Abstract
We have identified the membrane-active regions of the severe acute respiratory syndrome coronavirus (SARS CoV) spike glycoprotein by determining the effect on model membrane integrity of a 16/18-mer SARS CoV spike glycoprotein peptide library. By monitoring the effect of this peptide library on membrane leakage in model membranes, we have identified three regions on the SARS CoV spike glycoprotein with membrane-interacting capabilities: region 1, located immediately upstream of heptad repeat 1 (HR1) and suggested to be the fusion peptide; region 2, located between HR1 and HR2, which would be analogous to the loop domain of human immunodeficiency virus type 1; and region 3, which would correspond to the pretransmembrane region. The identification of these membrane-active regions, which are capable of modifying the biophysical properties of phospholipid membranes, supports their direct role in SARS CoV-mediated membrane fusion, as well as facilitating the future development of SARS CoV entry inhibitors.
An infectious disease, designated severe acute respiratory syndrome (SARS), broke out in China in late 2002 and quickly spread to several countries. The infectious agent responsible for this epidemic outbreak was identified as a previously unknown member of the family of coronaviruses (CoV), SARS CoV (10, 21, 30, 31). Its phylogenetic analysis showed that it was neither a mutant nor a recombinant of previously characterized CoV (35). These viruses are a diverse group of enveloped, positive-strand RNA viruses, with three or four proteins embedded in the envelope, that cause respiratory and enteric diseases in humans and other animals (10, 21, 30, 31). CoV infection, similarly to other envelope viruses, is achieved through fusion of the lipid bilayer of the viral envelope with the host cell membrane.
The fusion of viral and cellular membranes, the critical early events in viral infection, are mediated by envelope glycoproteins located on the outer surfaces of the viral membranes (11, 17). SARS CoV membrane fusion is mediated by the viral spike glycoprotein located on the viral envelope, which is synthesized as a 180-kDa precursor and displayed in ∼200 copies on the viral membrane in a trimeric or dimeric structure (10, 21, 30, 35, 47). In some CoV strains, the spike glycoprotein is cleaved by a protease to yield two noncovalently associated subunits, S1 and S2 (Fig. 1A), which have different functions (16, 40). However, cleavage is not an absolute requirement for the mechanism of fusion, and the available data suggest that the SARS CoV spike glycoprotein is not cleaved into two subunits (9, 18, 35). S1, which forms the globular portion of the spike, contains the receptor-binding site and thus defines the host range of the virus (42), while S2, more conserved than S1, forms the membrane-anchored stalk region and mediates the fusion between the viral and cellular membranes (35, 47).
FIG. 1.
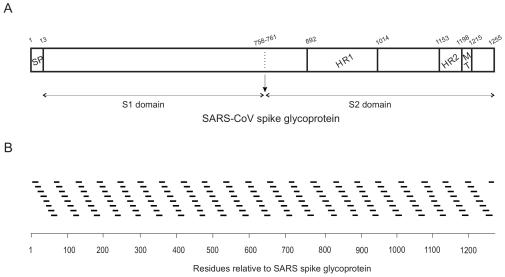
(A) Scheme of the structure of SARS-CoV spike glycoprotein S (amino acid residues 1 to 1255 for the full length), as well as the S1 and S2 domains, according to literature consensus. Boundaries between the different domains cannot be accurately identified at present but are indicated for the convenience of comparison with other coronaviruses. The relevant functional regions are highlighted: the N-terminal signal peptide (SP), the transmembrane domain (TM) and the predicted heptad repeat regions pertaining to the S2 domain, HR1 and HR2. (B) Sequence of the 169 16/18-mer peptides used in this study with respect to the sequence of the whole spike glycoprotein S. Maximum overlap between adjacent peptides is 10 amino acids.
S2 contains two predicted α-helical heptad repeat (HR) domains (HR1 and HR2) which form coiled-coil structures (5, 6, 20, 22, 35, 43, 50). These regions, separated by a stretch of 140 amino acid residues called the interhelical domain, are thought to play important roles in defining the oligomeric structure of the spike protein in its native state and its fusogenic ability (23). The presence of the HR regions, in conjunction with recent studies, indicates that CoV spike proteins can be classified as class 1 viral fusion proteins (5, 6, 20, 22, 43, 50). In the current paradigm of virus-host cell membrane fusion for class 1 viral fusion proteins, the HR domains form a six-helix bundle, where three HR1 helices fold into a central parallel triple-stranded α-helical coiled coil, and wrapped antiparallel on the outside of this core is an outer layer of three antiparallel HR2 α-helices, each HR1-HR2 pair connected by a loop that reverses the polypeptide chain (5, 15, 17, 20, 22, 43, 50). The HR1 and HR2 regions are believed to be important domains in this process and show different conformations in different fusion states (11, 49). Under the current model, there are at least three conformational states of the envelope fusion protein, the prefusion native state, the prehairpin intermediate state, and the postfusion hairpin state (11, 49). This trimeric helical hairpin structure is thought to form at a late stage during the membrane fusion process (15, 17). Formation of the six-helix coiled-coil bundle brings into close proximity the fusion peptide (FP) and the pretransmembrane (PTM) and transmembrane (TM) domains, thereby driving the viral and host cell membranes into close contact, making possible the formation of the fusion pore (11, 15, 17). In class 1 viral fusion proteins, the FP invariably occurs upstream of the HR1 region; however, no FP has been experimentally identified in any CoV spike protein, although a hydrophobic region has been predicted recently at the N terminus of the HR1 region (5).
Although much information has been gathered in recent years, we do not yet know the exact mechanism of membrane fusion and the processes which are behind it. The mechanism by which proteins facilitate the formation of fusion intermediates is a complex process involving several segments of fusion proteins (15, 32). These regions, either directly or indirectly, might interact with biological membranes, contributing to the viral envelope and cell membrane merging. Even though the detailed structures of different segments of the SARS CoV spike glycoprotein have been elucidated, there are still many questions to be answered regarding its mode of action in accelerating membrane fusion. Moreover, SARS CoV entry is an attractive target for anti-SARS therapy. To investigate the structural basis of SARS CoV membrane fusion and identify new fusion inhibitors, we carried out the analysis of the different regions of the SARS CoV spike glycoprotein that might interact with phospholipid membranes, using an approach similar to that used for studying the human immunodeficiency virus (HIV) gp41 ectodomain (25), i.e., the identification of membrane-active regions of SARS CoV spike glycoprotein by determining the effect on membrane integrity of a 16/18-mer spike glycoprotein-derived peptide library. By monitoring the effect of this peptide library on membrane integrity, i.e., leakage, we have identified different regions on the SARS CoV spike glycoprotein with membrane-interacting capabilities, which supports their direct role in membrane fusion and therefore might help in understanding the molecular mechanism of membrane merging, as well as making possible the future development of SARS entry inhibitors, which may lead to new vaccine strategies.
MATERIALS AND METHODS
Materials and reagents.
Egg l-α-phosphatidylcholine (EPC), egg sphingomyelin (SM), and cholesterol (Chol), were obtained from Avanti Polar Lipids (Alabaster, Ala.). 5-Carboxyfluorescein (CF) (>95% by high-performance liquid chromatography) was from Sigma-Aldrich (Madrid, Spain). A set of 169 peptides 16 or 18 amino acids in length derived from the SARS spike glycoprotein, with 10-amino-acid overlap between sequential peptides, was obtained through the AIDS Research and Reference Reagent Program (Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Md.). Porcine lungs were obtained from a local slaughterhouse. Plasma membranes from lung tissue pneumocytes were obtained according to the method of Müller et al. (26), and lipid extraction from porcine lungs was performed according to the procedure of Bligh and Dyer using a ratio of 1:1:0.9 (vol/vol/vol) between chloroform-methanol and the corresponding aqueous sample (4). All other reagents used were of analytical grade and were obtained from Merck (Darmstad, Germany). Water was deionized, distilled twice, and passed through a Milli-Q apparatus (Millipore Ibérica, Madrid, Spain) to a resistivity better than 18 MΩ/cm.
Sample preparation.
Aliquots containing the appropriate amount of lipid in chloroform-methanol (2:1 [vol/vol]) were placed in a test tube, the solvents were removed by evaporation under a stream of O2-free nitrogen, and finally, traces of solvents were eliminated under vacuum in the dark for >3 h. After that, 1 ml of buffer containing 10 mM Tris, 20 mM NaCl, pH 7.4, and CF at a concentration of 40 mM was added, and multilamellar vesicles were obtained. Large unilamellar vesicles (LUV) with a mean diameter of 90 nm were prepared from multilamellar vesicles by the extrusion method (19), using polycarbonate filters with a pore size of 0.1 μm (Nuclepore Corp., Cambridge, Calif.). Breakdown of the vesicle membrane leads to leakage of the contents, i.e., CF fluorescence. Nonencapsulated CF was separated from the vesicle suspension through a Sephadex G-75 filtration column (Pharmacia, Uppsala, Sweden) eluted with buffer containing 10 mM Tris-0.1 M NaCl-1 mM EDTA, pH 7.4.
Leakage measurement.
Leakage of intraliposomal CF was assayed by treating the probe-loaded liposomes (final lipid concentration, 0.125 mM) with the appropriate amounts of peptide on microtiter plates using a microplate reader (FLUOstar; BMG Labtech, Offenburg, Germany), each well containing a final volume of 170 μl stabilized at 25°C. The medium in the microtiter plates was continuously stirred to allow the rapid mixing of peptide and vesicles. Leakage was measured at approximate peptide-to-lipid ratios of 1:15, 1:10, and 1:5. Changes in fluorescence intensity were recorded with excitation and emission wavelengths set at 492 and 517 nm, respectively. One hundred percent release was achieved by adding Triton X-100 to the microtiter plates to a final concentration of 0.5% (wt/wt). Fluorescence measurements were made initially with probe-loaded liposomes, then by adding peptide solution, and finally by adding Triton X-100 to obtain 100% leakage. Leakage was quantified on a percentage basis according to the following equation: % release = (Ff − F0)/(F100 − F0) × 100, where Ff is the equilibrium value of fluorescence after peptide addition, F0 is the initial fluorescence of the vesicle suspension, and F100 is the fluorescence value after the addition of Triton X-100. The phospholipid concentration was measured by methods described previously (7).
Hydrophobic moments, hydrophobicity, and interfaciality.
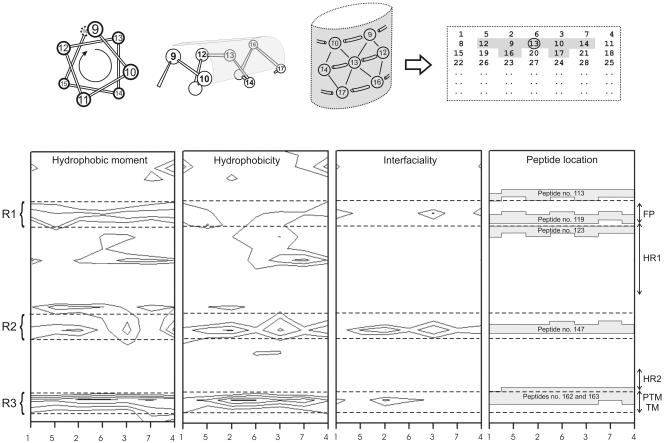
The hydrophobic-moment calculations were carried out according to the method of Eisenberg et al. (12, 13), and the scale for calculating hydrophobic moments was taken from Engelman et al. (14). Hydrophobicity and interfacial values, i.e., whole residue scales for the transfer of an amino acid of an unfolded chain into the membrane hydrocarbon palisade and the membrane interface, respectively, were obtained from http://blanco.biomol.uci.edu/hydrophobicity_scales.html (45, 46). Two-dimensional plots of the hydrophobic moments, hydrophobicity, and interfaciality were obtained using a window of seven amino acids, taking into consideration the arrangement of the amino acids in the space and assuming an α-helical structure (see Fig. 5). Each specific value in the two-dimensional plot represents the mean of the values pertaining to the hydrophobic moment, hydrophobicity, and interfaciality of the amino acid at that position and its neighbors. Positive values correspond to positive bilayer-to-water transfer free-energy values, and therefore, the higher the value, the greater the probability to interact with the membrane surface and/or the hydrophobic core (45, 46).
FIG. 5.
(Top) Helical-wheel diagram indicating the amino acids which lie on the surface of a helix and their representation on a two-dimensional array to identify an amino acid position as a surface-interacting point. Each value in the two-dimensional plot represents the mean of the values pertaining to the hydrophobic moment, hydrophobicity, and interfaciality of the amino acid at that position plus those of its neighbors (window, seven amino acids). (Bottom) Hydrophobic moment, hydrophobicity, interfaciality distribution, and relative positions of peptides 113, 119, 123, 147, 162, and 163 along the SARS CoV spike S2 domain, assuming it forms an α-helical wheel, as shown above, and its correlation with the S2 different functional regions. Only positive bilayer-to-water transfer free-energy values are depicted (solid lines). The dashed lines identify three different domains (R1 to R3) with highly positive values along the helix. The suggested FP, heptad repeats HR1 and HR2, and the PTM and TM domains are indicated (see the text for details). Column numbers define amino acid positions as in the diagram above, upper row.
RESULTS
The SARS CoV spike glycoprotein consists of an extracellular domain, a TM domain, and an intracellular domain (Fig. 1A), and it can be classified as a class I viral fusion protein (5, 6, 20, 22, 43, 50). Although it was previously thought that the FP of class I proteins was the only factor responsible for the cell membrane interaction leading to membrane fusion, it has been shown recently that, apart from the FP, other regions of viral fusion proteins bind and interact with membranes and experience conformational changes which all combine to make possible the fusion of the viral and cell membranes (8, 15, 25, 32). To explore the structural basis of SARS CoV membrane fusion, we have carried out the analysis of the different regions of the full SARS CoV spike glycoprotein that might interact with phospholipid membranes by using a peptide library derived from the SARS CoV spike glycoprotein. This peptide library was composed of 169 peptides 16 and 18 amino acids in length, and by using an approach similar to that used for studying the HIV gp41 ectodomain (25), we studied their effects on the release of an encapsulated fluorophore, CF, using the experimental setup described in Materials and Methods. The 169 peptides we used in this study and their correlation with the SARS CoV spike glycoprotein sequence are shown in Fig. 1B. As can be observed, the 16/18-mer peptide library encompasses the whole sequence of the SARS CoV spike glycoprotein, i.e., from the signal peptide to the sequence that follows the TM domain, including the recently identified HR1 and HR2 domains.
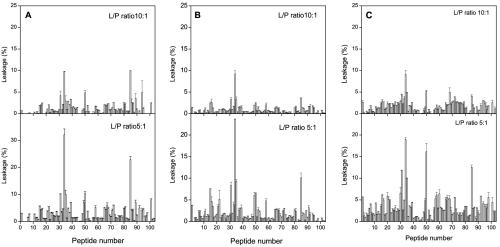
The S1 domain of the SARS CoV spike glycoprotein forms the globular portion of the spike and mediates binding to the host cells (16, 40, 42), being the receptor-binding domain localized in the amino acid sequence 318 to 510 (9, 47). Although the proposed role for the S1 domain is the attachment of the protein to its functional receptor, angiotensin-converting enzyme 2, several hydrophobic patches have been identified, which might be important not only for protein-protein binding but also for membrane interaction, since, as was mentioned above, S1 probably remains covalently attached to the S2 domain during the fusion process (9, 34, 35, 47). Figure 2A and B shows the effect of the S1 domain-derived 16/18-mer peptides on membrane integrity, i.e., leakage, at different peptide-to-lipid ratios and two different liposome compositions, namely, EPC-SM-Chol at a molar ratio of 70:15:15 and EPC-Chol at a molar ratio of 5:1. For liposomes composed of EPC-SM-Chol (Fig. 2A), it is clearly evident that some peptides exerted hardly any effect, but other peptides showed notable effects on liposome leakage in comparison. The most notable effects were observed for peptides 34 and 85, which produced leakage of ∼33 and 22%, respectively, at the highest peptide-to-lipid ratio used. Similar to what was found for EPC-SM-Chol liposomes, the same peptides, i.e., peptides 34 and 85, were the ones which interacted with liposomes composed of EPC-Chol (Fig. 2B). For EPC-Chol liposomes, the leakage values for peptides 34 and 85 were ∼22 and 10%, i.e., slightly lower than those found for EPC-SM-Chol liposomes. The other peptides from the S1 domain, apart from peptides 34 and 85, exerted no significant effect on liposome leakage.
FIG. 2.
Effects of the S1 domain derived 16/18-mer peptides on the release of LUV contents for different lipid compositions. Leakage data for LUV composed of (A) EPC-SM-Chol at a phospholipid molar ratio of 70:15:15, (B) EPC-Chol at a phospholipid molar ratio of 5:1, and (C) a lipidic extract of lung tissue pneumocytes at different peptide (P)-to-lipid (L) ratios as indicated. Experimental conditions are described in the text. The error bars indicate standard deviations of the mean for quadruplicate samples.
In order to compare leakage values obtained in phospholipid model membranes with those obtained in biologically derived model membranes, we also studied liposome leakage produced by peptides derived from the S1 domain of the SARS CoV spike glycoprotein in model membranes whose lipids were obtained from a lipidic extract of lung tissue pneumocytes (Fig. 2C). It can be observed that the extent of leakage was slightly reduced compared to the EPC-SM-Chol and EPC-Chol liposome systems mentioned above. However, patterns of leakage similar to those described above can be discerned, since peptides 34 and 85 again showed relatively high leakage values compared to the leakage values exhibited by the other peptides pertaining to the S1 domain. It is worth noting that, for this liposome composition, peptide 50 also showed a similar leakage value.
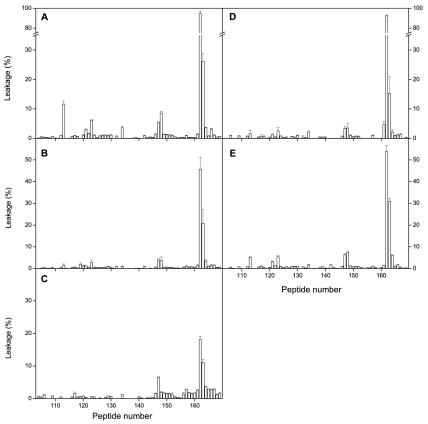
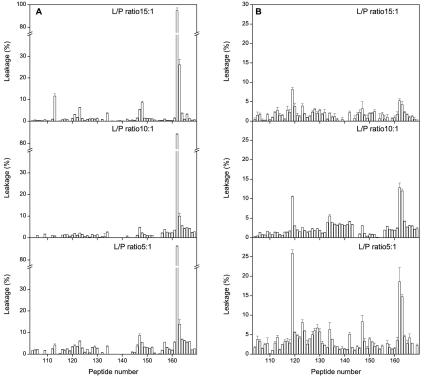
We studied the interaction of the 16/18-mer S2 spike glycoprotein library with LUV model membranes having variable Chol and SM compositions, namely, EPC-SM-Chol at molar ratios of 70:15:15, 52:18:30, and 37:18:45 (Fig. 3A, B, and C, respectively). These model membranes, showing the coexistence of liquid-ordered and liquid-disordered phases, have a high probability of raft formation (2); as has been documented, the presence of laterally segregated membrane microdomains or lipid rafts is important for membrane fusion (1, 3, 28, 33, 37, 38, 44). For liposomes composed of EPC-SM-Chol at a molar ratio of 70:15:15 (Fig. 3A), it can be clearly observed that, whereas the vast majority of peptides exerted hardly any effect on membrane leakage, peptide 162 showed a dramatic effect, i.e., ca. 100% leakage for peptide (compare the ordinate scales of Fig. 3A with those in Fig. 2). Other peptides that showed a significant effect, although not as high as peptide 162, were peptide 163 and, to a lesser extent, peptides 113, 123, 147, and 148. It is worth noting that peptide 163 did have a significant effect, not as great as that of peptide 162 but higher than the leakage values observed for peptides 113, 123, 147, and 148 (Fig. 3A). Similar to what was found for liposomes containing EPC-SM-Chol at a molar ratio of 70:15:15, some peptides interacted with liposomes composed of EPC-SM-Chol at molar ratios of 52:18:30 and 37:18:45 (Fig. 3B and C). The most notable effects were again observed for peptides 162 and 163, the former presenting the higher leakage values. However, the leakage values were reduced compared to those observed for liposomes containing EPC-SM-Chol at a molar ratio of 70:15:15. The major difference in the compositions of these liposomes is the PC-Chol ratio. In EPC-SM-Chol at molar ratios of 70:15:15, 52:18:30,and 37:18:45, the proportion of SM remains the same, whereas the PC-Chol ratio decreases, i.e., leakage values decrease as Chol contents increase. When liposomes composed of PC-SM at a molar ratio of 5:1 were studied, peptides 162 and 163 were again the ones which presented the most significant extents of leakage (Fig. 3D). Similar results were obtained for liposomes composed of PC-Chol at a molar ratio of 5:1 (Fig. 3E). Other peptides that showed positive leakage values, although not as significant as peptides 162 and 163, were peptides 113, 123, 147, and 148, similar to what was found earlier. However, it is interesting that EPC-Chol liposomes presented a lower extent of leakage than EPC-SM liposomes, stressing the fact that, as mentioned above, the Chol content might be responsible for the lower extent of leakage rather than the presence of both SM and Chol.
FIG. 3.
Effects of the S2 domain-derived 16/18-mer peptides on the release of LUV contents for different lipid compositions at a peptide-to-lipid ratio of 1:15. Leakage data for LUV composed of (A) EPC-SM-Chol at a molar ratio of 70:15:15, (B) EPC-SM-Chol at a molar ratio of 52:18:30, (C) EPC-SM-Chol at a molar ratio of 37:18:45, (D) EPC-SM at a molar ratio of 5:1, and (E) EPC-Chol at a molar ratio of 5:1. The error bars indicate standard deviations of the mean of quadruplicate samples.
We also studied the effects of different lipid-to-peptide ratios on leakage for liposomes composed of EPC-SM-Chol at a phospholipid molar ratio of 70:15:15 (Fig. 4A). It is clearly evident that peptide 162 was again the one which showed a dramatic effect, since nearly complete rupture of the liposomes was observed (ca. 100% leakage for peptide) at all lipid-to-peptide ratios tested. It is worth noting that, even at the lowest lipid-to-peptide ratio used, peptide 163 was the only one, apart from peptide 162, that had a relatively significant effect, although not as great as that of peptide 162 (Fig. 4A). As described above, other peptides that showed positive leakage values, although not as significant as peptide 162 or even peptide 163, were peptides 113, 123, 147, and 148.
FIG. 4.
Effects of the S2 domain-derived 16/18-mer peptides on the release of LUV contents for (A) EPC-SM-Chol at a phospholipid-molar ratio of 70:15:15 and (B) lipidic extract of lung tissue pneumocytes at different peptide (P)-to-lipid (L) ratios as indicated. The error bars indicate standard deviations of the mean for quadruplicate samples.
In order to compare leakage values observed in model membranes with those obtained in biologically derived model membranes, we studied liposome leakage produced by peptides derived from the S2 domain in model membranes whose lipids were obtained from a lipidic extract of lung tissue pneumocytes at different lipid-to-peptide ratios (Fig. 4B; note the increase in leakage scale compared with Fig. 4A). It can be observed that the extent of leakage was greatly reduced compared to the model systems studied before. However, two patterns of leakage can be discerned, i.e., regions defined by peptide 119 and by peptides 162 and 163 (Fig. 4B).
DISCUSSION
Recent studies point out that CoV spike envelope glycoproteins can be classified as class I viral fusion proteins, and functional and biochemical analyses of the SARS CoV spike glycoprotein show that SARS CoV uses a membrane fusion mechanism that is similar to that of class I viral fusion proteins (5, 6, 20, 22, 43, 50). The SARS CoV spike glycoprotein consists of an extracellular domain, a TM domain, and an intracellular domain (Fig. 1A). SARS CoV spike glycoprotein does not contain a typical proteolytic cleavage site, and the boundary between the S1 and S2 domains cannot be accurately identified at present (22). In addition, the available data suggest that the SARS CoV spike glycoprotein is not cleaved into two subunits, so most likely the S1 and S2 domains remain covalently bonded during the fusion process (9, 35). A signal peptide at the amino terminus has been recognized, and analogous to other class I viral fusion proteins, two α-helical heptad repeat domains, HR1 and HR2, have been identified (Fig. 1A). Although no obvious fusogenic peptide sequence has been experimentally identified, it is clear that viral and cellular membrane fusion is mediated by an internal FP in the SARS CoV spike glycoprotein. Nevertheless, a hydrophobic region located upstream of the HR1 region has been recently predicted to be the FP (5).
Several lines of evidence indicate that, in addition to classical FPs, different regions of viral envelope glycoproteins are essential for membrane fusion to occur. In the case of the HIV type 1 (HIV-1) gp41 ectodomain, we have recently shown that, apart from the FP, three different regions of this protein, namely the 15- to 20-residue peptide segment that follows the FP at the N terminus, the immunodominant loop, and the 10- to 15-residue peptide segment that precedes the TM domain at the C terminus, i.e., the PTM domain, are capable of modifying the biophysical properties of phospholipid membranes, suggesting a direct role in membrane fusion (25). In the case of the SARS CoV spike glycoprotein, both the S1 and S2 subunits probably remain attached to each other during the fusion process; however, the most probable role for the S1 subunit is the attachment to its specific receptor, whereas the S2 subunit is responsible for the fusion between the viral and cellular membranes (35, 47). As mentioned above, peptides 34, 50, and 85, which belong to the S1 domain, showed relatively significant membranotropic activity (Fig. 2). Similar to the HIV gp120 inner-domain regions interacting with gp41 (48), the regions of the S1 subunit where these peptides reside could associate with the S2 domain by specific hydrophobic interactions before the S1 subunit itself interacts with its receptors. As described above, these hydrophobic patches might be important not only for protein-protein binding but also for membrane interaction, since S1 probably remains covalently attached to the S2 domain during the fusion process. In fact, we have found that several peptides derived from the HIV gp120 region suggested to associate with its receptors have a significant effect on membrane leakage (unpublished data).
Hydrophobic moments measure the periodicity of residue distribution along a secondary-structure element (12, 13). The preferential orientation of the hydrophobic moments toward one face of that element (Fig. 5, top) has been proposed to favor hydrophobic interactions between proteins and/or between proteins and membranes. In order to detect surfaces along the S2 domain of the SARS CoV spike glycoprotein that might be identified as membrane-partitioning and/or membrane-interacting zones, we plotted the average surface hydrophobic moments, hydrophobicity, and interfaciality versus the S2 domain amino acid sequence, assuming it adopts an α-helical structure along the whole sequence (Fig. 5, bottom). It is readily evident that there are three different regions with high positive values covering the surface of the helix and along it: one of these regions is located immediately upstream of the HR1 region (region 1 [R1]), another one is located between the heptad repeats HR1 and HR2 (R2), and the last is located at the end of the S2 domain sequence (R3). The last region partially matches the proposed TM domain of the spike glycoprotein. These regions, having positive bilayer-to-water transfer free-energy values, might show a tendency to partition into and interact with membrane surfaces or proteins. The adoption of a specific conformation can generate rich hydrophobic surfaces along the structure and emphasize that the actual distribution of hydrophobicity and interfaciality, i.e., structure-related factors, along the S2 domain sequence might affect the biological function of these sequences (36).
As mentioned above, peptides 113, 119, 123, 147, and 162-163 were the peptides that exerted the most significant effect on model membranes, with peptide 162 the one that showed the most dramatic effects, since nearly 100% leakage was observed in many of the different compositions studied (Fig. 3 and 4). The extents of leakage observed for the other peptides were not as high as for peptide 162, but they were significantly higher than for the rest of the S2-derived peptides tested. Interestingly, peptide 119 showed the most important effect on pneumocyte-derived model membranes. We do not know the lipid compositions of these model membranes, but they must be very complex; nevertheless, two regions, defined by peptides 119 and 162-163, can be discerned (three if we count peptide 147 [Fig. 4B]). Actually, some peptides that are very active in model membrane systems are much less active in the biological membrane system, and the contrary is also true. However, it should be noted that membrane systems having different lipid compositions have diverse properties which make them behave in significantly different ways (24). What is outstanding is that peptides 113 (ARDLICAQKFNGLTVL828-843) and 123 (VLYENQKQIANQFNKAI897-913) are located at the boundaries of R1, peptide 119 (GAALQIPFAMQMAYRF873-888) is located inside R1, peptide 147 (FVFNGTSWFITQRNFF1077-1092) is located inside R2, and peptides 162 and 163 (LGKYEQYIKWPWYVWLGF1185-1202 and KWPWYVWLGFIAGLIAIV1193-1210) are located inside R3; all these regions represent surfaces with high bilayer-to-water transfer free-energy values (Fig. 5, bottom).
As mentioned above, the FP, a stretch of hydrophobic amino acids, is an essential factor in viral fusion proteins; however, it has not been identified, although it was recently predicted that the sequence comprising residues 858 to 886 from the S2 domain of the SARS CoV spike glycoprotein is indeed the FP domain (5). Peptide 119, which has a significant experimental membranotropic effect, as shown here, belongs to R1, as depicted in Fig. 5, bottom, and comprises residues 873 to 888 of the S2 domain of the SARS CoV spike glycoprotein. Since this peptide overlaps the theoretically predicted FP sequence 858 to 886 (5), the sequence is therefore a candidate to be the FP domain of the SARS CoV spike glycoprotein. Inspection of the sequence where this peptide is located reveals that it has a high content of alanine and glycine, characteristic of viral fusion peptides (15), and a high degree of interfaciality and hydrophobicity, essential properties for membrane-interacting sequences in proteins (Fig. 6). Its location immediately upstream of HR1, as found for other class I viral fusion proteins, is appropriate, since the formation of the six-helix coiled-coil bundle would bring into close proximity the FP and the PTM and TM domains (11, 15, 17).
FIG. 6.
Analysis of the interfacial and hydrophobic distribution using a window of 15 amino acids according to the scales of Wimley and White (46) along the proposed FP, PTM, and TM sequences of the SARS CoV spike glycoprotein S2 domain. Values above the 0 level correspond to positive bilayer-to-water transfer free-energy values.
Another fundamental structural and functional sequence for a class I viral fusion mechanism is the PTM, and one of the most thoroughly studied PTM sequences is that belonging to HIV-1 gp41 (27, 39, 41). The PTM is a region immediately adjacent to the membrane-spanning domain of class I fusion proteins containing highly conserved hydrophobic residues and unusually rich in tryptophan residues. In addition, the PTM domain of HIV-1 gp41 shows a significant tendency to partition into membranes and is highly fusogenic (41). Peptides 162 and 163 from the SARS CoV S2 domain exert a dramatic effect on leakage for different model membranes (Fig. 3 and 4), and a careful inspection of the sequences reveals that they have characteristics similar to those of the PTM domain of HIV-1 gp41, notably, high tryptophan content and an interfacial stretch immediately following a hydrophobic one (Fig. 6). It is likely, then, that peptides 162 and 163 comprise the PTM domain of the SARS CoV spike glycoprotein S2 subunit and, similar to the PTM domain of HIV-1 gp41, this sequence of the SARS CoV S2 subunit might be involved in the promotion of the membrane destabilization required for fusion, as well as in fusion pore formation and enlargement (32, 36).
Not only are the FP and PTM domains of HIV-1 gp41 essential for the perturbation of the membrane, but also, other regions of the protein interact and destabilize the viral and host membranes; this is what has been suggested for the HIV-1 gp41 immunodominant loop, which could play an essential role in the viral fusion process (8, 25, 29). One of the main characteristics of the gp41 loop domain, located between the HR1 and HR2 regions and instrumental in the formation of the gp41 central parallel triple-stranded α-helical coiled coil, is its tendency to partition into and interact with membrane surfaces (8, 25, 29). We have found a highly hydrophobic and interfacial domain flanked by SARS CoV HR1 and HR2 domains (R2) (Fig. 5, bottom), and remarkably, peptide 147, which has shown a notable leakage effect, is located in it. Therefore, R2 could play a role similar to that of the HIV-1 gp41 loop domain, i.e., destabilization of both viral and cellular membranes, favoring the formation of the fusion pore as well as its stabilization and therefore facilitating, along with the other regions, membrane fusion.
In conclusion, our results demonstrate that peptides originating from three different regions of the SARS CoV spike glycoprotein (Fig. 5, bottom)—R1, which corresponds to the 15- to 20-residue peptide segment immediately upstream of HR1; R2, which corresponds to a sequence between HR1 and HR2; and R3, which corresponds to the 10- to 15-residue peptide segment that immediately precedes the TM domain at the C terminus—are capable of modifying the biophysical properties of phospholipid membranes, a property which could provide an additional driving force for the merging of the viral and target cell membranes, suggesting that they have a direct role in SARS CoV-mediated membrane fusion and therefore might be necessary for assistance and enhancement of the viral and cell fusion process. These results should also facilitate the development of SARS CoV entry inhibitors, which might lead to new vaccine strategies, an important focus for clinical intervention.
Acknowledgments
This work was supported by grant BMC2002-00158 from MCYT, Madrid, Spain (J.V.). M. R. Moreno and A. J. Pérez-Berná are recipients of predoctoral fellowships from Ministerio de Educación y Ciencia and Generalidad Valenciana, Spain, respectively.
We are especially grateful to Maite Garzón for her excellent assistance, as well as to the National Institutes of Health AIDS Research and Reference Reagent Program for making available the peptides used in this work.
REFERENCES
- 1.Alfsen, A., and M. Bomsel. 2002. HIV-1 gp41 envelope residues 650-685 exposed on native virus acs as a lectin to bind epithelial cell galactosyl ceramide. J. Biol. Chem. 277:25649-25659. [DOI] [PubMed] [Google Scholar]
- 2.Almeida, R. F., A. Fedorov, and M. Prieto. 2003. Sphingomyelin-phosphatidylcholine-cholesterol phase diagram: boundaries and composition of lipid rafts. Biophys. J. 85:2406-2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn, A., D. L. Gibbons, and M. Kielian. 2002. The fusion peptide of Semliki Forest virus associates with sterol-rich membrane domain. J. Virol. 76:3267-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 3:911-917. [DOI] [PubMed] [Google Scholar]
- 5.Bosch, B. J., B. E. Martina, R. Van Der Zee, J. Lepault, B. J. Haijema, C. Versluis, A. J. Heck, R. De Groot, A. D. Osterhaus, and P. J. Rottier. 2004. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. USA 101:8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosch, B. J., R. van der Zee, C. A. de Haan, and P. J. Rottier. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77:8801-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Böttcher, C. S. F., C. M. Van Gent, and C. Fries. 1961. A rapid and sensitive submicro phosphorous determination. Anal. Chim. Acta 1061:297-303. [Google Scholar]
- 8.Contreras, L. M., F. J. Aranda, F. Gavilanes, J. M. González-Ros, and J. Villalaín. 2001. Structure and interaction with membrane model systems of a peptide derived from the major epitope region of HIV protein gp41. Implications on viral fusion mechanism. Biochemistry 40:3196-3207. [DOI] [PubMed] [Google Scholar]
- 9.Dimitrov, D. S. 2003. The secret life of ACE2 as a receptor for the SARS virus. Cell 115:652-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 11.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 12.Eisenberg, D., E. Schwarz, M. Komaromy, and R. Wall. 1984. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 179:125-142. [DOI] [PubMed] [Google Scholar]
- 13.Eisenberg, D., R. M. Weiss, and T. C. Terwilliger. 1982. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature 299:371-374. [DOI] [PubMed] [Google Scholar]
- 14.Engelman, D. M., T. A. Steitz, and A. Goldman. 1986. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu. Rev. Biophys. Chem. 15:321-353. [DOI] [PubMed] [Google Scholar]
- 15.Epand, R. M. 2003. Fusion peptides and the mechanism of viral fusion. Biochim. Biophys. Acta 1614:116-121. [DOI] [PubMed] [Google Scholar]
- 16.Frana, M. F., J. N. Behnke, L. S. Sturman, and K. V. Holmes. 1985. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell fusion. J. Virol. 56:912-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallo, S. A., C. M. Finnegan, M. Viard, Y. F. Raviv, A. Dimitrov, S. S. Rawat, A. Puri, S. Durell, and R. Blumenthal. 2003. The HIV Env-mediated fusion reaction. Biochim. Biophys. Acta 1614:36-50. [DOI] [PubMed] [Google Scholar]
- 18.de Haan, C. A., K. Stadler, G. J. Godeke, B. J. Bosch, and P. J. Rottier. 2004. Cleavage inhibition of the murine coronavirus spike protein by a furin-like enzyme affects cell-cell but not virus-cell fusion. J. Virol. 78:6048-6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hope, M. J., M. B. Bally, G. Webb, and P. R. Cullis. 1985. Production of large unilamellar vesicles by a rapid extrusion procedure. Characterization of size distribution, trapped volume and ability to maintain a membrane potential. Biochim. Biophys. Acta 812:55-65. [DOI] [PubMed] [Google Scholar]
- 20.Ingallinella, P., E. Bianchi, M. Finotto, G. Cantoni, D. M. Eckert, V. M. Supekar, C. Bruckmann, A. Carfi, and A. Pessi. 2004. Structural characterization of the fusion-active complex of severe acute respiratory syndrome (SARS) coronavirus. Proc. Natl. Acad. Sci. USA 101:8709-8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 22.Liu, S., G. Xiao, Y. Chen, Y. He, J. Niu, C. R. Escalante, H. Xiong, J. Farmar, A. K. Debnath, P. Tien, and S. Jiang. 2004. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet 363:938-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo, Z., A. M. Matthews, and S. R. Weiss. 1999. Amino acid substitutions within the leucine zipper domain of the murine coronavirus spike protein cause defects in oligomerization and the ability to induce cell-to-cell fusion. J. Virol. 73:8152-8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McIntosh, T. J. 2004. The 2004 Biophysical Society-Avanti Award in Lipids: roles of bilayer structure and elastic properties in peptide localization in membranes. Chem. Phys. Lipids 130:83-98. [DOI] [PubMed] [Google Scholar]
- 25.Moreno, M. R., R. Pascual, and J. Villalaín. 2004. Identification of membrane-active regions of the HIV-1 envelope glycoprotein gp41 using a 15-mer gp41-peptide scan. Biochim. Biophys. Acta 1661:97-105. [DOI] [PubMed] [Google Scholar]
- 26.Müller, M., C. Meijer, G. J. Zaman, P. Borst, R. J. Scheper, N. H. Mulder, E. G. de Vries, and P. L. Jansen. 1994. Overexpression of the gene encoding the multidrug resistance-associated protein results in increased ATP-dependent glutathione S-conjugate transport. Proc. Natl. Acad. Sci. USA 91:13033-13037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muñoz-Barroso, I., K. Salzwedel, E. Hunter, and R. Blumenthal. 1999. Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J. Virol. 73:6089-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen, D. H., and J. E. K. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pascual, R., M. R. Moreno, and J. Villalaín. A peptide pertaining to the loop segment of HIV gp41 binds and interacts with model biomembranes. Implications for the fusion mechanism. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 30.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peiris, J. S., K. Y. Yuen, A. D. Osterhaus, and K. Stohr. 2003. The severe acute respiratory syndrome. N. Engl. J. Med. 349:2431-2441. [DOI] [PubMed] [Google Scholar]
- 32.Peisajovich, S. G., and Y. Shai. 2003. Viral fusion proteins: multiple regions contribute to membrane fusion. Biochim. Biophys. Acta 1614:122-129. [DOI] [PubMed] [Google Scholar]
- 33.Phalen, T., and M. Kielian. 1991. Cholesterol is required for infection by Semliki Forest Virus. J. Cell Biol. 112:615-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prabakaran, P., X. Xiao, and D. S. Dimitrov. 2004. A model of the ACE2 structure and function as a SARS-CoV receptor. Biochem. Biophys. Res. Commun. 314:235-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 36.Sáez-Cirión, A., J. L. Arrondo, M. J. Gomara, M. Lorizate, I. Iloro, G. Melikyan, and J. L. Nieva. 2003. Structural and functional roles of HIV-1 gp41 pretransmembrane sequence segmentation. Biophys. J. 85:3769-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sáez-Cirión, A., M. J. Gómara, A. Agirre, and J. L. Nieva. 2003. Pre-transmembrane sequence of Ebola glycoprotein interfacial hydrophobicity distribution and interaction with membranes. FEBS Lett. 533:47-53. [DOI] [PubMed] [Google Scholar]
- 38.Sáez-Cirión, A., S. Nir, M. Lorizate, A. Aguirre, A. Cruz, J. Pérez-Gil, and J. L. Nieva. 2002. Sphingomyelin and cholesterol promote HIV-1 gp41 pretransmembrane sequence surface aggregation and membrane restructuring. J. Biol. Chem. 277:21776-21785. [DOI] [PubMed] [Google Scholar]
- 39.Salzwedel, K., J. T. West, and E. Hunter. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 73:2469-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sturman, L. S., C. S. Ricard, and K. V. Holmes. 1985. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: activation of cell-fusing activity of virions by trypsin and separation of two different 90K cleavage fragments. J. Virol. 56:904-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suárez, T., S. Nir, F. M. Goñi, A. Sáez-Cirión, and J. L. Nieva. 2000. The pre-transmembrane region of the human immunodeficiency virus type-1 glycoprotein: a novel fusogenic sequence. FEBS Lett. 477:145-149. [DOI] [PubMed] [Google Scholar]
- 42.Taguchi, F. 1995. The S2 subunit of the murine coronavirus spike protein is not involved in receptor binding. J. Virol. 69:7260-7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tripet, B., M. W. Howard, M. Jobling, R. K. Holmes, K. V. Holmes, and R. S. Hodges. 2004. Structural characterization of the SARS-coronavirus spike S fusion protein core. J. Biol. Chem. 279:20836-20849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vincenta, N., C. Genina, and E. Malvoisina. 2002. Identification of a conserved domain of the HIV-1 transmembrane protein gp41 which interacts with cholesteryl groups. Biochim. Biophys. Acta 1567:157-164. [DOI] [PubMed] [Google Scholar]
- 45.White, S. H., and W. C. Wimley. 1999. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 28:319-365. [DOI] [PubMed] [Google Scholar]
- 46.Wimley, W. C., and S. H. White. 1996. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3:842-848. [DOI] [PubMed] [Google Scholar]
- 47.Xiao, X., S. Chakraborti, A. S. Dimitrov, K. Gramatikoff, and D. S. Dimitrov. 2003. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 312:1159-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.York, J., and J. H. Nunberg. 2004. Role of hydrophobic residues in the central ectodomain of gp41 in maintaining the association between human immunodeficiency virus type 1 envelope glycoprotein subunits gp120 and gp41. J. Virol. 78:4921-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zelus, B. D., J. H. Schickli, D. M. Blau, S. R. Weiss, and K.V. Holmes. 2003. Conformational changes in the spike glycoprotein of murine coronavirus are induced at 37°C either by soluble murine CEACAM1 receptors or by pH 8. J. Virol. 77:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu, J., G. Xiao, Y. Xu, F. Yuan, C. Zheng, Y. Liu., H. Yan, D. K. Cole, J. I. Bell, Z. Rao, P. Tien, and G. F. Gao. 2004. Following the rule: formation of the 6-helix bundle of the fusion core from severe acute respiratory syndrome coronavirus spike protein and identification of potent peptide inhibitors. Biochem. Biophys. Res. Commun. 319:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]