Abstract
EGF-like domain 6 (EGFL6), a member of the epidermal growth factor (EGF) repeat protein superfamily, is a secreted protein that promotes endothelial cell migration and angiogenesis. The current study investigated the association between the clinicopathological characteristics and plasma level of EGFL6 in patients with oral squamous cell carcinoma (OSCC). We measured the plasma EGFL6 levels of 392 OSCC patients by using a commercial enzyme-linked immunosorbent assay. We also analyzed EGFL6 mRNA levels of 328 OSCC patients from The Cancer Genome Atlas (TCGA) dataset. The results showed that plasma EGFL6 levels were significantly higher in patients with OSCC than in healthy controls (p < 0.001). Similar results were observed for the TCGA bioinformatics database. Moreover, plasma EGFL6 levels were significantly higher in the patients with advanced T status (p = 0.002), distant metastasis (p = 0.001), and higher TNM stage (p=0.033). In conclusion, our results suggest that plasma level of EGFL6 may be useful to assess disease progression, and especially advanced T status and higher TNM stage in patients with OSCC.
Keywords: EGFL6, oral squamous cell carcinoma, biomarker.
Introduction
Both genetic and environmental factors contribute to the development of oral cancer, and major risk factors include the use of tobacco products, betel nut chewing, and alcohol consumption 1-3. Approximately 90% of oral cancers are squamous cell carcinomas, and oral squamous cell carcinoma (OSCC) is the tenth most common cancer worldwide and the fourth most common cancer among men in Taiwan 4-6. It is also the most common malignancy of the head and neck region, accounting for 2-4% of all cancer cases in Western countries and more than 10% in some areas of Asia 7, 8. The 5-year relative survival rate of OSCC is unfavorable even with aggressive interventions, because invasion of the neighboring tissues and metastasis to the neck lymph nodes are common 9. Identifying new biomarkers that can predict the risk of OSCC progression, especially local invasion and metastasis, are needed to improve the control of this deadly cancer.
Tumor invasion and metastasis are related to a series of complex processes, including cell adhesion, migration, invasion, angiogenesis, and anchorage-independent growth 10-15. In addition, degradation of the extracellular matrix (ECM) giving cancer cells access to blood vessels and lymphatics is also a key process. The epidermal growth factor (EGF) repeat superfamily features a series of conserved cysteines and glycines positioned in a domain of 30 to 40 residues 16. EGF-like proteins are characterized by their multiple EGF repeats 17. EGF-like repeat family members are predominantly secreted as cell surface molecules, and are often involved in the regulation of the cell cycle, proliferation, and developmental processes 18, 19. The binding of EGF-like proteins to their receptors triggers a wide range of biological functions, including proliferation, differentiation, apoptosis, adhesion, and migration 17. EGF motif-containing molecules have been previously linked to the progression of various cancers 20, 21, and the expression of EGF-like domain 6 (EGFL6) in tumors suggests that it may also be linked to cancer 22-25.
The EGFL6 protein is a member of the EGF repeat superfamily which is secreted and then promotes endothelial cell migration and angiogenesis 26. EGFL6 has been shown to be expressed in fetal tissues and pancreatic, lung, ovarian and breast tumors 20, 27-29. In microarray-based detection and expression analysis of ECM proteins in drug-resistant ovarian cancer cell lines, the over-expression of EGFL6 has been observed in the WITR cell line 28. Since EGFL6 is expressed specifically in certain tumors but not in normal adult tissues, the EGFL6 gene product represents a potential marker of malignancy 20. However, the potential expression and role of EGFL6 in patients with OSCC have yet to be elucidated. In this study, we investigated the association between the clinicopathological characteristics and plasma level of EGFL6 in patients with OSCC.
Materials and Methods
Subjects and specimen collection
We recruited 392 patients with OSCC (mean age 55.33±10.93 years) at Chung Shan Medical University Hospital in Taichung and Changhua Christian Hospital in Changhua, Taiwan between 2008 and 2015. OSCC were clinically staged at the time of diagnosis according to the TNM staging system of the American Joint Committee on Cancer (AJCC) Staging Manual, seventh edition. Medical information of the OSCC patients including TNM clinical staging, primary tumor size, lymph node involvement, and histological grade was obtained from their medical records. Whole blood samples were collected from the patients and placed in tubes containing ethylene diamine tetraacetic acid. After immediate centrifugation at 3000 rpm, the supernatants were stored at -80°C. Before conducting this study, approval from the Institutional Review Board of Chung Shan Medical University Hospital (CSMUH No: CS13214-1; CSMUH No: CS15150) and informed written consent to participate in the study were obtained from each patient.
Quantitative analysis of plasma EGFL6 level
The plasma EGFL6 concentration was determined quantitatively using an enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's instructions (Human EGFL6 Immunoassay E01E0401; BlueGene Biotech, Shanghai, China). One hundred microliters of plasma sample (100-fold dilution), standard control sample and internal quality control were placed into microtiter plates coated with a monoclonal antibody against EGFL6 and incubated for 2 hours at room temperature on a horizontal orbital shaker at 200 rpm. The absorbance was measured at 450 nm by using a microtest plate spectrophotometer (BioTek Instruments, Vemont, USA). EGFL6 levels were quantified with a calibration curve using human EGFL6 as the standard.
Statistical analysis
The demographic data are presented as number (%) and mean ± standard deviation (SD). Significances of differences between means were calculated using the Student's t-test. In addition, gender, smoking status, alcohol consumption status and betel nuts chewing status were analyzed using the x2 test. A p value less than 0.05 was considered to be statistically significant. All analyses were performed using SPSS version 16.0 statistical software (SPSS Inc., Chicago, IL, USA).
Results
Patient characteristics
Three hundred and ninety-two patients with OSCC were included in the analysis. Table 1 presents the demographic data, and shows that 88.3% of the patients were smokers, 51.5% consumed alcohol, and 78.3% chewed betel nuts. The TNM status and types of tumor cell differentiation of the patients are also shown in Table 1.
Table 1.
Demographic characteristics and clinical features of OSCC patients.
| Variables | OSCC (n = 392) |
|---|---|
| Age (years) | 55.33 ± 10.93 |
| Gender: male (%) | 385 (98.2%) |
| Smoking status | |
| No | 46 (11.7%) |
| Yes | 346 (88.3%) |
| Drinking status | |
| No | 190 (48.5%) |
| Yes | 202 (51.5%) |
| Betel nuts chewing | |
| No | 85 (21.7%) |
| Yes | 307 (78.3%) |
| EGFL6 (pg/mL) | 304.48 ± 194.55 |
| Stage | |
| I | 103 (26.3%) |
| II | 67 (17.1%) |
| III | 42 (10.7%) |
| IV | 180 (45.9%) |
| Tumor T status | |
| T1 | 124 (31.6%) |
| T2 | 109 (27.8%) |
| T3 | 33 (8.4%) |
| T4 | 126 (32.1%) |
| Lymph node status | |
| N0 | 253 (64.5%) |
| N1 | 46 (11.7%) |
| N2 | 90 (23.0%) |
| N3 | 3 (0.8%) |
| Metastasis | |
| M0 | 390 (99.5%) |
| M1 | 2 (0.5%) |
| Cell differentiation | |
| Well differentiated | 57 (14.5%) |
| Moderately or poorly differentiated | 335 (85.2%) |
Correlation between plasma EGFL6 levels and clinicopathological characteristics of the patients
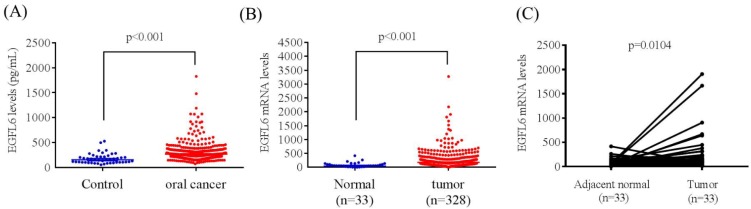
The mean plasma EGFL6 level was significantly higher in patients with OSCC than in controls (304.48 ± 194.55 pg/mL vs. 178.69 ± 102.96 pg/mL; p < 0.001) (Figure 1A). To verify our findings, TCGA OSCC database was used in this study. Samples filtered involved only six oral cancer subtypes (alveolar ridge, base of tongue, buccal mucosa, floor of mouth, oral cavity, oral tongue; filtered oral cancer dataset size: n = 328). The EGFL6 mRNA levels of OSCC and normal tissues were evaluated. Figure 1B shows the EGFL6 mRNA levels were also significantly higher in patients with OSCC tissues than in normal tissues (Figure 1B). Moreover, the EGFL6 expression was also significantly increased in cancer tissue compared with that in the normal parts in OSCC of the TCGA database (Figure 1C).
Figure 1.
ELISA-determined plasma EGFL6 level of OSCC patients. (A) EGFL6 levels were compared according to normal control and OSCC patients. (B) EGFL6 mRNA expressions were compared according to normal tissue and OSCC patients' tissue from TCGA database. (C). Relative expression of EGFL6 in 33 pairs of oral squamous cell carcinoma tumor tissues and their corresponding adjacent non-cancerous tissues.
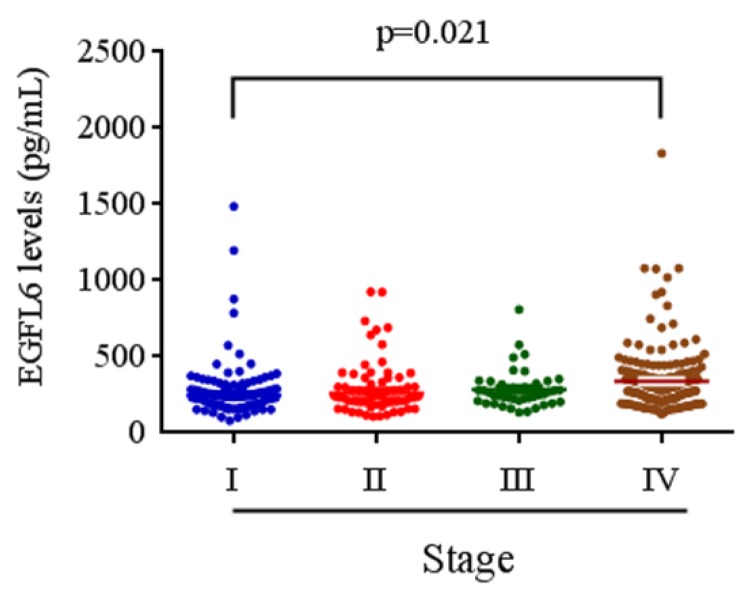
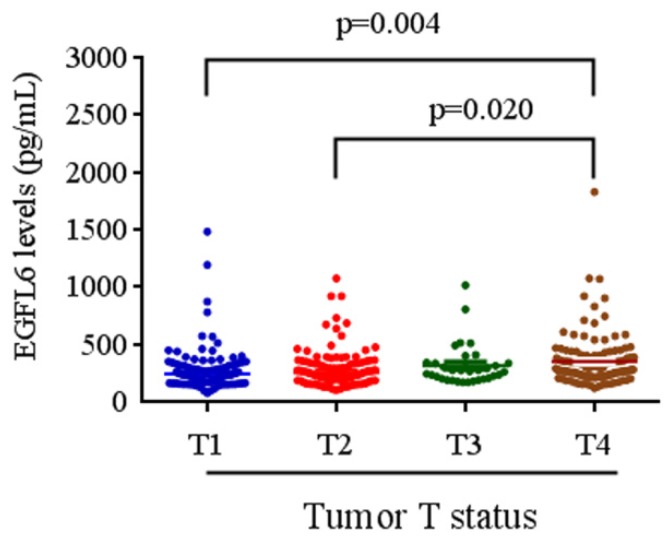
The relationships between plasma EGFL6 levels and various clinicopathological parameters of the patients are summarized in Table 2. Plasma levels of EGFL6 protein were not correlated with age, gender, smoking, drinking, betel nuts chewing, lymph node metastasis or cell differentiation. However, they were significantly higher in the patients with higher TNM stage (stage III + stage IV; p = 0.033), advanced T status (T3+T4; p = 0.002) and distant metastasis (M1; p = 0.001). Detailed comparisons of plasma EGFL6 levels between the patients with different disease severity are illustrated in Figure 2. With regards to TNM stage, the levels of EGFL6 were significantly higher in the patients with stage IV (332.83 pg/mL) compared to those with an early stage (stage I: 274.22 pg/mL) (Figure 2). The levels of EGFL6 were significantly higher in the patients with advanced tumor T status (T4: 347.73 pg/mL) compared to those with early T status (T1: 273.67 pg/mL and T2: 286.64 pg/mL; p=0.004 and p=0.020) (Figure 3).
Table 2.
Correlation between plasma levels of EGFL6 and clinicopathological parameters in 392 OSCC patients.
| No. of case (%) | EGFL6 level | p value | |
|---|---|---|---|
| Variables | n = 392 | Mean ± S.D. (pg/mL) | |
| Age (years) | |||
| <55 | 182 (46.4%) | 303.25 ± 192.82 | 0.907 |
| ≥55 | 210 (53.6%) | 305.55 ± 196.49 | |
| Gender | |||
| male | 385 (98.2%) | 304.81 ± 195.85 | 0.802 |
| female | 7 (1.8%) | 286.17 ± 105.22 | |
| Smoking status | |||
| No | 46 (11.7%) | 283.02 ± 135.87 | 0.427 |
| Yes | 346 (88.3%) | 307.33 ± 201.04 | |
| Drinking status | |||
| No | 190 (48.5%) | 314.45 ± 191.09 | 0.326 |
| Yes | 202 (51.5%) | 295.10 ± 197.76 | |
| Betel nuts chewing | |||
| No | 85 (21.7%) | 275.86 ± 140.22 | 0.126 |
| Yes | 307 (78.3%) | 312.40 ± 206.58 | |
| Stage | |||
| I+II | 170 (43.4%) | 280.63 ± 186.88 | 0.033* |
| III+IV | 222 (56.6%) | 322.74 ± 198.71 | |
| Tumor T status | |||
| T1+ T2 | 233 (59.4%) | 279.74 ± 175.98 | 0.002* |
| T3+T4 | 159 (40.6%) | 340.73 ± 214.39 | |
| Lymph node status | |||
| N0 | 253 (64.5%) | 297.42 ± 185.82 | 0.333 |
| N1+N2+N3 | 139 (35.5%) | 317.34 ± 209.59 | |
| Metastasis | |||
| M0 | 390 (99.5%) | 302.19 ± 191.46 | 0.001* |
| M1 | 2 (0.5%) | 751.21 ± 371.22 | |
| Cell differentiation | |||
| Well differentiated | 57 (14.5%) | 338.69 ± 39.65 | 0.151 |
| Moderately or poorly differentiated | 335 (85.2%) | 298.66 ± 170.44 |
*p<0.05.
Figure 2.
EGFL6 levels were compared according to stage. The levels of EGFL6 were significantly higher in the patients with stage IV (332.83 pg/mL) compared to those with an early stage (stage I: 274.22 pg/mL).
Figure 3.
EGFL6 levels were compared according to tumor T status. The levels of EGFL6 were significantly higher in the patients with advanced tumor T status (T4: 347.73 pg/mL) compared to those with early T status (T1: 273.67 pg/mL and T2: 286.64 pg/mL; p=0.004 and p=0.020).
Discussion
The advantage of using plasma tumor markers is that plasma can easily be obtained before treatment and at any time during follow-up without the necessity to invasively acquire tissue. It is clinically invaluable to have biomarkers which can provide information regarding the likelihood of a high TNM stage. This is particularly important in the context of patients with OSCC because their outcomes are closely related to disease progression and prognosis. Reliable tumor markers for these patients would therefore assist in assessing the prognosis and prediction of tumor behavior and in planning adequate therapy.
In this study, we investigated the levels of EGFL6 in 392 OSCC patients, and found that elevated plasma levels of EGFL6 were correlated with advanced T status, distant metastasis and high TNM stage. EGFL6 protein levels were significantly higher in patients with advanced T status (T3+T4; p = 0.002), distant metastasis (M1; p = 0.001), and higher TNM stage (stage III + stage IV; p=0.033). Previous reports have shown that tumor invasion and metastasis are related to cell adhesion, migration, invasion, angiogenesis, and anchorage-independent growth 10, 30-34. In addition, the EGFL6 protein has been reported to induce cell migration and angiogenesis of endothelial cells 26, 35-38. These findings may explain our results, and suggest that EGFL6 may promote OSCC tumor invasion and metastasis by promoting cell migration and angiogenesis. Our results also suggest that EGFL6 may play an important role in the carcinogenesis of OSCC.
Several EGF-like superfamily members have been identified, including EGFL2, EGFL3, EGFL5, EGFL6, EGFL7, EGFL8, and EGFL9. EGFL2, EGFL5 and EGFL9 contain transmembrane domains, however EGFL3, EGFL6, EGFL7and EGFL8 lack transmembrane domains and are secreted as proteins 26. The EGFL6 gene maps to the human Xp22 chromosome and encodes a secreted protein containing multiple EGF repeat motifs, which is highly expressed in certain tumors and fetal tissues, suggesting a role as a growth factor 27, 39. Using RNA in situ hybridization, the expression of EGFL6 has been detected in several sites, including all of the dermatome derivatives including the dermis of the trunk, hair follicles, and mesenchyme of the cranio-facial region 40.
Previous studies have reported that the EGFL6 protein induces migration and angiogenesis of endothelial cells 35-38, but that endothelial cells themselves do not express EGFL6 26. Several signaling pathways during angiogenesis have been reported to be potentially activated, such as the integrin/FAK-mediated pathway, MAPK pathway, and the PIK3/Akt pathway 35, 41, 42. Chim et al 26 reported that extracellular signal-regulated kinase (ERK) is activated by the EGFL6 protein, and that inhibition of the ERK signaling pathway blocks EGFL6-induced ERK activation and endothelial cell migration. They further validated that EGFL6 promotes endothelial cell migration and angiogenesis via activation of the ERK pathway 26.
In addition to oral cancer, the overexpression of plasma EGFL6 has been observed in several tumors including brain, lung, ovarian, and breast tumors, but generally not in normal adult tissues 27-29. EGFL6 has also been proposed to be a new target for diagnostic and therapeutic interventions in patients with breast cancer, which shows promise for new areas of basic research in tumor biology 29. Combined with our results, the plasma level of the EGFL6 protein appears to be a likely candidate biomarker for various human cancers.
To the best of our knowledge, this is the first report to examine the association between plasma EGFL6 level and clinicopathological characteristics for patients with OSCC with regards to the possible application of this molecule as a tumor marker. We suggest that EGFL6 may play an important role in the carcinogenesis of OSCC, and that this may have an important implication in the treatment of patients with OSCC. The detection of the EGFL6 protein in the plasma may serve as tumor marker to predict the likelihood of OSCC in patients without the disease.
In summary, we found that a substantial increase in the plasma level of EGFL6 by ELISA is useful to assess disease progression, especially in patients with OSCC with an advanced T status and higher TNM stage. As a secreted protein, EGFL6 may not only play an important role in the carcinogenesis of OSCC, but also find clinical applications as a biomarker for disease diagnosis and in planning therapy for patients with OSCC.
Acknowledgments
This work was supported in part by grants from the Ministry of Science and Technology, Taiwan (MOST-105-2320-B-040-001-MY2). This study was also supported by a grant from Chung Shan Medical University Hospital, Taiwan (CSH-2013-C-033; CSH-2014-C-021).
References
- 1.Ko YC, Huang YL, Lee CH, Chen MJ, Lin LM, Tsai CC. Betel quid chewing, cigarette smoking and alcohol consumption related to oral cancer in Taiwan. J Oral Pathol Med. 1995;24:450–453. doi: 10.1111/j.1600-0714.1995.tb01132.x. [DOI] [PubMed] [Google Scholar]
- 2.Proia NK, Paszkiewicz GM, Nasca MA, Franke GE, Pauly JL. Smoking and smokeless tobacco-associated human buccal cell mutations and their association with oral cancer-a review. Cancer Epidemiol Biomarkers Prev. 2006;15:1061–1077. doi: 10.1158/1055-9965.EPI-05-0983. [DOI] [PubMed] [Google Scholar]
- 3.Chou YE, Hsieh MJ, Hsin CH, Chiang WL, Lai YC, Lee YH, Huang SC, Yang SF, Lin CW. CD44 gene polymorphisms and environmental factors on oral cancer susceptibility in Taiwan. PLoS One. 2014;9:e93692. doi: 10.1371/journal.pone.0093692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chuang CY, Sung WW, Wang L, Lin WL, Yeh KT, Su MC, Hsin CH, Lee SY, Wu BC, Cheng YW, Lee H. Differential impact of IL-10 expression on survival and relapse between HPV16-positive and -negative oral squamous cell carcinomas. PLoS One. 2012;7:e47541. doi: 10.1371/journal.pone.0047541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 6.Su SC, Lin CW, Liu YF, Fan WL, Chen MK, Yu CP, Yang WE, Su CW, Chuang CY, Li WH, Chung WH, Yang SF. Exome Sequencing of Oral Squamous Cell Carcinoma Reveals Molecular Subgroups and Novel Therapeutic Opportunities. Theranostics. 2017;7:1088–1099. doi: 10.7150/thno.18551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsin CH, Chen MK, Tang CH, Lin HP, Chou MY, Lin CW, Yang SF. High level of plasma matrix metalloproteinase-11 is associated with clinicopathological characteristics in patients with oral squamous cell carcinoma. PLoS One. 2014;9:e113129. doi: 10.1371/journal.pone.0113129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 9.Zini A, Czerninski R, Sgan-Cohen HD. Oral cancer over four decades: epidemiology, trends, histology, and survival by anatomical sites. J Oral Pathol Med. 2010;39:299–305. doi: 10.1111/j.1600-0714.2009.00845.x. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Chien MH, Lin CW, Cheng CW, Wen YC, Yang SF. Matrix metalloproteinase-2 as a target for head and neck cancer therapy. Expert Opin Ther Targets. 2013;17:203–216. doi: 10.1517/14728222.2013.740012. [DOI] [PubMed] [Google Scholar]
- 12.Su SC, Lin CW, Yang WE, Fan WL, Yang SF. The urokinase-type plasminogen activator (uPA) system as a biomarker and therapeutic target in human malignancies. Expert Opin Ther Targets. 2016;20:551–566. doi: 10.1517/14728222.2016.1113260. [DOI] [PubMed] [Google Scholar]
- 13.Yang JS, Lin CW, Su SC, Yang SF. Pharmacodynamic considerations in the use of matrix metalloproteinase inhibitors in cancer treatment. Expert Opin Drug Metab Toxicol. 2016;12:191–200. doi: 10.1517/17425255.2016.1131820. [DOI] [PubMed] [Google Scholar]
- 14.Ho HY, Lin CW, Chien MH, Reiter RJ, Su SC, Hsieh YH, Yang SF. Melatonin suppresses TPA-induced metastasis by downregulating matrix metalloproteinase-9 expression through JNK/SP-1 signaling in nasopharyngeal carcinoma. J Pineal Res. 2016;61:479–492. doi: 10.1111/jpi.12365. [DOI] [PubMed] [Google Scholar]
- 15.Su SC, Hsieh MJ, Yang WE, Chung WH, Reiter RJ, Yang SF. Cancer metastasis: Mechanisms of inhibition by melatonin. J Pineal Res; 2017. p. 62. doi: 10.1111/jpi.12370. [DOI] [PubMed] [Google Scholar]
- 16.Davis CG. The many faces of epidermal growth factor repeats. New Biol. 1990;2:410–419. [PubMed] [Google Scholar]
- 17.Singh AB, Harris RC. Autocrine, paracrine and juxtacrine signaling by EGFR ligands. Cell Signal. 2005;17:1183–1193. doi: 10.1016/j.cellsig.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 18.Carter TH, Kung HJ. Tissue-specific transformation by oncogenic mutants of epidermal growth factor receptor. Crit Rev Oncog. 1994;5:389–428. doi: 10.1615/critrevoncog.v5.i4.40. [DOI] [PubMed] [Google Scholar]
- 19.Rusch V, Mendelsohn J, Dmitrovsky E. The epidermal growth factor receptor and its ligands as therapeutic targets in human tumors. Cytokine Growth Factor Rev. 1996;7:133–141. doi: 10.1016/1359-6101(96)00016-0. [DOI] [PubMed] [Google Scholar]
- 20.Birk D, Gansauge F, Gansauge S, Formentini A, Lucht A, Beger HG. Serum and correspondent tissue measurements of epidermal growth factor (EGF) and epidermal growth factor receptor (EGF-R). Clinical relevance in pancreatic cancer and chronic pancreatitis. Int J Pancreatol. 1999;25:89–96. doi: 10.1385/IJGC:25:2:89. [DOI] [PubMed] [Google Scholar]
- 21.Panin VM, Papayannopoulos V, Wilson R, Irvine KD. Fringe modulates Notch-ligand interactions. Nature. 1997;387:908–912. doi: 10.1038/43191. [DOI] [PubMed] [Google Scholar]
- 22.Bai S, Ingram P, Chen YC, Deng N, Pearson A, Niknafs Y, O'Hayer P, Wang Y, Zhang ZY, Boscolo E, Bischoff J, Yoon E, Buckanovich RJ. EGFL6 Regulates the Asymmetric Division, Maintenance, and Metastasis of ALDH+ Ovarian Cancer Cells. Cancer Res. 2016;76:6396–6409. doi: 10.1158/0008-5472.CAN-16-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larimer BM, Deutscher SL. Identification of a Peptide from In vivo Bacteriophage Display with Homology to EGFL6: A Candidate Tumor Vasculature Ligand in Breast Cancer. J Mol Biomark Diagn; 2014. p. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Gong Y, Wang D, Xie Q, Zheng M, Zhou Y, Li Q, Yang Z, Tang H, Li Y, Hu R, Chen X, Mao Y. Analysis of gene expression profiling in meningioma: deregulated signaling pathways associated with meningioma and EGFL6 overexpression in benign meningioma tissue and serum. PLoS One. 2012;7:e52707. doi: 10.1371/journal.pone.0052707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buckanovich RJ, Sasaroli D, O'Brien-Jenkins A, Botbyl J, Hammond R, Katsaros D, Sandaltzopoulos R, Liotta LA, Gimotty PA, Coukos G. Tumor vascular proteins as biomarkers in ovarian cancer. J Clin Oncol. 2007;25:852–861. doi: 10.1200/JCO.2006.08.8583. [DOI] [PubMed] [Google Scholar]
- 26.Chim SM, Qin A, Tickner J, Pavlos N, Davey T, Wang H, Guo Y, Zheng MH, Xu J. EGFL6 promotes endothelial cell migration and angiogenesis through the activation of extracellular signal-regulated kinase. J Biol Chem. 2011;286:22035–22046. doi: 10.1074/jbc.M110.187633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeung G, Mulero JJ, Berntsen RP, Loeb DB, Drmanac R, Ford JE. Cloning of a novel epidermal growth factor repeat containing gene EGFL6: expressed in tumor and fetal tissues. Genomics. 1999;62:304–307. doi: 10.1006/geno.1999.6011. [DOI] [PubMed] [Google Scholar]
- 28.Januchowski R, Zawierucha P, Rucinski M, Zabel M. Microarray-based detection and expression analysis of extracellular matrix proteins in drugresistant ovarian cancer cell lines. Oncol Rep. 2014;32:1981–1990. doi: 10.3892/or.2014.3468. [DOI] [PubMed] [Google Scholar]
- 29.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 30.Buck E, Eyzaguirre A, Barr S, Thompson S, Sennello R, Young D, Iwata KK, Gibson NW, Cagnoni P, Haley JD. Loss of homotypic cell adhesion by epithelial-mesenchymal transition or mutation limits sensitivity to epidermal growth factor receptor inhibition. Mol Cancer Ther. 2007;6:532–541. doi: 10.1158/1535-7163.MCT-06-0462. [DOI] [PubMed] [Google Scholar]
- 31.Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol. 2010;188:11–19. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gocheva V, Wang HW, Gadea BB, Shree T, Hunter KE, Garfall AL, Berman T, Joyce JA. IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev. 2010;24:241–255. doi: 10.1101/gad.1874010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bergers G, Benjamin LE. Tumorigenesis and the angiogenic switch. Nat Rev Cancer. 2003;3:401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- 34.Hanahan D, Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86:353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 35.Mehta VB, Besner GE. HB-EGF promotes angiogenesis in endothelial cells via PI3-kinase and MAPK signaling pathways. Growth Factors. 2007;25:253–263. doi: 10.1080/08977190701773070. [DOI] [PubMed] [Google Scholar]
- 36.Parker LH, Schmidt M, Jin SW, Gray AM, Beis D, Pham T, Frantz G, Palmieri S, Hillan K, Stainier DY, De Sauvage FJ, Ye W. The endothelial-cell-derived secreted factor Egfl7 regulates vascular tube formation. Nature. 2004;428:754–758. doi: 10.1038/nature02416. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt M, Paes K, De Maziere A, Smyczek T, Yang S, Gray A, French D, Kasman I, Klumperman J, Rice DS, Ye W. EGFL7 regulates the collective migration of endothelial cells by restricting their spatial distribution. Development. 2007;134:2913–2923. doi: 10.1242/dev.002576. [DOI] [PubMed] [Google Scholar]
- 38.Campagnolo L, Leahy A, Chitnis S, Koschnick S, Fitch MJ, Fallon JT, Loskutoff D, Taubman MB, Stuhlmann H. EGFL7 is a chemoattractant for endothelial cells and is up-regulated in angiogenesis and arterial injury. Am J Pathol. 2005;167:275–284. doi: 10.1016/S0002-9440(10)62972-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buchner G, Orfanelli U, Quaderi N, Bassi MT, Andolfi G, Ballabio A, Franco B. Identification of a new EGF-repeat-containing gene from human Xp22: a candidate for developmental disorders. Genomics. 2000;65:16–23. doi: 10.1006/geno.2000.6146. [DOI] [PubMed] [Google Scholar]
- 40.Osada A, Kiyozumi D, Tsutsui K, Ono Y, Weber CN, Sugimoto N, Imai T, Okada A, Sekiguchi K. Expression of MAEG, a novel basement membrane protein, in mouse hair follicle morphogenesis. Exp Cell Res. 2005;303:148–159. doi: 10.1016/j.yexcr.2004.04.053. [DOI] [PubMed] [Google Scholar]
- 41.Kim HS, Shin HS, Kwak HJ, Cho CH, Lee CO, Koh GY. Betacellulin induces angiogenesis through activation of mitogen-activated protein kinase and phosphatidylinositol 3'-kinase in endothelial cell. Faseb j. 2003;17:318–320. doi: 10.1096/fj.02-0570fje. [DOI] [PubMed] [Google Scholar]
- 42.Lamalice L, Le Boeuf F, Huot J. Endothelial cell migration during angiogenesis. Circ Res. 2007;100:782–794. doi: 10.1161/01.RES.0000259593.07661.1e. [DOI] [PubMed] [Google Scholar]