Abstract
The interferon (IFN) response is the first line of defense against viral infections, and the majority of viruses have developed different strategies to counteract IFN responses in order to ensure their survival in an infected host. In this study, the abilities to inhibit IFN signaling of two closely related West Nile viruses, the New York 99 strain (NY99) and Kunjin virus (KUN), strain MRM61C, were analyzed using reporter plasmid assays, as well as immunofluorescence and Western blot analyses. We have demonstrated that infections with both NY99 and KUN, as well as transient or stable transfections with their replicon RNAs, inhibited the signaling of both alpha/beta IFN (IFN-α/β) and gamma IFN (IFN-γ) by blocking the phosphorylation of STAT1 and its translocation to the nucleus. In addition, the phosphorylation of STAT2 and its translocation to the nucleus were also blocked by KUN, NY99, and their replicons in response to treatment with IFN-α. IFN-α signaling and STAT2 translocation to the nucleus was inhibited when the KUN nonstructural proteins NS2A, NS2B, NS3, NS4A, and NS4B, but not NS1 and NS5, were expressed individually from the pcDNA3 vector. The results clearly demonstrate that both NY99 and KUN inhibit IFN signaling by preventing STAT1 and STAT2 phosphorylation and identify nonstructural proteins responsible for this inhibition.
The interferons (IFNs) are a large family of multifunctional secreted cytokines involved in antiviral defense, cell growth regulation, and immune activation. IFNs are produced by the majority of cells and include 14 different species of alpha IFN (IFN-α) and one species of beta IFN (IFN-β); these IFNs are involved primarily in antiviral and antiproliferative responses (7, 16, 17, 28). Gamma IFN (IFN-γ) is IFN that is usually produced by specific cells of the immune system, including CD8+ T cells, and has potent antiviral and immunomodulating activities (7, 16, 17, 28). The binding of IFNs to corresponding receptors on cell surfaces triggers a cascade of different signaling pathways that eventually lead to the transcriptional activation of a large number of IFN-stimulated genes (ISGs), which can establish antiviral, antiproliferative, and/or immunoregulatory states in host cells. The best-studied IFN signaling pathways are based on IFN receptor-Janus Kinase (JAK)/signal transducer and activator of transcription (STAT) activation (7, 16). The binding of IFN-α and IFN-β to the IFN-α/β receptor, which consists of IFNAR1 and IFNAR2 molecules, leads to the activation of JAK1 and Tyk-2 kinases via tyrosine phosphorylation. Activated Tyk-2 phosphorylates IFNAR1, which then serves as a binding site for STAT2. STAT2 is then phosphorylated by Tyk-2 and serves as a binding site for STAT1, which is subsequently phosphorylated by JAK1. The phosphorylated STAT2-STAT1 heterodimers then dissociate from the receptor and associate with p48/IRF-9 to form an ISGF3 complex that translocates to the nucleus, where it initiates the transcription of ISGs via binding to the IFN-stimulated response element (ISRE). The binding of IFN-γ to the IFN-γ receptor, which consists of two IFNGR1 and IFNGR2 molecules, leads to the activation of JAK1 and JAK2 kinases via tyrosine phosphorylation. Activated JAK1 phosphorylates the IFNGRα chain, which serves as a binding site for STAT1, which is subsequently phosphorylated by JAK2. Two phosphorylated STAT1 molecules form a homodimer, which dissociates from the receptor and migrates to the nucleus, where it initiates the transcription of ISGs via binding to gamma-activated sequence (GAS) (7, 16).
Many viruses have developed different strategies to counteract IFN responses in order to ensure their survival in an infected host. A number of comprehensive recent reviews discuss this in great detail (7, 17, 25, 28). The examples of RNA viruses interfering with IFN induction and signaling pathways include influenza virus, Ebola virus, Sendai virus, simian virus 5 (SV5), bovine respiratory syncytial virus, parainfluenza virus, hepatitis C virus, bovine viral diarrhea virus (7, 25, 28), and, most recently, dengue (DEN) virus (26). Viruses counter IFN responses by three means of inhibition: (i) inhibition of IFN production by sequestering double-stranded RNA or inhibition of the activation of the double-stranded RNA-dependent protein kinase R (PKR), NF-κB, and other IFN regulatory factors, e.g., IRF-1 and IRF-3; (ii) inhibition of IFN signaling at different levels (i.e., signaling of IFN receptors, JAK/STAT activation, and signaling of p48 and ISGF3 transcriptional factors); and (iii) inhibition of IFN-induced antiviral enzymes, such as the PKR, 2′-5′ oligoadenylate synthetase-RNase L. Some viruses, e.g., SV5 and hepatitis C virus, were shown to inhibit both IFN induction and IFN signaling (1, 5, 12, 13, 27).
Several members of the family Flaviviridae, including West Nile (WN) virus, DEN virus, yellow fever virus, tick-borne encephalitis virus, Murray valley encephalitis virus, Japanese encephalitis (JE) virus, and hepatitis C virus, are major pathogens of humans (14). Studies with WN virus, DEN virus, yellow fever virus, and Murray valley encephalitis virus demonstrated that pretreatment with IFNs inhibited flavivirus infection in cell culture and in animals, with IFN-α and -β apparently inhibiting virus infection by preventing the translation and replication of viral RNA. IFN-γ affects virus replication probably via the generation of proinflammatory and antiviral molecules, including nitric oxide (reviewed in reference 2). However, the antiviral effect of IFNs were minimal if the treatment was performed as early as 4 h after infection (3), indicating that flaviviruses are able to overcome the antiviral activities of IFNs once they establish replication. Similarly, a recent study with WN virus showed that the virus was able to overcome the host antiviral response despite demonstrated induction of IFN-β transcription via the IRF-3 activation pathway (6). The mechanism by which flaviviruses acquire resistance to the antiviral activity of IFN is not well understood; however, recent studies with DEN and JE viruses suggest that one or more nonstructural proteins may be involved in preventing the activation of the STAT1 and STAT2 proteins, most likely by blocking the phosphorylation of Tyk-2 kinase (21, 26).
An Australian flavivirus, the Kunjin virus (KUN), recently classified as a subtype of WN virus (14), appears to be naturally attenuated and does not cause an overt disease in humans (10). In contrast, the New York 99 strain (NY99) of WN virus, which is genetically closely related to KUN (more than 98% homology in amino acid sequence [20, 22, 30]), has already caused ∼500 deaths and over 20,000 registered infections since its emergence in North America in 1999, including 262 deaths and 9,858 infections in 2003 alone (http://www.cdc.gov/ncidod/dvbid/westnile/surv&controlCaseCount03_detailed.htm). In this study we analyzed the ability of these two viruses, KUN and NY99, to inhibit cellular antiviral responses. We showed that KUN and NY99 as well as their replicon RNAs inhibited IFN signaling to ISRE and GAS promoters by preventing the phosphorylation of STAT1 and/or STAT2 and their subsequent transport to the nucleus. We also showed that the inhibition of IFN signaling and STAT2 phosphorylation can be achieved by the expression of individual nonstructural proteins.
KUN replicon RNA replication is resistant to treatment with IFN-α and IFN-γ.
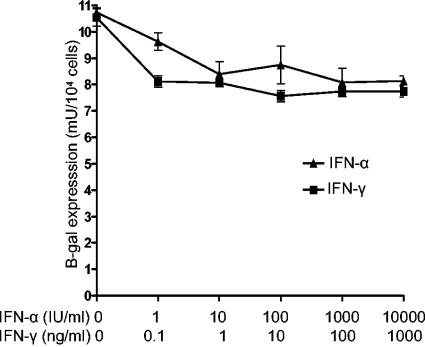
A previous study with HeLa cells stably transfected with KUN replicon RNA showed that treatment with up to 160 IU of IFN-α per ml did not significantly inhibit the replication of KUN replicon RNA (8). Here we employed HEp2 cells stably transfected with KUN replicon RNA expressing the β-galactosidase reporter gene to examine whether higher concentrations of IFN-α or treatment with IFN-γ would affect KUN RNA replication. HEp2 cells stably expressing KUN replicon RNA (H-KUNrep cells) were generated by propagating HEp2 cells transfected with replicon RNA repPAC/β-gal (23) in medium containing 1 μg of puromycin (Sigma, St. Louis, Mo.) per ml. Treatment of H-KUNrep cells with 1 and 10 IU of IFN-α per ml resulted in ∼10 and ∼20% decreases in the levels of β-galactosidase expression, respectively, while a further increase in IFN-α concentration from 10 to 10,000 IU/ml did not produce additional inhibition of β-galactosidase expression (Fig. 1). Similarly, an ∼10% decrease in β-galactosidase expression was observed after treatment with 0.1 ng of IFN-γ/ml, while no further inhibition of β-galactosidase expression was observed with increasing concentrations of IFN-γ up to 1,000 ng/ml (Fig. 1). The results (i) confirmed a previous finding of the relative resistance of KUN replicon RNA replication to treatment with IFN-α (8), (ii) extended the range of IFN-α concentrations that do not substantially affect KUN RNA replication to 10,000 IU/ml, and (iii) demonstrated the relative resistance of KUN replicon RNA replication to treatment with up to a 1,000 ng of IFN-γ/ml. Our results correlate well with the results of others showing the relative resistance of DEN virus replication to treatment with IFN-α, IFN-β, and IFN-γ when the IFN treatment was initiated after virus replication was established (3).
FIG. 1.
IFN-α and IFN-γ did not inhibit KUN replicon RNA replication and expression. HEp2 cells stably expressing the KUN replicon RNA repPAC/β-gal (23) in a 96-well plate were treated with different concentrations of IFN-α2A (Roferon-A; Roche, Basel, Switzerland) and IFN-γ (I-1520; Sigma) as indicated. Forty-eight hours after IFN treatment, cells were lysed and analyzed for β-galactosidase expression to compare the replication and expression efficiencies of KUN replicon RNAs. The values of β-galactosidase expression are averages from triplicate samples, and the error bars show standard deviations.
IFN signaling is blocked in Vero cells stably or transiently transfected with KUN or NY99 replicon RNAs.
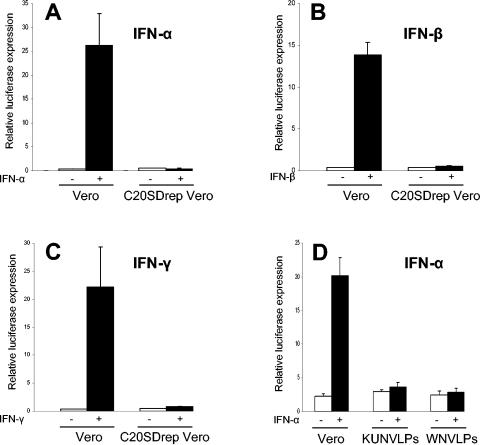
We next analyzed whether the resistance of KUN RNA replication to IFN treatment is mediated by the inhibition of IFN signaling using reporter assays with plasmids encoding the luciferase reporter gene under the control of IFN-α/β (ISRE)- or IFN-γ (GAS)-responsive elements. Luciferase expression from the ISRE or GAS promoter in response to treatment with IFN-α and IFN-β or IFN-γ, respectively, was severely inhibited in Vero cells stably transfected with KUN replicon RNA (C20SDrepVero) (24). In contrast, 75-, 35-, and 55-fold induction of luciferase expression in response to IFN-α, IFN-β, and IFN-γ treatments, respectively, were observed in normal Vero cells (Fig. 2A to C). In a separate experiment, IFN-α signaling was also inhibited in HEp2 cells stably expressing KUN replicon RNA (data not shown). Further examination of IFN-α signaling in Vero cells that were previously infected with virus-like particles (VLPs) containing KUN or NY99 replicon RNA showed that both KUN and NY99 replicon RNA replication blocked the induction of luciferase expression in response to IFN-α treatment with similar efficiencies (Fig. 2D). Fluorescence-activated cell sorter analysis of VLP-infected cells with cross-reacting KUN anti-NS3 antibodies showed that the efficiencies of infection were similar for KUN and NY99 replicon VLPs (data not shown). Thus, both the stable and transient replication of KUN and NY99 replicon RNAs blocked IFN signaling.
FIG. 2.
IFN signaling is inhibited in Vero cells stably (A-C) or transiently (D) transfected with KUN (KUNrep) or NY99 (WNrep) replicon RNAs. (A-C) Normal Vero cells and Vero cells stably transfected with KUN replicon RNA (C20SDrepVero) (24) in 24-well plates (four wells from each transfection mixture) were cotransfected with the control plasmid pCMV-β (Clontech, Palo Alto, Calif.) and either the IFN-α/β-responsive (ISRE) luciferase reporter plasmid p(9-27)4thΔ(−39)Lucter (18) or the IFN-γ-responsive (GAS) luciferase reporter plasmid p(IRF-1*GAS)6tkΔ(−39)Lucter (18). Forty hours after transfection, cells were treated with 1,000 U of IFN-αA (I-4276; Sigma) per ml (A), 1,000 IU of IFN-β (I-4151; Sigma) per ml (B), or 50 ng of IFN-γ (I-1520; Sigma) per ml (C) for 14 h and then assayed for luciferase and β-galactosidase expression. (D) Vero cells were infected with VLPs containing KUN replicon RNA (C20DXHDVrep, derivative of C20UbHDVrep) (31) or NY99 replicon RNA (29) at an MOI of 10. Replicon VLPs were produced by transfection of replicon RNAs into the tetKUNCprME packaging cell line as described previously (11). Four hours after infection, cells were cotransfected with plasmid p(9-27)4thΔ(−39)Lucter and the control plasmid pCMV-β. Forty hours after transfection, cells were treated with 1,000 U of IFN-αA (I-4276; Sigma) per ml for 14 h and assayed for luciferase and β-galactosidase expression. The luciferase activity in panels A to D is expressed in relative light units normalized to the expression of β-galactosidase from the cotransfected pCMV-β plasmid and represent average values from duplicate samples. The error bars represent standard deviations.
KUN and NY99 infection and the stable replication of their replicon RNAs prevent the translocation of STAT1 and STAT2 from the cytoplasm to the nucleus in response to IFN treatment.
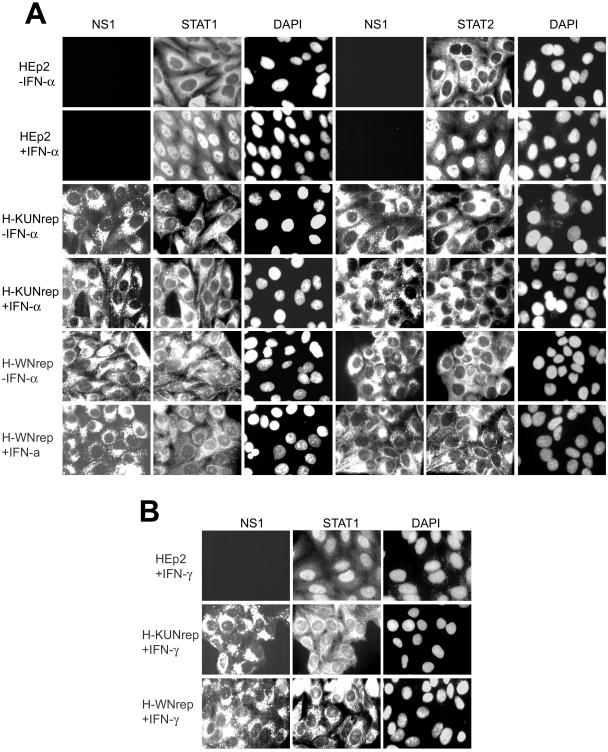
It was recently shown that infection with DEN virus inhibited the activation of STAT1 and its translocation to the nucleus in response to IFN-β and IFN-γ treatment (26). In our experiments, we examined the activation and translocation into the nucleus of STAT1 and STAT2 in HEp2 cells stably transfected with KUN (H-KUNrep) or NY99 (H-WNrep) replicon RNAs. H-WNrep cells stably transfected with NY99 replicon RNAs were generated by propagating HEp2 cells transfected with NY99 replicon (NeoRep) RNA expressing the neomycin gene (29) RNA in medium containing 0.5 mg of G418 (Sigma) per ml. Immunofluorescence analysis of H-KUNrep and H-WNrep cells with anti-STAT1 and anti-STAT2 antibodies after 30 min of treatment with 1,000 U of IFN-α per ml clearly showed no STAT1 or STAT2 in the nucleus (Fig. 3A). In contrast, treatment of normal HEp2 cells with IFN-α resulted in the translocation of STAT1 and STAT2 from the cytoplasm to the nucleus (Fig. 3A). Similarly, the translocation of STAT1 into the nuclei of KUN and NY99 replicon-expressing cells was blocked in response to treatment with 50 ng of IFN-γ per ml (Fig. 3B).
FIG. 3.
The nuclear translocation of STAT1 and STAT2 in response to IFN treatment is blocked in HEp2 cells stably transfected with KUN (KUNrep) and WN (WNrep) replicon RNAs. (A) Normal HEp2 cells and HEp2 cells stably transfected with KUN (23) or NY99 (29) replicon RNAs were treated with 1,000 U of IFN-αA (I-4276; Sigma) per ml for 30 min at 37°C. Cells were fixed in 4% formaldehyde-phosphate-buffered saline for 10 min at room temperature, permeabilized with ice-cold acetone-methanol (1:1) for 30 min at −20°C, and stained sequentially with KUN NS1 antibodies (9) and with STAT1 (SC-345; Santa Cruz Biotechnology, Santa Cruz, Calif.) or STAT2 (SC-476; Santa Cruz Biotechnology) antibodies at concentrations of 1 μg/ml essentially as described by the manufacturer. (B) Block in nuclear translocation of STAT1 in KUN or WN replicon cells in response to treatment with 50 ng of IFN-γ/ml.
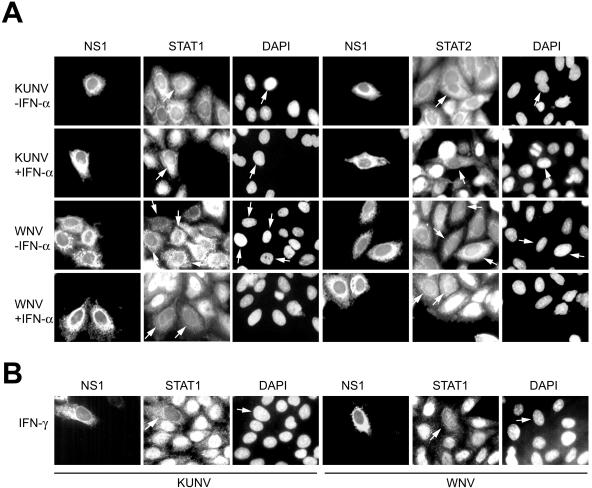
After obtaining the results with replicon-expressing cells, we performed the same experiment on HEp2 cells infected with KUN or NY99. KUN (MRM61C strain [32]) and NY99 (NY99-4132 strain, obtained from Roy Hall, University of Queensland, Brisbane, Australia) stocks were prepared and titrated on Vero cells. Similar to what occurred with the replicon cells, infections of HEp2 cells with both viruses blocked the translocation into the nucleus of both STAT1 and STAT2 in response to treatment with IFN-α (Fig. 4A) and of STAT1 in response to treatment with IFN-γ (Fig. 4B). Unlike with replicon-expressing cells, all of which produced and expressed replicon RNA, the low multiplicity of infection (MOI) of virus infections (MOI = 0.01) resulted in the expression of viral RNAs in only a limited number of cells (Fig. 4). The mixed population of infected and uninfected cells allowed clear visualization of STAT1 and STAT2 staining in the cytoplasm of infected cells and in the nuclei of neighboring uninfected cells (Fig. 4). The intensity of anti-STAT1 fluorescence in virus-infected cells also appeared to be noticeably lower than that in noninfected cells (Fig. 4). The inability of STAT1 and STAT2 to translocate to the nucleus in response to IFN treatment in virus-infected or replicon-expressing cells as detected by immunofluorescence analysis presented the first indication that activation (i.e., phosphorylation) of these proteins is inhibited by KUN and NY99 or replicon RNA replication. The lower viral intensity of STAT1 staining in KUN- and NY99-infected cells suggested that these viruses may down regulate STAT1 expression.
FIG. 4.
The nuclear translocation of STAT1 and STAT2 in response to IFN treatment is blocked in HEp2 cells infected with KUN and NY99. HEp2 cells on coverslips in 24-well plates were infected with KUN and NY99 at an MOI of 0.01. KUN (MRM61C strain) (32) and NY99 (NY99-4132 strain, provided by Roy Hall, University of Queensland) were grown and titrated on Vero cells. Seventy-two hours after infection, cells were treated for 30 min with 1,000 IU of IFN-αA (I-4276; Sigma) per ml (A) or 50 ng of IFN-γ (I-1520; Sigma) per ml (B), fixed, permeabilized with acetone-methanol, and stained sequentially with KUN NS1, STAT1, and STAT2 antibodies as described for Fig. 3. Arrows show cells positively infected with either KUN or NY99.
Stably replicating KUN and NY99 replicon RNAs inhibit STAT1 and STAT2 phosphorylation in response to IFN treatment and down regulate STAT1 expression.
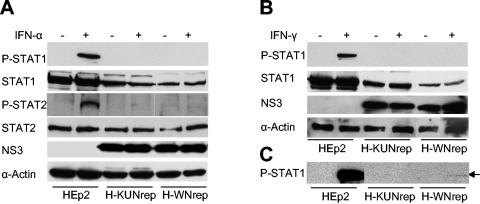
In order to confirm the immunofluorescence results and to establish whether the phosphorylation of both STAT2 and STAT1 was blocked by KUN and NY99 RNA replication, we performed a Western blot analysis of KUN and NY99 replicon-expressing cells using antibodies to phosphorylated (activated) forms of STAT1 (monoclonal mouse antibodies) and STAT2 (polyclonal rabbit serum). The results showed that no phosphorylated STAT1 was present in KUN or NY99 replicon-expressing cells in response to IFN-α treatment (Fig. 5A). Longer exposure of the membrane to X-ray film did not reveal any phosphorylated STAT1 in any of the replicon-expressing cells treated with IFN-α (results not shown). A very faint band of a size similar to that of phosphorylated STAT2 was visible in KUN replicon- and NY99 replicon-expressing cells either treated or not treated with IFN-α, suggesting some nonspecific binding of STAT2 polyclonal antibodies (Fig. 5A). It is therefore reasonable to conclude that STAT2 phosphorylation was blocked in KUN replicon- and NY99 replicon-expressing cells in response to IFN-α treatment. No detectable phosphorylated STAT1 was found in KUN or NY99 replicon-expressing cells in response to IFN-γ treatment when the membrane was exposed to X-ray film for a short (1-min) period (Fig. 5B). However, a very faint band of phosphorylated STAT1 was detected in NY99 replicon-expressing, but not in KUN replicon-expressing, cells and only in response to IFN-γ treatment when the membrane was exposed to X-ray film for a longer (10-min) period (Fig. 5C). The results suggest that the IFN-γ-induced phosphorylation of STAT1 was not completely blocked in cells stably expressing NY99 replicon RNA.
FIG. 5.
Western blot analysis of STAT1 and STAT2 expression and phosphorylation in HEp2 cells stably transfected with KUN and NY99 replicon RNAs. H-KUNrep and H-WNrep cells were treated with IFN-αA (I-4276; Sigma) (A) and IFN-γ (I-1520; Sigma) (B) as described for Fig. 4, and cell lysates were used for the detection of STAT1 and STAT2 expression and phosphorylation by Western blot analysis with antibodies recognizing nonphosphorylated and phosphorylated (P) forms of STAT1 and STAT2, respectively (anti-P-STAT1 antibody 550428; Pharmingen, San Diego, Calif.; anti-P-STAT2 antibody SC-21689 and the anti-STAT1 and anti-STAT2 antibodies described in the legend to Fig. 3; Santa Cruz Biotechnology). Controls included the detection of replicon RNA expression by cross-reacting KUN anti-NS3 antibodies and of host cell protein expression by anti-α-actin antibodies. The detection of reacted proteins on the blotted membrane was performed with an ECL Plus chemiluminescence kit (Amersham). In panels A and B, the membrane was exposed to X-ray film for 1 to 2 min, and in panel C, the membrane was exposed for 10 min.
In view of the demonstrated inhibition of STAT1 and STAT2 phosphorylation (Fig. 3 to 5) and the inhibition of IFN signaling to ISRE and GAS promoter elements (Fig. 2), the reason for the observed 10 to 20% decrease in β-galactosidase expression from stably transfected replicon RNA after the addition of IFNs (Fig. 1) is not clear. It is possible that a small amount of STAT2 and/or STAT1 was phosphorylated and provided sufficient signaling to induce the transcription of small amounts of antiviral genes whose products inhibited KUN RNA replication and translation. Alternatively, other STAT1/STAT2-independent IFN signaling pathways (16) may not be inhibited by KUN RNA replication, and thus IFN treatment may have resulted in a small reduction in KUN RNA replication and expression.
Western blot analysis of cell lysates from KUN and NY99 replicon-expressing HEp2 cells with antibodies to nonphosphorylated STAT1 and STAT2 proteins showed approximately two- to threefold less STAT1, but not STAT2, protein in replicon-expressing cells than in normal HEp2 cells (Fig. 5). Interestingly, slightly less STAT1 was detected in cells stably transfected with NY99 replicon RNA than in those stably transfected with a KUN replicon RNA. The significance if any of this down regulation of STAT1 expression in virus pathogenesis is not clear and requires further investigation. Since STAT1 expression itself is inducible by IFN, it is possible that the inhibition of signaling to IFN produced in response to replicon RNA replication results in the absence of the autocrine loop and thus reduces the level of STAT1.
In the IFN-α/β signaling pathway, STAT2 is phosphorylated by Tyk-2 after it binds to the Tyk-2-phosphorylated IFN-α/β receptor, which then leads to phosphorylation of STAT1 by JAK1 (16). In the IFN-γ signaling pathway, STAT1 is phosphorylated by JAK1 after binding to the JAK2-phosphorylated IFN-γ receptor (16). A recent study published after the submission of this paper has shown the inhibition of STAT1 and STAT2 phosphorylation by JE virus in response to IFN-α treatment and identified the phosphorylation of Tyk-2 as the primary step targeted by the virus for inhibition of IFN-α/β signaling (21). However, which step in IFN-γ signaling (where Tyk-2 is not involved) is inhibited by flaviviruses is still not clear. The abilities of KUN and NY99 replicon RNAs to block STAT1 phosphorylation in response to both IFN-α/β and IFN-γ and to block STAT2 phosphorylation in response to IFN-α/β suggest that there may be different components in the IFN-α/β and IFN-γ signaling pathways prior to the phosphorylation of STATs that are likely to be affected by the products of KUN and NY99 replicon RNA replication.
Inhibition of IFN-α/β signaling by the individually expressed KUN nonstructural proteins.
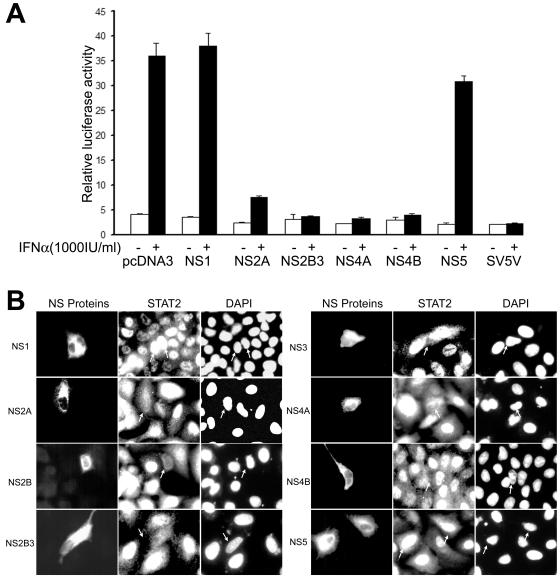
Recent studies with DEN virus showed that the individually expressed nonstructural protein NS4B and, to a lesser extent, NS4A and NS2A enhanced the replication of IFN-sensitive virus and down regulated the expression of IFN-β-stimulated genes (26). In addition, expression of DEN virus NS4B alone was shown to inhibit the phosphorylation of STAT1 and its translocation to the nucleus in response to IFN-β and IFN-γ treatments (26). We were interested in determining the effect of expression of KUN nonstructural proteins on ISRE-driven transcription and on STAT2 activation in response to IFN-α treatment (Fig. 6). For this purpose we constructed a set of plasmids expressing KUN nonstructural genes individually or as cassettes in the mammalian expression vector pcDNA3. For easier detection, some constructs incorporated the C-terminal C-myc tag (NS2A and NS4B in Fig. 6A and NS2A, NS2B, NS3, NS2B-3, NS4A, and NS4B in Fig. 6B) or the green fluorescence protein (GFP) fused either at the C terminus (NS2B-3 in Fig. 6A) or at the N terminus (NS4A in Fig. 6A) of the expressed gene product. In the first experiment, plasmids expressing KUN nonstructural proteins were cotransfected in Vero cells with the IFN-α/β-responsive ISRE-luciferase reporter plasmid and treated with IFN-α. Substantial inhibition of IFN-α-induced ISRE-driven luciferase expression was observed in cells expressing the NS2A, NS2B-3, NS4A, and NS4B genes but not in cells expressing the NS1 and NS5 genes (Fig. 6A). Noticeably, transfection of pcDNA3 or pcDNA3-GFP did not have any effect on IFN-α-induced ISRE-driven transcription (Fig. 6A for pcDNA3 and results not shown for pcDNA3-GFP). Interestingly, in contrast to the results with DEN virus showing no inhibition of IFN-β-induced ISRE-driven transcription by the NS2B and NS3 proteins (26), expression of the KUN NS2B-3 protein cassette strongly inhibited IFN-α-induced ISRE-driven transcription.
FIG. 6.
Inhibition of IFN-α signaling by KUN nonstructural proteins. (A) Inhibition of ISRE-driven transcription by KUN nonstructural proteins in response to IFN-α treatment. Vero cells were cotransfected with pcDNA plasmids expressing the indicated KUN nonstructural proteins, the ISRE reporter plasmid p(9-27)4thΔ(−39)Lucter (19), and the pCMV-β plasmid. The controls included pcDNA3 vector plasmid (for positive IFN stimulation) and a plasmid expressing a known inhibitor of IFN signaling, V protein of SV5 (4). Thirty-two hours after transfection, cells were treated or not treated with 1,000 U of IFN-αA (I-4276; Sigma) per ml for 14 h and assayed for luciferase and β-galactosidase expression. Luciferase activity, expressed in relative light units, was normalized to β-galactosidase activity. The error bars represent standard deviations. (B) The nuclear translocation of STAT2 in response to IFN-α treatment is blocked by KUN nonstructural (NS) proteins. A549 cells on coverslips in 24-well plates were transfected with pcDNA plasmids expressing the indicated KUN nonstructural proteins. Forty-eight hours after transfection, cells were treated with 800 IU of IFN-α2A (Roferon-A; Roche) per ml for 30 min at 37°C. Cells were fixed and stained sequentially with STAT2 antibody and with either KUN anti-NS1 and anti-NS5 antibodies or anti-C-myc antibodies (15).
Further confirmation of inhibition of IFN-α signaling by the KUN nonstructural proteins was sought by detection of the activation of STAT2 and its translocation to the nucleus in A549 cells after IFN-α treatment (Fig. 6B). Immunofluorescence analysis with anti-STAT2 antibodies showed no STAT2 in the nucleus after IFN-α treatment in the majority of A549 cells expressing individual KUN nonstructural proteins, NS2A, NS2B, NS3, NS4A, and NS4B, as well as the protein cassette NS2B-3 (Fig. 6B). In contrast, STAT2 clearly translocated to the nucleus in neighboring cells not expressing corresponding KUN nonstructural proteins or in cells expressing NS1 or NS5 protein (Fig. 6B). Thus, reporter assays and immunofluorescence analysis demonstrated that KUN nonstructural proteins NS2A, NS2B, NS3, NS4A, and NS4B, but not NS1 and NS5, were involved in the inhibition of IFN-α/β signaling. Given the very high homology of these nonstructural proteins between KUN and NY99 (four amino acid changes in NS2A, two in NS2B, five in NS3, four in NS4A, and three in NS4B) (22, 30), it is likely that the same proteins will be involved in the inhibition of IFN signaling by NY99. However, further experimental data are required to confirm this prediction.
The relative contribution of each of the identified nonstructural proteins in the inhibition of IFN signaling exhibited by KUN and other WN viruses is not clear and requires further investigation. It is likely, however, that the combined actions of all identified nonstructural proteins may be required for the most efficient inhibition of IFN signaling during virus infection or replicon RNA replication. Noticeably, a more-efficient inhibition of IFN-α/β signaling was observed when the DEN virus proteins NS2A, NS4A, and NS4B were expressed together than when they were expressed separately (26). The reasons why NS2B and NS3 are involved in the inhibition of IFN signaling by WN virus(es) but not by DEN virus are not clear. It is possible that flaviviruses from different subgroups have adapted different mechanisms for evading host innate immunity and that, as a result, they have different means of pathogenesis. For example, a recent study showed that JE virus (the prototype virus for the subgroup which also includes KUN and NY99) was much more efficient in inhibiting the antiviral activity of IFN-α than DEN virus (21). It is therefore possible that the addition of two more IFN antagonists, NS2B and NS3, may provide more-efficient protection against the antiviral activity of IFN for the members of the JE virus subgroup than that for the viruses from the DEN virus subgroup. Clearly, further studies are warranted to provide answers to these and many other questions in the rapidly expanding field of modulation of innate immune responses by flaviviruses.
In conclusion, we have extended the results of recent studies with DEN virus (26) and JE virus (21) by showing that two WN viruses, KUN and NY99, and their replicon RNAs block the activation of STAT1 and STAT2 and their translocation to the nucleus, and we have added the NS2B and NS3 proteins to the list of flavivirus IFN antagonists. Our results also showed that, apart from a slightly more efficient down regulation of STAT1 expression by NY99 replicon RNA, no other apparent differences in inhibition of IFN signaling by KUN and NY99 were observed. This finding implies that other factors not related to the inhibition of IFN signaling may be involved in determining the difference in the levels of pathogenicity of these two viral infections.
Acknowledgments
We are grateful to Steven Goodbourn for providing IFN-responsive reporter plasmids and for helpful discussions during preparation of the manuscript, to Roy Hall for providing NY99 stock, to Jason Mackenzie for providing plasmids expressing KUN NS2B-NS3-GFP and GFP-NS4A gene cassettes, and to Andreas Suhrbier for helpful suggestions during preparation of the manuscript.
This work was supported by the grants to A.A.K. from the National Health and Medical Research Council of Australia.
Footnotes
This is publication 208 from the Clinical Medical Virology Centre and the Sir Albert Sakzewski Virus Research Centre.
REFERENCES
- 1.Bossert, B., S. Marozin, and K. K. Conzelmann. 2003. Nonstructural proteins NS1 and NS2 of bovine respiratory syncytial virus block activation of interferon regulatory factor 3. J. Virol. 77:8661-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diamond, M. S. 2003. Evasion of innate and adaptive immunity by flaviviruses. Immunol. Cell Biol. 81:196-206. [DOI] [PubMed] [Google Scholar]
- 3.Diamond, M. S., T. G. Roberts, D. Edgil, B. Lu, J. Ernst, and E. Harris. 2000. Modulation of Dengue virus infection in human cells by alpha, beta, and gamma interferons. J. Virol. 74:4957-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 6.Fredericksen, B. L., M. Smith, M. G. Katze, P. Y. Shi, and M. Gale, Jr. 2004. The host response to West Nile virus infection limits viral spread through the activation of the interferon regulatory factor 3 pathway. J. Virol. 78:7737-7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 8.Guo, J. T., Q. Zhu, and C. Seeger. 2003. Cytopathic and noncytopathic interferon responses in cells expressing hepatitis C virus subgenomic replicons. J. Virol. 77:10769-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall, R. A., A. K. Broom, A. C. Hartnett, M. J. Howard, and J. S. Mackenzie. 1995. Immunodominant epitopes on the NS1 protein of MVE and KUN viruses serve as targets for a blocking ELISA to detect virus-specific antibodies in sentinel animal serum. J. Virol. Methods 51:201-210. [DOI] [PubMed] [Google Scholar]
- 10.Hall, R. A., A. K. Broom, D. W. Smith, and J. S. Mackenzie. 2002. The ecology and epidemiology of Kunjin virus. Curr. Top. Microbiol. Immunol. 267:253-269. [DOI] [PubMed] [Google Scholar]
- 11.Harvey, T. J., W. J. Liu, X. J. Wang, R. Linedale, M. Jacobs, A. Davidson, T. T. Le, I. Anraku, A. Suhrbier, P. Y. Shi, and A. A. Khromykh. 2004. Tetracycline-inducible packaging cell line for production of flavivirus replicon particles. J. Virol. 78:531-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He, B., R. G. Paterson, N. Stock, J. E. Durbin, R. K. Durbin, S. Goodbourn, R. E. Randall, and R. A. Lamb. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 303:15-32. [DOI] [PubMed] [Google Scholar]
- 13.Heim, M. H., D. Moradpour, and H. E. Blum. 1999. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J. Virol. 73:8469-8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heinz, F. X., M. S. Collet, R. H. Purcell, E. A. Gould, C. R. Howard, M. Houghton, R. J. M. Moormann, C. M. Rice, and H.-J. Thiel. 2000. Family Flaviviridae, p. 859-879. In M. H. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, et al. (ed.), Virus taxonomy, classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 15.Hilpert, K., G. Hansen, H. Wessner, G. Kuttner, K. Welfle, M. Seifert, and W. Hohne. 2001. Anti-c-myc antibody 9E10: epitope key positions and variability characterized using peptide spot synthesis on cellulose. Protein Eng. 14:803-806. [DOI] [PubMed] [Google Scholar]
- 16.Kalvakolanu, D. V. 2003. Alternate interferon signaling pathways. Pharmacol. Ther. 100:1-29. [DOI] [PubMed] [Google Scholar]
- 17.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-687. [DOI] [PubMed] [Google Scholar]
- 18.King, P., and S. Goodbourn. 1994. The beta-interferon promoter responds to priming through multiple independent regulatory elements. J. Biol. Chem. 269:30609-30615. [PubMed] [Google Scholar]
- 19.King, P., and S. Goodbourn. 1998. STAT1 is inactivated by a caspase. J. Biol. Chem. 273:8699-8704. [DOI] [PubMed] [Google Scholar]
- 20.Lanciotti, R. S., J. T. Roehrig, V. Deubel, J. Smith, M. Parker, K. Steele, B. Crise, K. E. Volpe, M. B. Crabtree, J. H. Scherret, R. A. Hall, J. S. MacKenzie, C. B. Cropp, B. Panigrahy, E. Ostlund, B. Schmitt, M. Malkinson, C. Banet, J. Weissman, N. Komar, H. M. Savage, W. Stone, T. McNamara, and D. J. Gubler. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286:2333-2337. [DOI] [PubMed] [Google Scholar]
- 21.Lin, R. J., C. L. Liao, E. Lin, and Y. L. Lin. 2004. Blocking of the alpha interferon-induced Jak-Stat signaling pathway by Japanese encephalitis virus infection. J. Virol. 78:9285-9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, W. J., H. B. Chen, and A. A. Khromykh. 2003. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. J. Virol. 77:7804-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, W. J., P. L. Sedlak, N. Kondratieva, and A. A. Khromykh. 2002. Complementation analysis of the flavivirus Kunjin NS3 and NS5 proteins defines the minimal regions essential for formation of a replication complex and shows a requirement of NS3 in cis for virus assembly. J. Virol. 76:10766-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackenzie, J. M., A. A. Khromykh, and E. G. Westaway. 2001. Stable expression of noncytopathic Kunjin replicons simulates both ultrastructural and biochemical characteristics observed during replication of Kunjin virus. Virology 279:161-172. [DOI] [PubMed] [Google Scholar]
- 25.Mahalingam, S., J. Meanger, P. S. Foster, and B. A. Lidbury. 2002. The viral manipulation of the host cellular and immune environments to enhance propagation and survival: a focus on RNA viruses. J. Leukoc. Biol. 72:429-439. [PubMed] [Google Scholar]
- 26.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 100:14333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poole, E., B. He, R. A. Lamb, R. E. Randall, and S. Goodbourn. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 303:33-46. [DOI] [PubMed] [Google Scholar]
- 28.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 29.Shi, P. Y., M. Tilgner, and M. K. Lo. 2002. Construction and characterization of subgenomic replicons of New York strain of West Nile virus. Virology 296:219-233. [DOI] [PubMed] [Google Scholar]
- 30.Shi, P. Y., M. Tilgner, M. K. Lo, K. A. Kent, and K. A. Bernard. 2002. Infectious cDNA clone of the epidemic West Nile virus from New York City. J. Virol. 76:5847-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varnavski, A. N., P. R. Young, and A. A. Khromykh. 2000. Stable high-level expression of heterologous genes in vitro and in vivo by noncytopathic DNA-based Kunjin virus replicon vectors. J. Virol. 74:4394-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westaway, E. G. 1968. Antibody responses in rabbits to the group B arbovirus Kumjin: serologic activity of the fractionated immunoglobulins in homologous and heterologous reactions. J. Immunol. 100:569-580. [PubMed] [Google Scholar]