Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) can be driven into the lytic cycle in vitro by phorbol esters and sodium butyrate. This report begins to analyze the process by which butyrate activates the promoter of KSHV open reading frame 50 (ORF50), the key viral regulator of the KSHV latency to lytic cycle switch. A short fragment of the promoter, 134 nucleotides upstream of the translational start of ORF50, retained basal uninduced activity and conferred maximal responsiveness to sodium butyrate. The butyrate response element was mapped to a consensus Sp1-binding site. By means of electrophoretic mobility shift assays, both Sp1 and Sp3 were shown to form complexes in vitro with the ORF50 promoter at the Sp1 site. Butyrate induced the formation of a group of novel complexes, including several Sp3-containing complexes, one Sp1-containing complex, and several other complexes that were not identified with antibodies to Sp1 or Sp3. Formation of all butyrate-induced DNA-protein complexes was mediated by the consensus Sp1 site. In insect and mammalian cell lines, Sp1 significantly activated the ORF50 promoter linked to luciferase. Chromatin immunoprecipitation experiments in a PEL cell line showed that butyrate induced Sp1, CBP, and p300 binding to the ORF50 promoter in vivo in an on-off manner. The results suggest that induction of the KSHV lytic cycle by butyrate is mediated through interactions at the Sp1/Sp3 site located 103 to 112 nucleotides upstream of the translational initiation of ORF50 presumably by enhancing the binding of Sp1 to this site.
The classic form of Kaposi's sarcoma (KS) was first described as “idiopathic multiple pigmented sarcoma” of the skin by Moritz Kaposi in 1872 (73). The causative agent for the disease was identified more than a century later as a DNA virus belonging to the gammaherpesvirus family with substantial sequence similarity to other oncogenic primate gammaherpesviruses, including herpesvirus saimiri, rhesus rhadinovirus, and Epstein-Barr virus (EBV) (27, 46, 54, 67). Although Kaposi's patients were elderly, otherwise healthy, men, KS now occurs primarily in immunosuppressed patients, such as those with AIDS and recipients of organ transplantation (1, 8, 11). KS is also endemic to sub-Saharan Africa (12, 13). KS-associated herpesvirus (KSHV) also causes lymphoproliferative disorders, such as body cavity-based primary effusion lymphomas (PELs) and multicentric Castleman's disease, primarily in patients with AIDS (3, 70).
The KSHV genome contains more than 85 open reading frames (ORF) that include genes encoding homologues of cellular genes, such as cyclin, G protein-coupled receptor, chemokines, and cytokines (9, 50). These viral genes influence many aspects of cellular physiology to regulate cell proliferation, apoptosis, and immunity. Immediately after infection, KSHV establishes a latent state during which only a few KSHV genes are expressed. These include ORF K1 (an integral membrane protein), K12 (kaposin A), K15 (transmembrane proteins), ORF 73 (a latency-associated nuclear antigen), ORF72 (a viral homologue of cyclin D), and ORF K13 (vFLIP, a homolog of a cellular inhibitor of apoptosis) (16, 58, 68, 74, 87). It is not clear what triggers KHSV to switch from latency to lytic replication in vivo; however, chemicals, such as phorbol ester and butyric acid, have induced the KSHV lytic cycle in cultured cells derived from PELs (47, 85). In these cell culture systems, genes expressed during the lytic cycle have been classified into three categories: immediate early, delayed early, and late. Early genes are expressed prior to and late genes are expressed after viral DNA replication. The immediate-early group refers to those genes whose transcripts can be induced without de novo protein synthesis. Several regions giving rise to immediate-early transcripts have been identified in the KSHV genome, including ORF50, ORF 48/29b, ORF45, ORFK4.2, and ORF K8 (66, 86).
Both latent and lytic viral gene products play essential roles in oncogenesis by KSHV. For example, latency-associated nuclear antigen has been shown to interact with the tumor suppressor p53 and inhibit p53 transcriptional activity (21). Although the majority of cells in KSHV-associated tumors are latently infected and exhibit a restricted pattern of viral gene expression, some monocytic cells and spindle cells in KS lesions also express lytic genes (49, 72). Lytic viral replication is vital for viral transmission between cells and among individuals. Ganciclovir, which functions by inhibiting viral DNA replication, markedly reduces KS incidence in human immunodeficiency virus type 1-infected patients (42). KSHV lytic genes may also play important direct roles in KSHV-mediated oncogenesis. For example viral interleukin 6 promotes cell proliferation and prevents apoptosis (28, 43). vFLIP, a latent product, and vBcl-2, a lytic product, may both be important for tumorigenesis and tumor maintenance as they block apoptosis (24).
Although the detailed mechanism of how KSHV undergoes the switch from latency to lytic replication is not fully understood, studies of ORF50 have demonstrated that its product plays a key role in this process. ORF50 (also called KSHV replication and transcription activator, or Rta) encodes a protein of 691 amino acids that shares significant homology with ORF50a of herpesvirus saimiri and Rta (R transactivator) encoded by BRLF1 of EBV (56, 76, 83). The function of these proteins as immediate-early transactivators appears to be conserved among all gammaherpesviruses. ORF50 mRNA is detected within 2 to 4 h after chemical induction of PEL-derived cell lines in the presence of cycloheximide, an inhibitor of protein synthesis (77). ORF50 has been shown to increase the expression of KSHV early and late genes, such as viral interleukin 6, polyadenylated nuclear RNA, and small viral capsid protein, encoded by ORF65; it has also been shown to drive the KSHV lytic cycle into completion in HH-B2 cells (23, 39, 76). These experiments suggest that ORF50 expression is sufficient to activate the entire KSHV lytic replication cascade. A mutant form of ORF50 that lacks the carboxyl terminal transactivation domain acts as a repressor to block KSHV replication induced by wild-type ORF50 or chemicals, suggesting that ORF50 is not only sufficient but required for KSHV lytic induction (38).
ORF50 protein has been shown to stimulate the expression of KSHV lytic genes by several distinct mechanisms. ORF50 activates the expression of polyadenylated nuclear RNA and K12 directly, by binding to specific elements in their promoters (10, 69). ORF50 interacts with RBP-Jκ and, through this interaction, indirectly activates the expression of two other viral genes, mRNA transcript accumulation (ORF57) and major single-stranded-DNA-binding protein (SSB, ORF6) (34).
Most reported studies deal with the question of how the ORF50 protein regulates expression of other viral genes. The essential question of how ORF50 gene expression is regulated remains largely unanswered. Wang et al. have shown that the phorbol ester tetradecanoyl phorbol acetate (TPA) leads to enhanced binding of AP1 and CCAAT/enhancer-binding protein α (C/EBPα) to the ORF50 promoter and rapid phosphorylation of c-JUN (81, 82). It has also been shown that ORF50 activates itself, and octomer-binding protein 1 mediates this autostimulation (65). However, autoactivation can only be used to amplify ORF50 expression and does not provide the mechanism for the initial step of induction of ORF50 expression in response to environmental stimuli. In the studies presented here, we address the question of how ORF50 is regulated by cellular genes. We identified an element in the ORF50 promoter that is essential for its constitutive activity and mediates induction by butyrate. We further showed that this element is a bona fide stimulating protein 1 (Sp1) binding site and demonstrated that Sp1 activated the ORF50 promoter by luciferase reporter assays with insect and human cells. Moreover, we found that, following butyrate treatment of PEL cell lines, Sp1 and its associated coactivators were transiently induced to bind to the ORF50 promoter in vivo.
MATERIALS AND METHODS
Cell lines.
PEL cell lines HH-B2, BC1, and BC3 were maintained in RPMI 1640 supplemented with 15% fetal calf serum, penicillin (50 U/ml), streptomycin (50 μg/ml), and amphotericin B (1 μg/ml). Cells were grown at 37°C under 5% CO2. Drosophila melanogaster Schneider cells (S2) were grown in Schneider's Drosophila media (Invitrogen) supplemented with 10% fetal calf serum and antibiotics (50 U of penicillin/ml, 50 μg of streptomycin) at 26°C. HKB5/B5 cells, a somatic cell hybrid between 293 human embryonic kidney cells and HH514-16 Burkitt lymphoma cells were cultured in RPMI 1640 supplemented with 5% fetal calf serum, penicillin (50 U/ml), streptomycin (50 μg/ml), and amphotericin B (1 μg/ml).
Reagents and antibodies.
Phorbol-12-myristate-13-acetate (PMA) purchased from Sigma was used at 20 ng/ml. Sodium butyrate purchased from Sigma was used at 3 mM. Trichostatin A (TSA) from Wako was used at 5 μM. Antibodies against Sp1, Sp3, E2F-1, Ku-70, CBP, and p300 were purchased from Santa Cruz Biotechnology. Horseradish peroxidase-conjugated goat anti-rabbit antibody was purchased from Amersham. Rabbit polyclonal antibodies against ORF50 were made in our laboratory by using the bacterially expressed carboxyl-terminal half (amino acids 333 to 691) of the ORF50 protein.
Plasmids.
pLuc was made by ligating firefly (Phontinus pyralis) luciferase cDNA into the pcDNA3.1 vector (Invitrogen). Luciferase cDNA was amplified from pGL2 (Promega) by PCR with Pfu polymerase. A BamHI recognition sequence was included in the 5′ primer (GTTGGATCCATGGAAGACGCCAAAAACATAAAG) and an XhoI site was included in the 3′ primer (TGTCTCGAGTTACAATTTGGACTTTCCGCCCTT) to facilitate subcloning of luciferase cDNA into the pcDNA3.1 vector linearized with BamHI and XhoI. The resulting construct (pcDNA-Luc) was digested with BglII and HindIII to delete a 900-bp fragment that includes the cytomegalovirus promoter and enhancer sequences, the ends were filled, and the plasmid was religated to make the pLuc control reporter. ORF50 promoter sequences of different lengths were amplified from BC1 genomic DNA by PCR with Pfu polymerase. The same 3′ primer (TTGAAGCTTTTTTGTGGCTGCCTGGACAGTAT) and different 5′ primers were employed (2,500 bp, ACAAGATCTATCCACACTAAACGCCAGGCCAG; 950 bp, TCAAGATCTCTGCCCATGGGCGGGTGGGTG; 134 bp, AAAAGATCTGCAGCTACCGGCGACTCATTAAG; 115 bp, GCGAGATCTTAAGCCCCGCCCAGAAACCAGT; 95 bp, CAGAGATCTGTAGCTGGGTGGCAATGACACG; 75 bp, GCAAGATCTCGTCCCCTTTAAAAAGTCAACCTT; 69 bp, ACAAGATCTCTTTAAAAAGTCAACCTTACTCCG; 60 bp, TTTAGATCTGTCAACCTTACTCCGCAAGGGG). A BglII recognition sequence was included in the 5′ primer, and a HindIII site was included in the 3′ primer. PCR products were digested with BglII and HindIII and ligated into the pcDNA-Luc vector linearized with the same enzymes to generate the p2500Luc, p950Luc, p134Luc, p115Luc, p95Luc, p75Luc, p69Luc, and p60Luc reporters, respectively. Each plasmid construct was confirmed by restriction digestion and DNA sequencing. Linker insertion mutants of p134Luc shown in Fig. 3A were made with the QuikChange site-directed mutagenesis kit (Stratagene).
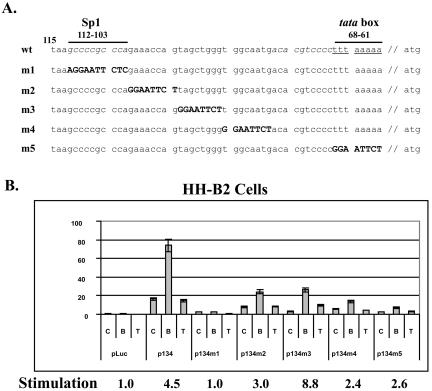
FIG. 3.
An Sp1 site confers butyrate responsiveness to the ORF50 promoter. (A) Schematic diagram of sequence replacement mutants in the ORF50 promoter. Numbers indicate the corresponding positions of nucleotides in the ORF50 promoter map upstream of the translation start site. The putative Sp1 site is located at 103 to 112; the putative TATA box is from 61 to 68. The sequence in boldface type represents linker DNA used to replace various regions in the ORF50 promoter to generate mutant constructs (m1 to m5). The putative Sp1 site is in italics; the putative TATA element is underlined. Wt, wild type; m, mutant. (B) Luciferase assay of ORF50 promoter mutants. HH-B2 cells were transfected and treated with drugs as described in the legend to Fig. 2. C, control (unreated); B, butyrate; T, TSA. The numbers below the bars indicate the induction (n-fold) of reporter activity in butyrate-treated cells compared to untreated HH-B2 cells.
The full-length Sp1 clone was constructed by two-stage PCR from cDNA clones. A PCR product of amino acids 1 to 102 of Sp1 (PCR-A) was generated from plasmid pOTB7 (ATCC 9406919; purchased from the American Type Culture Collection). A second PCR product of amino acids 83 to 779 (PCR-B) was generated from plasmid pEVR2. PCR-A was made by using primers GCGCGAATTCGGATGATGAAATGACAGCTGTG and GATCTGCCAGCCATTGCCACC. PCR-B was generated by using primers ACCAGGTGAGCTTGACCTCACA and GCGCCTCGAGTCAGAAGCCATTGCCACT. Purified PCR-A and PCR-B products were mixed and used as template for a second-step PCR (PCR-C) by using primers CGCGAATTCGGATGGATGAAATGACAGCTGTG and GCGCCTCGAGTCAGAAGCCATTGCCACT. An aliquot of the PCR-C reaction was run on a 0.75% agarose gel. The remaining PCR was phenol-chloroform extracted once, followed by one chloroform extraction. The DNA was ethyl alcohol (EtOH) precipitated, and the pellet was washed once with 70% EtOH and dried. The pellet was resuspended in restriction digest mix and digested with EcoRI and XhoI. The DNA was purified and ligated to the EcoRI/XhoI-digested pET22 vector. A positive clone was confirmed by sequencing. This clone was used as the source of full-length Sp1 for cloning into pCMVHA. A full-length Sp1 PCR product was generated by using primers GCGCGAATTCGGGATGAAATGACAGCTGTG and GCGCCTCGAGTCAGAAGCCATTGCCACT. The product was purified, digested, and ligated into the vector pCMVHA cut with EcoRI/XhoI. Transformants were screened by restriction enzyme digestion, and a positive clone (pCMVHA-Sp1) was confirmed by sequencing. The full-length Sp1 cDNA was in frame with the hemagglutinin tag.
Transfection and luciferase reporter assay.
HH-B2 cells were transfected with 8 μg of DNA by electroporation. Cells were harvested and resuspended in growth media at 3.25 × 107/ml. An aliquot of 0.4 ml of cells (about 1.5 × 107) was transferred into an electroporation cuvette (0.4 cm; Bio-Rad) and was electroporated at 250 V and 960 μF. Cells were then seeded in 15 ml of fresh media and grown for 48 h before being harvested. The transfection efficiency of HH-B2 cells was about 1.8%, as assessed by the green fluorescent protein expression vector pGreenLantern (Invitrogen). This efficiency was highly reproducible in different transfections. Drosophila Schneider cells (S2) were transfected with Lipofectin (Invitrogen) according to the manufacturer's recommended protocol. Cells were resuspended in Schneider's Drosophila medium (Invitrogen) at a concentration of 5 × 106/ml and aliquoted into six-well plates. One microgram of DNA and 6 μl of Lipofectin reagent (2 mg/ml) were diluted into 100 μl of Schneider's Drosophila medium, mixed, and incubated at room temperature for 30 min. The DNA-Lipofectin mixture was added to 1 ml of cells and incubated for 3 to 4 h. Serum was added to a final concentration of 10%. Cells were incubated for 48 h and collected for a luciferase activity assay. HKB5/B5 cells were transfected with DMRIE-C reagent (Invitrogen). Four micrograms of DNA and 12 μl of DMRIE-C reagent (2 mg/ml) were diluted in 0.5 ml of OPTI-MEM1 reduced serum medium (Invitrogen) and mixed, incubated at room temperature for 30 min. HKB5/B5 cells (2 × 106) in 0.2 ml of OPTI-MEM1 medium were added to the mixture in each well of a six-well plate. Cells were incubated at 37°C in a CO2 incubator for 4 h before the addition of serum to a final concentration of 5%. Cells were furthered cultured for 20 h and then harvested for luciferase assay. All transfection and luciferase reporter assays were repeated at least three times.
RNA isolation and RT-PCR.
Total RNA was isolated from 1.5 × 107 HH-B2 cells by using an RNeasy mini kit (Qiagen). Each RNA sample was treated with 30 U of DNase I by using the RNase-free DNase set (Qiagen). RNA was quantified by UV spectrometry. Total RNA (0.5 μg) was used as a template for reverse transcription (RT)-PCR to amplify an ∼350-bp fragment from ORF50 mRNA by using the SuperScript one-step RT-PCR system (Invitrogen). The primer pair (forward, GCAGCCACAAAAATGGCGCAAGATG; reverse, CATCCCAAGGCATTATTCGGATCCT) spans the only intron of the ORF50 gene so that the PCR product could be differentiated from amplifications of residual genomic DNA (∼1.3 kbp). PCR products were resolved by 2% agarose gel electrophoresis. A multi-intron-spanning primer pair (forward, TGAAGGTCGGAGTCAACGGATTTGG; reverse, GACTGTGGTCATGAGTCCTTCCACG) was used to amplify an ∼520-bp fragment from the human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA.
Western blot.
Polypeptides in a cell lysate were resolved on a sodium dodecyl sulfate (SDS)-10% polyacrylamide gel and transferred to a nitrocellulose filter membrane by using the Trans-Blot SD semidry transfer cell (Bio-Rad Laboratories). The membrane was blocked in Tris-buffered saline (TBS; 20 mM Tris-Cl [pH 7.5], 150 mM NaCl) with 5% nonfat milk for 1 h at room temperature. The blocked membrane was then incubated with the first antibody for 1 h at room temperature or overnight at 4°C. The membrane was then washed three times, for 5 min each wash, with TBS plus 0.2% Triton X-100, one time with TBS plus 1% Triton X-100 and 12% urea, and one time with TBS. The membrane was incubated with a 1:20,000 dilution of horseradish peroxidase-conjugated secondary antibody (Amersham) for 1 h, and washes were repeated. Protein bands were detected by enhanced chemiluminescence fluorography.
EMSA.
About 1.5 × 107 HH-B2 cells were collected by low-speed centrifugation (500 × g, 5 min) and washed once with 15 ml of ice-cold phosphate-buffered saline. Cells were incubated in NP-40 lysis buffer (10 mM Tris-Cl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40) for 5 min on ice. Nuclei were collected by low-speed centrifugation (500 × g, 5 min) and washed twice with NP-40 lysis buffer. Nuclear extract was prepared by treatment with high-salt buffer (20 mM HEPES [pH 7.5], 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and 2 μg of aprotinin/ml). The protein concentration of nuclear extract was measured by Bradford assay (Bio-Rad Laboratories). Five micrograms of nuclear extract protein was used per reaction in electrophoretic mobility shift assay (EMSA) experiments. The DNA probes were end-labeled with [γ-32P]ATP and incubated with nuclear extract in DNA-binding buffer (10 mM HEPES [pH 7.5], 50 mM NaCl, 2 mM MgCl2, 2.5 mM ZnSO4, 0.5 mM EDTA, 1 mM dithiothreitol, and 15% glycerol). Five micrograms of poly(dI-dC) was included per reaction to reduce nonspecific DNA binding. When applicable, cold unlabeled duplex oligonucleotide competitors were added before the addition of DNA probe. Supershift antibodies were added 5 min after mixing nuclear extract and the probe. After 25 min at room temperature, the reaction mixture was loaded onto a 6% polyacrylamide gel to resolve the DNA-protein complexes by electrophoresis. After electrophoresis, gels were fixed in 10% methanol and 10% acetic acid and dried onto Whatman 3M filter paper in a gel dryer (Bio-Rad). DNA-protein complexes were visualized by X-ray autoradiography.
ChIP.
HH-B2 cells were treated with 1% formaldehyde for 10 min and then with 125 mM glycine for 5 min at room temperature. Cells were pelleted and washed once with ice-cold phosphate-buffered saline. To prepare chromatin lysate, cells were resuspended in SDS lysis buffer (1% SDS, 50 mM Tris-Cl [pH 8.0], 10 mM EDTA, and 1× protease inhibitor cocktail [Roche Biochemistry]). The lysates were sonicated three times for 10 s to shear chromatin DNA to an average of 200 to 500 bp and centrifuged at ∼10,000 × g for 15 min at 4°C. The supernatant was transferred to a fresh tube and was measured for absorbance at 260 nm. An equivalent of 1 ml of supernatant at an optical density at 260 nm of 1 was diluted in chromatin immunoprecipitation (ChIP) dilution buffer (0.1% SDS, 1% Triton X-100, 15 mM Tris-Cl [pH 8.0], 1 mM EDTA, 150 mM NaCl, and 1× protease inhibitor cocktail). To preclear the lysate, 5 μl of protein G agarose beads was added to the diluted chromatin lysate and incubated for 1 h at 4°C in a rotator. The protein G beads were removed by brief centrifugation. Ten micrograms of control immunoglobulin G or specific antibodies and 5 μl of protein G beads was added to the precleared lysate and incubated overnight at 4°C in a rotator. Immune complexes (antibody-protein-DNA) were washed sequentially with the following buffers: buffer 1, 0.1% SDS, 1% Triton X-100, 20 mM Tris-Cl (pH 8.0), 2 mM EDTA, 150 mM NaCl, and 0.1 μg of aprotinin/ml; buffer 2, 0.1% SDS, 1% Triton X-100, 20 mM Tris-Cl (pH 8.0), 2 mM EDTA, 500 mM NaCl, and 0.1 μg of aprotinin/ml; buffer 3, 250 mM LiCl, 1% NP-40, 1% sodium deoxycholate, 10 mM Tris-Cl (pH 8.0), and 1 mM EDTA; buffer 4, 10 mM Tris-Cl (pH 8.0) and 1 mM EDTA. Immune complexes were then eluted with 250 μl of 1% SDS and 100 mM NaHCO3 for 10 min at 65°C. NaCl was added to the eluate to a final concentration of 200 mM, and the eluate was incubated at 65°C for 5 h to reverse cross-links. Protein and RNA were digested with proteinase K (0.2 mg/ml) and RNase A (0.2 mg/ml) for 30 min at 37°C. DNA was extracted sequentially with phenol, phenol-chloroform, and chloroform and precipitated with 2 volumes of EtOH at −20°C overnight. The DNA pellet was collected by centrifugation, washed with 70% EtOH, and dissolved in 50 μl of sterile double-distilled H2O. Two microliters of DNA solution was used for PCR analysis to detect an ORF50 promoter-specific sequence (5′ primer, GCAGCTACCGGCGACTCATTAAG; 3′ primer, TGCCTGGACAGTATTCTCACAACAG; product size, 125 bp) or a control DNA fragment from the ORF K8 coding region (5′ primer, GCTCGCTGTTGTCAACCTACGTAG; 3′ primer, GTGTGGTCCTCTTGGTGGTGTGTG; product size, 197 bp).
RESULTS
Sodium butyrate reproducibly induces ORF50 gene expression.
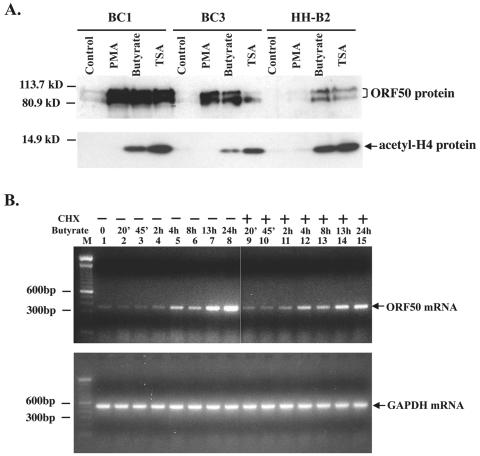
Sodium butyrate and TSA potently activated KSHV ORF50 protein expression in three PEL cell lines (Fig. 1A). Both drugs are inhibitors of histone deacetylase and lead to hyperacetylation of the tails of histones H4 in these cell lines (Fig. 1A). However, TSA, which was a more potent inducer of H4 hyperacetylation, was a less effective activator of ORF50 protein expression. In some PEL cell backgrounds, such as BC-1 and BC-3, phorbol esters were as potent as histone deacetylase inhibitors in inducing ORF50 protein expression, but in others, such as HH-B2 cells, phorbol esters were minimally active. Phorbol esters did not cause hyperacetylation of H4 (Fig. 1A). Thus, treatment with butyrate allows exploration of the lytic cycle induction pathway upstream of ORF50 gene expression in several KSHV-infected PEL cell lines.
FIG. 1.
Butyrate induces ORF50 expression. (A) ORF50 protein expression in three PEL cell lines treated with PMA, butyrate, or TSA. Western blot analysis shows the expression of ORF50 protein in BC1, BC3, and HH-B2 cells 24 h after chemical treatment. The same immunoblot was probed with antibody to acetylated histone H4. (B) Kinetics of induction of ORF50 mRNA expression by butyrate. HH-B2 cells were treated with butyrate in the presence (+) or absence (−) of cycloheximide; cells were harvested at intervals. ORF50 mRNA was detected by an RT-PCR assay. Lanes: M, DNA length marker; 1, untreated samples; 2 to 8, butyrate-treated samples; 9 to 15, butyrate- and cycloheximide (CHX)-treated samples; 2 and 9, 20 min; 3 and 10, 45 min; 4 and 11, 2 h; 5 and 12, 4 h; 6 and 13, 8 h; 7 and 14, 13 h; 8 and 15, 24 h.
Within 4 h after treatment with butyrate, there was a reproducible increase in ORF50 mRNA abundance detectable by an RT-PCR assay with HH-B2 cells (Fig. 1B). This increase in ORF50 mRNA was not inhibited by cycloheximide. By 13 and 24 h after butyrate treatment, there was a further increase in the abundance of ORF50 mRNA which was slightly, but not completely, dampened by cycloheximide treatment. Thus, the initial phase of ORF50 gene expression induced by butyrate occurs rapidly and is independent of de novo protein synthesis. A later phase, possibly due to autostimulation, involves protein synthesis.
Identification of a butyrate-responsive element in the promoter of ORF50.
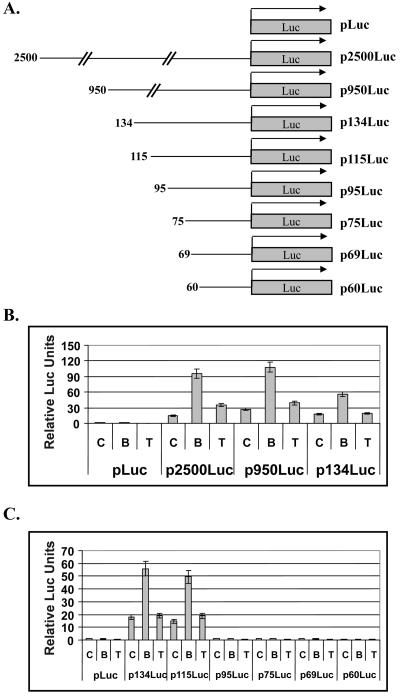
By means of a series of ORF50 promoter truncations linked to a firefly luciferase reporter, a butyrate-responsive region of the ORF50 promoter was found to be located between 95 and 115 nucleotides (nt) upstream of the ATG start codon of ORF50 (Fig. 2B and C). Deletions downstream of 115 nt reduced basal ORF50 promoter activity and eliminated its responsiveness to butyrate (Fig. 2C). The response of ORF50-luc to butyrate was observed in several KSHV-positive and KSHV-negative B cell lines (data not shown). While treatment with TSA induced expression of ORF50 protein (Fig. 1A) and ORF50 mRNA (data not shown), in KSHV-infected HH-B2 cells, TSA exerted only minimal effects on expression of the ORF50p-luciferase reporters (Fig. 2B and C).
FIG. 2.
Identification of the butyrate-responsive region of the ORF50 promoter. (A) Schematic diagram of ORF50 promoter-luciferase reporter constructs. Numbers indicate the length of the ORF50 promoter in base pairs upstream of the ATG translation initiation codon. (B and C) Luciferase reporter assays. HH-B2 cells (1.5 × 107) were transfected by electroporation with equal molar amounts of the indicated luciferase constructs. Transfected cells were incubated at 37°C for 24 h under 5% CO2 before the addition of the indicated drugs. Cells were harvested 24 h after drug treatment and analyzed for luciferase activity. C, control (untreated); B, butyrate; T, TSA.
A putative Sp1 binding site is required for butyrate responsiveness of the ORF50 promoter.
The butyrate-responsive clones ORF50p134luc and ORF50p115luc contain a putative Sp1 response element located 103 to 112 nt upstream of the ATG of ORF50 (−103 to −112). To determine whether this Sp1 site mediated the response to butyrate, a series of linker-scanning insertion mutants were placed in the ORF50p134Luc construct (Fig. 3). Spontaneous transcriptional activity was decreased in all five linker-scanning mutants in the region from −61 to −112; however, only one mutant, ORF50p134m1, lost responsiveness to butyrate. This mutant destroyed the Sp1 site. The other four linker-scanning mutants retained a 2.4-fold to 8.8-fold response to butyrate. Replacement of the putative TATA box effectively annulled the spontaneous activity of the ORF50 promoter, but this mutant, ORF50p134m5, still maintained a weak response to butyrate (Fig. 3B). This result indicated that the putative TATA box is required for the ORF50 promoter to be fully active, but TATA is not required for the response to butyrate.
Sp1 markedly activates the ORF50 promoter in insect and mammalian cells.
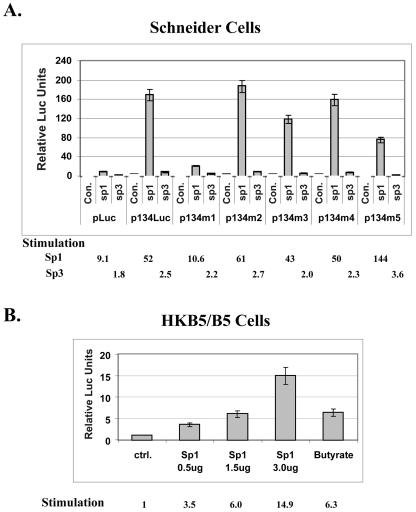
The next experiments were designed to determine whether Sp1 was competent to activate the ORF50 promoter and to determine whether the Sp1 site within the ORF50 promoter that had been identified as the butyrate response element was responsive to Sp1. The reporter ORF50p134luc and its mutants were transfected into Drosophila Schneider 2 cells, where endogenous Sp1 activity is absent (Fig. 4A). Cotransfection of Sp1 strongly stimulated ORF50p/luc activity (52-fold). Transfection of Sp3 exerted only minor effects. The response to Sp1 of mutant p134m1, in which the Sp1 site was replaced with linker DNA, was reduced fivefold. The other mutants still responded strongly to Sp1, including mutant p134m5, in which the putative TATA element was replaced by a linker. These experiments confirmed the functional importance of the Sp1 site in the ORF50 promoter and showed that the response to Sp1 was independent of the TATA element. A eukaryotic vector that overexpressed Sp1 transfected into HKB5/B5 human cells also stimulated the ORF50p/luc reporter in a dose-dependent manner (Fig. 4B). The maximal dose of Sp1 expression plasmid used stimulated the reporter 14.9-fold above the background; butyrate stimulated the reporter 6.3-fold in the same experiment.
FIG. 4.
Sp1 activates the ORF50 promoter in Drosophila Schneider 2 cells and HKB5/B5 cells. (A) Schneider cells were transfected with equal molar amounts of ORF50 promoter luciferase reporter (p134Luc) or mutant reporters (p134m1 to p134m5) in the presence or absence of an Sp1 or Sp3 expression construct. p134m1 to p134m5 contain the same mutations shown in Fig. 3A. Cells were harvested 2 days after transfection and assayed for luciferase activity. Con, control (transfected with salmon sperm carrier DNA). (B) HKB/B5 cells were transfected with p2500Luc and different amounts of pCMVHA-Sp1. Cells were harvested 1 day after transfection and assayed for luciferase activity.
Sp1 and Sp3 form complexes with ORF50 promoter in vitro.
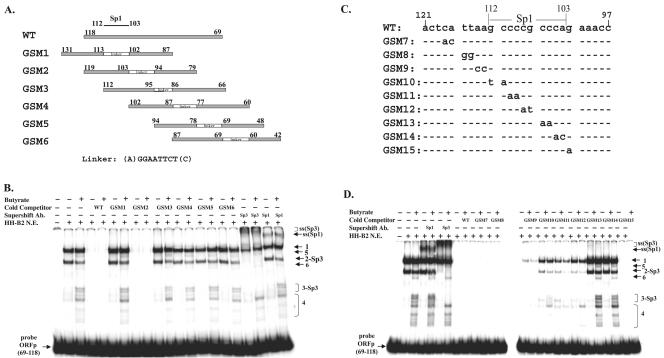
To determine whether Sp1 and Sp3 bound ORF50 promoter DNA in vitro, a 50-bp DNA fragment encompassing nt 69 to 118 upstream of the ORF50 ATG was used as a probe in an EMSA (Fig. 5). Nuclear extracts of HH-B2 cells that were untreated or treated with butyrate were tested for DNA binding activity (Fig. 5B). Two abundant DNA-protein complexes (designated 1 and 2) were observed in untreated cells. Complex 2 reproducibly decreased in abundance in butyrate-treated cells. Several less-abundant complexes (designated 3, 4, 5, and 6) appeared only in nuclear extracts from butyrate-treated cells. On the basis of supershifts with antibody to Sp3, complexes 2 and 3 were identified as containing Sp3 (Fig. 5B, lanes 18 and 19). Antibody to Sp1 also induced a supershift of complexes formed on untreated and butyrate-treated cells, possibly derived from complex 1 (Fig. 5B, lanes 20 and 21). Anti-Sp1 also eliminated complex 5, which was specific to butyrate-treated cells (Fig. 5B, lane 21, and D, lane 5). Antibodies to Ku-70 and E2F1 did not produce any supershift or eliminate any complexes (data not shown).
FIG. 5.
Sp1 and Sp3 bind to the ORF50 promoter in vitro. (A) Schematic diagram showing the region of ORF50 promoter sequences used as wild-type (WT) and linker-scanning mutant competitors in the EMSA illustrated in panel B. The numbers indicate the positions of the nucleotides upstream of the ATG site. For example, mutant 1 (GSM1) contains sequences ranging from 87 to 102 and 113 to 131. (B) EMSA with nuclear extracts (N.E.) from HH-B2 cells that were untreated or treated with sodium butyrate for 24 h. Lanes 4 to 17 represent competitions with a 50-fold excess of cold oligonucleotides. Lanes 18 to 21 represent supershifts (ss) with antibodies (Ab) to Sp3 and Sp1. (C) DNA sequences of point mutant competitors used in the EMSA illustrated in panel D. (D) EMSA with nuclear extracts of untreated or butyrate-treated HH-B2 cells. Lanes 4 to 7, supershifts with antibodies to Sp1 and Sp3; lanes 8 to 27, competition with a 50-fold molar excess of wild-type or mutant oligonucleotide from the ORF50 promoter (−69 to −118). +, present; −, absent.
Duplex oligonucleotides containing linker insertions and encompassing nt −42 to −118 of ORF50p were used as cold competitors in an EMSA (Fig. 5A and B). The only mutant oligonucleotide that competed effectively (GSM2) contained an Sp1 site and flanking sequences. Oligonucleotides lacking an Sp1 site, either as the result of linker replacement or deletion, i.e., GSM1, GSM4, GSM5, and GSM6, failed to compete for any of the complexes. Mutant GSM3, which contained an Sp1 site without flanking sequences, also did not compete. These experiments showed that Sp1 and Sp3 bound to the ORF50 promoter in vitro and that novel Sp1- and Sp3-related complexes appeared after butyrate treatment.
Effect of point mutants on ORF50p complex formation.
The next experiment used point mutagenesis to determine whether the Sp1 site in ORF50p was required for formation of all the observed complexes. To address this question, a 25-bp DNA fragment from ORF50p (encompassing nt 97 to 121 upstream of ORF50 ATG) was used as a cold competitor for formation of complexes on ORF50p (−69 to −118). Nine mutants (GSM7 to GSM15) were generated from the competitor fragment by replacing 1 or 2 nt (Fig. 5C). The 25-bp (−97 to −121) competitor completely inhibited all probe-protein complex formation (Fig. 5D, lanes 8 and 9). Thus, all complexes observed on the probe from −69 to −118 are attributable to sequences from −97 to −118. Mutants GSM7, GSM8, and GSM15, containing nucleotide substitutions outside the Sp1 site, were as active competitors as duplex oligonucleotide from −97 to −121. Thus, nt 115 to 118 and 102 upstream of the ATG are not important in mediating DNA-protein complex formation. All of the remaining substitutions in the competitor oligonucleotide, from 2 nt upstream of the Sp1 site (GSM9) and including the Sp1 site, impaired competition but to varying degrees. Substitution of four nucleotides in the 3′ end of the Sp1 site abolished binding of all complexes, indicating that the last 4 nt, CCCA, are most important for formation of the Sp1 site in the ORF50p. Substitution of 6 nt in the 5′ end of the Sp1 site reduced but did not eliminate binding of all complexes. Mutant GSM9, with two nucleotide substitutions 5′ of the Sp1 site (113 to 114), was slightly compromised in the competition assay. Some of complex 1 remained, but all of the other complexes were eliminated. These results strongly suggested that the Sp1 consensus site was responsible for all of the protein-DNA complexes formed on the region from −69 to −118 of ORF50p.
Expression of Sp1 and Sp3 in untreated and butyrate-treated cells.
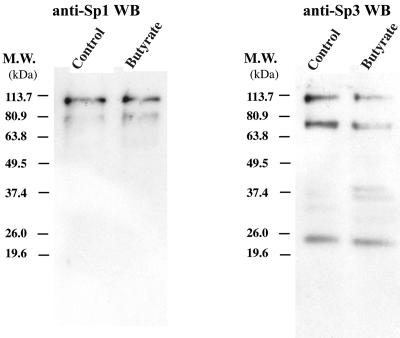
The EMSA experiments suggested that new complexes related to Sp1 and Sp3 were formed following butyrate treatment of HH-B2 cells. A Western blot with antibody to Sp1 detected two forms of Sp1, ∼80 and 110 kDa, of approximately equal abundance in butyrate-treated and untreated cells (Fig. 6). Antibody to Sp3 detected three forms: 110, 70, and 23 kDa in untreated and butyrate-treated cells (Fig. 6). In butyrate-treated cells, Sp3 antibody revealed two new polypeptides of ∼36 and 40 kDa (Fig. 6).
FIG. 6.
Butyrate does not alter the abundance of Sp1 or Sp3. Immunoblots of extracts of untreated or butyrate-treated HH-B2 cells reacted with antibody to Sp1 and Sp3 are shown. M.W., molecular mass.
Butyrate induces dynamic binding of Sp1, CBP, and p300 to the ORF50 promoter in vivo.
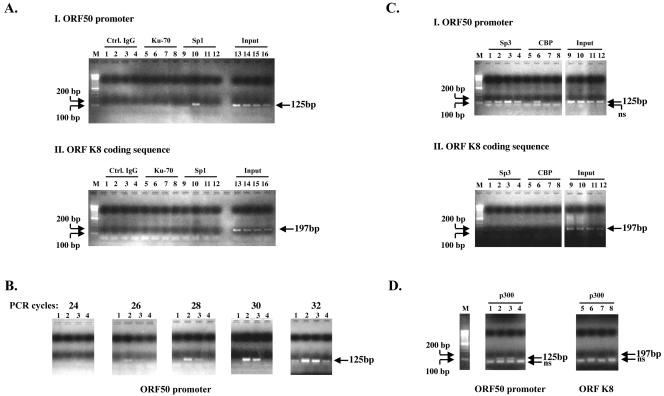
Most cellular signaling pathways utilize feedback control mechanisms to achieve tight regulation. We reasoned that the butyrate effect may also be temporally regulated. We used ChIP to study whether butyrate treatment affects the kinetics of association of Sp1 with ORF50p in living HH-B2 cells (Fig. 7A and B). There is an increase in ORF50 mRNA by 4 h after butyrate treatment (Fig. 1B). The level of Sp1 associated with ORF50p in vivo also increased at 4 h, diminished at 11 h after butyrate treatment, and returned to baseline levels at 24 h (Fig. 7AI, lane 10, and B, lanes 2 and 3). By varying the number of PCR cycles, Sp1 could be detected in the 4-h sample at 26 cycles, in the 11-h sample at 28 cycles, and in the untreated and 24-h butyrate-treated samples at 32 cycles (Fig. 7B). Sp1 did not bind to the ORFK8 coding sequence (Fig.7AII). The K8 coding sequence was used as a negative control for the ORF50 promoter, since this stretch of DNA does not contain any consensus Sp1 binding site. Ku-70 could not be immunoprecipitated to ORF50p or to ORF K8. Antibody to Ku-70, which binds nonspecifically to the ends of DNA, was used as a negative control for antibody to Sp1. Thus, butyrate induction of Sp1 binding to ORF50p was specific and temporally regulated. Sp3 was constitutively associated with ORF50p and increased slightly 11 h after butyrate treatment (Fig.7CI, lane 3). Thus, the kinetics of association of Sp1 and Sp3 with ORF50p were different. In a ChIP assay with antibody to Sp3, a band smaller than the expected size was also amplified (Fig.7CI). This is likely to be due to nonspecific amplification. Sp3 was also found to associate weakly and nonspecifically with the K8 coding sequence (Fig.7CII).
FIG. 7.
Butyrate induces transient association of Sp1 and CBP/p300 with the ORF50 promoter. (A) ChIP experiments exploring the temporal association of Sp1, CBP, and p300 with the ORF50 promoter in HH-B2 cells after butyrate induction. Immunoprecipitation was performed with normal goat immunoglobulin G (control [Ctrl]) (lanes 1 to 4), goat antibody to Ku-70 (lanes 5 to 8), goat antibody to Sp1 (lanes 9 to 12), or input DNA representing 5% of the sample (lanes 13 to 16). Cells were harvested at 0 h (lanes 1, 5, 9, 13), 4 h (lanes 2, 6, 10, 14), 11 h (lanes 3, 7, 11, 15), or 24 h (lanes 4, 8, 12, 16) after butyrate treatment. PCR primers detected the ORF50 promoter (I) or the K8 coding sequence (II). (B) The samples immunoprecipitated with antibody to Sp1 were analyzed with primers from the ORF50 promoter by using increasing numbers of PCR cycles. Samples were obtained 0 h (lane 1), 4 h (lane 2) 11 h (lane 3), or 24 h (lane 4) after butyrate treatment. (C) ChIP experiment exploring the association of Sp3 and CBP with the ORF50 promoter. M, marker; lanes 1 to 4, rabbit anti-Sp3; lanes 5 to 8, rabbit anti-CBP; lanes 9 to 12, 5% input. Samples were harvested at 0 h (lanes 1, 5, 9), 4 h (lanes 2, 6, 10), 11 h (lanes 3, 7, 11) and 24 h (lanes 4, 8, 12) after butyrate treatment. Primers specific for ORF50 promoters (I) or the K8 coding sequence (II) were used. (D) Transient association of p300 with the ORF50 promoter. ChIP experiment with antibody to p300. Samples were taken 0, 4, 11, and 24 h (lanes 1 to 4, respectively) after butyrate treatment.
Several transcriptional coactivators, including histone acetylases, CBP/p300, and TATA-associated factors, associate with Sp1 (71, 78). We detected a markedly enhanced association of CBP with ORF50p 4 h after butyrate treatment (Fig. 7C, lane 6) and with p300 (Fig. 7D, lanes 2 and 3) at 4 and 11 h after butyrate treatment. Neither CBP nor p300 associated with ORF K8 sequences. The kinetics of association of CBP and p300 with ORF50p following butyrate treatment was remarkably similar to that of Sp1. This result suggests that, following butyrate treatment, Sp1 may recruit these coactivators to the ORF50p.
DISCUSSION
Experiments directed at understanding regulation of the latency to lytic replication switch not only address a fundamental unanswered problem in KSHV biology but they may illuminate the behavior of many viruses and control of cellular gene expression. The principal advantage of using the gammaherpesvirus system to address this phenomenon is that it is possible to manipulate the transition between latency and lytic cycle viral gene expression in cultured cells and to examine its molecular mechanism. Although butyrate has been known for many years to induce both EBV and KSHV lytic gene expression, its mechanism of action remains obscure (37, 48, 85). Since butyrate is a histone deacetylase inhibitor that leads to global hyperacetylation of the tails of histones H3 and H4, it has been assumed that open chromatin formation, on the promoters of viral lytic cycle activator genes, such as the EBV ZEBRA and Rta genes, and the KSHV ORF50 gene, is sufficient to activate gene expression (10, 37). This report emphasizes several novel aspects of butyrate action on the ORF50 promoter. The butyrate response element was shown to be colinear with an Sp1/Sp3 binding site. Using in vitro DNA binding assays, we demonstrated that, while Sp1 and Sp3 constitutively bound this site, other DNA binding proteins related to Sp1 and Sp3 were induced by butyrate. We proved that the butyrate-responsive element in the ORF50 promoter was a bona fide Sp1 binding site by showing that Sp1 protein activated the ORF50 promoter in Schneider insect cells, where there is very little endogenous Sp1 activity, and also, when overexpressed, in mammalian cells. Finally, Sp1 and its coactivators, CBP and p300, were found to be recruited to the ORF50 promoter within 4 h after butyrate treatment. Enhanced occupancy of the ORF50 promoter by Sp1 was transient, while Sp3 abundance at this promoter did not change detectably over 24 h. Taken together, the results of this study indicate that a principal downstream effect of butyrate treatment is to enhance the occupancy of the ORF50 promoter by Sp1. The results suggest that Sp1 may play a critical role in activation of ORF50 expression.
The butyrate response element is an Sp1/Sp3 site.
Although a very short fragment of the ORF50 promoter, located 134 bp upstream of the ORF50 ATG, contains most of the constitutive activity and confers most of the response to butyrate induction, we found that the reporters p2500Luc and p950Luc were slightly more active than p134Luc in cells treated with butyrate (Fig. 2B). This result indicates that there may be other weak butyrate-responding elements located between 134 and 950 bp upstream of the ATG. p2500Luc and p950Luc weakly respond to TSA, suggesting that there may also exist weak TSA responsive elements in this region that are not present in p134Luc (Fig. 2B). ORF50 has been shown to up-regulate its own promoter in a variety of cell lines; however, the induction of the ORF50 promoter by butyrate does not depend on autostimulation, as we observed a similar level of induction by butyrate in cell lines that do not contain the KSHV genome (Fig. 4B). The principal butyrate-responsive element was mapped to the consensus Sp1-binding site located between nt 103 and 112 upstream of ATG on the ORF50 promoter. This site was shown to be essential for both basal promoter activity and butyrate induction (Fig. 3).
About two decades ago, the Sp1 protein was first described by Dynan and Tjian as a sequence-specific transcriptional activator that stimulated gene expression from a group of promoters including the simian virus 40 early promoter (18). Dynan and Tijan showed that Sp1 bound to the DNA sequences within the tandem 21-bp repeats about 70 to 110 bp upstream of the transcription start site of the simian virus 40 early promoter (19). Later on, they and others demonstrated that Sp1 binds to a GC-rich sequence and thereafter activates many genes that are involved in different aspects of cellular function (17, 29, 33). Although Sp1 can be detected in most cell types, cells have evolved complicated mechanisms to regulate the expression level and activity of Sp1 protein to ensure precise control of cellular physiology. The Sp1 level varies among different cell types during development; the lowest level is found in fully differentiated cells (64). Sp1 has also been shown to undergo alternative splicing to generate an amino-terminal truncated form that lacks transcriptional activation activity (53). This truncated form of Sp1 may act as a repressor or as a coactivator for full-length Sp1 or other related proteins of the Sp family in different cell backgrounds. In addition, Sp1 has been shown to undergo proteosome-dependent and caspase-mediated partial degradation to generate amino-terminal or carboxyl-terminal deletion forms that may modulate full-length Sp1 activity or have unique functions (61, 75).
Sp1 also undergoes posttranslational modifications, including phosphorylation and glycosylation (4, 5, 20, 31, 41, 44, 55, 60, 63, 84). So far, there is no evidence supporting regulation of Sp1 activity through acetylation (22). However, in view of the known action of butyrate as a histone deacetylase inhibitor, it will be important to examine the effect of butyrate on Sp1 acetylation. Glycosylation and phosphorylation have been shown to regulate Sp1 activity both positively and negatively under different circumstances. Glycosylation may regulate the cellular localization and stability of Sp1 protein, which indirectly affects Sp1-mediated transcription (26). Many cellular kinases have been shown to phosphorylate Sp1 at different serine and threonine residues. These kinases include DNA-dependent protein kinase, casein kinase II, protein kinase C-γ, cyclin A-dependent kinase, mitogen-activated protein kinase/extracellular signal-regulated kinase, and protein kinase A (2, 25, 30, 44, 55, 62).
Possible role of Sp3.
Sp3, another member of the Sp family of transcription factors, mainly plays a negative role in regulation of gene expression, although it may also activate certain promoters (15, 40). Using EMSA, we found that Sp3 also binds to the ORF50 promoter. Several novel Sp3-containing complexes were formed after butyrate induction (Fig. 5B and D). We also noted that butyrate treatment decreased the intensity of the Sp3-containing complex 2, indicating that butyrate may induce partial degradation of Sp3. Western blot analysis confirmed that several faster-migrating Sp3 species were induced after butyrate treatment (Fig. 6). We also found, by using ChIP, that the level of Sp3 associated with the ORF50 promoter in vivo did not change significantly after butyrate treatment (Fig. 7C). This result suggests that the presumably truncated forms of Sp3 identified by EMSA do not differ much from the long form in terms of DNA binding capacity. However, it is not clear whether these short forms of Sp3 are altered in their properties as transcription factors, e.g., whether they change Sp3 from a transcriptional repressor to a coactivator on the ORF50 promoter.
Butyrate also induces several novel DNA-protein complexes that are not disrupted or supershifted by Sp1 or Sp3 antibodies. We found that formation of these complexes also depends on the integrity of the consensus Sp1 site, suggesting that they may contain Sp-related proteins. It is possible that the Sp1 and Sp3 antibodies used in our assay do not recognize all Sp1 or Sp3 isoforms. However, it is also possible that these complexes contain other proteins not related to Sp1 or Sp3.
Butyrate activates some cellular genes via Sp1 sites in their promoters.
Butyrate is a short-chain fatty acid that was originally identified in butter (52). It is also produced in the colon by microbial fermentation of dietary fiber (59, 80). Since butyrate is known to promote growth arrest and cellular differentiation, its role as a regulator of cellular gene expression, particularly in cancer cells, has recently received considerable attention. In cells derived from colon carcinoma, butyrate has been found to induce the p21WAF1/CIP1 gene and the mitochondrial 3-hydroxy-3-methylglutaryl coenzyme A synthase gene via Sp1 sites in their promoters (7, 51). The human insulin-like growth factor binding protein 3 promoter is activated via Sp1 sites in cells from breast cancer (79). In murine and rat cells, the Sp1 site has been found to be important in induction of the promoters of the gamma glutamyl transferase, nonmuscle myosin heavy chain II-3, and galectin genes (6, 36, 45). Butyrate is known to cause hyperacetylation of the tails of histones H3 and H4 in many cell types, including Friend leukemia cells, HeLa cells from cervical carcinoma, and CoCa from colon carcinoma (14). Yet there is limited information that directly links this activity to the role of Sp1 in butyrate induction of gene expression. A single recent report suggests that histone hyperacetylation is not sufficient to activate the promoter of the p21WAF1/CIP1 gene (32). A serine/threonine kinase inhibitor, H7, suppressed the capacity of butyrate to induce p21WAF1/CIP1 mRNA but did not influence the capacity of butyrate to cause hyperacetylation of H3 and H4 (32).
Hypotheses that may link induction of ORF50 expression by butyrate to activity of Sp1.
We have considered the following five hypotheses attempting to relate butyrate action to the activity of Sp1. Our experimental data address some of these possibilities. (i) Butyrate may alter the level of expression of Sp1. Since ORF50 is an immediate-early gene and expression of ORF50 occurs when protein synthesis is inhibited, it is unlikely that the earliest effects of butyrate result from enhanced Sp1 expression per se. Moreover, our data (Fig. 6) indicate that, 24 h after butyrate treatment, the level of Sp1 does not change significantly. (ii) Butyrate may enhance the nuclear localization of Sp1. However, nuclear extracts prepared from cells with and without butyrate treatment contained equal amounts of Sp1 (Fig. 6). (iii) By altering histone acetylation, butyrate may lead to chromatin remodeling that permits Sp1 to bind to the ORF50 promoter. This is the hypothesis favored by a report that appeared while our work was in progress (35). Our data (Fig. 7) describing the kinetics of association of Sp1 with the ORF50 promoter in vivo show that Sp1 eventually falls off the ORF50 promoter. These data favor the idea that binding of Sp1 to the ORF50 promoter is regulated. If so, chromatin remodeling induced by butyrate must also be regulated. (iv) A likely hypothesis in our view is that butyrate treatment somehow alters Sp1 itself, allowing the protein to bind to ORF50 promoter DNA. The DNA binding activity of Sp1 is known to be regulated by phosphorylation. It is conceivable that butyrate treatment activates a kinase cascade that leads to phosphorylation of Sp1 in a manner that enhances its DNA binding activity. (v) Butyrate treatment may act by removing an inhibitor that prevents Sp1 from binding to DNA. This latter hypothesis raises the possibility that butyrate may induce a variety of posttranslational modifications that influence the capacity of Sp1 to interact with other proteins, either transcriptional coactivators or corepressors. In this context, the capacity of Sp1 to interact with coactivators such as CBP and corepressors such as histone deacetylases may be relevant.
Some problems for further investigation.
One unanswered question is whether all KSHV lytic cycle-inducing stimuli converge on Sp1. Some KSHV-positive PEL cell lines (e.g., BC1 and BC3) are also induced to undergo lytic viral replication by the phorbol ester PMA (Fig. 1). Yet, in these cell lines, PMA treatment does not cause any detectable change of global histone H4 acetylation (Fig. 1). Although the global chromatin acetylation level is not significantly different between untreated and PMA-treated cells, it is possible that PMA induces the KSHV lytic cycle through selective opening of chromatin at some KSHV genomes. TSA is a more potent inhibitor of histone deacetylase than is butyrate; nonetheless, TSA is less active in inducing the KSHV lytic cycle (Fig. 1) and only weakly activates ORF50 promoter luciferase reporter constructs (Fig. 2 and 3). Butyrate is a more pleiotropic agent than TSA and may possess important additional effects. Histone hyperacetylation may not be sufficient for KSHV to switch from latency into the lytic cycle.
It will be very interesting to find out whether the consensus Sp1 site is also important for the activation of the ORF50 promoter in BC1 and BC3 cells by PMA and whether Sp1 protein is also induced to bind to the ORF50 promoter in these cells in vivo. An important unanswered question is whether different inducers of KSHV lytic replication use different cellular signaling pathways or converge on the same cell signaling component to activate ORF50 expression. Wang et al. found that C/EBPα activates the ORF50 promoter in transient reporter assay and that C/EBPα binds to the ORF50 promoter in JSC1 PEL cells after 40 h of TPA treatment (82); furthermore, they demonstrated that short-term TPA treatment induces cJUN phosphorylation and binding to ORF50 promoter, suggesting that AP1 activity may mediate the early phase of ORF50 induction after TPA treatment (81). Future experiments should investigate whether butyrate induction is associated with enhanced activity of AP1 and C/EBPα.
Another question for further study is whether butyrate induction of EBV and other related gammaherpesvirus lytic gene expression also operates via a linkage to Sp1. There are several Sp1-binding sites in the promoter of EBV Rta, an immediate-early gene which is a homologue of KSHV ORF50. While these Sp1 sites are important for autoactivation of the Rta promoter, their role in butyrate activation of the EBV lytic cycle has not yet been investigated (57).
Our studies provoke the question of whether environmental triggers of viral lytic cycle activation may explain some of the perplexing epidemiology of KSHV infection. One of the unsolved mysteries of KSHV epidemiology is the marked geographic variation in the frequency of KSHV infection and its associated diseases. One possibility that merits further inquiry is that variations in components of the diet that generate butyrate may lead to differences in lytic cycle activation of KSHV. High levels of KSHV lytic replication may predispose people to a high frequency of KS and other KSHV-associated diseases.
Acknowledgments
This work was supported by grants CA70036 and CA16038 from the NIH to G.M. J.Y. was supported by the Interdisciplinary Immunology Training Program, NIH AI07019-27 and -28.
REFERENCES
- 1.Alkan, S., D. S. Karcher, A. Ortiz, S. Khalil, M. Akhtar, and M. A. Ali. 1997. Human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus in organ transplant patients with immunosuppression. Br. J. Haematol. 96:412-414. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, S. A., D. A. Barry, R. W. Leggett, and C. R. Mueller. 1997. Casein kinase II-mediated phosphorylation of the C terminus of Sp1 decreases its DNA binding activity. J. Biol. Chem. 272:13489-13495. [DOI] [PubMed] [Google Scholar]
- 3.Ascoli, V., C. M. Mastroianni, V. Galati, M. C. Sirianni, A. Fruscalzo, A. Pistilli, and C. F. Lo. 1998. Primary effusion lymphoma containing human herpesvirus 8 DNA in two AIDS patients with Kaposi's sarcoma. Haematologica 83:8-12. [PubMed] [Google Scholar]
- 4.Benasciutti, E., G. Pages, O. Kenzior, W. Folk, F. Blasi, and M. P. Crippa. 2004. MAPK and JNK transduction pathways can phosphorylate Sp1 to activate the uPA minimal promoter elements and endogenous gene transcription. Blood 104:256-262. [DOI] [PubMed] [Google Scholar]
- 5.Bouwman, P., and S. Philipsen. 2002. Regulation of the activity of Sp1-related transcription factors. Mol. Cell. Endocrinol. 195:27-38. [DOI] [PubMed] [Google Scholar]
- 6.Buxton, D. B., E. Golomb, and R. S. Adelstein. 2003. Induction of nonmuscle myosin heavy chain II-C by butyrate in RAW 264.7 mouse macrophages. J. Biol. Chem. 278:15449-15455. [DOI] [PubMed] [Google Scholar]
- 7.Camarero, N., A. Nadal, M. J. Barrero, D. Haro, and P. F. Marrero. 2003. Histone deacetylase inhibitors stimulate mitochondrial HMG-CoA synthase gene expression via a promoter proximal Sp1 site. Nucleic Acids Res. 31:1693-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 9.Cesarman, E., R. G. Nador, F. Bai, R. A. Bohenzky, J. J. Russo, P. S. Moore, Y. Chang, and D. M. Knowles. 1996. Kaposi's sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi's sarcoma and malignant lymphoma. J. Virol. 70:8218-8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, P. J., D. Shedd, L. Gradoville, M. S. Cho, L. W. Chen, J. Chang, and G. Miller. 2002. Open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus directly activates the viral PAN and K12 genes by binding to related response elements. J. Virol. 76:3168-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 12.Chow, J. W., and S. B. Lucas. 1990. Endemic and atypical Kaposi's sarcoma in Africa-histopathological aspects. Clin. Exp. Dermatol. 15:253-259. [DOI] [PubMed] [Google Scholar]
- 13.Cook-Mozaffari, P., R. Newton, V. Beral, and D. P. Burkitt. 1998. The geographical distribution of Kaposi's sarcoma and of lymphomas in Africa before the AIDS epidemic. Br. J. Cancer 78:1521-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davie, J. R. 2003. Inhibition of histone deacetylase activity by butyrate. J. Nutr. 133:2485S-2493S. [DOI] [PubMed] [Google Scholar]
- 15.De Luca, P., B. Majello, and L. Lania. 1996. Sp3 represses transcription when tethered to promoter DNA or targeted to promoter proximal RNA. J. Biol. Chem. 271:8533-8536. [DOI] [PubMed] [Google Scholar]
- 16.Dittmer, D., M. Lagunoff, R. Renne, K. Staskus, A. Haase, and D. Ganem. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dynan, W. S., S. Sazer, R. Tjian, and R. T. Schimke. 1986. Transcription factor Sp1 recognizes a DNA sequence in the mouse dihydrofolate reductase promoter. Nature 319:246-248. [DOI] [PubMed] [Google Scholar]
- 18.Dynan, W. S., and R. Tjian. 1983. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell 32:669-680. [DOI] [PubMed] [Google Scholar]
- 19.Dynan, W. S., and R. Tjian. 1983. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell 35:79-87. [DOI] [PubMed] [Google Scholar]
- 20.Fojas de Borja, P., N. K. Collins, P. Du, J. Azizkhan-Clifford, and M. Mudryj. 2001. Cyclin A-CDK phosphorylates Sp1 and enhances Sp1-mediated transcription. EMBO J. 20:5737-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 22.Furia, B., L. Deng, K. Wu, S. Baylor, K. Kehn, H. Li, R. Donnelly, T. Coleman, and F. Kashanchi. 2002. Enhancement of nuclear factor-kappa B acetylation by coactivator p300 and HIV-1 Tat proteins. J. Biol. Chem. 277:4973-4980. [DOI] [PubMed] [Google Scholar]
- 23.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guasparri, I., S. A. Keller, and E. Cesarman. 2004. KSHV vFLIP is essential for the survival of infected lymphoma cells. J. Exp. Med. 199:993-1003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Haidweger, E., M. Novy, and H. Rotheneder. 2001. Modulation of Sp1 activity by a cyclin A/CDK complex. J. Mol. Biol. 306:201-212. [DOI] [PubMed] [Google Scholar]
- 26.Han, I., and J. E. Kudlow. 1997. Reduced O glycosylation of Sp1 is associated with increased proteasome susceptibility. Mol. Cell. Biol. 17:2550-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henle, W., and G. Henle. 1970. Evidence for a relation of Epstein-Barr virus to Burkitt's lymphoma and nasopharyngeal carcinoma. Bibl. Haematol. 36:706-713. [DOI] [PubMed] [Google Scholar]
- 28.Hideshima, T., D. Chauhan, G. Teoh, N. Raje, S. P. Treon, Y. T. Tai, Y. Shima, and K. C. Anderson. 2000. Characterization of signaling cascades triggered by human interleukin-6 versus Kaposi's sarcoma-associated herpes virus-encoded viral interleukin 6. Clin. Cancer Res. 6:1180-1189. [PubMed] [Google Scholar]
- 29.Ishii, S., J. T. Kadonaga, R. Tjian, J. N. Brady, G. T. Merlino, and I. Pastan. 1986. Binding of the Sp1 transcription factor by the human Harvey ras1 proto-oncogene promoter. Science 232:1410-1413. [DOI] [PubMed] [Google Scholar]
- 30.Jackson, S., T. Gottlieb, and K. Hartley. 1993. Phosphorylation of transcription factor Sp1 by the DNA-dependent protein kinase. Adv. Second Messenger Phosphoprotein Res. 28:279-286. [PubMed] [Google Scholar]
- 31.Kang, H. T., J. W. Ju, J. W. Cho, and E. S. Hwang. 2003. Down-regulation of Sp1 activity through modulation of O-glycosylation by treatment with a low glucose mimetic, 2-deoxyglucose. J. Biol. Chem. 278:51223-51231. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi, H., E. M. Tan, and S. E. Fleming. 2004. Acetylation of histones associated with the p21WAF1/CIP1 gene by butyrate is not sufficient for p21WAF1/CIP1 gene transcription in human colorectal adenocarcinoma cells. Int. J. Cancer. 109:207-213. [DOI] [PubMed] [Google Scholar]
- 33.Lee, W., A. Haslinger, M. Karin, and R. Tjian. 1987. Activation of transcription by two factors that bind promoter and enhancer sequences of the human metallothionein gene and SV40. Nature 325:368-372. [DOI] [PubMed] [Google Scholar]
- 34.Liang, Y., J. Chang, S. J. Lynch, D. M. Lukac, and D. Ganem. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway. Genes Dev. 16:1977-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu, F., J. Zhou, A. Wiedmer, K. Madden, Y. Yuan, and P. M. Lieberman. 2003. Chromatin remodeling of the Kaposi's sarcoma-associated herpesvirus ORF50 promoter correlates with reactivation from latency. J. Virol. 77:11425-11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu, Y., and R. Lotan. 1999. Transcriptional regulation by butyrate of mouse galectin-1 gene in embryonal carcinoma cells. Biochim. Biophys. Acta 1444:85-91. [DOI] [PubMed] [Google Scholar]
- 37.Luka, J., B. Kallin, and G. Klein. 1979. Induction of the Epstein-Barr virus (EBV) cycle in latently infected cells by n-butyrate. Virology 94:228-231. [DOI] [PubMed] [Google Scholar]
- 38.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 40.Majello, B., P. De Luca, and L. Lania. 1997. Sp3 is a bifunctional transcription regulator with modular independent activation and repression domains. J. Biol. Chem. 272:4021-4026. [DOI] [PubMed] [Google Scholar]
- 41.Majumdar, G., A. Harmon, R. Candelaria, A. Martinez-Hernandez, R. Raghow, and S. S. Solomon. 2003. O-glycosylation of Sp1 and transcriptional regulation of the calmodulin gene by insulin and glucagon. Am. J. Physiol Endocrinol. Metab. 285:E584-E591. [DOI] [PubMed] [Google Scholar]
- 42.Martin, D. F., B. D. Kuppermann, R. A. Wolitz, A. G. Palestine, H. Li, and C. A. Robinson. 1999. Oral ganciclovir for patients with cytomegalovirus retinitis treated with a ganciclovir implant. Roche Ganciclovir Study Group. N. Engl. J. Med. 340:1063-1070. [DOI] [PubMed] [Google Scholar]
- 43.Menke, D. M., A. Chadbum, E. Cesarman, E. Green, J. Berenson, J. Said, M. Tiemann, R. Parwaresch, and S. D. Thome. 2002. Analysis of the human herpesvirus 8 (HHV-8) genome and HHV-8 vIL-6 expression in archival cases of castleman disease at low risk for HIV infection. Am. J. Clin. Pathol. 117:268-275. [DOI] [PubMed] [Google Scholar]
- 44.Merchant, J. L., M. Du, and A. Todisco. 1999. Sp1 phosphorylation by Erk 2 stimulates DNA binding. Biochem. Biophys. Res. Commun. 254:454-461. [DOI] [PubMed] [Google Scholar]
- 45.Mikkelsen, I. M., N. E. Huseby, A. Visvikis, and U. Moens. 2002. Activation of the gamma-glutamyltransferase promoter 2 in the rat colon carcinoma cell line CC531 by histone deacetylase inhibitors is mediated through the Sp1 binding motif. Biochem. Pharmacol. 64:307-315. [DOI] [PubMed] [Google Scholar]
- 46.Miller, G. 1971. Human lymphoblastoid cell lines and Epstein-Barr virus: a review of their interrelationships and their relevance to the etiology of leukoproliferative states in man. Yale J. Biol. Med. 43:358-384. [PMC free article] [PubMed] [Google Scholar]
- 47.Miller, G., L. Heston, E. Grogan, L. Gradoville, M. Rigsby, R. Sun, D. Shedd, V. M. Kushnaryov, S. Grossberg, and Y. Chang. 1997. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J. Virol. 71:314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller, G., M. O. Rigsby, L. Heston, E. Grogan, R. Sun, C. Metroka, J. A. Levy, S. J. Gao, Y. Chang, and P. Moore. 1996. Antibodies to butyrate-inducible antigens of Kaposi's sarcoma-associated herpesvirus in patients with HIV-1 infection. N. Engl. J. Med. 334:1292-1297. [DOI] [PubMed] [Google Scholar]
- 49.Monini, P., S. Colombini, M. Sturzl, D. Goletti, A. Cafaro, C. Sgadari, S. Butto, M. Franco, P. Leone, S. Fais, P. Leone, G. Melucci-Vigo, C. Chiozzini, F. Carlini, G. Ascherl, E. Cornali, C. Zietz, E. Ramazzotti, F. Ensoli, M. Andreoni, P. Pezzotti, G. Rezza, R. Yarchoan, R. C. Gallo, and B. Ensoli. 1999. Reactivation and persistence of human herpesvirus-8 infection in B cells and monocytes by Th-1 cytokines increased in Kaposi's sarcoma. Blood 93:4044-4058. [PubMed] [Google Scholar]
- 50.Moore, P. S., C. Boshoff, R. A. Weiss, and Y. Chang. 1996. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 274:1739-1744. [DOI] [PubMed] [Google Scholar]
- 51.Nakano, K., T. Mizuno, Y. Sowa, T. Orita, T. Yoshino, Y. Okuyama, T. Fujita, N. Ohtani-Fujita, Y. Matsukawa, T. Tokino, H. Yamagishi, T. Oka, H. Nomura, and T. Sakai. 1997. Butyrate activates the WAF1/Cip1 gene promoter through Sp1 sites in a p53-negative human colon cancer cell line. J. Biol. Chem. 272:22199-22206. [DOI] [PubMed] [Google Scholar]
- 52.Parodi, P. W. 1997. Cows' milk fat components as potential anticarcinogenic agents. J. Nutr. 127:1055-1060. [DOI] [PubMed] [Google Scholar]
- 53.Persengiev, S. P., J. D. Saffer, and D. L. Kilpatrick. 1995. An alternatively spliced form of the transcription factor Sp1 containing only a single glutamine-rich transactivation domain. Proc. Natl. Acad. Sci. USA 92:9107-9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rabin, H., R. H. Neubauer, G. R. Pearson, J. L. Cicmanec, W. C. Wallen, W. F. Loeb, and M. G. Valerio. 1975. Spontaneous lymphoma associated with herpesvirus saimiri in owl monkeys. J. Natl. Cancer Inst. 54:499-502. [PubMed] [Google Scholar]
- 55.Rafty, L. A., and L. M. Khachigian. 2001. Sp1 phosphorylation regulates inducible expression of platelet-derived growth factor B-chain gene via atypical protein kinase C-zeta. Nucleic Acids Res. 29:1027-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ragoczy, T., L. Heston, and G. Miller. 1998. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol. 72:7978-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ragoczy, T., and G. Miller. 2001. Autostimulation of the Epstein-Barr virus BRLF1 promoter is mediated through consensus Sp1 and Sp3 binding sites. J. Virol. 75:5240-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S. J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rechkemmer, G., K. Ronnau, and W. von Engelhardt. 1988. Fermentation of polysaccharides and absorption of short chain fatty acids in the mammalian hindgut. Comp. Biochem. Physiol. A 90:563-568. [DOI] [PubMed] [Google Scholar]
- 60.Reisinger, K., R. Kaufmann, and J. Gille. 2003. Increased Sp1 phosphorylation as a mechanism of hepatocyte growth factor (HGF/SF)-induced vascular endothelial growth factor (VEGF/VPF) transcription. J. Cell Sci. 116:225-238. [DOI] [PubMed] [Google Scholar]
- 61.Rickers, A., N. Peters, V. Badock, R. Beyaert, P. Vandenabeele, B. Dorken, and K. Bommert. 1999. Cleavage of transcription factor SP1 by caspases during anti-IgM-induced B-cell apoptosis. Eur. J. Biochem. 261:269-274. [DOI] [PubMed] [Google Scholar]
- 62.Rohlff, C., S. Ahmad, F. Borellini, J. Lei, and R. I. Glazer. 1997. Modulation of transcription factor Sp1 by cAMP-dependent protein kinase. J. Biol. Chem. 272:21137-21141. [DOI] [PubMed] [Google Scholar]
- 63.Roos, M. D., K. Su, J. R. Baker, and J. E. Kudlow. 1997. O glycosylation of an Sp1-derived peptide blocks known Sp1 protein interactions. Mol. Cell. Biol. 17:6472-6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saffer, J. D., S. P. Jackson, and M. B. Annarella. 1991. Developmental expression of Sp1 in the mouse. Mol. Cell. Biol. 11:2189-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75:6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saveliev, A., F. Zhu, and Y. Yuan. 2002. Transcription mapping and expression patterns of genes in the major immediate-early region of Kaposi's sarcoma-associated herpesvirus. Virology 299:301-314. [DOI] [PubMed] [Google Scholar]
- 67.Searles, R. P., E. P. Bergquam, M. K. Axthelm, and S. W. Wong. 1999. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 73:3040-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharp, T. V., H. W. Wang, A. Koumi, D. Hollyman, Y. Endo, H. Ye, M. Q. Du, and C. Boshoff. 2002. K15 protein of Kaposi's sarcoma-associated herpesvirus is latently expressed and binds to HAX-1, a protein with antiapoptotic function. J. Virol. 76:802-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Song, M. J., X. Li, H. J. Brown, and R. Sun. 2002. Characterization of interactions between RTA and the promoter of polyadenylated nuclear RNA in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 76:5000-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 71.Soutoglou, E., B. Viollet, M. Vaxillaire, M. Yaniv, M. Pontoglio, and I. Talianidis. 2001. Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity. EMBO J. 20:1984-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Staskus, K. A., W. Zhong, K. Gebhard, B. Herndier, H. Wang, R. Renne, J. Beneke, J. Pudney, D. J. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sternbach, G., and J. Varon. 1995. Moritz Kaposi: idiopathic pigmented sarcoma of the skin. J. Emerg. Med. 13:671-674. [DOI] [PubMed] [Google Scholar]
- 74.Sturzl, M., C. Hohenadl, C. Zietz, E. Castanos-Velez, A. Wunderlich, G. Ascherl, P. Biberfeld, P. Monini, P. J. Browning, and B. Ensoli. 1999. Expression of K13/v-FLIP gene of human herpesvirus 8 and apoptosis in Kaposi's sarcoma spindle cells. J. Natl. Cancer Inst. 91:1725-1733. [DOI] [PubMed] [Google Scholar]
- 75.Su, K., M. D. Roos, X. Yang, I. Han, A. J. Paterson, and J. E. Kudlow. 1999. An N-terminal region of Sp1 targets its proteasome-dependent degradation in vitro. J. Biol. Chem. 274:15194-15202. [DOI] [PubMed] [Google Scholar]
- 76.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Suzuki, T., S. Muto, S. Miyamoto, K. Aizawa, M. Horikoshi, and R. Nagai. 2003. Functional interaction of the DNA-binding transcription factor Sp1 through its DNA-binding domain with the histone chaperone TAF-I. J. Biol. Chem. 278:28758-28764. [DOI] [PubMed] [Google Scholar]
- 79.Tsubaki, J., V. Hwa, S. M. Twigg, and R. G. Rosenfeld. 2002. Differential activation of the IGF binding protein-3 promoter by butyrate in prostate cancer cells. Endocrinology 143:1778-1788. [DOI] [PubMed] [Google Scholar]
- 80.von Engelhardt, W., J. Bartels, S. Kirschberger, H. D. Meyer zu Duttingdorf, and R. Busche. 1998. Role of short-chain fatty acids in the hind gut. Vet. Q. 20(Suppl. 3):S52-S59. [PubMed] [Google Scholar]
- 81.Wang, S. E., F. Y. Wu, H. Chen, M. Shamay, Q. Zheng, and G. S. Hayward. 2004. Early activation of the Kaposi's sarcoma-associated herpesvirus RTA, RAP, and MTA promoters by the tetradecanoyl phorbol acetate-induced AP1 pathway. J. Virol. 78:4248-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang, S. E., F. Y. Wu, Y. Yu, and G. S. Hayward. 2003. CCAAT/enhancer-binding protein-alpha is induced during the early stages of Kaposi's sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters. J. Virol. 77:9590-9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Whitehouse, A., I. M. Carr, J. C. Griffiths, and D. M. Meredith. 1997. The herpesvirus saimiri ORF50 gene, encoding a transcriptional activator homologous to the Epstein-Barr virus R protein, is transcribed from two distinct promoters of different temporal phases. J. Virol. 71:2550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang, X., K. Su, M. D. Roos, Q. Chang, A. J. Paterson, and J. E. Kudlow. 2001. O-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability. Proc. Natl. Acad. Sci. USA 98:6611-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu, Y., J. B. Black, C. S. Goldsmith, P. J. Browning, K. Bhalla, and M. K. Offermann. 1999. Induction of human herpesvirus-8 DNA replication and transcription by butyrate and TPA in BCBL-1 cells. J. Gen. Virol. 80(Pt 1):83-90. [DOI] [PubMed] [Google Scholar]
- 86.Zhu, F. X., T. Cusano, and Y. Yuan. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zong, J. C., D. M. Ciufo, D. J. Alcendor, X. Wan, J. Nicholas, P. J. Browning, P. L. Rady, S. K. Tyring, J. M. Orenstein, C. S. Rabkin, I. J. Su, K. F. Powell, M. Croxson, K. E. Foreman, B. J. Nickoloff, S. Alkan, and G. S. Hayward. 1999. High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi's sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. J. Virol. 73:4156-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]