and R5G15X6+ phenotypes constituted 74% of all HIV-2 variants. At least some of these variants are likely to use even more coreceptors, as indicated by the fact that the majority (63%) of 27 R5G15X6 and R5G15X6+ variants tested could also use CX3CR1.
The R5X6G15 and R5X6G15+ HIV-2 phenotypes were the only two phenotypes observed in aviremic HIV-2-infected individuals. In the group of individuals with viremia a higher number of different phenotypes was observed, among which were X4 variants and R5-using variants with a more restricted coreceptor use. Despite the apparent high level of phenotypic variation in the group of viremic individuals, generally no more than two different types of variants were observed per patient, which is similar to the variation in aviremic patients (Table 3). The presence of variants with different phenotypes in aviremic individuals may suggest a low level of ongoing virus replication and evolution, despite the undetectable levels of virus in their plasma.
Analysis of coreceptor use of HIV-2 variants in U87 cells.
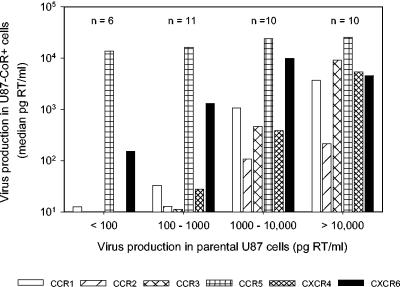
The observation that CCR1, CCR2b, and CCR3 did not generally function as HIV-2 coreceptors in GHOST cells, even in individuals with progressive infection, was unanticipated and contrasted with previous observations (7, 29, 38, 42). To verify the data we obtained with the GHOST cells, we did an additional coreceptor analysis in a panel of U87 cells for 38 of the variants (Table 6). Most HIV-2 variants tested could infect a broad range of U87 cells, in agreement with previous observations (29, 37, 38). However, 37 (97%) of the variants also infected the parental cell line, often with high efficiency, indicating that these cells express a major HIV-2 coreceptor. The appearance of promiscuity of coreceptor usage increased, along with increasing efficiency in infecting the parental cell line (Fig. 5), suggesting that virus production in all cell lines is at least partially due to entry via the same unidentified coreceptor. When virus production in the parental cells was taken as background production in the coreceptor-transduced cells, coreceptor usage appeared generally the same or more restricted than that based on GHOST cells (Table 6).
TABLE 6.
Infection of U87 cells
| Virus | GHOSTb | RT concna
|
||||||
|---|---|---|---|---|---|---|---|---|
| Parental | CCR1 | CCR2b | CCR3 | CCR5 | CXCR4 | CXCR6 | ||
| Aviremic individuals | ||||||||
| RH2-3 3B3 | R5G15X6 | 15,274 | 3,795 | 216 | 3,946 | 16,092 | 3,059 | 6,143 |
| RH2-3 3C3 | R5G15X6 | 31,055 | 4,965 | 477 | >31,055 | >31,055 | 7,683 | 16,701 |
| RH2-3 5B1 | R5G15X6 | 7,394 | 1,718 | 324 | 5,876 | >31,055 | 1,829 | 1,272 |
| RH2-3 8A5 | R5G15X6 | >31,055 | 2,013 | 303 | 2,823 | 17,001 | 1,834 | 10,598 |
| RH2-3 8A3 | R5G15X6 | 13,886 | 3,523 | 202 | 6,105 | 22,182 | 3,071 | 2,008 |
| RH2-13 1D6 | R5G15X6 | >31,055 | 3,363 | 341 | 21,154 | 27,143 | >31,055 | 1,859 |
| RH2-13 5C1 | R5G15X6 | >31,055 | 3,595 | 116 | 8,169 | >31,055 | 16,336 | 2,927 |
| RH2-13 7C3 | R5G15X6 | 27,631 | 826 | 100 | 143 | 26,904 | 678 | 1,959 |
| RH2-13 6A2 | R5G15X6p | >31,055 | 4,827 | 212 | 10,103 | >31,055 | 18,746 | 1,387 |
| RH2-13 6B1 | R5G15X6p | − | − | − | − | >31,055 | NT | 11 |
| RH2-14 1C3 | R5G15X6 | 1,444 | 1,426 | 100 | 498 | 29,452 | 519 | 9,996 |
| RH2-14 1D1 | R5G15X6 | 5,484 | 865 | 113 | 431 | 9,902 | 675 | 13,341 |
| RH2-14 2A2 | R5G15X6 | 4,165 | 3,529 | 389 | 639 | >31,055 | 448 | >31,055 |
| RH2-14 2A5 | R5G15X6 | 4,867 | 3,582 | 156 | 823 | 27,631 | 330 | 9,098 |
| RH2-14 2D5 | R5G15X6 | 1,921 | 646 | 103 | 71 | 18,281 | 102 | 9,809 |
| RH2-22 2B2 | R5G15X6 | 617 | 42 | − | 19 | 14,658 | 16 | 1,302 |
| RH2-22 2C4 | R5G15X6 | 365 | 33 | 13 | − | 8,349 | 39 | 1,323 |
| RH2-22 3A1 | R5G15X6 | 226 | − | − | 11 | 19,186 | 16 | 310 |
| RH2-22 2A4 | R5G15X6p | 639 | 39 | 15 | 32 | >31,055 | 68 | 4,304 |
| RH2-22 1B1 | R1R5G15X6p | 438 | 71 | 19 | 45 | 7,652 | 68 | 1,649 |
| RH2-22 1B2 | R1R5G15X6p | 555 | 26 | − | 11 | 10,837 | 21 | 2,152 |
| RH2-22 2C2 | R5G15X6p | 166 | 29 | 16 | 49 | 5,829 | 28 | 724 |
| RH2-8 3D5 | R5G15X6p | 48 | 16 | − | − | 12,388 | − | 52 |
| RH2-4 3B1 | R5G15X6p | 6,806 | 1,255 | 449 | 1,168 | 20,467 | 962 | 15,786 |
| RH2-4 4D1 | R5G15X6 | 758 | 375 | 107 | 273 | 27,083 | 131 | 3,988 |
| Viremic individuals | ||||||||
| Asymptomatic | ||||||||
| RH2-15 B9 | R5G15X6 | 81 | 12 | − | − | 11,685 | 11 | 888 |
| RH2-15 E5 | R5G15 | 52 | − | − | − | 21,404 | − | 252 |
| RH2-15 A9 | − | 56 | − | 19 | − | 31 | − | 48 |
| RH2-15 E10 | R3R5G15X6 | 39 | 162 | 42 | 18,450 | 15,117 | 20,669 | 14,314 |
| Progressor | ||||||||
| RH2-1 A9 | R5G15 | 136 | 21 | − | − | 25,645 | − | 100 |
| RH2-1 C7 | R5 | 892 | 31 | − | − | 17,690 | − | − |
| RH2-1 D9 | R5G15 | 42 | 13 | − | 10 | 10,700 | − | 30 |
| RH2-1 G7 | R5G15 | 6,585 | 162 | − | 18 | 17,786 | 176 | 527 |
| RH2-5 2D11 | R5G15X6 | 9,793 | − | − | − | >31,055 | − | 661 |
| RH2-5 2A12 | R5G15X6 | 927 | 42 | 37 | − | 24,838 | 212 | 324 |
| RH2-5 2F10 | R5G15X6p | 30,078 | 10,889 | 206 | 20,467 | 15,301 | 1,379 | 11,875 |
| RH2-7 E4 | R5G15X6 | 19,935 | 107 | 10 | − | 9,809 | 220 | 9,809 |
| RH2-7 B3 | R5G15X6p | 27,385 | 24,733 | 2,208 | 14,509 | 23,918 | 25,380 | >31,055 |
Shown are concentrations of RT (pg/ ml) in culture supernatants after 14 days of culture. Negative cutoff of the assay is 10 pg/ml. Values below cutoff are considered negative (−). Values in the coreceptor-transduced cell lines that are at least 1.5 times higher than values in parental cells are underlined. NT, not tested.
Coreceptor usage in GHOST cells as deduced from tables 3 and 5. R1, CCR1; R5, CCR5; G15, GPR15; X6, CXCR6; p, parental infection. Note that GPR15 was not tested in U87 cells.
FIG. 5.
Infection in parental versus coreceptor-transduced U87 cells. Thirty-seven HIV-2 variants were tested on all U87 cell lines (Table 6), and these variants were grouped based on the level of virus production in the parental cells. The resulting four groups are specified on the x axis, with the number of HIV-2 variants in each group above the bars. Shown are the group median levels of RT production in the coreceptor-transduced U87 cells, with each bar representing a cell line as indicated.
Four HIV-2 variants previously demonstrated to be capable of using a broad range of coreceptors but incapable of infecting the parental cell lines (29, 37) were included in the present study. In our hands, these variants infected the whole range of U87 cells, including the parental U87 cells (Table 7). Taking into account the level of replication in parental cells, previous conclusions regarding coreceptor usage could largely be confirmed for two of these variants, but not for two others.
TABLE 7.
Infection of U87 cells: control variants
| Variant | Referenceb | Phenotypec | RT concna
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Parental | CCR1 | CCR2b | CCR3 | CCR5 | CXCR4 | CXCR6 | |||
| HIV-2 | |||||||||
| MIR | 37 | R1R2R3R5X4 | 78 | 287 | 108 | 47,267 | 10,059 | >61,730 | 9,412 |
| prCBL20 | 37 | R1R2R3R5X4 | 205 | 94 | 78 | 287 | 189 | >61,730 | 39,175 |
| PH2-1 E6 | 29 | R1R3R5X4 | 41,785 | 49,719 | 6,748 | 33,023 | 48,376 | >61,730 | 17,696 |
| RH2-5 E4 | 29 | R1R3R5X4 | 28,740 | >61,730 | 667 | >61,730 | >61,730 | 17,763 | >61,730 |
| HIV-1 | |||||||||
| ACH337 9.2F4 | 45 | R5 | 48 | − | − | − | 7,581 | − | − |
| ACH583 9.2A2 | 45 | R5 | 541 | − | − | − | 25,480 | − | − |
| ACH39 *H4 | 55 | R3X4 | − | − | − | 25,209 | − | 30,508 | − |
| ACH208 *4C8 | 55 | X4 | − | − | − | − | − | >31,055 | − |
See legend to Table 6.
Study previously describing coreceptor use of the variants indicated.
Phenotypes of the variants as established in the studies indicated; the abbreviations R1, R2, R3, R5, and X4 represent the different chemokine receptors used.
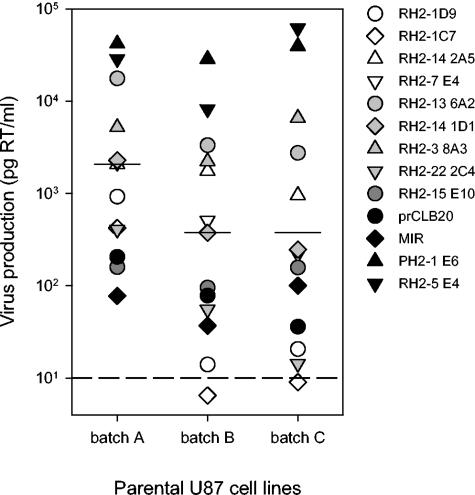
To exclude the possibility that the permissiveness to HIV-2 infection was a property specific to our parental U87 cells, we compared the susceptibility of this cell line (batch A) with that of parental cells freshly obtained via an AIDS reagent distribution program (batch B) and a batch of parental cells that were of the same lineage as the cells previously used by McKnight et al. (37) (batch C). All three batches of cells supported HIV-2 infection (Fig. 6), although in a paired analysis the cells of batches B and C scored, on average, lower than batch A (mean ranks, 1.8 [C] and 1.5 [B] versus 2.8 [A]; P = 0.002; Kendal W test). Importantly, in each cell batch the relative tropisms of HIV-2 variants for parental U87 cells were similar, i.e., variants with relatively high virus production in one batch of cells also had relatively high virus production in the other two batches. This is confirmed by the correlation between the levels of virus production in different batches of cells (A versus B, rs = 0.82 and P = 0.001; A versus C, rs = 0.80 and P = 0.001) (data not shown).
FIG. 6.
Infection of parental U87 cells. The susceptibilities to HIV-2 infection of three batches of parental cells with different origins and culture histories were compared. Shown is the virus production by 13 HIV-2 variants, with each symbol representing a variant. The horizontal bars represent median levels of virus production in each cell batch. The four control HIV-2 variants are indicated by black symbols. The relative susceptibilities of the batches of parental U87 cells were analyzed by assigning ranks based on the level of RT production for each virus individually and comparing the average ranks using the Kendal W test. The dashed line indicates the lower cutoff value of the assay (10 pg/ml).
Notably, one HIV-2 variant (RH2-15 E10) appeared capable of infecting U87-CXCR4 but not GHOST-CXCR4 cells. This variant could infect CCR5−/− PBMC, which could be partially inhibited by T22 (not shown). Although the HIV-2-infected individual from whom this variant was isolated had detectable levels of virus in the plasma, there were no signs of disease progression for 4 years after the first detection of a variant with discordant CXCR4 use.
DISCUSSION
The present study is the first to describe the coreceptor usage of HIV-2 variants from individuals who had undetectable levels of viral RNA in their plasma and high and stable CD4 counts for prolonged periods. Of the eight chemokine receptors tested in GHOST cells, these HIV-2 variants used only CCR5, GPR15, and CXCR6 with high efficiency. Most HIV-2 variants isolated from individuals with progressive HIV-2 infection and/or viremia were restricted to the same chemokine receptors, while some variants from half of these individuals used CXCR4 instead. Use of CCR1, CCR2b, and CCR3 was rare.
Almost a quarter of all tested HIV-2 variants could infect the parental GHOST cells, albeit inefficiently. This could be partially explained by the presence of CXCR4, which is known to be endogenously expressed by HOS cells, from which GHOST cells are derived (11, 47). Every GHOST cell line contained a fraction of CXCR4-expressing cells, the magnitude of which correlated with the level of virus produced by X4-restricted HIV variants. Discrepancies exist in the literature regarding infection of parental GHOST cells by X4 HIV variants (8, 42, 57, 58, 60, 61). This may suggest that different batches of parental cell lines have different levels of CXCR4 expression and consequently different susceptibilities to X4 HIV infection. Other variants infected parental GHOST cells independently of CXCR4, as evidenced by their insensitivity to T22, relatively low capacity to infect GHOST-CXCR4 cells, and inability to infect CCR5−/− PBMC. One can speculate that such variants are able to infect CD4-positive cells independently of a coreceptor. However, if it exists, coreceptor-independent infection occurs only under specific conditions, since these variants could not infect CCR5−/− PBMC. Alternatively, GHOST cells may express a second, still unidentified HIV-2 coreceptor. If so, the low susceptibility of parental GHOST cells to only a minority of HIV-2 variants indicates either that this unidentified GHOST coreceptor is not a major HIV-2 coreceptor or that expression levels are low, allowing only variants with the highest affinity for the receptor to enter the cells. A candidate might be CXCR6, which is expressed by HOS cells (3), but as we show here, at levels too low to be detectable by FACS analysis. However, the lack of an association between virus production in GHOST-CXCR6 cells and that in parental cells argues against CXCR6 being the second GHOST coreceptor. An alternative candidate may be CXCR5/BLR1 (31).
Parental U87 cells appeared highly susceptible to infection with HIV-2, although the efficiency of infection varied among virus variants. This was observed for three different batches of parental U87 cells, among which were cells freshly obtained from an AIDS reagent distribution program and cells closely related to those used in a previous study, and is in agreement with previous studies demonstrating infection of U87.CD4 cells by HIV-2 (14, 23, 51). The high frequency with which parental cells are infected and the high levels of virus produced indicate the presence of a major HIV-2 coreceptor on the surfaces of U87 cells. Although U87 cells express CXCR6 mRNA (25, 48), we could not detect CXCR6 on the cell surface of any of the U87 derivatives other than that of the U87-CXCR6 cell line. Given the high virus production in all U87 cell lines, it is unlikely that CXCR6 represents the unknown U87 coreceptor. Alternative candidates are GPR1 and RDC1, both of which are expressed by U87 cells (25, 26, 48, 49) and both of which confer HIV-2 susceptibility on the otherwise nonpermissive human glioma cell line NP-2 when coexpressed with CD4 (48, 49). Further studies are needed to reveal the identity of this HIV-2 coreceptor.
In previous coreceptor studies, parental U87 cells were not susceptible to infection by all of the 33 primary HIV-2 isolates tested (29, 37, 38). Four of these variants were included in our study and appeared capable of infecting three batches of parental U87 cells tested. Our results indicate that variable susceptibilities of parental cell batches used, possibly in combination with the degree of U87.CD4 tropism of HIV-2 variants tested, are responsible for different findings regarding infection of parental U87 cells. The variable susceptibility of parental U87 cells may be due to variable expression levels of the endogenous HIV-2 coreceptor and/or of transduced CD4. In support of the latter, parental cells of batch B had lower levels of CD4+ cells than cells of batch C, and parental cells we used in pilot experiments appeared to have even lower levels of CD4+ cells. In addition, retrospective analysis of an old batch of parental cells stored in our laboratory since 1998 revealed that these cells had no detectable levels of CD4, and the cells were not susceptible to HIV-2 infection (data not shown).
In the present study, use of CCR1, CCR2b, and CCR3 by HIV-2 was rarely detected using GHOST cells. This contrasts particularly with previous studies using U87 cells, which demonstrated frequent use of CCR1, CCR2b, and CCR3, albeit with different efficiencies in different studies (29, 37, 38). In line with our data, with GHOST cells, use of these coreceptors was less frequently detected, and if detected, was mainly associated with use of CXCR4 (42, 60). It is conceivable that GHOST cells are less sensitive than U87 cells and that the differences between studies and different systems reflect weak use of these coreceptors. In our hands, virus production levels at least 100 times lower than that in CCR5-expressing GHOST cells could still have been detected for most variants, indicating that if CCR1, CCR2b, and CCR3 are used, then it is only with relatively low efficiency. Inefficient use of these coreceptors might be revealed using more sensitive cell-to-cell fusion assays, as was previously demonstrated for the HIV-2ROD strain (7).
Alternatively, underestimation of nonspecific entry in coreceptor-transduced GHOST and U87 cells, concealed due to the relative lack of susceptibility of parental cells, may have resulted in overestimation of coreceptor use. In support of this, our conclusions based on U87 cells are similar to those based on GHOST cells when virus production in the parental U87 cells is considered to represent nonspecific infection in the other cell lines. Nevertheless, analysis in U87 cells confirmed efficient use of a broad spectrum of coreceptors for two out of four previously tested variants, suggesting that at least some of the discrepancies between previous studies and our own reflect true differences in virus properties.
The use of CXCR6 and/or GPR15 appeared to be a common property of HIV-2 and has also been observed by others (9, 38, 60). HIV-2 shares this property with SIV (9, 56, 60), which differentiates these two viruses from HIV-1 (8, 55, 59, 61). The capacity to use these two chemokine receptors, and also the use of the unidentified U87 coreceptor, may explain the broader tropism of HIV-2 than HIV-1 observed in vitro (14, 36). The demonstration that HIV-2 variants isolated from individuals with undetectable levels of virus in their plasma efficiently use these chemokine receptors argues against a role in enhancing HIV virulence. The paradox of broad in vitro tropism and low in vivo virus production may be explained by the absence of significant numbers of target cells expressing both CD4 and sufficient levels of the chemokine receptors in vivo. Both CXCR6 and GPR15 are expressed by activated T cells (20, 26, 32, 48, 53), but none of the HIV-2 variants analyzed here could infect CCR5−/− PBMC in vitro independently of CXCR4. However, others did observe low levels of infection of CCR5−/− PBMC by HIV-2 variants that cannot use CXCR4. The inconsistencies may partially result from the different genetic background and CXCR6 and GPR15 expression levels of the donor of the CCR5−/− PBMC (9, 39).
In contrast to CXCR6 and GPR15, use of CXCR4 was observed only in variants isolated from individuals with viremia, three of whom had overt progressive disease. The association of HIV-2 variants capable of using CXCR4 with progressive infection is reminiscent of what is known for HIV-1 (16,33). Variants from one asymptomatic individual with viremia could infect some, but not all, CXCR4-positive cell types, which may suggest that X4 HIV-2 variants with different CXCR4 binding properties and different pathogenic potentials exist. In analogy to HIV-1 infection, X4 variants were not observed in all individuals with progressive infection, indicating that although the capacity to use CXCR4 is likely a determinant of HIV-2 virulence, it is not the only one.
In conclusion, we find that HIV-2 variants from nonprogressing individuals have a broader coreceptor repertoire than HIV-1. Moreover, we find that the number of coreceptors used per HIV-2 variant does not increase with disease progression. These findings establish for the first time that promiscuous coreceptor usage per se is not a determinant of HIV pathogenicity.
Acknowledgments
This study was financially supported by Zon-MW (grant number 906-02-007) and approved by the Ethical Medical Committee (no. 196.454/2000/221).
We thank the HIV-2-infected participants of the cohort, as well as the donor of the CCR5−/− PBMC, for their participation. We are very grateful to R. P. van Rij for helpful discussions and pivotal suggestions, to C. A. van Baalen for critically reading the manuscript, to D. Kwa for supplying HIV-1 control variants, to Á. McKnight and D. Marchant for advice and for donating HIV-2 isolates and parental U87 cells, and to P. Marx and T. Williams for donating SIVrcm. GHOST cells were provided by D. R. Littman and V. N. Kewalramani via the EU program EVA/MRC Centralised Facility for AIDS Reagents, NIBSC, United Kingdom (grant numbers QLKZ-CT-1999-00609 and GP828102). U87 cells were provided by D. R. Littman and H. Deng via the National Institutes of Health AIDS Research and Reference Reagent Program (U87-CCR3, U87-CCR5, and U87.CD4) and via the EU program EVA/MRC Centralised Facility for AIDS Reagents (U87-CCR1, U87-CCR2, U87-CXCR4, and U87.CD4).
REFERENCES
- 1.Albert, J., A. Naucler, B. Bottiger, P. A. Broliden, P. Albino, S. A. Ouattara, C. Bjorkegren, A. Valentin, G. Biberfeld, and E. M. Fenyo. 1990. Replicative capacity of HIV-2, like HIV-1, correlates with severity of immunodeficiency. AIDS 4:291-295. [DOI] [PubMed] [Google Scholar]
- 2.Berkowitz, R. D., S. Alexander, C. Bare, V. Linquist-Stepps, M. Bogan, M. E. Moreno, L. Gibson, E. D. Wieder, J. Kosek, C. A. Stoddart, and J. M. McCune. 1998. CCR5- and CXCR4-utilizing strains of human immunodeficiency virus type 1 exhibit differential tropism and pathogenesis in vivo. J. Virol. 72:10108-10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berry, N., K. Ariyoshi, S. Jaffar, S. Sabally, T. Corrah, R. Tedder, and H. Whittle. 1998. Low peripheral blood viral HIV-2 RNA in individuals with high CD4 percentage differentiates HIV-2 from HIV-1 infection. J. Hum. Virol. 1:457-468. [PubMed] [Google Scholar]
- 4.Björndal, A., H. Deng, M. Jansson, J. R. Fiore, C. Colognesi, A. Karlsson, J. Albert, G. Scarlatti, D. R. Littman, and E. M. Fenyö. 1997. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 71:7478-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaak, H., P. H. Boers, M. Schutten, M. E. Van Der Ende, and A. D. Osterhaus. 2004. HIV-2-infected individuals with undetectable plasma viremia carry replication-competent virus in peripheral blood lymphocytes. J. Acquir. Immun. Defic. Syndr. 36:777-782. [DOI] [PubMed] [Google Scholar]
- 6.Blaak, H., A. B. van't Wout, M. Brouwer, B. Hooibrink, E. Hovenkamp, and H. Schuitemaker. 2000. In vivo HIV-1 infection of CD45RA+CD4+ T cells is established primarily by syncytium-inducing variants and correlates with the rate of CD4+ T cell decline. Proc. Natl. Acad. Sci. USA 97:1269-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bron, R., P. J. Klasse, D. Wilkinson, P. R. Clapham, A. Pelchen-Matthews, C. Power, T. N. Wells, J. Kim, S. C. Peiper, J. A. Hoxie, and M. Marsh. 1997. Promiscuous use of CC and CXC chemokine receptors in cell-to-cell fusion mediated by a human immunodeficiency virus type 2 envelope protein. J. Virol. 71:8405-8415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cecilia, D., V. N. Kewalramani, J. O'Leary, B. Volsky, P. Nyambi, S. Burda, S. Xu, D. R. Littman, and S. Zolla-Pazner. 1998. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J. Virol. 72:6988-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, Z., A. Gettie, D. D. Ho, and P. A. Marx. 1998. Primary SIVsm isolates use the CCR5 coreceptor from sooty mangabeys naturally infected in West Africa: a comparison of coreceptor usage of primary SIVsm, HIV-2, and SIVmac. Virology 246:113-124. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Z., D. Kwon, Z. Jin, S. Monard, P. Telfer, M. S. Jones, C. Y. Lu, R. F. Aguilar, D. D. Ho, and P. A. Marx. 1998. Natural infection of a homozygous delta24 CCR5 red-capped mangabey with an R2b-tropic simian immunodeficiency virus. J. Exp Med. 188:2057-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho, M. W., M. K. Lee, M. C. Carney, J. F. Berson, R. W. Doms, and M. A. Martin. 1998. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J. Virol. 72:2509-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choe, H., M. Farzan, M. Konkel, K. Martin, Y. Sun, L. Marcon, M. Cayabyab, M. Berman, M. E. Dorf, N. Gerard, C. Gerard, and J. Sodroski. 1998. The orphan seven-transmembrane receptor apj supports the entry of primary T-cell-line-tropic and dualtropic human immunodeficiency virus type 1. J. Virol. 72:6113-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 14.Clapham, P. R., D. Blanc, and R. A. Weiss. 1991. Specific cell surface requirements for the infection of CD4-positive cells by human immunodeficiency virus types 1 and 2 and by simian immunodeficiency virus. Virology 181:703-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Combadiere, C., K. Salzwedel, E. D. Smith, H. L. Tiffany, E. A. Berger, and P. M. Murphy. 1998. Identification of CX3CR1. A chemotactic receptor for the human CX3C chemokine fractalkine and a fusion coreceptor for HIV-1. J. Biol. Chem. 273:23799-23804. [DOI] [PubMed] [Google Scholar]
- 16.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damond, F., M. Gueudin, S. Pueyo, I. Farfara, D. L. Robertson, D. Descamps, G. Chene, S. Matheron, P. Campa, F. Brun-Vezinet, and F. Simon. 2002. Plasma RNA viral load in human immunodeficiency virus type 2 subtype A and subtype B infections. J. Clin. Microbiol. 40:3654-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Cock, K. M., G. Adjorlolo, E. Ekpini, T. Sibailly, J. Kouadio, M. Maran, K. Brattegaard, K. M. Vetter, R. Doorly, and H. D. Gayle. 1993. Epidemiology and transmission of HIV-2. Why there is no HIV-2 pandemic. JAMA 270:2083-2086. [DOI] [PubMed] [Google Scholar]
- 19.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 20.Deng, H. K., D. Unutmaz, V. N. Kewalramani, and D. R. Littman. 1997. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature 388:296-300. [DOI] [PubMed] [Google Scholar]
- 21.Dittmar, M. T., L. Zekeng, L. Kaptue, J. Eberle, H. G. Krausslich, and L. Gurtler. 1999. Coreceptor requirements of primary HIV type 1 group O isolates from Cameroon. AIDS Res. Hum. Retrovir. 15:707-712. [DOI] [PubMed] [Google Scholar]
- 22.Doranz, B. J., J. Rucker, Y. Yi, R. J. Smyth, M. Samson, S. C. Peiper, M. Parmentier, R. G. Collman, and R. W. Doms. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149-1158. [DOI] [PubMed] [Google Scholar]
- 23.Dragic, T., and M. Alizon. 1993. Different requirements for membrane fusion mediated by the envelopes of human immunodeficiency virus types 1 and 2. J. Virol. 67:2355-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. Huang, K. A. Nagashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667-673. [DOI] [PubMed] [Google Scholar]
- 25.Edinger, A. L., T. L. Hoffman, M. Sharron, B. Lee, B. O'Dowd, and R. W. Doms. 1998. Use of GPR1, GPR15, and STRL33 as coreceptors by diverse human immunodeficiency virus type 1 and simian immunodeficiency virus envelope proteins. Virology 249:367-378. [DOI] [PubMed] [Google Scholar]
- 26.Farzan, M., H. Choe, K. Martin, L. Marcon, W. Hofmann, G. Karlsson, Y. Sun, P. Barrett, N. Marchand, N. Sullivan, N. Gerard, C. Gerard, and J. Sodroski. 1997. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J. Exp. Med. 186:405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 28.Gottlieb, G. S., P. S. Sow, S. E. Hawes, I. Ndoye, M. Redman, A. M. Coll-Seck, M. A. Faye-Niang, A. Diop, J. M. Kuypers, C. W. Critchlow, R. Respess, J. I. Mullins, and N. B. Kiviat. 2002. Equal plasma viral loads predict a similar rate of CD4+ T cell decline in human immunodeficiency virus (HIV) type 1- and HIV-2-infected individuals from Senegal, West Africa. J. Infect. Dis. 185:905-914. [DOI] [PubMed] [Google Scholar]
- 29.Guillon, C., M. E. van der Ende, P. H. Boers, R. A. Gruters, M. Schutten, and A. D. Osterhaus. 1998. Coreceptor usage of human immunodeficiency virus type 2 primary isolates and biological clones is broad and does not correlate with their syncytium-inducing capacities. J. Virol. 72:6260-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho, W. Z., R. Cherukuri, S. D. Ge, J. R. Cutilli, L. Song, S. Whitko, and S. D. Douglas. 1993. Centrifugal enhancement of human immunodeficiency virus type 1 infection and human cytomegalovirus gene expression in human primary monocyte/macrophages in vitro. J. Leukoc. Biol. 53:208-212. [DOI] [PubMed] [Google Scholar]
- 31.Kanbe, K., N. Shimizu, Y. Soda, K. Takagishi, and H. Hoshino. 1999. A CXC chemokine receptor, CXCR5/BLR1, is a novel and specific coreceptor for human immunodeficiency virus type 2. Virology 265:264-273. [DOI] [PubMed] [Google Scholar]
- 32.Kim, C. H., E. J. Kunkel, J. Boisvert, B. Johnston, J. J. Campbell, M. C. Genovese, H. B. Greenberg, and E. C. Butcher. 2001. Bonzo/CXCR6 expression defines type 1-polarized T-cell subsets with extralymphoid tissue homing potential. J. Clin. Investig. 107:595-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koot, M., I. P. Keet, A. H. Vos, R. E. de Goede, M. T. Roos, R. A. Coutinho, F. Miedema, P. T. Schellekens, and M. Tersmette. 1993. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann. Intern. Med. 118:681-688. [DOI] [PubMed] [Google Scholar]
- 34.Liao, F., G. Alkhatib, K. W. Peden, G. Sharma, E. A. Berger, and J. M. Farber. 1997. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J. Exp. Med. 185:2015-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marlink, R., P. Kanki, I. Thior, K. Travers, G. Eisen, T. Siby, I. Traore, C. C. Hsieh, M. C. Dia, E. H. Gueye, J. Hellinger, A. Guèye-Ndiaye, J. L. Sankalé, I. Ndoye, S. Mboup, and M. Essex. 1994. Reduced rate of disease development after HIV-2 infection as compared to HIV-1. Science 265:1587-1590. [DOI] [PubMed] [Google Scholar]
- 36.McKnight, A., P. R. Clapham, and R. A. Weiss. 1994. HIV-2 and SIV infection of nonprimate cell lines expressing human CD4: restrictions to replication at distinct stages. Virology 201:8-18. [DOI] [PubMed] [Google Scholar]
- 37.McKnight, A., M. T. Dittmar, J. Moniz-Periera, K. Ariyoshi, J. D. Reeves, S. Hibbitts, D. Whitby, E. Aarons, A. E. Proudfoot, H. Whittle, and P. R. Clapham. 1998. A broad range of chemokine receptors are used by primary isolates of human immunodeficiency virus type 2 as coreceptors with CD4. J. Virol. 72:4065-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mörner, A., A. Björndal, J. Albert, V. N. Kewalramani, D. R. Littman, R. Inoue, R. Thorstensson, E. M. Fenyö, and E. Björling. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 73:2343-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mörner, A., A. Björndal, A. C. Leandersson, J. Albert, E. Björling, and M. Jansson. 2002. CCR5 or CXCR4 is required for efficient infection of peripheral blood mononuclear cells by promiscuous human immunodeficiency virus type 2 primary isolates. AIDS Res. Hum. Retrovir. 18:193-200. [DOI] [PubMed] [Google Scholar]
- 40.Murakami, T., T. Nakajima, Y. Koyanagi, K. Tachibana, N. Fujii, H. Tamamura, N. Yoshida, M. Waki, A. Matsumoto, O. Yoshie, T. Kishimoto, N. Yamamoto, and T. Nagasawa. 1997. A small molecule CXCR4 inhibitor that blocks T cell line-tropic HIV-1 infection. J. Exp. Med. 186:1389-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakami, T., T. Y. Zhang, Y. Koyanagi, Y. Tanaka, J. Kim, Y. Suzuki, S. Minoguchi, H. Tamamura, M. Waki, A. Matsumoto, N. Fujii, H. Shida, J. A. Hoxie, S. C. Peiper, and N. Yamamoto. 1999. Inhibitory mechanism of the CXCR4 antagonist T22 against human immunodeficiency virus type 1 infection. J. Virol. 73:7489-7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Owen, S. M., D. Ellenberger, M. Rayfield, S. Wiktor, P. Michel, M. H. Grieco, F. Gao, B. H. Hahn, and R. B. Lal. 1998. Genetically divergent strains of human immunodeficiency virus type 2 use multiple coreceptors for viral entry. J. Virol. 72:5425-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pietroboni, G. R., G. B. Harnett, and M. R. Bucens. 1989. Centrifugal enhancement of human immunodeficiency virus (HIV) and human herpesvirus type 6 (HHV-6) infection in vitro. J. Virol. Methods 24:85-90. [DOI] [PubMed] [Google Scholar]
- 44.Popper, S. J., A. D. Sarr, K. U. Travers, A. Gueye-Ndiaye, S. Mboup, M. E. Essex, and P. J. Kanki. 1999. Lower human immunodeficiency virus (HIV) type 2 viral load reflects the difference in pathogenicity of HIV-1 and HIV-2. J. Infect. Dis. 180:1116-1121. [DOI] [PubMed] [Google Scholar]
- 45.Roda Husman, A. M., R. P. van Rij, H. Blaak, S. Broersen, and H. Schuitemaker. 1999. Adaptation to promiscuous usage of chemokine receptors is not a prerequisite for human immunodeficiency virus type 1 disease progression. J. Infect. Dis. 180:1106-1115. [DOI] [PubMed] [Google Scholar]
- 46.Rucker, J., A. L. Edinger, M. Sharron, M. Samson, B. Lee, J. F. Berson, Y. Yi, B. Margulies, R. G. Collman, B. J. Doranz, M. Parmentier, and R. W. Doms. 1997. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J. Virol. 71:8999-9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schols, D., J. A. Este, C. Cabrera, and E. De Clercq. 1998. T-cell-line-tropic human immunodeficiency virus type 1 that is made resistant to stromal cell-derived factor 1α contains mutations in the envelope gp120 but does not show a switch in coreceptor use. J. Virol. 72:4032-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimizu, N., Y. Soda, K. Kanbe, H. Y. Liu, A. Jinno, T. Kitamura, and H. Hoshino. 1999. An orphan G protein-coupled receptor, GPR1, acts as a coreceptor to allow replication of human immunodeficiency virus types 1 and 2 in brain-derived cells. J. Virol. 73:5231-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimizu, N., Y. Soda, K. Kanbe, H. Y. Liu, R. Mukai, T. Kitamura, and H. Hoshino. 2000. A putative G protein-coupled receptor, RDC1, is a novel coreceptor for human and simian immunodeficiency viruses. J. Virol. 74:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simon, F., S. Matheron, C. Tamalet, I. Loussert-Ajaka, S. Bartczak, J. M. Pepin, C. Dhiver, E. Gamba, C. Elbim, and J. A. Gastaut. 1993. Cellular and plasma viral load in patients infected with HIV-2. AIDS 7:1411-1417. [DOI] [PubMed] [Google Scholar]
- 51.Sol, N., F. Ferchal, J. Braun, O. Pleskoff, C. Treboute, I. Ansart, and M. Alizon. 1997. Usage of the coreceptors CCR-5, CCR-3, and CXCR-4 by primary and cell line-adapted human immunodeficiency virus type 2. J. Virol. 71:8237-8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tersmette, M., R. E. de Goede, B. J. Al, I. N. Winkel, R. A. Gruters, H. T. Cuypers, H. G. Huisman, and F. Miedema. 1988. Differential syncytium-inducing capacity of human immunodeficiency virus isolates: frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J. Virol. 62:2026-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Unutmaz, D., W. Xiang, M. J. Sunshine, J. Campbell, E. Butcher, and D. R. Littman. 2000. The primate lentiviral receptor Bonzo/STRL33 is coordinately regulated with CCR5 and its expression pattern is conserved between human and mouse. J. Immunol. 165:3284-3292. [DOI] [PubMed] [Google Scholar]
- 54.van der Ende, M. E., M. Schutten, T. D. Ly, R. A. Gruters, and A. D. Osterhaus. 1996. HIV-2 infection in 12 European residents: virus characteristics and disease progression. AIDS 10:1649-1655. [DOI] [PubMed] [Google Scholar]
- 55.van Rij, R. P., H. Blaak, J. A. Visser, M. Brouwer, R. Rientsma, S. Broersen, A. M. Roda Husman, and H. Schuitemaker. 2000. Differential coreceptor expression allows for independent evolution of non-syncytium-inducing and syncytium-inducing HIV-1. J. Clin. Investig. 106:1039-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vödrös, D., R. Thorstensson, G. Biberfeld, D. Schols, E. De Clercq, and E. M. Fenyö. 2001. Coreceptor usage of sequential isolates from cynomolgus monkeys experimentally Infected with simian immunodeficiency virus (SIVsm). Virology 291:12-21. [DOI] [PubMed] [Google Scholar]
- 57.Vödrös, D., C. Tscherning-Casper, L. Navea, D. Schols, E. De Clercq, and E. M. Fenyö. 2001. Quantitative evaluation of HIV-1 coreceptor use in the GHOST3 cell assay. Virology 291:1-11. [DOI] [PubMed] [Google Scholar]
- 58.Xiao, L., D. L. Rudolph, S. M. Owen, T. J. Spira, and R. B. Lal. 1998. Adaptation to promiscuous usage of CC and CXC-chemokine coreceptors in vivo correlates with HIV-1 disease progression. AIDS 12:F137-F143. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, L., T. He, Y. Huang, Z. Chen, Y. Guo, S. Wu, K. J. Kunstman, R. C. Brown, J. P. Phair, A. U. Neumann, D. D. Ho, and S. M. Wolinsky. 1998. Chemokine coreceptor usage by diverse primary isolates of human immunodeficiency virus type 1. J. Virol. 72:9307-9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, Y., B. Lou, R. B. Lal, A. Gettie, P. A. Marx, and J. P. Moore. 2000. Use of inhibitors to evaluate coreceptor usage by simian and simian/human immunodeficiency viruses and human immunodeficiency virus type 2 in primary cells. J. Virol. 74:6893-6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, Y. J., T. Dragic, Y. Cao, L. Kostrikis, D. S. Kwon, D. R. Littman, V. N. Kewalramani, and J. P. Moore. 1998. Use of coreceptors other than CCR5 by non-syncytium-inducing adult and pediatric isolates of human immunodeficiency virus type 1 is rare in vitro. J. Virol. 72:9337-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]