Abstract
Rearrangements of the JC virus (JCV) regulatory region (RR) are consistently found in the brains of patients with progressive multifocal leukoencephalopathy (PML), whereas the archetype RR is present in their kidneys. In addition, the C terminus of the large T antigen (T-Ag) shows greater variability in PML than does the rest of the coding region. To determine whether similar changes in simian virus 40 (SV40) are necessary for disease induction in monkeys, we sequenced the SV40 RR and the C terminus of the T-Ag from the brain of simian/human immunodeficiency virus (SHIV)-infected monkey 18429, which presented spontaneously with an SV40-associated PML-like disease, as well as from the peripheral blood mononuclear cells (PBMC), kidneys, and brains of SV40-seronegative, SHIV-infected monkeys 21289 and 21306, which were inoculated with the 18429 brain SV40 isolate. These animals developed both SV40-associated PML and meningoencephalitis. Thirteen types of SV40 RR were characterized. Compared to the SV40 archetype, we identified RRs with variable deletions in either the origin of replication, the 21-bp repeat elements, or the late promoter, as well as deletions or duplications of the 72-bp enhancer. The archetype was the most prominent RR in the brain of monkey 18429. Shortly after inoculation, a wide range of RRs could be found in the PBMC of monkeys 21289 and 21306. However, the archetype RR became the predominant type in their blood, kidneys, and brains at the time of sacrifice. On the contrary, the T-Ag C termini remained identical in all compartments of the three animals. These results indicate that unlike JCV in humans, rearrangements of SV40 RR are not required for brain disease induction in immunosuppressed monkeys.
Progressive multifocal leukoencephalopathy (PML) is emerging as a persistent cause of death in human immunodeficiency virus-infected patients, despite the availability of highly active antiretroviral therapy, and survival is limited to an average of 11 months (1). This disease is the consequence of a lytic infection of oligodendrocytes by the polyomavirus JC (JCV). Since there is no specific treatment for PML, the development of an animal model of PML would be a tremendous asset for studying its pathophysiology and devising therapeutic interventions. However, earlier attempts to create a useful animal model of PML were unsuccessful since the host range of JCV is restricted to humans. Indeed, this virus may cause a variety of tumors in animals but it does not replicate in glial cells of rodents or nonhuman primates (9, 19, 23), probably because of a block at the transcriptional level (3).
Since JCV cannot be used directly to create an animal model of PML in immunosuppressed monkeys, we hypothesized that a closely related simian polyomavirus could be used to this effect. Simian polyomavirus 40 (SV40) has 69% sequence homology with JCV, and a few cases of PML-like disease have been reported in rhesus monkeys infected with SV40, with detection of polyomavirus-like virions or SV40 nucleic acid and large T antigen (T-Ag) in the lesions (5-7). In a previous report, we described the transmission of an SV40-induced PML-like disease in two simian/human immunodeficiency virus (SHIV) 89.6P-immunosuppressed rhesus monkeys (2). These animals were inoculated intravenously with an SV40 strain isolated from the brain of an SHIV-infected monkey that presented spontaneously with PML.
The determinants of JCV neurotropism and neurovirulence are thought to be located in the regulatory region (RR) of this virus. Indeed, the type of JCV RR present in the kidneys of healthy and immunosuppressed individuals alike has been called “the archetype,” as it is assumed that it is the form of RR from which all others have evolved (14, 24). Indeed, rearranged JCV RRs, including forms with a tandem repeat pattern, are usually found in the blood, cerebrospinal fluid (CSF), and brains of patients with PML (15, 18). On the contrary, the coding region of JCV is extremely conserved, except for the host range domain (HRD), located in the C terminus of the T-Ag, which has a higher mutation rate (4, 8).
Whether the SV40 RR and the C terminus of the T-Ag are also implicated in the development of PML in monkeys is unclear. Indeed, only a few retrospective studies have investigated the presence of SV40 sequence variation in its natural host (8, 11, 16, 20), and most of the animals tested did not have PML (11, 16). Furthermore, the dynamic evolution of SV40 sequences preceding disease onset in those animals that developed PML was not investigated (8, 20). In the present study, we characterized the SV40 RR and the T-Ag C terminus over time in the blood of the previously described animals prior to the development of PML, as well as in their kidneys and brains at the time of sacrifice (2). Our results show a stable C-terminal T-Ag pattern and the initial presence of multiple SV40 RR types in the blood of these animals, leading to the emergence of a predominant type similar to the SV40 archetype in all of the compartments tested by the time of PML onset.
MATERIALS AND METHODS
Explant supernatant was used to infect primary rhesus macaque fetal fibroblasts. Virus recovered from fibroblast cultures was subjected to three rounds of plaque purification in fetal fibroblasts. Plaque-purified virus was propagated in fetal fibroblasts and concentrated by ultracentrifugation at 100,000 × g for 2 h onto a 30% sucrose cushion. The primary rhesus macaque fetal fibroblasts used have been extensively characterized and found not to contain adventitious viruses common to rhesus macaques, including type D oncornaviruses, type C oncornaviruses, lentiviruses, spumaretroviruses, SV40 or lymphotropic polyomaviruses, adenoviruses, alphaherpesviruses (B virus), betaherpesviruses (cytomegalovirus and human herpesvirus 6-like agents), or gammaherpesvirus (lymphocryptovirus, rhadinovirus, and the paramyxoviruses SV5 and respiratory syncytial virus). Further, specific-pathogen-free macaques, including the macaques in this study, do not seroconvert to these agents after receiving virus inocula propagated in these cells.
Peripheral blood mononuclear cell (PBMC), plasma, brain, and kidney samples were obtained from rhesus macaques 18429, 21289, and 21306 (2). DNA extraction was performed by standard methods (10). PCR amplification of the SV40 RR was performed with primers SRP-5.0 (5′-GACCTAGAAGGTCCATTAGCTGC-3′ [positions 5109 to 5131]) and SRP-6.0 (5′-CTACGAACCTTAACGGAGGCC-3′ [positions 381 to 361]) as previously described (16). These primers amplify a 516-bp fragment of the RR of SV40 reference strain 776. PCR amplification of the SV40 HRD was performed with primers HRD S2573 (5′-ACCACAACTAGAATGCAGTGAAA-3′ [positions 2573 to 2595]) and HRD R3069 (5′-ACCTGTGGCTGAGTTTGCTCA-3′ [positions 3069 to 3049]), which amplify a 497-bp fragment of SV40 776. PCR was performed in a PE 9700 thermal cycler with 1 μg of brain, kidney, or PBMC DNA; 25 pmol of each primer; 1.5 mM MgCl2; and 1 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, Calif.) in a final volume of 25 μl. The amplification protocol consisted of 94°C for 10 min, followed by 40 cycles of 94°C for 30 s, 56°C for 1 min, and 72°C for 1 min and 15 min of elongation at 72°C.
PCR products were cloned with a TA TOPO cloning kit (Invitrogen, Carlsbad, Calif.). They were categorized into different groups according to size on a 2% agarose gel. RR types of similar lengths (less than 20-bp difference in size) were combined together as one group. A total of 55 clones were sequenced, including up to 5 clones of each group of PCR products.
The cycle sequencing reaction was performed in accordance with the manufacturer's instructions with the ABI PRISM BigDye Terminator Cycle Sequencing Ready Kit (Applied Biosystems, Weiterstadt, Germany) with 400 ng of double-stranded DNA and 3.2 pmol of sequencing primer. Fluorescence-based DNA sequencing was performed with an ABI 377 DNA sequencer (Applied Biosystems). Sequence analysis was performed with Lasergene Software for PC, MegAlign 3.12 (DNASTAR Inc., Madison, Wis.) comparing the sequences obtained to that of SV40 reference strain 776 (GenBank accession no. J02400, GI 965480).
RESULTS
The pathology of SV40-induced disease in animals 21289 and 21306 has been described in detail elsewhere (2). Briefly, both animals presented with neurologic syndromes and were euthanized at 63 and 112 days post SV40 infection, respectively.
At necropsy, both animals had CSF pleocytosis with an elevated total protein concentration. Attempts to isolate bacterial agents from CSF yielded negative results. Demyelinating lesions were present in small focal areas in the subcortical white matter, optic tract, corpus callosum, and periventricular white matter. Lesions were characterized by the presence of large, bizarre astrocytes and oligodendrocytes with enlarged nuclei, which stained positively for SV40 DNA by in situ hybridization, as well as abundant large foamy macrophages containing phagocytized myelin debris. In addition to PML-like lesions, SV40 meningoencephalitis (ME) was also present in both animals (2).
RR.
The RR of SV40 reference strain 776 contains the origin of replication, three 21-bp repeats, and two 72-bp enhancer elements (Fig. 1). However, most of the SV40 isolates found in naturally infected animals have only one 72-bp element, and this form of RR was considered the archetype (8, 11, 22).
FIG. 1.
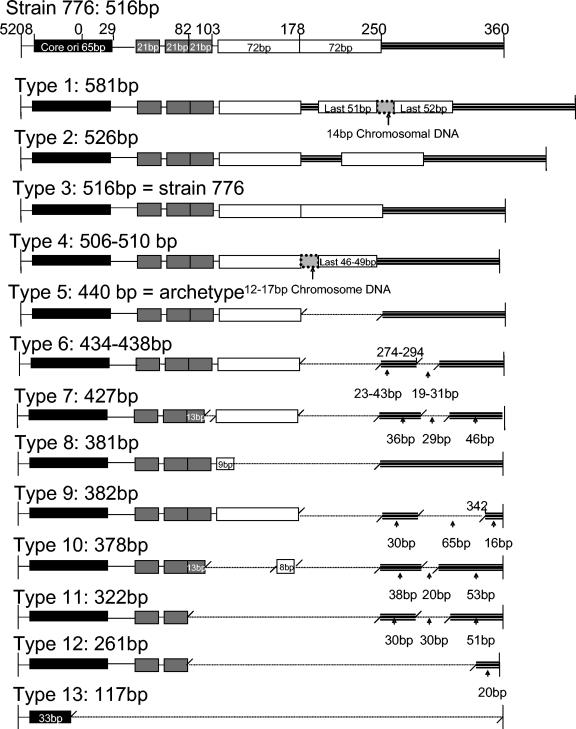
Diversity of SV40 RR sequences. At the top is a representation of the RR of SV40 reference strain 776. The nucleotide numbers are based on this sequence. Thirteen different subtypes of RR were obtained from the brain of monkey 18429 and the PBMCs, brains, and kidneys of monkeys 21289 and 21306. They are displayed in the order of the sizes of the amplified PCR products, from the longest to the shortest. The thick triple line following the 72-bp element represents the late promoter sequence. Deletions are indicated by a single dotted line. Types 1 and 4 contain fragments of nonviral chromosomal DNA. Type 3 is identical to reference strain 776. Type 5 is identical to the archetype RR and only contains one 72-bp element.
To explore the role of the SV40 RR and the T-Ag C terminus in the pathogenesis of PML, we studied the composition of these sequences in three monkeys that developed a PML-like disease. As reported previously (2), rhesus macaque 18429 was an SHIV-KB9-infected animal that presented with PML caused by SV40 reactivation more than 2 years after SHIV infection. Histological examination of the brain of this animal showed multifocal areas of demyelination harboring numerous oligodendroglial cells with enlarged nuclei, gemistocytic astrocytes, and foamy macrophages containing myelin debris. In situ hybridization demonstrated that glial cells in the periphery of areas of demyelination were infected with SV40. Brain explant cultures from this animal were established, and the 18429 isolate was grown on primary fetal monkey fibroblasts. This isolate was then inoculated intravenously into SV40-negative, SHIV-89.6P-immunosuppressed rhesus macaques 21289 and 21306. These two animals presented with an SV40-associated PML-like disease similar to that seen in animal 18429, as well as ME, after 63 and 112 days, respectively (2).
A total of 55 SV40 RR clones were sequenced from the PBMCs, kidneys, and brains of these animals. Thirty different forms of SV40 RR were characterized. These RRs are displayed according to size from the longest to the shortest in Fig. 1. Among these 30 SV40 RRs, 18 subtypes have a small variable deletion in the late promoter sequence (from nucleotide [nt] 250 to nt 360). The only differences between them are the beginning position and the length of the deletion. For this reason, we grouped these 18 different SV40 RRs together in type 6 (Fig. 1). Accordingly, a total of 13 types of SV40 RR were identified.
The highest variability within the RR was found in the late promoter sequence, with deletions ranging from 20 bp to its entirety in types 7 and 9 to 13. Of all types, only 2 and 3 had two intact 72-bp elements, similar to SV40 776. Types 5 to 7 and 9 had a single 72-bp element, as is the case for the SV40 RR found in naturally infected monkeys (11, 13), while other types had either a partial duplication in a 72-bp element (1 and 4), a truncated 72-bp element (8 and 9), or no 72-bp element at all (11 to 13).
In addition, types 1 and 4 had a variable insertion of 12 to 17 bp of nonviral DNA between duplications of the 72-bp element. These sequences were very similar to each other and included a conserved fragment (5′-CTAACTGACACACA-3′) that did not match any particular sequences in the GenBank database. Since these sequences were not reported in any other SV40 isolate, we hypothesize that they were acquired by direct recombination with chromosomal DNA. Finally, the three 21-bp repeat sequences were the most conserved of all of the types, although partial or total deletion of one or more repeats was present in types 7 and 10 to 13. Finally, the 65 bp containing the origin of replication was entirely conserved in all of the types, except for a truncation in type 13.
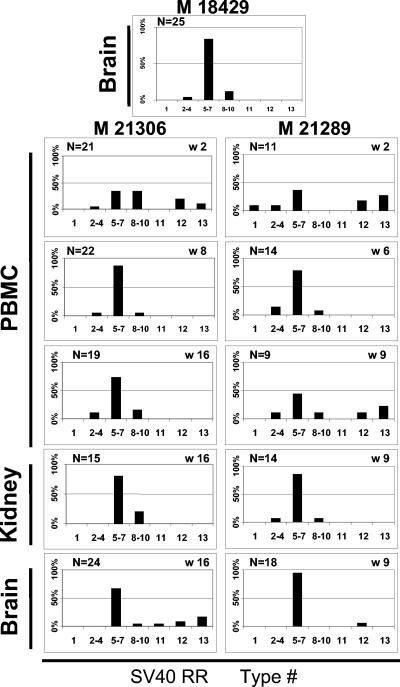
The frequency and distribution of each RR type in various compartments over time are shown in Fig. 2. RR types were classified on the basis of the length of the amplified product. Since the lengths of RR types 5 to 7 are similar and impossible to differentiate on an agarose gel, these were grouped together. This was also the case for RR types 8 to 10. Analysis of the RR from the brain isolate of monkey 18429 showed types 5 to 7 in >80% of the clones (Fig. 2). Type 5 is identical to the archetype RR. Type 6 is the archetype with a variable 19- to 31-bp deletion at nt 274 to 294 in the late promoter, and type 7 is a minor form with an 8-bp deletion in the third 21-bp repeat (Fig. 1). Two weeks after infection of monkeys 21306 and 21289, none of these RR types seemed to be predominant. Five different groups of RR could be readily detected in the PBMCs of each animal, representing all of the 13 types described in Fig. 1, except for type 11. Surprisingly, short RR types 12 and 13 were also well represented in the PBMCs of both animals at this time point. Type 12 lacks one 21-bp repeat and the 72-bp enhancer and has a large deletion in the late promoter, whereas type 13 solely contains a truncated origin of replication and lacks all other elements.
FIG. 2.
Frequency and distribution of SV40 RRs over time in monkeys 21289 and 21306 compared with the SV40 RR obtained from the brain of monkey 18429. The SV40 RR types are numbered 1 to 13 in the order of their sizes as indicated in Fig. 1. Types 5 to 7 and 8 to 10 could not be differentiated by size and are considered together. N indicates the numbers of positive clones analyzed from each sample. W is the number of weeks post SV40 infection. Samples of the brains and kidneys of monkeys 21289 and 21306 were only available at the time of sacrifice. Types 5 to 7 are the most common forms of SV40 RR in the brain of monkey 18429 and gradually became the predominant types in the blood of monkeys 21289 and 21306 over time, as well as in their kidneys and brains at autopsy. Within this group, the type 5 RR is identical to the archetype SV40 RR.
At 6 and 8 weeks after infection, RR types 5 to 7 became predominant in the PBMCs of both animals (>60% of the total RR variants). As was the case in the brain of monkey 18429, types 2 to 4 and 8 to 10, containing duplications and deletions of the 72-bp enhancer, were also identified in a limited number of clones, while the truncated RRs from types 12 and 13 disappeared. However, these truncated RR types reappeared 3 weeks later in monkey 21289, coincident with the onset of neurological disease. At that time, SV40 RR types 5 to 7 were also the most frequently represented in the brains and kidneys of these animals. As was the case in their PBMCs, minor RR types 2 to 4 and 8 to 10 were also found in their kidneys. However, truncated RR types 12 and 13, which had been initially identified in the PBMCs, appeared within the brain as well. Finally, RR type 11, which lacks one 21-bp repeat and the 72-bp element and has a deletion in the late promoter region, was found for the first time in the brain of one animal (Fig. 2).
Besides the SV40 RR, another variable region has been identified in the C terminus of the T-Ag, which includes the HRD. We sequenced at least two clones per PBMC sample at each time point and from the kidneys and brains of these animals at the time of sacrifice. A single pattern was found in all samples. Analysis of the deduced amino acid sequence showed a duplication of the amino acids SQS and deletion of the amino acids HN compared to the HRD of SV40 776. Some single amino acid mutations were also found in the C terminus of the T-Ag (Fig. 3). Each amino acid mutation has been reported separately in SV40 isolates (8, 16) but never in combination. Our results showed that the HRDs from different compartments of all of the animals studied were identical. In addition, our PCR primers, which amplify a fragment from nt 2573 to nt 3069 of reference strain SV40 776, allowed us to examine the intergenic sequence (nt 2594 to 2690) located between the T and VP1 genes. This region was also identical in all samples and showed a 3-bp deletion starting at nt 2677 compared to SV40 776.
FIG. 3.
HRD sequence of SV40-infected monkeys. Alignment of the amino acid sequences of the SV40 strain 776 T-Ag C terminus and that found in all compartments of monkeys 18429, 21289, and 21306 (mm). These include the HRD (amino acids 682 to 708). Dots indicate identity with SV40 strain 776, and dashes indicate deletions. The numbers correspond to the amino acid sequence of SV40 strain 776.
DISCUSSION
Central to the neurotropism and neurovirulence of JCV in humans are rearrangements of its RR found in the blood, CSF, and brains of patients with PML (14, 18, 21). Understanding whether such mutations also occur in the SV40 RR during induction of a PML-like disease in rhesus monkeys is a crucial step toward the creation of an animal model of PML in this species.
Indeed, SV40 naturally infects rhesus monkeys in the wild, and as is the case with JCV in humans, this virus has not been associated with any disease in immunocompetent hosts. However, molecular events occurring at the time of primary SV40 infection in its natural host and those associated with latency and pathogenesis have never been prospectively studied.
Our data indicate that RR rearrangements were not necessary for reactivation of SV40 and development of PML in monkey 18429, since the archetype SV40 RR was the predominant type in the brain of this animal. The archetype SV40 RR is also the predominant RR of the input virus inoculum from monkey 18429. Of 24 clones screened by restriction digest analysis, 23 were types 5 to 7. Of five clones sequenced, two were type 5 (archetype) and two were type 6. We also found a new type of SV40 RR that contained an insertion between two 72-bp repeats. The insertion consisted of nt 251 to 285 of the late promoter, followed by nt 98 to 108 of the 21-bp repeat. This RR form was never found in any of the animals. In addition, we also identified minor RR types containing deletions and duplications of the 72-bp elements. These include one type of RR identical to that of SV40 prototype strain 776, which has two 72-bp elements. To our knowledge, this is the first time that such an RR was found in naturally infected monkeys, indicating that duplication of the 72-bp element is not necessarily a tissue culture artifact.
Surprisingly, a large variability of RR types was found in the blood of monkeys 21289 and 21306 shortly after primary infection with the isolate from the brain of monkey 18429. These include largely truncated types 12 and 13, which would not be expected to grow in tissue culture since they lack the 72-bp enhancer. In addition, type 13 lacks the three 21-bp repeats, which contain the GC boxes and SP1 binding sites and promote replication of viral DNA. Moreover, this type also lacks most of the origin of replication. Since these truncated types were not detected in the brain of monkey 18429, the possibility that they arose during in vitro culture of this isolate cannot be entirely ruled out. Rearrangements of the SV40 RR have been observed when the virus is grown in transformed cell lines (reviewed in reference 12). For this study, we cultured the 18429 SV40 brain isolate on primary rhesus monkey fetal fibroblasts. According to our experience, SV40 grows better in fetal fibroblast than in other cell lines, and primate lentiviruses do not replicate in these cells, which provides a means of differentiating SV40 from the SHIV given to the animal experimentally. In any event, these truncated RR types cleared from the blood of both monkeys 21289 and 21306 by weeks 6 to 8 after inoculation. However, they were again found at the onset of neurological disease in the blood of monkey 21289 and in the brains of both animals. This finding largely rules out an in vitro culture artifact and indicates that these truncated types actually arise in vivo in immunosuppressed hosts. Whether these minor types spread to the brain at the time of intravenous inoculation with SV40 or only later, concomitant with the onset of neurologic symptoms, cannot be determined by this study. However, this question is largely irrelevant since the predominant RR type in the brain was the archetype, indicating that unlike that of JCV, the SV40 archetype RR is the pathogenic type in both SV40-associated PML and ME.
Another interesting and unexpected finding in this study was the presence of short sequences of chromosomal DNA between partial duplications of the 72-bp enhancer in RR types 1 and 4 (Fig. 1). These sequences may have been acquired by direct recombination with chromosomal DNA. Indeed, it is likely that the nonviral sequences were adjacent to the SV40 integration site in chromosomal DNA and remained attached to the viral genome when it went back to its original episomal form. Our findings indicate that a small chromosomal fragment can become inserted into the RR of the SV40 genome in vivo (25).
Our model differs from most cases of PML in humans, since JCV is thought to be initially acquired by the urine-oral route during childhood and remains quiescent in the kidneys thereafter. In the setting of immunosuppression, JCV can be found at higher frequency in the blood (10) and may seed the CNS via the hematogenous route. Therefore, intravenous inoculation of SV40 in our model reproduces this viremic phase. Furthermore, as is the case in humans who become immunosuppressed after being infected with JCV, reactivation of endogenous SV40 is rare and has only been observed in 2.6% of simian immunodeficiency virus-infected animals after a long period of latency (20), which would clearly not be adequate for an animal model. Our model more resembles cases of PML occurring in immunosuppressed children in the context of primary JCV infection. In one such case, multiple JCV archetype RR variants were present in different tissues (17). Nevertheless, regardless of the route of infection, and of the timing of SHIV immunosuppression with regard to SV40 infection, the archetype SV40 RR appears to be the pathogenic type associated with CNS disease in immunosuppressed monkeys, including both those with PML and those with ME. Our results are consistent with those of other studies. Archetype or archetype-like sequences containing variable deletions in the late promoter region (type 6, Fig. 1) have been reported by other investigators in the brains and kidneys of simian immunodeficiency virus-infected monkeys with or without SV40-associated ME or PML, as well as in the kidneys of an immunocompetent animal (8, 11, 16, 20). Although the transcription factor binding sites in the RRs of JCV and SV40 differ, this model will allow us to study the immune response to SV40, test medications, and devise immunotherapeutic interventions for this incurable disease.
In contrast to the high variability of the RR, the T-Ag C terminus, containing the HRD, was conserved in all of the compartments of these three animals at all time points. We detected few mutations, compared to prototype strain 776, corresponding to changes in the deduced amino acid sequence. Most of these mutations had been previously reported separately in both the kidneys and brains of monkeys with SV40-associated ME or PML and in the kidneys of an immunocompetent monkey, although never in the exact combination found in our animals (8, 11, 16, 20). These results indicate that the T-Ag C terminus is not subject to mutations after primary SV40 infection of immunosuppressed animals and does not contain determinants of SV40 neurotropism.
These data shed new light on the mechanisms of brain disease induction by SV40 in its natural host. The availability of this animal model of PML in a species closely related to humans will be a tremendous asset for studying disease pathogenesis and testing therapeutic inventions for this incurable disease.
Acknowledgments
This study was supported by NIH grant R21 046243 and development pilot project grant P30-AI28691 to I.J.K., NIH grant U24 018107 to M.K.A., and NIH grant P51 RR00163 supporting the Oregon National Primate Research Center.
REFERENCES
- 1.Astrom, K. E., E. L. Mancall, and E. P. Richardson, Jr. 1958. Progressive multifocal leuko-encephalopathy; a hitherto unrecognized complication of chronic lymphatic leukaemia and Hodgkin's disease. Brain 81:93-111. [DOI] [PubMed] [Google Scholar]
- 2.Axthelm, M. K., I. J. Koralnik, X. Dang, C. Wuthrich, D. Rohne, I. E. Stillman, and N. L. Letvin. 2004. Meningoencephalitis and demyelination are pathologic manifestations of primary polyomavirus infection in immunosuppressed rhesus monkeys. J. Neuropathol. Exp. Neurol. 63:750-758. [DOI] [PubMed] [Google Scholar]
- 3.Daniel, A. M., J. J. Swenson, R. P. Mayreddy, K. Khalili, and R. J. Frisque. 1996. Sequences within the early and late promoters of archetype JC virus restrict viral DNA replication and infectivity. Virology 216:90-101. [DOI] [PubMed] [Google Scholar]
- 4.Frisque, R. J., G. L. Bream, and M. T. Cannella. 1984. Human polyomavirus JC virus genome. J. Virol. 51:458-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gribble, D. H., C. C. Haden, L. W. Schwartz, and R. V. Henrickson. 1975. Spontaneous progressive multifocal leukoencephalopathy (PML) in macaques. Nature 254:602-604. [DOI] [PubMed] [Google Scholar]
- 6.Holmberg, C. A., D. H. Gribble, K. K. Takemoto, P. M. Howley, C. Espana, and B. I. Osburn. 1977. Isolation of simian virus 40 from rhesus monkeys (Macaca mulatta) with spontaneous progressive multifocal leukoencephalopathy. J. Infect. Dis. 136:593-596. [DOI] [PubMed] [Google Scholar]
- 7.Horvath, C. J., M. A. Simon, D. J. Bergsagel, D. R. Pauley, N. W. King, R. L. Garcea, and D. J. Ringler. 1992. Simian virus 40-induced disease in rhesus monkeys with simian acquired immunodeficiency syndrome. Am. J. Pathol. 140:1431-1440. [PMC free article] [PubMed] [Google Scholar]
- 8.Ilyinskii, P. O., M. D. Daniel, C. J. Horvath, and R. C. Desrosiers. 1992. Genetic analysis of simian virus 40 from brains and kidneys of macaque monkeys. J. Virol. 66:6353-6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King, N. W., R. D. Hunt, and N. L. Letvin. 1983. Histopathologic changes in macaques with an acquired immunodeficiency syndrome (AIDS). Am. J. Pathol. 113:382-388. [PMC free article] [PubMed] [Google Scholar]
- 10.Koralnik, I. J., D. Boden, V. X. Mai, C. I. Lord, and N. L. Letvin. 1999. JC virus DNA load in patients with and without progressive multifocal leukoencephalopathy. Neurology 52:253-260. [DOI] [PubMed] [Google Scholar]
- 11.Lednicky, J. A., A. S. Arrington, A. R. Stewart, X. M. Dai, C. Wong, S. Jafar, M. Murphey-Corb, and J. S. Butel. 1998. Natural isolates of simian virus 40 from immunocompromised monkeys display extensive genetic heterogeneity: new implications for polyomavirus disease. J. Virol. 72:3980-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lednicky, J. A., and J. S. Butel. 1997. Tissue culture adaptation of natural isolates of simian virus 40: changes occur in viral regulatory region but not in carboxy-terminal domain of large T-antigen. J. Gen. Virol. 78(Pt. 7):1697-1705. [DOI] [PubMed] [Google Scholar]
- 13.Lednicky, J. A., R. L. Garcea, D. J. Bergsagel, and J. S. Butel. 1995. Natural simian virus 40 strains are present in human choroid plexus and ependymoma tumors. Virology 212:710-717. [DOI] [PubMed] [Google Scholar]
- 14.Major, E. O., K. Amemiya, C. S. Tornatore, S. A. Houff, and J. R. Berger. 1992. Pathogenesis and molecular biology of progressive multifocal leukoencephalopathy, the JC virus-induced demyelinating disease of the human brain. Clin. Microbiol. Rev. 5:49-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin, J. D., D. M. King, J. M. Slauch, and R. J. Frisque. 1985. Differences in regulatory sequences of naturally occurring JC virus variants. J. Virol. 53:306-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman, J. S., G. B. Baskin, and R. J. Frisque. 1998. Identification of SV40 in brain, kidney and urine of healthy and SIV-infected rhesus monkeys. J. Neurovirol. 4:394-406. [DOI] [PubMed] [Google Scholar]
- 17.Newman, J. T., and R. J. Frisque. 1997. Detection of archetype and rearranged variants of JC virus in multiple tissues from a pediatric PML patient. J. Med. Virol. 52:243-252. [DOI] [PubMed] [Google Scholar]
- 18.Pfister, L. A., N. L. Letvin, and I. J. Koralnik. 2001. JC virus regulatory region tandem repeats in plasma and central nervous system isolates correlate with poor clinical outcome in patients with progressive multifocal leukoencephalopathy. J. Virol. 75:5672-5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reimann, K. A., J. T. Li, R. Veazey, M. Halloran, I.-W. Park, G. B. Karlsson, J. Sodroski, and N. L. Letvin. 1996. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J. Virol. 70:6922-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simon, M. A., P. O. Ilyinskii, G. B. Baskin, H. Y. Knight, D. R. Pauley, and A. A. Lackner. 1999. Association of simian virus 40 with a central nervous system lesion distinct from progressive multifocal leukoencephalopathy in macaques with AIDS. Am. J. Pathol. 154:437-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tornatore, C., J. R. Berger, S. A. Houff, B. Curfman, K. Meyers, D. Winfield, and E. O. Major. 1992. Detection of JC virus DNA in peripheral lymphocytes from patients with and without progressive multifocal leukoencephalopathy. Ann. Neurol. 31:454-462. [DOI] [PubMed] [Google Scholar]
- 22.Vacante, D. A., R. Traub, and E. O. Major. 1989. Extension of JC virus host range to monkey cells by insertion of a simian virus 40 enhancer into the JC virus regulatory region. Virology 170:353-361. [DOI] [PubMed] [Google Scholar]
- 23.Walker, D. L., B. L. Padgett, G. M. ZuRhein, A. E. Albert, and R. F. Marsh. 1973. Human papovavirus (JC): induction of brain tumors in hamsters. Science 181:674-676. [DOI] [PubMed] [Google Scholar]
- 24.Yogo, Y., T. Kitamura, C. Sugimoto, T. Ueki, Y. Aso, K. Hara, and F. Taguchi. 1990. Isolation of a possible archetypal JC virus DNA sequence from nonimmunocompromised individuals. J. Virol. 64:3139-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, S., and G. Magnusson. 2003. Cellular mobile genetic elements in the regulatory region of the pneumotropic mouse polyomavirus genome: structure and function in viral gene expression and DNA replication. J. Virol. 77:3477-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]