Abstract
Coronaviruses are enveloped, positive-stranded RNA viruses with a genome of approximately 30 kb. Based on genetic similarities, coronaviruses are classified into three groups. Two group 2 coronaviruses, human coronavirus OC43 (HCoV-OC43) and bovine coronavirus (BCoV), show remarkable antigenic and genetic similarities. In this study, we report the first complete genome sequence (30,738 nucleotides) of the prototype HCoV-OC43 strain (ATCC VR759). Complete genome and open reading frame (ORF) analyses were performed in comparison to the BCoV genome. In the region between the spike and membrane protein genes, a 290-nucleotide deletion is present, corresponding to the absence of BCoV ORFs ns4.9 and ns4.8. Nucleotide and amino acid similarity percentages were determined for the major HCoV-OC43 ORFs and for those of other group 2 coronaviruses. The highest degree of similarity is demonstrated between HCoV-OC43 and BCoV in all ORFs with the exception of the E gene. Molecular clock analysis of the spike gene sequences of BCoV and HCoV-OC43 suggests a relatively recent zoonotic transmission event and dates their most recent common ancestor to around 1890. An evolutionary rate in the order of 4 × 10−4 nucleotide changes per site per year was estimated. This is the first animal-human zoonotic pair of coronaviruses that can be analyzed in order to gain insights into the processes of adaptation of a nonhuman coronavirus to a human host, which is important for understanding the interspecies transmission events that led to the origin of the severe acute respiratory syndrome outbreak.
Coronaviruses are large (120- to 160-nm), roughly spherical particles with a linear, nonsegmented, capped, and polyadenylated positive-sense single-stranded RNA genome that is encapsidated in a helical nucleocapsid. The envelope is derived from intracellular membranes and contains a characteristic crown of widely spaced club-shaped spikes that are 12 to 24 nm long. The genus Coronavirus (International Committee on the Taxonomy of Viruses database [ICTVdb], virus code 03.019.0.1) belongs to the family Coronaviridae in the order Nidovirales (7, 8).
Before the 2002-to-2003 severe acute respiratory syndrome (SARS) epidemic, coronaviruses were somewhat neglected in human medicine, but they have always been of considerable importance in animal health. Coronaviruses infect a variety of livestock, poultry, and companion animals, in whom they can cause serious and often fatal respiratory, enteric, cardiovascular, and neurologic diseases (25). Most of our understanding about the molecular pathogenic properties of coronaviruses has been achieved by the veterinary virology community.
The coronaviruses are classified into three groups based on genetic and serological relationships (19). Group 1 contains the porcine epidemic diarrhea virus (PEDV), porcine transmissible gastroenteritis virus (TGEV), canine coronavirus (CCoV), feline infectious peritonitis virus (FIPV), human coronavirus 229E (HCoV-229E), and the recently identified human coronavirus NL63 (HCoV-NL63). Group 2 contains the murine hepatitis virus (MHV), bovine coronavirus (BCoV), human coronavirus OC43 (HCoV-OC43), rat sialodacryoadenitis virus (SDAV), porcine hemagglutinating encephalomyelitis virus (PHEV), canine respiratory coronavirus (CRCoV), and equine coronavirus (ECoV). Group 3 contains the avian infectious bronchitis virus (IBV) and turkey coronavirus (TCoV). The SARS coronavirus (SARS-CoV) is not assigned to any of these groups but is most closely related to group 2 coronaviruses (21, 54).
HCoV-OC43 (ICTVdb code 19.0.1.0.006) and HCoV-229E (ICTVdb code 19.0.1.0.005) were isolated in 1967 from volunteers at the Common Cold Unit in Salisbury, United Kingdom. HCoV-OC43 was initially propagated on ciliated human embryonic tracheal and nasal organ cultures (42). HCoV-OC43 and HCoV-229E are responsible for 10 to 30% of all common colds, and infections occur mainly during the winter and early spring (38). The incubation period is 2 to 4 days. During the 2002-to-2003 winter season, a new human coronavirus, HCoV-NL63, was isolated from a 7-month-old child suffering from bronchiolitis and conjunctivitis in The Netherlands (61). Seven additional HCoV-NL63-infected individuals, both infants and adults, were identified, indicating that HCoV-NL63 can be considered an important new etiologic agent in respiratory tract infections. Coronaviruses infect all age groups, and reinfections are common. The infection can be subclinical and is usually mild, but there have been reports of more-severe lower respiratory tract involvement in infants and elderly people (17, 60). Human coronaviruses can induce a demyelinating disease in rodents and can infect primary cultures of human astrocytes and microglia. A possible etiological role for HCoV-OC43 and HCoV-229E in multiple sclerosis is being debated (4, 13, 15).
The coronavirus genomes are the largest of the known RNA viruses (27 to 31.5 kb) and are polycistronic, generating a nested set of subgenomic RNAs with common 5′ and 3′ sequences (35). The 5′ two-thirds of the genome consists of two large replicase open reading frames (ORFs), ORF1a and ORF1b. The ORF1a polyprotein (pp1a) can be extended with ORF1b-encoded sequences via a −1 ribosomal frameshift at a conserved slippery site (6), generating the >7,000-amino-acid polyprotein pp1ab, which includes the putative RNA-dependent RNA polymerase (RdRp) and RNA helicase (HEL) activity (20, 39). The polyproteins pp1a and pp1ab are autocatalytically processed by two or three different viral proteases encoded by ORF1a: one or two papain-like proteases (PLP1 and PLP2) and a 3C-like protease (3CLpro) (39, 67, 68). Other putative domains presumably associated with a 3′-to-5′ exonuclease (ExoN) activity, a poly(U)-specific endo-RNase (XendoU) activity, and a 2′-O-methyltransferase (2′-O-MT) activity are predicted in pp1ab (27, 54). The 3′ end of a coronavirus genome includes several structural and accessory protein genes: an envelope-associated hemagglutinin esterase (HE) glycoprotein gene, present only in group 2 coronaviruses; a spike (S) glycoprotein gene; an envelope (E) protein gene; a matrix (M) glycoprotein gene; a nucleocapsid (N) phosphoprotein gene; and several ORFs that encode putative nonstructural (ns) proteins (35).
Coronaviruses are well equipped to adapt rapidly to changing ecological niches by the high mutation rate of their RNA genome (about 10−4 nucleotide substitution/site/year) and high recombination frequencies (51). Many animal coronaviruses cause long-term or persistent enzootic infections. Long periods of coronavirus infection combined with a high mutation and recombination rate increase the probability that a virus mutant with an extended host range might arise.
The current emergence of the SARS-CoV is an example of a crossing of the animal-human species barrier. It is likely that the SARS-CoV was enzootic in an unknown animal or bird species before suddenly emerging as a virulent virus for humans. Chinese scientists found that six masked palm civets (Paguma larvata) and a racoon dog (Nyctereutes procyonoides) for sale in an exotic food market in Shenzhen, in the Guangdong province in Southern China, were harboring a virus very similar to the SARS-CoV (1). Thirteen percent of the civet merchants tested at markets in Guangdong also had SARS antibodies. Sequence analysis showed that the animal version of the SARS-CoV contained an extra stretch of 29 bases (22). It is still not clear whether the civets were a reservoir for the virus or were infected by another species.
HCoV-OC43 and BCoV (ICTVdb code 03.019.0.01.002) show remarkable antigenic and genetic similarities (23, 29, 36, 44, 52, 63, 65). They both have hemagglutinating activity by attaching to the N-acetyl-9-O-acetylneuraminic acid moiety on red blood cells (33). BCoV causes severe diarrhea in newborn calves. The complete nucleotide sequences of different BCoV strains are known, but only fragments of the HCoV-OC43 genome had been determined previously. In this paper, we report the complete HCoV-OC43 sequence (30,738 bases) and the comparative characterization and evolutionary relationship of the BCoV-HCoV-OC43 pair. This is the first animal-human zoonotic pair of coronaviruses that can be analyzed in order to gain insights into the processes of adaptation of a nonhuman coronavirus to a human host.
MATERIALS AND METHODS
Preparation of HCoV-OC43 RNA.
An HCoV-OC43 strain (VR759) was obtained from the American Type Culture Collection (ATCC). The ATCC VR759 strain originated from a volunteer with a common cold-like illness at the Common Cold Unit in Salisbury, United Kingdom (42). HCoV-OC43 was propagated in a human rhabdomyosarcoma (RD) cell line, obtained from the European Collection of Cell Cultures (ECACC). The supernatant was harvested after 7 days of incubation at 33°C, and RNA was isolated by using the QIAamp viral RNA kit (QIAGEN, Westburg, The Netherlands). A real-time quantitative reverse transcription PCR (RT-PCR) (Taqman; Perkin-Elmer Applied Biosystems, Foster City, Calif.) was developed to determine the number of RNA copies present in the supernatant.
Sequencing of the HCoV-OC43 genome.
To determine the HCoV-OC43 genomic sequence, a set of overlapping RT-PCR products (average size, 1.5 kb) encompassing the entire genome was generated. For both RT-PCR and sequencing, oligonucleotide primers were designed in regions that were conserved between the BCoV and MHV genomes. The forward PCR primer in the 5′-terminal sequence (OC43F1 [5′-GATTGTGAGCGATTTGC-3′]) was based on the HCoV-OC43 5′ untranslated region partial sequence (H. Y. Wu, J. S. Guy, D. Yoo, R. Vlasak, and D. A. Brian, unpublished data; GenBank accession number AF523847). To generate RT-PCR products containing the exact 3′-terminal sequence, we used oligonucleotide OC43R74 (5′-TTTTTTTTTTGTGATTCTTCCA-3′) based on the conserved 3′-end sequence of all known group 2 coronaviruses. By using 150 sequencing primers, sequencing in both directions was performed on an ABI Prism 3100 genetic analyzer (Perkin-Elmer Applied Biosystems) using the BigDye terminator cycle sequencing kit (version 3.1). Chromatogram sequencing files were inspected with Chromas 2.2 (Technelysium, Helensvale, Australia), and contigs were prepared by using SeqMan II (DNASTAR, Madison, Wis.).
DNA and protein sequence analyses.
ORF analysis was performed by using the NCBI ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Potential 3C-like protease cleavage sites were identified by using the NetCorona 1.0 server (30). DNA and protein similarity searches were performed using the NCBI WWW-BLAST (basic local alignment search tool) server on the GenBank DNA database, release 118.0 (2). Pairwise nucleotide and protein sequence alignments were performed by using FASTA algorithms in the ALIGN program on the GENESTREAM network server (http://www2.igh.cnrs.fr) at the Institut de Génétique Humaine in Montpellier, France (47). Maizel-Lenk dot matrix plots were calculated using the pairwise FLAG 1.0 (fast local alignment for gigabases) algorithm at the server of the Biomedical Engineering Center of the Industrial Technology Research Institute in Hsinchu City, Taiwan (http://bioinformatics.itri.org.tw/prflag/prflag.php). Multiple sequence alignments were prepared by using CLUSTALW (58) and CLUSTALX, version 1.82 (59) and were manually edited in GENEDOC (46). Phylogenetic analyses were conducted by using MEGA, version 2.1 (34).
Evolutionary rate analyses and timing of the most recent common ancestor.
The relationship between isolation date and genetic divergence was investigated using a linear regression, based on a maximum-likelihood tree, as implemented in the Path-O-Gen software, kindly provided by Andrew Rambaut (University of Oxford, Oxford, United Kingdom). Evolutionary rates and divergence times were estimated by using maximum likelihood in the TipDate software package, version 1.2 (49), and Bayesian inference in BEAST, version 1.03 (kindly made available by A. J. Drummond and A. Rambaut, University of Oxford; http://evolve.zoo.ox.ac.uk/beast/). The molecular clock hypothesis was tested by using the likelihood ratio test.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper were deposited in GenBank under accession number AY391777 by using the National Center for Biotechnology Information (NCBI; Bethesda, Md.) BankIt v3.0 submission tool (http://www3.ncbi.nlm.nih.gov/BankIt/).
RESULTS
HCoV-OC43 complete genomic sequence.
We report here the complete nucleotide sequence of the prototype HCoV-OC43 strain (VR759), isolated in 1967 from an adult with common cold-like symptoms (42). The HCoV-OC43 genome encompasses 30,738 nucleotides [excluding the 3′ poly(A) tail] and was deposited in the GenBank database under accession number AY391777. The HCoV-OC43 genome has a GC-content of 36.9%.
ORF organization of HCoV-OC43.
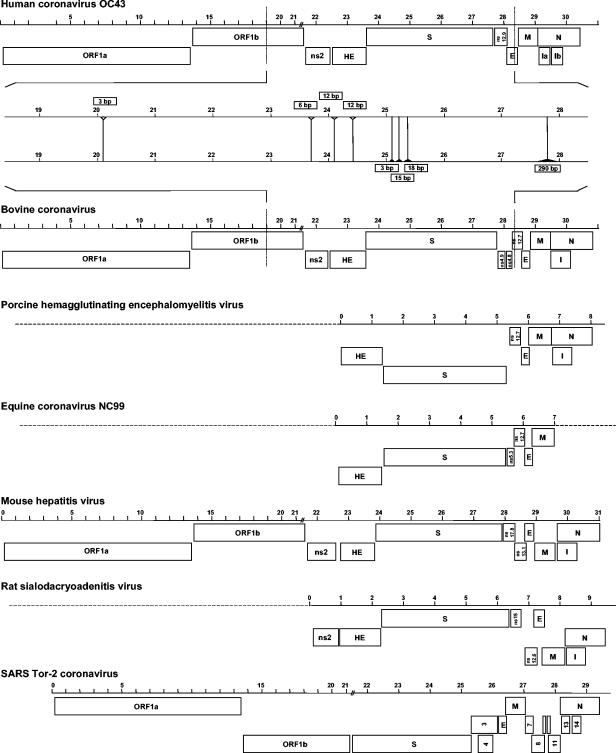
The HCoV-OC43 genome contains 11 major ORFs flanked by 5′ and 3′ untranslated regions of 211 and 288 nucleotides, respectively. A linear representation of the major ORFs of HCoV-OC43, other group 2 coronaviruses, and SARS-CoV is given in Fig. 1. Table 1 shows a comparison of the positions of the major ORFs of HCoV-OC43 and BCoV strain Mebus.
FIG. 1.
Linear representation of the ORFs of the group 2 coronaviruses and SARS-CoV. Nucleotide insertions (open arrowheads) and deletions (solid arrowheads) in the HCoV-OC43 genome compared to BCoV are shown.
TABLE 1.
Positions of the major ORFs of HCoV-OC43 (ATCC VR759) and BCoV (Mebus strain)
| ORF | Nucleotide position
|
No. of bases | No. of amino acids | |||
|---|---|---|---|---|---|---|
| Virus | Start ORF | First ATG | Stop codon | |||
| ORF1a | HCoV-OC43 | 205 | 211 | 13362 | 13,158 | 4,385 |
| BCoV | 205 | 211 | 13362 | 13,158 | 4,385 | |
| ORF1b | HCoV-OC43 | 13332 | 13566 | 21497 | 8,166 | 2,721 |
| BCoV | 13332 | 13566 | 21494 | 8,163 | 2,720 | |
| ns2 | HCoV-OC43 | 21498 | 21507 | 22343 | 846 | 281 |
| BCoV | 21495 | 21504 | 22340 | 846 | 281 | |
| HE | HCoV-OC43 | 22334 | 22355 | 23629 | 1,296 | 431 |
| BCoV | 22331 | 22352 | 23626 | 1,296 | 431 | |
| S | HCoV-OC43 | 23623 | 23644 | 27729 | 4,107 | 1,368 |
| BCoV | 23620 | 23641 | 27732 | 4,113 | 1,370 | |
| ns4.9 | HCoV-OC43 | NAa | NA | NA | NA | NA |
| BCoV | 27811 | 27889 | 28026 | 216 | 71 | |
| ns12.9 | HCoV-OC43 | 27802 | 27817 | 28146 | 345 | 114 |
| BCoV | 28095 | 28110 | 28439 | 345 | 114 | |
| E | HCoV-OC43 | 28133 | 28133 | 28387 | 255 | 84 |
| BCoV | 28426 | 28426 | 28680 | 255 | 84 | |
| M | HCoV-OC43 | 28381 | 28402 | 29094 | 714 | 237 |
| BCoV | 28674 | 28695 | 29387 | 714 | 237 | |
| N | HCoV-OC43 | 29095 | 29104 | 30450 | 1,356 | 451 |
| BCoV | 29388 | 29397 | 30743 | 1,356 | 451 | |
| Ia | HCoV-OC43 | 29102 | 29165 | 29347 | 246 | 81 |
| Ib | HCoV-OC43 | 29348 | 29441 | 29788 | 441 | 146 |
| I | BCoV | 29395 | 29458 | 30081 | 687 | 228 |
NA, not applicable.
The first two-thirds of the genome consists of two large replicase ORFs, ORF1a and ORF1b. ORF1a is 4,383 codons long and overlaps with ORF1b, which consists of 2,721 codons. The coronavirus replicase polyprotein of 7,095 amino acids (aa) is synthesized by a −1 ribosomal frameshift at a conserved slippery site (UUUAAAC, nucleotides 13335 to 13341). In this polyprotein (GenBank accession number AAR01012) numerous putative functional domains are predicted: PLP1 and PLP2 (aa 852 to 2750), 3CLpro (aa 3247 to 3549), RdRp (aa 4370 to 5297), HEL (aa 5298 to 5900) (20, 39), and several putative nidovirus homologs of cellular RNA-processing enzymes, such as ExoN (aa 5901 to 6421), XendoU (aa 6422 to 6796), and a 2′-O-MT domain (aa 6797 to 7095) (Fig. 2) (27, 54). The amino-terminal part of the polyprotein is predicted to be cleaved by the papain-like proteases (68), while the carboxy-terminal part is putatively processed by the main coronavirus protease, 3CLpro (67).
FIG. 2.
Overview of the putative domain organization and potential proteolytic cleavage sites of the HCoV-OC43 replicase polyprotein pp1ab. Cleavage sites that are predicted to be processed by 3C-like protease are indicated by black arrowheads, while potential papain-like protease cleavage sites are indicated by white arrowheads. The following predicted domains are shown: papain-like proteases 1 and 2 (PLP1 and PLP2), X domain (X), putative transmembrane domains 1, 2, and 3 (TM1, TM2, and TM3), 3C-like protease (3CL), growth factor-like domain (GFL), RdRp, metal ion-binding domain (MB), HEL, ATPase, putative 3′-to-5′ exonuclease (ExoN), putative poly(U)-specific endo-RNase (XendoU), and a putative S-adenosylmethionine-dependent ribose 2′-O-methyltransferase (MT).
The 3′-proximal part of the HCoV-OC43 genome contains several ORFs, which encode a variety of structural and accessory proteins. Downstream of ORF1b, a nonstructural protein gene (ns2) of 837 nucleotides, which is a group 2-specific gene, is present. Although these group 2-specific genes are not essential for viral growth, recent work has shown that deletion of MHV ns2 leads to a significant attenuation of the virus when inoculated into mice (12). The HE gene, another group 2-specific gene, consists of 1,275 nucleotides and encodes a protein of 424 aa. The S gene is located immediately downstream of the HE gene and has 4,086 nucleotides. The S protein, which consists of 1,361 aa, plays an important role in the attachment of the virus to cell surface receptors and induces the fusion of the viral and cellular membranes (5, 55). In the genomic region between the S gene and the membrane glycoprotein (M) gene, two ORFs can be identified: the ns12.9 gene encodes a putative nonstructural protein of 12.9 kDa and is 330 nucleotides long (43), while the E gene, 255 nucleotides long, codes for the E protein of approximately 9.5 kDa (43). At the 3′ end of the HCoV-OC43 genome, four major ORFs are present. The M gene is 693 nucleotides long and encodes a polypeptide of 230 aa with a molecular size of approximately 26 kDa (24). The membrane glycoprotein is anchored in the viral membrane with only a short amino-terminal domain exposed to the exterior of the viral envelope. The nucleocapsid protein (N) gene, consisting of 1,347 nucleotides, lies at the 3′ end of the genome and encodes a 448-aa protein, which is associated with the RNA genome to form the nucleocapsid inside the viral envelope. In the 5′ part of the HCoV-OC43 N region, two small internal (I) ORFs can be identified (Ia and Ib). In BCoV, this region is uninterrupted and contains a single I gene which has the capacity to code for a 207-aa protein (37).
HCoV-OC43 sequence similarity to other group 2 coronaviruses.
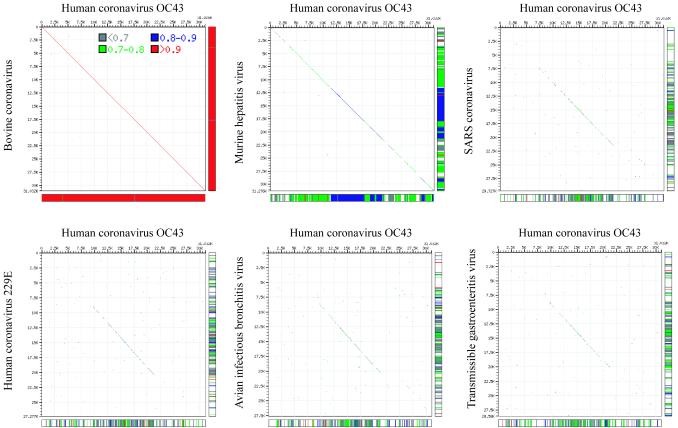
The sequence similarity among HCoV-OC43, BCoV, CRCoV, PHEV, ECoV, MHV, and SDAV was investigated by pairwise alignments of the corresponding ORFs and their proteins (Table 2). HCoV-OC43 showed the highest percentage of similarity to BCoV in all ORFs except for the HCoV-OC43 E gene, which showed 99.6% identity on the nucleotide level and 98.8% identity on the protein level to the PHEV E gene. Maizel-Lenk dot matrix plots illustrate the similarity between HCoV-OC43 and BCoV (Fig. 3).
TABLE 2.
Nucleotide and amino acid similarities of the major HCoV-OC43 (ATCC VR759) ORFs with the ORFs of BCoV, CRCoV, PHEV, ECoV, MHV, and SDAV
| HCoV-OC43 ORF | % Nucleotide (amino acid) similarity
|
|||||
|---|---|---|---|---|---|---|
| BCoV | CRCoV | PHEV | ECoV | MHV | SDAV | |
| ORF1a | 97.4 (97.0) | NAa | NA | NA | 69.3 (65.9) | NA |
| ORF1b | 97.8 (98.6) | NA | NA | NA | 82.7 (87.1) | NA |
| ns2 | 95.1 (95.0) | NA | NA | NA | 59.0 (59.7) | 60.3 (51.5) |
| HE | 96.7 (95.3) | NA | 89.8 (88.7) | 73.7 (72.9) | 60.8 (58.1)b | 64.2 (59.5) |
| S | 93.5 (91.4) | 92.8 (89.9) | 81.7 (81.0) | 79.3 (79.2) | 66.5 (62.9) | 67.0 (64.4) |
| ns12.9 | 96.1 (93.6) | NA | 89.7 (86.2) | 88.8 (81.7) | 55.0 (47.4) | 59.8 (51.4) |
| E | 98.0 (96.4) | NA | 99.6 (98.8) | 96.1 (91.7) | 72.5 (65.9) | 69.2 (66.7) |
| M | 94.8 (94.3) | NA | 92.1 (93.0) | 91.2 (88.7) | 77.9 (83.5) | 77.5 (82.3) |
| N | 96.8 (96.4) | NA | 94.3 (94.0) | NA | 71.8 (69.6) | 72.0 (69.9) |
| Ia/Ibc | 97.1 (NDd) | NA | 95.2 (ND) | NA | 72.3 (ND) | 71.4 (ND) |
NA, not applicable; the corresponding sequence is not available in the GenBank database.
The 5′ end of the MHV HE ORF is missing due to a frameshift mutation or sequencing error in NC_001846.
The OC43 (ATCC VR759 strain) internal ORF (I) coding region contains a stop codon at position 29345, resulting in two potential coding regions of 60 aa (Ia) and 115 aa (Ib). This stop codon is not present in BCoV, which has the capacity to code for a 207-aa protein. This stop codon is also absent in PHEV, MHV, and SADV. The percentage of nucleotide similarity is calculated for the continuous Ia/Ib region.
ND, not done.
FIG. 3.
Maizel-Lenk dot matrix plots: the complete genome sequence of HCoV-OC43 is compared to the complete genome sequences of BCoV, MHV, SARS-CoV, HCoV-229E, IBV, and TGEV, respectively. Sequence identities are indicated by a dot.
Phylogenetic analysis.
A neighbor-joining phylogenetic tree of HCoV-OC43 and 11 other coronaviruses was constructed based on an alignment of ORF1b replicase amino acid sequences (Fig. 4). As an outgroup, we used the equine torovirus (EToV; accession number X52374), belonging to the genus Torovirus in the family Coronaviridae. Three phylogenetic clusters, corresponding to the three coronavirus groups, can be demonstrated. The SARS-CoV forms a separate branch, although there is strong support for monophyly of SARS-CoV with the group 2 coronaviruses, such as HCoV-OC43 and BCoV.
FIG. 4.
Phylogenetic analysis of the coronavirus ORF1b replicase amino acid sequences. The HCoV-OC43 ORF1b protein (GenBank accession number AY391777) was compared to other coronaviruses and to an equine torovirus as an outgroup. Group 1, HCoV-229E (accession number AF304460), HCoV-NL63 (AY567487), PEDV strain CV777 (AF353511), and TGEV strain Purdue (AJ271965). Group 2, BCoV strain Mebus (U00735), MHV type 2 (MHV-2; AF201929), MHV strain Penn 97-1 (AF208066), and MHV-A59 (X51939). Group 3, IBV strain Beaudette (M95169), IBV strain LX4 (AY338732), IBV strain BJ (AY319651). SARS-CoV strain Frankfurt-1 (AY291315) is not classified in any of these groups but is most closely related to group 2 coronaviruses. Outgroup, equine Berne torovirus (EToV; X52374). Regions that were poorly conserved in the manually edited multiple protein sequence alignment were deleted from the alignment. All columns containing gaps were removed. The resulting alignment included 2,083 characters (1,122 being parsimony informative) and contained the meld of the following HCoV-OC43 fragments: 13686-13721, 13737-13793, 13797-13820, 13857-13889, 13869-13994, 14013-14090, 14127-14174, 14247-14390, 14397-14594, 14598-14756, 14766-14855, 14859-15230, 15243-15443, 15480-15674, 15684-15719, 15729-15764, 15786-15854, 15864-15989, 16023-16358, 16374-16715, 16719-16898, 16902-17093, 17115-17258, 17268-17336, 17340-17363, 17379-17501, 17535-17561, 17568-17825, 17925-18008, 18018-18032, 18069-18101, 18207-18440, 18450-18527, 18531-18563, 18576-18602, 18612-18929, 18942-19010, 19,026-19139, 19143-19259, 19284-19466, 19479-19625, 19686-19793, 20289-20318, 20370-20603, 20625-20708, 20718-20762, 20769-20885, 20907-21008, 21045-21125, 21135-21233, 21252-21296, 21309-21431, and 21438-21476. The frequencies of occurrence of particular bifurcations (percentage of 10,000 bootstrap replicate calculations) are indicated at the nodes.
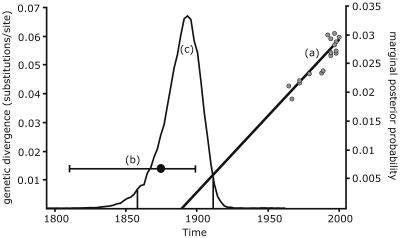
Based on the nucleotide sequence coding for the spike protein, a maximum-likelihood phylogenetic tree was constructed for HCoV-OC43 and several BCoV strains for which the date of isolation was known (Table 3; Fig. 5). HEC4408, a coronavirus isolated in 1988 from a child with acute diarrhea, was also included in the analysis and has actually been shown to be a BCoV (66). The time to the most recent common ancestor (TMRCA) of HCoV-OC43 and BCoV was dated by three methods (Fig. 6). Linear regression of root-to-tip divergence versus sampling time situates the TMRCA of HCoV-OC43 and BCoV in 1891. The maximum-likelihood estimate for TMRCA is 1873, with a 95% confidence interval of 1815 to 1899. The Bayesian coalescent approach dates TMRCA around 1890 (95% highest posterior density interval, 1859 to 1912). This estimate was highly consistent under different demographic models, including an exponential-growth model, which resulted in a TMRCA around 1893 (95% confidence interval, 1866 to 1918). The evolutionary rate of BCoV was also calculated by these three methods (Table 4). A maximum-likelihood evolutionary rate of 4.3 × 10−4 substitutions per site per year was estimated (95% confidence interval, 2.7 × 10−4 to 6.0 × 10−4). A likelihood ratio test indicated that the molecular clock hypothesis could not be rejected (P = 0.10).
TABLE 3.
Date and area of isolation of bovine and human coronaviruses used to calculate TMRCA
| Strain | Isolation date | Isolation area | GenBank accession no. | Reference |
|---|---|---|---|---|
| BCoV-LY138 | 1965 | Utah | AF058942 | 64 |
| BCoV Mebus | 1972 | Quebec, Canada | U00735 | 18 |
| BCoV Quebec | 1972 | Quebec, Canada | AF220295 | 32 |
| BCoV-BECS | 1979 | France | D00731 | 64 |
| BCoV-COBAAA | 1989 | Giessen, Germany | M80844 | 66 |
| BCoV-BCQ7373 | 1992 | Quebec, Canada | AF239306 | 18 |
| BCoV-BCQ1523 | 1994 | Quebec, Canada | AF239307 | 18 |
| BCoV-OK05143 | 1996 | Kansas | AF058944 | 18 |
| BCoV-LSU94 | 1994 | Louisiana | AF058943 | 9 |
| BCoV-BCO44175 | 1997 | Ontario, Canada | AF239309 | 18 |
| BCoV-OntarioBCO43277 | 1997 | Ontario, Canada | AH010241 | 18 |
| BCoV-BCQ3994 | 1998 | Quebec, Canada | AF339836 | 18 |
| BCoV-ENT | 1998 | Texas | AF391541 | 10, 57 |
| BCoV-LUN | 1998 | Texas | AF391542 | 57 |
| HEC4408 | 1988 | Laubach-Wetterfeld, Germany | L07748 | William Herbst, personal communication |
| HCoV-OC43 (VR759) | 1967 | Salisbury, United Kingdom | AY391777 | 42 |
FIG. 5.
Maximum-likelihood phylogenetic tree of spike gene nucleotide sequences of HCoV-OC43 and several BCoV strains for which the date of isolation was known.
FIG. 6.
Results of the evolutionary rate analysis. Line a, linear regression of root-to-tip divergence (y axis) versus sampling time (x axis). The point at which the regression line crosses the time axis indicates the TMRCA (1891). Line b, maximum-likelihood estimate (1873) with 95% confidence intervals (1815 to 1899) for the TMRCA. Curve c, marginal posterior probability (right y axis) for the TMRCA obtained by using the Bayesian coalescent approach. The vertical bars in the distribution represent the 95% highest posterior density interval. Dates of isolation of HCoV-OC43 and BCoV strains are indicated by grey dots.
TABLE 4.
Evolutionary rate estimations of the BCoV-HCoV-OC43 pair
| Method | Substitutions/site/yr (10−4) | 95% Confidence interval (10−4) |
|---|---|---|
| Linear regression | 5.0 | |
| Maximum likelihood | 4.3 | 2.7-6.0 |
| Bayesian inference | ||
| 1st codon position | 4.7 | 3.1-6.5 |
| 2nd codon position | 3.6 | 2.2-4.8 |
| 3rd codon position | 7.8 | 5.4-10.4 |
DISCUSSION
We report in this paper the first complete genome sequence (30,738 nucleotides) of the prototype HCoV-OC43 strain (VR759). Until now, only partial sequence fragments of the structural protein genes of HCoV-OC43 were available in GenBank, leaving the greater 5′ part of the genome to be determined. The recent discovery of a new human coronavirus, SARS-CoV, necessitates a better understanding of the genomic structure and evolution of other known coronaviruses in order to gain insights in how this new type could have emerged.
The prototype HCoV-OC43 strain (ATCC VR759) is a laboratory strain that, since its isolation in 1967, has been passaged 7 times in human embryonic tracheal organ culture, followed by 15 passages in suckling mouse brain cells and an unknown number of passages in human rectal tumor HRT-18 cells and/or Vero cells. During the passage history, it is likely that a number of mutations have accumulated. It would be interesting to analyze the complete nucleotide sequence of contemporary HCoV-OC43 strains that are free from in vitro expansion mutations.
Nucleotide and amino acid similarity percentages were determined for the major HCoV-OC43 ORFs and those of other group 2 coronaviruses (BCoV, CRCoV, PHEV, ECoV, MHV, and SDAV). For all HCoV-OC43 ORFs, the highest similarity demonstrated was that to the corresponding BCoV ORFs, except for the HCoV-OC43 E gene, which showed 99.6% identity on the nucleotide level and 98.8% identity on the amino acid level with the PHEV E gene. Based on the high similarity between HCoV-OC43 and PHEV in E, and between HCoV-OC43 and BCoV in all the other major ORFs, some hypotheses concerning the origin of HCoV-OC43 can be put forward. Adaptation of BCoV to a human host and a recombination event between BCoV and PHEV leading to a new type of coronavirus with a different species specificity could both have been responsible for the emergence of a new human coronavirus.
Phylogenetic analysis of coronavirus ORF1b replicase protein sequences confirms the presence of three coronavirus group clusters and a separate branch for SARS-CoV, which seems to be most closely related to group 2 coronaviruses (21, 54). HCoV-OC43 and BCoV cluster together, demonstrating the close relationship between the two viruses. There is in fact more divergence between the different MHV strains or between the different IBV strains than between HCoV-OC43 and BCoV. The close relationship between HCoV-OC43 and BCoV on the genetic level has also been shown to correspond to a close antigenic relationship: by using monoclonal antibodies directed against the BCoV S protein, common antigenic determinants for BCoV, HCoV-OC43, and PHEV have been demonstrated (62, 63). A phylogenetic tree was also constructed for the spike gene of HCoV-OC43 and several BCoV isolates for which the date of isolation could be traced. Different molecular clock calculations situate the most recent common ancestor of HCoV-OC43 and the different BCoV isolates around 1890. We suggest that around 1890, BCoV might have jumped the species barrier and became able to infect humans, resulting in the emergence of a new type of human coronavirus (HCoV-OC43), a scenario similar to the origin of the SARS outbreak. Indisputable evidence for the bovine-to-human direction of the interspecies transmission event, instead of a human-to-bovine direction, is not available. However, we consider the occurrence of a 290-nucleotide deletion (corresponding to the absence of BCoV ns4.9 and ns4.8) in HCoV-OC43 relative to the BCoV genome to be a potential supporting argument, as this additional sequence fragment in BCoV is also present in MHV and SDAV. Consequently, we assume that a deletion from BCoV to HCoV-OC43 rather than an insertion in the opposite direction took place during evolution, and thus, we hypothesize that the interspecies transmission event occurred from bovines to humans.
Nevertheless, it is possible that two other group 2 coronaviruses, CRCoV and PHEV, might have played a role in the emergence of HCoV-OC43. CRCoV appears to be very closely related to BCoV and HCoV-OC43 (16), and for the HCoV-OC43 E gene, the highest percentage of similarity was found with the PHEV E gene, suggesting a possible recombination event. To elucidate the evolutionary relationship of HCoV-OC43 and BCoV with CRCoV and PHEV, complete genome sequence data of CRCoV and PHEV would be required. Molecular dating has frequently been used to investigate the origin of viral epidemics (31, 40, 48). The reliability of such an analysis is dependent on the validity of the molecular clock hypothesis, which assumes that the evolutionary rate is roughly constant in the lineages of a phylogenetic tree. Although this assumption is frequently violated for viral sequence data (28), a molecular clock test indicated that this hypothesis could not be rejected for the coronavirus data set investigated here.
In the second half of the nineteenth century, a highly infectious respiratory disease with a high mortality rate affected cattle herds around the world (11, 41). The same disease, or a similar disease, is now known as contagious bovine pleuropneumonia (CBPP) and is caused by Mycoplasma mycoides mycoides. In the nineteenth century, the clinical symptoms of CBPP would have been difficult to distinguish from those of BCoV pneumonia, and it can be hypothesized that the bovine respiratory disease in the second half of the nineteenth century might have been similar to the coronavirus-associated shipping fever disease (56). Most industrialized countries mounted massive culling operations in the period between 1870 and 1890 (11) and were able to eradicate the disease by the beginning of the twentieth century. During the slaughtering of CBPP-affected herds, there was ample opportunity for the culling personnel to come into contact with bovine respiratory secretions. These respiratory secretions could have contained BCoV, either as the causal agent or as a coinfecting agent.
Interestingly, around the period in which the BCoV interspecies transmission would probably have taken place, a human epidemic ascribed to influenza was spreading around the world. The 1889-1890 pandemic probably originated in Central Asia (3) and was characterized by malaise, fever, and pronounced central nervous system symptoms (53). A significant increase in case fatality with increasing age was observed. Absolute evidence that an influenza virus was the causative agent of this epidemic was never obtained, due to the lack of tissue samples from that period. However, postepidemic analysis in 1957 of the influenza antibody pattern in sera of people who were 50 to 100 years old indicated that H2N2 influenza antibodies might have originated from the 1889-1890 pandemic (45). However, it is tempting to speculate about an alternative hypothesis, that the 1889-1890 pandemic may have been the result of interspecies transmission of bovine coronaviruses to humans, resulting in the subsequent emergence of HCoV-OC43. The dating of the most recent common ancestor of BCoV and HCoV-OC43 to around 1890 is one argument. Another argument is the fact that central nervous system symptoms were more pronounced during the 1889-1890 epidemic than in other influenza outbreaks. It has been shown that HCoV-OC43 has neurotropism and can be neuroinvasive (4).
Maximum-likelihood phylogenetic analysis of the spike gene of HCoV-OC43 and several BCoV strains for which the date of isolation is known indicates that these strains evolved according to a molecular clock. An evolutionary rate on the order of 4 × 10−4 nucleotide change per site per year was estimated, and this rate was highly consistent across the different methods used. This rate falls within the range reported for other RNA viruses, including SARS-CoV (14, 50, 51).
This study provides evidence for viral promiscuity, a phenomenon that has already been reported for several animal coronaviruses, including BCoV, for which the potential to infect other species, including humans, has already been described (26, 66). The isolation of the SARS-CoV from masked palm civets and raccoon dogs indicates that this new type of coronavirus was also enzootic in an animal species before suddenly emerging as a virulent virus for humans. The characterization of the BCoV-HCoV-OC43 pair presented in this study provides insights into the process of adaptation of a nonhuman coronavirus to a human host, which is important for understanding the interspecies transmission events that led to the origin of the SARS outbreak.
Acknowledgments
We thank all our colleagues at the Laboratory of Clinical and Epidemiological Virology, Department of Microbiology and Immunology, Rega Institute for Medical Research, University of Leuven, Leuven, Belgium, for helpful comments and discussions.
This work was supported by a fellowship from the Flemish Fonds voor Wetenschappelijk Onderzoek (FWO) to L.V. and by FWO grant G.0288.01.
REFERENCES
- 1.Abbott, A. 2003. Pet theory comes to the fore in fight against SARS. Nature 423:576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous. 1958. Influenza 1889 and 1957. Lancet i:833-835. [PubMed] [Google Scholar]
- 4.Arbour, N., R. Day, J. Newcombe, and P. J. Talbot. 2000. Neuroinvasion by human respiratory coronaviruses. J. Virol. 74:8913-8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch, B. J., R. van der Zee, C. A. de Haan, and P. J. Rottier. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77:8801-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brierley, I., M. E. Boursnell, M. M. Binns, B. Bilimoria, V. C. Blok, T. D. Brown, and S. C. Inglis. 1987. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 6:3779-3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Büchen-Osmond, C. 2003. ICTVdB: the Universal Virus Database of the International Committee on Taxonomy of Viruses, version 3, 03.019.0.1 (Coronaviridae). Biomedical Informatics Core, Northeast Biodefense Center, Columbia University, New York, N.Y.
- 8.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 9.Chouljenko, V. N., K. G. Kousoulas, X. Q. Lin, and J. Storz. 1998. Nucleotide and predicted amino acid sequences of all genes encoded by the 3′ genomic portion (9.5 kb) of respiratory bovine coronaviruses and comparisons among respiratory and enteric coronaviruses. Virus Genes 17:33-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chouljenko, V. N., X. Q. Lin, J. Storz, K. G. Kousoulas, and A. E. Gorbalenya. 2001. Comparison of genomic and predicted amino acid sequences of respiratory and enteric bovine coronaviruses isolated from the same animal with fatal shipping pneumonia. J. Gen. Virol. 82:2927-2933. [DOI] [PubMed] [Google Scholar]
- 11.Crookshank, E. M. 1897. Infectious pleuro-pneumonia, p. 239-248. In E. M. Crookshank (ed.), A textbook of bacteriology including the etiology and prevention of infective diseases. W. B. Saunders, Philadelphia, Pa.
- 12.de Haan, C. A., P. S. Masters, X. Shen, S. Weiss, and P. J. Rottier. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296:177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dessau, R. B., G. Lisby, and J. L. Frederiksen. 2001. Coronaviruses in brain tissue from patients with multiple sclerosis. Acta Neuropathol. (Berlin) 101:601-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domingo, E., and J. J. Holland. 1988. High error rates, population equilibrium and evolution of RNA replication systems, p. 3-36. In E. Domingo, J. J. Holland, and P. Ahlquist (ed.), RNA genetics, vol. 3. CRC Press, Boca Raton, Fla. [Google Scholar]
- 15.Edwards, J. A., F. Denis, and P. J. Talbot. 2000. Activation of glial cells by human coronavirus OC43 infection. J. Neuroimmunol. 108:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erles, K., C. Toomey, H. W. Brooks, and J. Brownlie. 2003. Detection of a group 2 coronavirus in dogs with canine infectious respiratory disease. Virology 310:216-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gagneur, A., J. Sizun, S. Vallet, M. C. Legr, B. Picard, and P. J. Talbot. 2002. Coronavirus-related nosocomial viral respiratory infections in a neonatal and paediatric intensive care unit: a prospective study. J. Hosp. Infect. 51:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gelinas, A. M., M. Boutin, A. M. Sasseville, and S. Dea. 2001. Bovine coronaviruses associated with enteric and respiratory diseases in Canadian dairy cattle display different reactivities to anti-HE monoclonal antibodies and distinct amino acid changes in their HE, S and ns4.9 protein. Virus Res. 76:43-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez, J. M., P. Gomez-Puertas, D. Cavanagh, A. E. Gorbalenya, and L. Enjuanes. 2003. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 148:2207-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 17:4847-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorbalenya, A. E., E. J. Snijder, and W. J. Spaan. 2004. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 78:7863-7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. M. Peiris, and L. L. M. Poon. 2003. Isolation and characterisation of viruses related to the SARS coronavirus from animals in Southern China. Science 302:276-278. [DOI] [PubMed] [Google Scholar]
- 23.Hogue, B. G., B. King, and D. A. Brian. 1984. Antigenic relationships among proteins of bovine coronavirus, human respiratory coronavirus OC43, and mouse hepatitis coronavirus A59. J. Virol. 51:384-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hogue, B. G., and D. A. Brian. 1986. Structural proteins of human respiratory coronavirus OC43. Virus Res. 5:131-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes, K. V., and M. M. Lai. 1996. Coronaviridae: the viruses and their replication, p. 1075-1093. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Raven Press, New York, N.Y.
- 26.Ismail, M. M., K. O. Cho, L. A. Ward, L. J. Saif, and Y. M. Saif. 2001. Experimental bovine coronavirus in turkey poults and young chickens. Avian Dis. 45:157-163. [PubMed] [Google Scholar]
- 27.Ivanov, K. A., V. Thiel, J. C. Dobbe, Y. van der Meer, E. J. Snijder, and J. Ziebuhr. 2004. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 78:5619-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenkins, G. M., A. Rambaut, O. G. Pybus, and E. C. Holmes. 2002. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J. Mol. Evol. 54:156-165. [DOI] [PubMed] [Google Scholar]
- 29.Kamahora, T., L. H. Soe, and M. M. Lai. 1989. Sequence analysis of nucleocapsid gene and leader RNA of human coronavirus OC43. Virus Res. 12:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiemer, L., O. Lund, S. Brunak, and N. Blom. 2004. Coronavirus 3CL-pro proteinase cleavage sites: possible relevance to SARS virus pathology. BMC Bioinformatics 5:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korber, B., M. Muldoon, J. Theiler, F. Gao, R. Gupta, A. Lapedes, B. H. Hahn, S. Wolinsky, and T. Bhattacharya. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789-1796. [DOI] [PubMed] [Google Scholar]
- 32.Kourtesis, A. B., A. M. Gelinas, and S. Dea. 2001. Genomic and antigenic variations of the HE glycoprotein of bovine coronaviruses associated with neonatal calf diarrhea and winter dysentery. Arch. Virol. 146:1219-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krempl, C., B. Schultze, and G. Herrler. 1995. Analysis of cellular receptors for human coronavirus OC43. Adv. Exp. Med. Biol. 380:371-374. [DOI] [PubMed] [Google Scholar]
- 34.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 35.Lai, M. M., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lapps, W., and D. A. Brian. 1985. Oligonucleotide fingerprints of antigenically related bovine coronavirus and human coronavirus OC43. Arch. Virol. 86:101-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lapps, W., B. G. Hogue, and D. A. Brian. 1987. Sequence analysis of the bovine coronavirus nucleocapsid and matrix protein gene. Virology 157:47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larson, H. E., S. E. Reed, and D. A. J. Tyrell. 1980. Isolation of rhinoviruses and coronaviruses from 38 colds in adults. J. Med. Virol. 5:221-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee, H. J., C. K. Shieh, A. E. Gorbalenya, E. V. Koonin, N. La Monica, J. Tuler, A. Bagdzhadzhyan, and M. M. Lai. 1991. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology 180:567-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lemey, P., O. G. Pybus, B. Wang, N. K. Saksena, M. Salemi, and A. M. Vandamme. 2003. Tracing the origin and history of the HIV-2 epidemic. Proc. Natl. Acad. Sci. USA 100:6588-6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McEachran, D. 1879. Extract report of the Minister of Agriculture for the Dominion of Canada for 1878: report of special investigation into existence of cattle disease in the United States, p. 206-209. In Thomas Walley (ed.), The four bovine scourges: pleuro-pneumonia, foot-and-mouth disease, cattle plague, tubercle (scrofula) with an appendix on the inspection of live animals and meat. MacLachlan and Stewart, Edinburgh, United Kingdom.
- 42.McIntosh, K., W. B. Becker, and R. M. Chanock. 1967. Growth in suckling mouse brain of “IBV-like” viruses from patients with upper respiratory tract disease. Proc. Natl. Acad. Sci. USA 58:2268-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mounir, S., and P. J. Talbot. 1993. Human coronavirus OC43 RNA 4 lacks two open reading frames located downstream of the S gene of bovine coronavirus. Virology 192:355-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mounir, S., P. Labonte, and P. J. Talbot. 1993. Characterization of the nonstructural and spike proteins of the human respiratory coronavirus OC43: comparison with bovine enteric coronavirus. Adv. Exp. Med. Biol. 342:61-67. [DOI] [PubMed] [Google Scholar]
- 45.Mulder, J., and N. Masurel. 1958. Pre-epidemic antibody against 1957 strain of Asiatic influenza in serum of older people living in The Netherlands. Lancet i:810-814. [DOI] [PubMed] [Google Scholar]
- 46.Nicholas, K. B., H. B. Nicholas, and D. W. Deerfield. 1997. GeneDoc: analysis and visualization of genetic variation. EMBnet News 4:14. [Google Scholar]
- 47.Pearson, W. R., T. Wood, Z. Zhang, and W. Miller. 1997. Comparison of DNA sequences with protein sequences. Genomics 46:24-36. [DOI] [PubMed] [Google Scholar]
- 48.Pybus, O. G., A. J. Drummond, T. Nakano, B. H. Robertson, and A. Rambaut. 2003. The epidemiology and iatrogenic transmission of hepatitis C virus in Egypt: a Bayesian coalescent approach. Mol. Biol. Evol. 20:381-387. [DOI] [PubMed] [Google Scholar]
- 49.Rambaut, A. 2000. Estimating the rate of molecular evolution: incorporating noncontemporaneous sequences into maximum likelihood phylogenies. Bioinformatics 16:395-399. [DOI] [PubMed] [Google Scholar]
- 50.Salemi, M., W. M. Fitch, M. Ciccozzi, M. J. Ruiz-Alvarez, G. Rezza, and M. J. Lewis. 2004. Severe acute respiratory syndrome coronavirus sequence characteristics and evolutionary rate estimate from maximum likelihood analysis. J. Virol. 78:1602-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanchez, C. M., F. Gebauer, C. Sune, A. Mendez, J. Dopazo, and L. Enjuanes. 1992. Genetic evolution and tropism of transmissible gastroenteritis coronaviruses. Virology 190:92-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sasseville, A. M., M. Boutin, A. M. Gelinas, and S. Dea. 2002. Sequence of the 3′-terminal end (8.1 kb) of the genome of porcine haemagglutinating encephalomyelitis virus: comparison with other haemagglutinating coronaviruses. J. Gen. Virol. 83:2411-2416. [DOI] [PubMed] [Google Scholar]
- 53.Sisley, R. 1891. The epidemic of 1889-1890. Bokhara. St. Petersburgh. Berlin, p. 47-53. In R. Sisley (ed.), Epidemic influenza: notes on its origin and method of spread. Longmans, Green, and Co., London, United Kingdom.
- 54.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. M. Poon, Y. Guan, M. Rozanov, W. J. M. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteonome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spaan, W., D. Cavanagh, and M. C. Horzinek. 1988. Coronaviruses: structure and genome expression. J. Gen. Virol. 69:2939-2952. [DOI] [PubMed] [Google Scholar]
- 56.Storz, J., L. Stine, A. Liem, and G. A. Anderson. 1996. Coronavirus isolation from nasal swab samples of cattle with signs of respiratory tract disease after shipping. J. Am. Vet. Med. Assoc. 208:1452-1456. [PubMed] [Google Scholar]
- 57.Storz, J., X. Lin, C. W. Purdy, V. N. Chouljenko, K. G. Kousoulas, F. M. Enright, W. C. Gilmore, R. E. Briggs, and R. W. Loan. 2000. Coronavirus and Pasteurella infections in bovine shipping fever pneumonia and Evans' criteria for causation. J. Clin. Microbiol. 38:3291-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vabret, A., T. Mourez, S. Gouarin, J. Petitjean, and F. Freymuth. 2003. An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin. Infect. Dis. 36:985-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. M. Berkhout, K. C. Wolthers, P. M. E. Wertheim-van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vautherot, J. F., and J. Laporte. 1983. Utilization of monoclonal antibodies for antigenic characterization of coronaviruses. Ann. Rech. Vet. 14:437-444. [PubMed] [Google Scholar]
- 63.Vieler, E., T. Schlapp, C. Anders, and W. Herbst. 1995. Genomic relationship of porcine hemagglutinating encephalomyelitis virus to bovine coronavirus and human coronavirus OC43 as studied by the use of bovine coronavirus S gene-specific probes. Arch. Virol. 140:1215-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, X., K. G. Kousoulas, and J. Storz. 1991. Comparison of the nucleotide and deduced amino acid sequences of the S genes specified by virulent and avirulent strains of bovine coronaviruses. Virology 183:397-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, X., K. G. Kousoulas, and J. Storz. 1992. The hemagglutinin/esterase gene of human coronavirus strain OC43: phylogenetic relationships to bovine and murine coronaviruses and influenza C virus. Virology 186:318-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, X. M., W. Herbst, K. G. Kousoulas, and J. Storz. 1994. Biological and genetic characterization of a hemagglutinating coronavirus isolated from a diarrhoeic child. J. Med. Virol. 44:152-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]
- 68.Ziebuhr, J., V. Thiel, and A. E. Gorbalenya. 2001. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 276:33220-33232. [DOI] [PMC free article] [PubMed] [Google Scholar]