Abstract
Accurately assessment of adipose tissue volume inside a human body plays an important role in predicting disease or cancer risk, diagnosis and prognosis. In order to overcome limitation of using only one subjectively selected CT image slice to estimate size of fat areas, this study aims to develop and test a computer-aided detection (CAD) scheme based on deep learning technique to automatically segment subcutaneous fat areas (SFA) and visceral fat areas (VFA) depicting on volumetric CT images. A retrospectively collected CT image dataset was divided into two independent training and testing groups. The proposed CAD framework consisted of two steps with two convolution neural networks (CNNs) namely, Selection-CNN and Segmentation-CNN. The first CNN was trained using 2,240 CT slices to select abdominal CT slices depicting SFA and VFA. The second CNN was trained with 84,000 pixel patches and applied to the selected CT slices to identify fat-related pixels and assign them into SFA and VFA classes. Comparing to the manual CT slice selection and fat pixel segmentation results, the accuracy of CT slice selection using the Selection-CNN yielded 95.8%, while the accuracy of fat pixel segmentation using the Segmentation-CNN was 96.8%. This study demonstrated the feasibility of applying a new deep learning based CAD scheme to automatically recognize abdominal section of human body from CT scans and segment SFA and VFA from volumetric CT data with high accuracy or agreement with the manual segmentation results.
Keywords: Computer-aided detection (CAD), Deep learning, Convolution neural network (CNN), Segmentation of adipose tissue, Subcutaneous fat area (SFA), Visceral fat area (VFA)
1. Introduction
Abdominal obesity is one of the most prevalent public health problems and over one third of adults were obese in the United States in recent years [1]. Obesity is strongly associated with many different diseases such as heart diseases, metabolic disorders, type 2 diabetes and certain types of cancers [1–3]. Inside a human body, there are subcutaneous fat areas (SFA) and visceral fat areas (VFA), which both contribute to the abdominal obesity. Studies have shown that in the clinical practice separate measurement or quantification of subtypes of adipose tissue in SFA and VFA is crucial for obesity assessment since visceral fat is more closely related to risk factors for hypertension, coronary artery disease, metabolic syndrome, and etc. [4, 5]. Other studies have also found that measurement of the total fat volume and/or the ratio between the VFA and SFA could generate useful clinical markers to assess response of cancer patients to the chemotherapies, in particular many antiangiogenic therapies [6–8].
Among different imaging techniques for adiposity tissue detection and measurement, computed tomography (CT) has been most widely adopted because of its higher accuracy and reproducibility [9]. Accurate segmentation and quantification of SFA and VFA from CT slices is important for clinical diagnosis and prediction of disease (or cancer) treatment efficacy. Currently, manual or semi-automated segmentation of SFA and VFA in a single subjectively chosen CT image slice has been adopted to determine fat areas and measure adiposity related features as demonstrated in the previous studies [7, 9]. However, this approach has a number of limitations including that 1) manual manipulations are time-consuming and cannot deal with large amount of data; 2) fat area measured from a single CT slice may not accurately correlate to the total fat volume of a human body; 3) measurement may also not be consistent due to the inter and intra reader variability in selecting CT slice and segmenting SFA and/or VFA areas. Therefore, developing a computer-aided detection (CAD) scheme for fully automated segmentation and quantification of SFA and VFA is necessary [10].
Recently, deep learning methods, especially deep convolutional neural networks (CNN), have gained extensive research interests and proven to be the state of art in a number of computer vision applications [11–15]. Unlike conventional machine learning methods where manual features design is crucial, deep learning models automatically learn hierarchical feature representations from raw inputs without knowledge of feature engineering [15]. With the availability of large amount of well-annotated datasets and high-speed parallel computing resources (i.e. Graphical Processing Units), deep learning models provide powerful tools for addressing the problems of object recognition, image segmentation and classification [16]. Following the tremendous applications of deep learning in computer vision area, there are a couple of previous works that successfully employed CNN methods to solve medical image analysis and CAD related problems [17–24]. For example, Roth et al [22] developed a multi-level deep CNN model for automated pancreas segmentation from CT scans; Brebisson et al [17] used a combination of 2D and 3D patches as the input of CNN for brain segmentation; Yan et al [24] applied a multi-instance deep learning framework to discover discriminative local anatomies for bodypart recognition; etc.
In this study, we developed a two-step CNN based CAD scheme for automated segmentation and quantification of SFA and VFA from abdominal CT scans. The new CAD scheme consists of two different CNNs, while the first one is used to automatically select and collect CT slices belonging to abdomen area from the whole CT scan series (i.e., the perfusion CT images acquired from ovarian cancer patients, which are scanned from lung to pelvis crossing the entire abdomen region), and the second CNN is used for automated segmentation of SFA and VFA in each single CT slice. While there has been a number of previously published studies that focused on automated quantification of visceral and subcutaneous adipose tissue by using combinations of traditional image processing techniques (e.g. labelling and morphological operation) [5, 8, 25–30], previous works have a number of limitations including that: 1) the selection of CT slice range of interest (i.e. abdomen area in this study) is either manually processed or not mentioned; 2) the optimal values of some parameters (e.g. morphological operation kernel size) in some of these models may not be consistent for different patients and thus human intervention might be necessary for tuning these parameters. Our proposed CNN based CAD scheme aims to overcome these limitations and achieve fully automated segmentation since (1) the first CNN was developed for automated selection of CT slices belonging to abdomen area, and (2) there is no parameters needed to be tuned in our CAD scheme after the two CNNs are sufficiently trained. The details of this study including the development of CAD scheme and performance evaluation are presented in the following sections.
2. Materials and Methods
2.1. Overview of the CAD framework
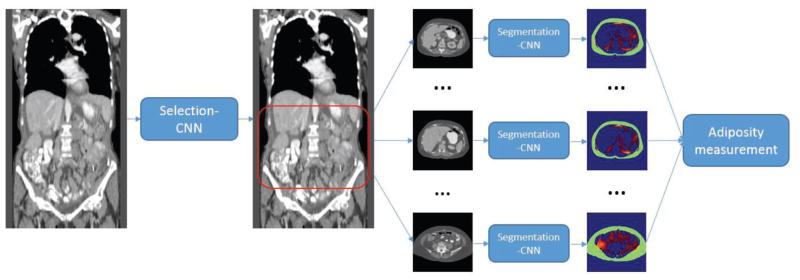
Our proposed CAD framework consisted of two steps with two CNNs, namely the Selection-CNN and Segmentation-CNN. The flowchart for demonstrating the whole process of this system is shown in Figure 1. The first Selection-CNN is applied to automatically select and collect CT slices of interest (i.e. abdomen area in this study) for each patient. The selected slices will then be used as the input of the second Segmentation-CNN for automated segmentation of SFA and VFA. The SFA and VFA volumes segmented from all CT slices of interest will be combined to compute and measure the adiposity characteristics of the particular patient at the very end step. The proposed CAD system is fully automated and no human intervention is needed. In the next two sections we will discuss the details of the CNN architectures, training process and evaluation methods for the two CNNs respectively.
Figure 1.
Overview of the two-step CNN based CAD scheme for adipose tissue quantification
2.2. A CT image dataset
Under an Institutional Review Board (IRB) approved image data collection and retrospective study protocol, we randomly assemble an image dataset, which consists of CT images acquired from 40 ovarian cancer patients who underwent cancer treatment in the Health Science Center of our University. The detailed image acquisition protocol has been reported in our previous publication [7, 8]. In brief, all CT scans were done using either a GE LightSpeed VCT 64-detector or a GE Discovery 600 16-detector CT machine. The X-ray power output was set at 120 kVp and a variable range from 100 to 600mA depending on patient body size. CT image slice thickness or spacing is 5mm and the images were reconstructed using a GE “Standard” image reconstruction kernel. Next, these 40 patients were randomly and equally divided into two groups namely, a training patient group and a testing patient group. CT image data from training patient group were used to train two CNNs used in the CAD scheme and the data from the testing patient group were used to evaluate the performance of the trained CAD scheme.
2.2.1. Training and testing dataset for Selection-CNN
In order to train Selection-CNN model to “learn” how to discriminate CT slices as belonging to abdomen area or not, an observer manually identified the abdomen area for each of the twenty patients in the training patient group. Specifically, an upper bound was subjectively placed just below the lung area and a lower bound was placed at the umbilicus level. All CT slices between the two bounds were labeled as positive (i.e. belong to slices of interest) and other slices were labeled as negative (i.e. not belong to slices of interest). By doing this, we can collect a sample of 2,240 CT slices as the training set of Selection-CNN. Among them, 757 are “positive slices” located inside the abdominal region and 1,483 are “negative slices” located outside the abdominal region. Although in order to optimally train a machine learning classifier, many previous studies chose to use “balanced” training datasets with the equal number of sample cases in two classes, the “balanced” approach has a limitation of potential sampling bias in selecting and removing part of sampling cases from the class with more samples. Thus, to maintain the diversity of all image slices, we used all 2,240 image slices in the training dataset although the number of “positive” and “negative” slices was different (or not balanced). Next, to verify the advantage of using all available training samples, we also conducted experiments to compare the classification accuracy between using “balanced” and “non-balanced” training datasets with 50 repeated training and validation tests in each training condition.
2.2.2. Training and testing dataset for Segmentation-CNN
Similar to Selection-CNN, Segmentation-CNN also needs some subjectively processed images as the training samples to train the classifier. For each of the twenty patients in the training patient group, 6 CT slices from the abdomen area were randomly selected for the training purpose (totally 120 CT images). A previously developed and tested CT image segmentation method was applied to remove the background (e.g. air and CT bed) and generate a body trunk mask. All the pixels inside the body trunk mask with CT number between the threshold −40 HU and −140 HU were defined as adipose tissue pixels [9]. An observer manually drew a boundary that contains the entire visceral area. The adipose tissue pixels inside the visceral area were labeled as VFA pixels whereas the adipose tissue pixels outside the visceral area were labeled as SFA pixels.
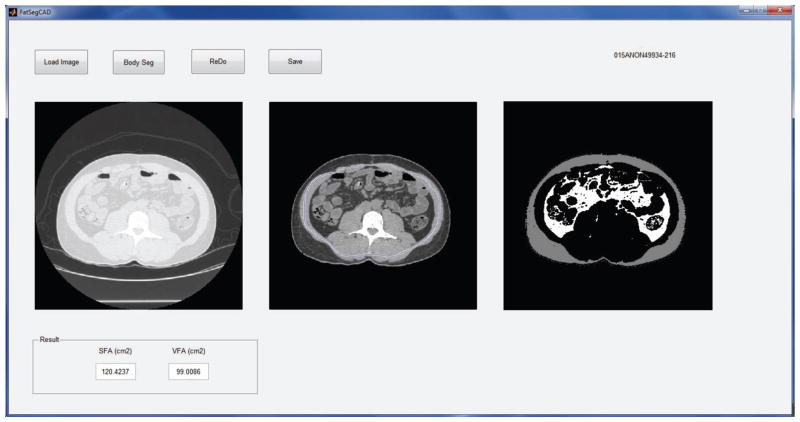
A Matlab based graphic user interface (GUI) program was developed for implementation of image pre-processing step and manual segmentation process. Figure 2 shows an example of the working procedure of the GUI. After loading a CT image from the local hard drive, the original image is shown in the left figure of the program. The “Body Seg” button has been designed to implement the body trunk segmentation algorithm and display the body trunk image in the middle figure of the program. Then, an observer can draw a boundary (the blue line in the middle image) that contains the entire visceral area. The segmented SFA/VFA result is then shown in the right figure of the GUI window.
Figure 2.
Demonstration of the Matlab GUI program for implementation of image preprocessing and manual segmentation process
Subsequently, 700 adipose tissue pixels (belonging to either VFA or SFA pixel class) were randomly selected from each of the 120 CT images for training Segmentation-CNN. By doing this, we can collect a sample of 84,000 adipose tissue pixels as the training set. Among them, 64,691 are labeled as SFA pixels and 19,309 are VFA pixels. The goal is to train the Segmentation-CNN to recognize or distinguish pixels with the CT number between −40 HU and −140 HU into SFA or VFA areas. By using the same segmentation criterion, 120 CT images from the 20 patients in the testing patient group were randomly selected and manually labelled for evaluation purpose.
2.3. CNN architectures
2.3.1. Selection-CNN
The task of Selection-CNN is to select the CT slices belonging to abdomen area, which will be collected for the computation of adipose tissue volume and features. We formulated this task as a binary classification problem. Specifically, we used each single CT slice image as the input of a classifier (Selection-CNN) and determine whether the CT slice belongs to the abdomen area or not. In this way, each CT slice was processed independently; the location and spatial consistency were not considered. In order to overcome this limitation, a simple post-processing step was performed to ensure the spatial consistency and dependence after obtaining the raw Selection-CNN output.
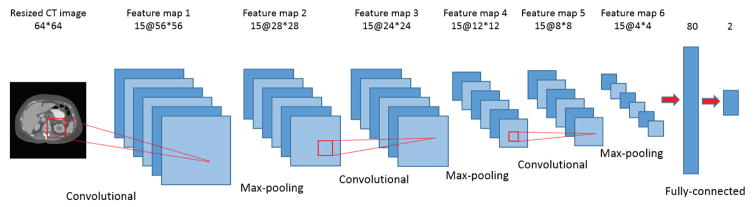
The architecture of Selection-CNN is shown in Figure 3 and it was developed based on LeNet, which was designed by LeCun et al. for image recognition [13]. First, the original 512×512 CT images were resized to 64×64 using an 8×8 averaging kernel. This step can be interpreted as either a down-sampling based image pre-processing step or a “pooling” layer in CNN architecture. By doing this, the input size and number of parameters of Selection-CNN was greatly reduced, which can potentially improve the training efficiency and reduce the risk of over-fitting. The down-sampled CT images were then used as input into a standard CNN architecture consisting of three convolutional-pooling layers, one fully connected layer, and one soft-max layer. Convolutional layers are the core part of CNN architecture. They consist of a number of rectangular convolutional filters; the parameters of these filters are randomly initialized and learnable during the training process. Each convolutional layer performs two-dimensional convolutional operations between the input image maps and the convolutional filters followed by a non-linear transformation. The convolutional layers can be interpreted as automatic feature extractors that are optimized from training data, and thus the outputs of convolutional layers are referred as “feature maps”.
Figure 3.
Architecture of Selection-CNN
Following convolutional layers, max-pooling layers were commonly performed in a number of CNN architectures [11, 13, 15]. The operation of max-pooling layer is to take the maximum values over sub-windows of feature maps, which can greatly reduce the spatial redundancy and the number of parameters. Several convolutional-pooling layer pairs can be stacked to get high-level feature representations. These features were then used as input into a standard Multi-Layer Perceptron (MLP) classifier, which consists of a fully connected hidden layer and a soft-max layer. The Selection-CNN developed in this study contained three convolutional-pooling layers. The numbers of feature maps were 15 for all the three layers and the filter sizes were 9×9, 5×5 and 5×5, respectively. A tanh function was applied for non-linear transformation. The size of max-pooling was 2×2 for all layers. The fully connected layer contained 80 hidden neurons and the soft-max layer contained 2 output neurons (i.e. positive or negative).
As a result, the Selection-CNN architecture mapped each 512×512 CT image slice to a vector of two continuous number between 0 and 1, indicating the probabilities of the input image belonging to positive or negative classes. Considering that the size of training set of Selection-CNN is relatively small compared to many of other computer vision datasets using deep learning (e.g. MNIST and ImageNet), we took following measures to avoid the potential over-fitting problem, which include that (1) the numbers of filters for each convolutional layer and hidden layer were set to be smaller than the commonly used CNNs, and (2) an L2 regularization term was adopted as a part of loss function.
2.3.2. Segmentation-CNN
After CT slices belonging to abdomen area were selected, a Segmentation-CNN based scheme was applied to segment SFA and VFA depicted on each single CT slice. This task was also formulated as a binary classification problem. Specifically, an image pre-processing step including a previously developed body trunk segmentation scheme [31] and a well-defined thresholding process for adipose tissue identification [10] was applied to identify all the pixels belonging to the fat area (namely adipose tissue pixels) in each CT image; a classifier (Segmentation-CNN in this study) was then trained and applied to classify each adipose tissue pixel as belonging to SFA or VFA by using their neighborhood pixels and location information as input.
There have been a couple of previous works that applied CNN methods for medical image segmentation tasks [17, 18, 20–22]. The basic method is a patch-wise classification process. Specifically, in order to classify each single pixel into its correct class, a 2-D rectangle patch centered at the specific pixel was extracted, resized and used as the input of a CNN classifier. The outputs of CNN indicated the probability of the center pixels belonging to each class. The advantage of patch-wise classification is that by extracting separate and possible overlapping image patches, we can collect a training set that is large enough for the requirement of deep CNN architectures. However, spatial consistency and pixel location information are lost to some degree in this basic method. Many approaches have been applied to overcome this limitation, including post-processing, increasing the size of image patches and using location information as part of the input of CNN, etc. [17, 18, 21, 22].
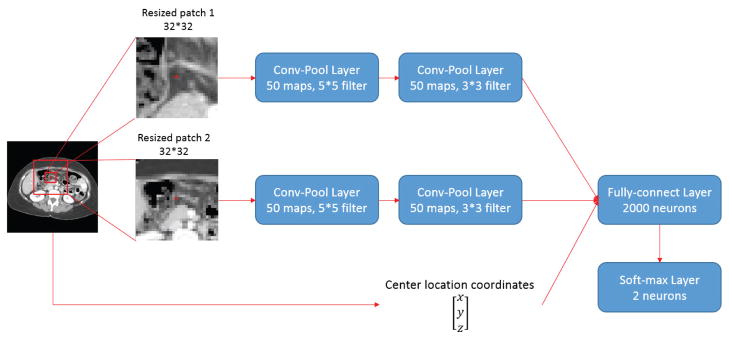
In this study, we developed the Segmentation-CNN architecture as shown in Figure 4. The objective of Segmentation-CNN is to classify an adipose tissue pixel as belonging to SFA or VFA. It is a 2-scale CNN architecture where the patches with smaller size are used to represent fine details around the target pixel and patches with larger size are used to maintain global consistency. The patch sizes of 64×64 and 128×128 were used because they yield better results than others. Multi-scale CNN architecture was not considered because it will generate larger network and take longer time to train and execute. In addition, it may also suffer from the over-fitting problems.
Figure 4.
Architecture of Segmentation-CNN
The input of Segmentation-CNN consists of three parts. The first part is a 32×32 rectangular patch obtained by down-sampling a 64×64 patch centered at the adipose tissue pixel depicted on the CT slice, which is the representation of fine details around the target adipose tissue pixel. Two convolutional-pooling layer pairs were stacked to get high-level image patch features. The numbers of feature maps were 50 for both two layers. The filter sizes were 5×5 and 3×3, respectively, while the sizes of max-pooling were 2×2. The second part of Segmentation-CNN input is a 32×32 rectangular patch obtained by down-sampling a 192×192 patch centered at the adipose tissue pixel, which was designed to preserve more spatial consistency. Same convolutional-pooling layer pairs were employed to get high-level features. The third part is a 3-D vector that contained the normalized and adjusted spatial location coordinates of the adipose tissue pixel. A fully connected layer with 2,000 hidden neurons was used to fuse the information and high-level features from the three parts of input. The dimension of the input of fully-connected layer is 50 (feature maps) × 6×6 (size of each feature map after two conv-pool layers) × 2 (two convolutional channel) + 3 (location coordinates). A soft-max layer was finally applied to generate the likelihood based prediction scores.
In addition, we also evaluated the performance of a baseline CNN for comparison. The baseline CNN only consists of one channel which is a resized 32×32 rectangular patch obtained from a 64×64 patch in the image. Two convolutional-pooling layer pairs and one fully-connected layer were stacked to build the network. The numbers of feature maps were 50 for both the convolutional layers. The filter sizes were 5×5 and 3×3 respectively and the fully-connected layer contained 1,000 hidden neurons.
2.4. Experiments and evaluation
2.4.1. Selection-CNN
The loss function of Selection-CNN is set to be the summation of a negative log-likelihood term and a L2 regularization term. Mini-batch stochastic gradient descent (SGD) methods were applied to minimize the loss function. SGD is one of the most popular and widely used training method in machine learning (especially in deep learning) applications [16]. It is more efficient for large-scale learning problems compared to some other training methods such as second-order approaches. Specifically, the training set was split into a number of batches; the gradient of loss function was estimated over each batch instead of the whole training set. By using mini-batch SGD, the parameters got more frequent updates and training efficiency was greatly improved. Here we set mini-batch size equal to 50 and iteratively trained the Selection-CNN for 50 epochs to get the optimal parameters.
After obtaining the raw Selection-CNN output, a post-processing step was performed to check and ensure the spatial consistency. Specifically, a one-dimensional median filter was performed to smooth the outputs (i.e. the probabilities of each CT slice belonging to positive or negative class). The longest consecutive CT slices with probabilities of being positive greater than 0.5 were predicted as positive and the remaining slices were predicted as negative.
For each CT case in the testing group, we compared the manually processed labels and the results generated by Selection-CNN with post-processing step. We computed the prediction accuracy, sensitivity and specificity for each testing case and averaged them over all 20 cases in the testing group for performance evaluation.
2.4.2. Segmentation-CNN
Similar to Selection-CNN, a mini-batch SGD was employed to train the Segmentation-CNN network to get optimal parameters. The loss function of Segmentation-CNN is solely a negative log-likelihood function since we have collected enough training samples. The size of mini-batch was set to 500 and the iteration time was set to 400 epochs.
For each input (i.e. a 512×512 CT image) in the testing dataset, CAD scheme firstly applied the body trunk segmentation algorithm [31] to remove the background. The pixels inside the body trunk were scanned one by one. If the pixel has a CT number between the threshold −140 HU and −40 HU, a neighborhood patch and location information were extracted and used as input of Segmentation-CNN. A likelihood score generated by Segmentation-CNN was used to label the pixel as belonging to either SFA or VFA. In order to evaluate the performance of this Segmentation-CNN based scheme, 120 CT images from the 20 patients in the testing patient group were randomly selected and manually labelled. For each CT image, the segmentation result generated subjectively and by Segmentation-CNN based scheme were compared. Pixel-wise prediction accuracy and dice coefficients (DC) of SFA/VFA were calculated for each image and averaged over the 120 CT images for performance evaluation. Dice coefficient is a similarity measurement index commonly used to evaluate the performance of image segmentation tasks. It calculates the ratio of overlapping volume between two segmented areas. The formula of DC with respect to two segmentation areas A and B is shown as below [21]:
3. Results
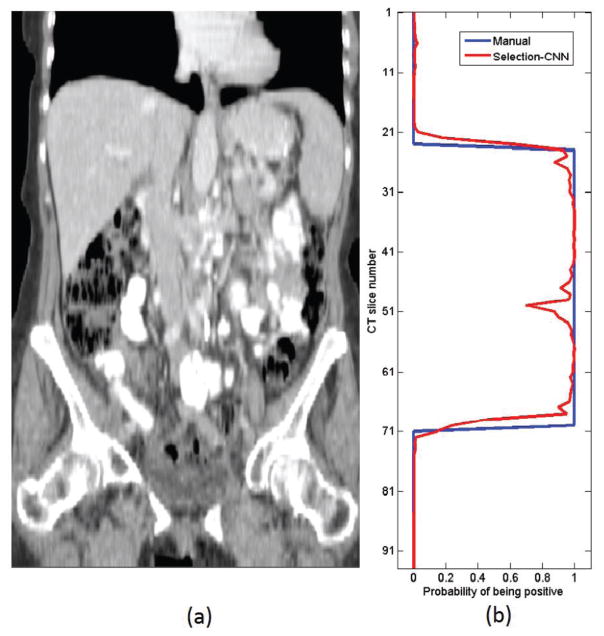
The training and evaluation process of CNNs were performed on a Dell T3610 workstation equipped with a quadcore 3.00GHz processor, 8 Gb RAM and a NVidia Quadro 600 GPU card. The models were implemented in Python using Theano library [32]. Figure 5 shows an example of the comparison between the abdomen area labelled subjectively by an observer and generated by the optimized Selection-CNN. Table 1 summarized and compared the quantitative performance evaluation results (i.e. accuracy and DC) of Selection-CNN with and without post-processing. Specifically, by using individual CT scans independently as the input of Selection-CNN, we can obtain averaged prediction accuracy and DC over 90%. Adding the post-processing steps enabled to further improve performance by taking the spatial consistence into account. Finally, the study results yielded a mean prediction accuracy equal to 0.9582 with standard deviation 0.0268, mean sensitivity equal to 0.9481 with standard deviation 0.0595 and mean specificity equal to 0.9625 with standard deviation 0.0521, respectively. The improvement is statistically significant by using paired t-tests (p < 0.005 for all three evaluation indices).
Figure 5.
Examples of abdomen area selected by Selection-CNN. (a) A patient CT scan from vertical view. (b) Comparison of abdomen area selected by an observer (blue line) and by optimized Selection-CNN (red line)
Table 1.
Summarization of quantitative performance evaluation of Selection-CNN without and with post-processing
| Prediction accuracy | Sensitivity | Specificity | |
|---|---|---|---|
| Selection-CNN | 0.9352 ± 0.0344 | 0.9287 ± 0.0690 | 0.9362 ± 0.0722 |
| Selection-CNN with post-processing | 0.9582 ± 0.0268 | 0.9481 ± 0.0595 | 0.9625 ± 0.0521 |
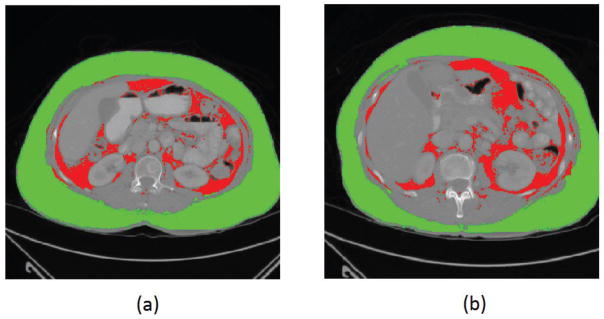
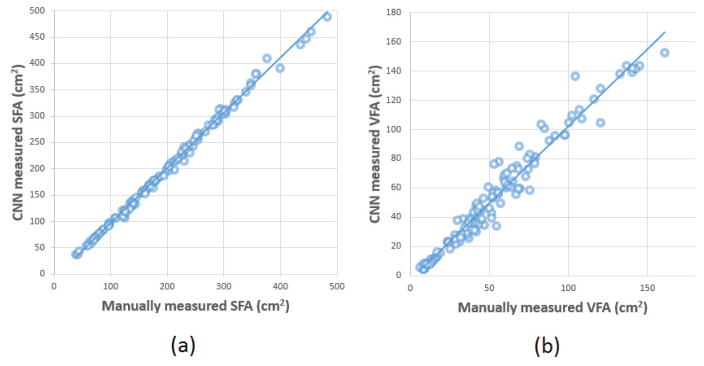
Table 2 summarized and compared the performance evaluation results of the proposed Segmentation-CNN and a baseline CNN architecture which only used the 64×64 neighborhood patches as input. It shows that the performance of Segmentation-CNN is statistically significantly better than the baseline CNN (p < 0.005 for all the three evaluation methods). Figure 6 shows two examples of segmentation results generated by Segmentation-CNN for segmenting SFA and VFA depicting on CT image slices. Figure 7 shows two scatter plots of the volumes of manually labelled SFA/VFA and CAD measured SFA/VFA among all the 120 testing CT images. The correlation coefficients are 0.9980 with 95% confidence interval (CI) (0.9972, 0.9986) for SFA and 0.9799 with 95% CI (0.9712, 0.9859) for VFA respectively.
Table 2.
Summarization of quantitative performance evaluation of baseline CNN and Segmentation-CNN
| Evaluation | Pixel-wise prediction accuracy | SFA Dice coefficient | VFA Dice coefficient |
|---|---|---|---|
| Method | |||
| A baseline CNN | 0.9535 ± 0.0253 | 0.9696 ± 0.0175 | 0.8890 ± 0.0563 |
| Segmentation-CNN | 0.9682 ± 0.0218 | 0.9797 ± 0.0145 | 0.9150 ± 0.0624 |
Figure 6.
Examples showing the segmentation of VFA and SFA generated by Segmentation-CNN in two CT image slices. In these two images, SFA is shown in green color and VFA is represented by red color.
Figure 7.
The scatter plots of the manually and automatically (i.e. by Segmentation-CNN) measured (a) SFA and (b) VFA volume.
To evaluate the stability of our CAD scheme, following are the results of our two experiments. First, by repeated training the Selection-CNN with different random initialized weights for 50 times, the Selection-CNNs without post-processing steps yielded a mean prediction accuracy of 0.9324, which is quite similar to the result reported in Table 1 (0.9352). The highest, median and lowest accuracies among the 50 experiments are 0.9456, 0.9328 and 0.9167, respectively. Second, the 50 pairs of repeated experiment that compared the performance of the trained Selection-CNN using balanced versus unbalanced datasets yielded an equal mean prediction accuracy of 0.9324. However, the standard deviations were 0.0119 and 0.0066 when using 757 pairs of “balanced” training samples and all available 2,240 “unbalanced” training samples, respectively, which indicates that using all available training samples increase diversity of training dataset and yielded more stable testing results.
4. Discussion
Identifying and computing new quantitative image markers for predicting efficacy of cancer treatment has been a focused research topic in our recent studies [33–35]. However, automatically segmenting SFA and VFA from volumetric CT image data is quite different from and/or more complicated than segmenting the targeted tumors from a few image slices. Thus, previous CAD schemes in this field are semi-automated schemes [7–9] that require manual selection of one targeted CT image slice and possible adjustment of placing segmentation seeds or boundary conditions. In this study, for the first time we developed a fully-automated CAD scheme and demonstrated its feasibility to segment volumetric SFA and VFA data without any human intervention. This study and the new CAD scheme have a number of unique characteristics. First, a Selection-CNN with post-processing step was developed for automated selection of CT slices belonging to abdomen areas. This CNN based process can not only overcome the limitation of manual selection in most previous studies which were quite difficult to deal with large-scale datasets, and also generated high segmentation accuracy as compared to manually processed results or “ground-truth” (i.e. yielding a prediction accuracy and DC greater than 0.95). Therefore, the Selection-CNN for automated selection of abdomen CT slices is reliable and can be used to replace manual selection, which provide the capability of managing large-scale dataset based medical data analysis studies with high efficiency.
Second, in most of the previously developed schemes, the segmentation of SFA and VFA was obtained by detecting visceral masks or abdomen wall masks using sequences of traditional image processing techniques such as morphological operations, pixel labelling and thresholding [5, 8, 25–30]. Therefore, the segmentation results might be sensitive to the selection of parameters (e.g. morphological operation kernels and distance thresholds); these parameters were mostly subjectively determined and the optimal values might be different for different patients. In this study, we developed a Segmentation-CNN based scheme to segment SFA and VFA, which was based on machine learning classifiers and thus provided a more “intelligent” way by considering the location coordinates and neighborhood information. After sufficiently trained and optimized, the CNN model is free of parameters and no human intervention is required for getting optimal segmentation results. Therefore, the Segmentation-CNN provided a reliable CAD scheme for fully-automated segmentation of SFA and VFA from single CT slices.
Third, the two tasks in this study (i.e. selection and segmentation) were both formulated as binary classification problems and Convolutional neural networks were employed as the classifier to solve the problems. In traditional machine learning classifiers (e.g. Support Vector Machines and random forests) based systems, how to design and select effective and discriminative features is a crucial but difficult task. The advantage of convolutional neural network is that it can automatically learn hierarchical feature representations from its raw input images and therefore, no manual feature extraction and selection process is needed [15]. Following the success of CNNs in many other computer vision and medical image analysis areas, this study demonstrated that CNN models are effective for recognizing CT slices that belong to abdomen areas and segmenting SFA and VFA from single CT slices.
Fourth, we further investigated and employed new approaches of adding post-processing and a 2-scale network to maintain global consistency and improve prediction accuracy. The study results showed that after using these new approaches CAD scheme enabled to yield significantly higher segmentation accuracy than the raw CNN outputs. Thus, this study provided an example of how to optimally apply CNN with consideration of spatial or location information in developing a deep learning based scheme. The stability of the deep learning based CAD scheme has also been tested and approved by the experiments using repeated random initializations and different training datasets with balanced and unbalanced training samples in two classes.
In addition, based on the experimental results, we also made several observations. For example, (1) although direct comparison of segmentation accuracy between using this new deep learning scheme (3D data) and previous semi-automated schemes (2D data) is difficult, the new automated scheme can achieve high segmentation accuracy as comparing to the general visual segmentation. (2) Unlike the conventional machine learning methods, which should be optimally trained using a balanced dataset with equal number of training samples in two classes, a deep learning based CAD scheme can be optimally trained without such a restriction. Thus, the deep learning scheme may have an advantage to build a more stable classification model using all available training samples with increased diversity.
Despite the encouraging results, this is a preliminary technology development study with a number of limitations. First, we one applied and evaluated a commonly used CNN architecture, activation functions, loss functions and the training methods in this study. The performance of this CAD system can be potentially improved by employing other advanced deep learning models and methodologies. Second, it took a relatively long time (i.e. a few minutes) to segment SFA and VFA in each single CT slice. This is because that each adipose pixel is used as an independent input of the deep network and the CAD scheme needs to scan all the pixels in the image. More research efforts should be devoted to investigate how to improve the computational efficiency of this system. For example, applying an optimal sampling and/or super-pixel concept might be helpful for reducing the amount of pixel-wise classification process in the system. Thus, at current stage, this deep learning based CAD scheme can only be used offline to process the images and segment SFA and VFA from the images. Last, the clinical potential of this new technology needs to be evaluated in the clinical studies to validate whether using volumetric adiposity-related image features can significantly improve accuracy in predicting disease prognosis (e.g. response of ovarian cancer patients to chemotherapies) as compared to previous manual or semi-automated methods that measured adiposity from one selected single CT slice.
In summary, in order to overcome the limitation of estimating SFA and VFA from one subjectively selected single CT image slice, we developed and tested a new CAD scheme for adipose tissue segmentation and quantification based on a sequential two-step process including (1) selecting CT slices belonging to abdomen areas and (2) segmenting SFA and VFA from each of the selected CT slices. We demonstrated that applying this new deep learning CNN based CAD scheme enabled to recognize specific body parts (abdomen in this study) from volumetric CT image data and segment SFA and VFA from each selected CT slice with high accuracy or agreement with manual segmentation results. As a result, this study provided researchers a new and reliable CAD tool to assist processing volumetric CT data and quantitatively computing a new adiposity related imaging marker in the future clinical practice.
Highlights.
A Selection-CNN was developed for automatically selecting abdomen part from CT slices.
A Segmentation-CNN was developed for automatic segmentation of SFA and VFA.
Both two schemes can generate high accuracy compared to manual selection/segmentation.
A fully automatic CAD scheme for quantifying adiposity tissue volumes was provided.
Acknowledgments
This work is supported in part by Grants of R01 CA197150 from the National Cancer Institute, National Institutes of Health and HR15-016 from Center for the Advancement of Science and Technology, State of Oklahoma. The authors would also like to acknowledge the support from the Peggy and Charles Stephenson Cancer Center, University of Oklahoma.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of childhood and adult obesity in the United States, 2011–2012. Jama. 2014;311:806–814. doi: 10.1001/jama.2014.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kissebah AH, VYDELINGUM N, MURRAY R, EVANS DJ, KALKHOFF RK, ADAMS PW. Relation of Body Fat Distribution to Metabolic Complications of Obesity*. The Journal of Clinical Endocrinology & Metabolism. 1982;54:254–260. doi: 10.1210/jcem-54-2-254. [DOI] [PubMed] [Google Scholar]
- 3.Mun EC, Blackburn GL, Matthews JB. Current status of medical and surgical therapy for obesity. Gastroenterology. 2001;120:669–681. doi: 10.1053/gast.2001.22430. [DOI] [PubMed] [Google Scholar]
- 4.Després JP, Lemieux I, Bergeron J, Pibarot P, Mathieu P, Larose E, Rodés-Cabau J, Bertrand OF, Poirier P. Abdominal obesity, the metabolic syndrome: contribution to global cardiometabolic risk. Arteriosclerosis, thrombosis, and vascular biology. 2008;28:1039–1049. doi: 10.1161/ATVBAHA.107.159228. [DOI] [PubMed] [Google Scholar]
- 5.Makrogiannis S, Caturegli G, Davatzikos C, Ferrucci L. Computer-aided assessment of regional abdominal fat with food residue removal in CT. Academic radiology. 2013;20:1413–1421. doi: 10.1016/j.acra.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guiu B, Petit JM, Bonnetain F, Ladoire S, Guiu S, Cercueil J-P, Krausé D, Hillon P, Borg C, Chauffert B. Visceral fat area is an independent predictive biomarker of outcome after first-line bevacizumab-based treatment in metastatic colorectal cancer. Gut. 2010;59:341–347. doi: 10.1136/gut.2009.188946. [DOI] [PubMed] [Google Scholar]
- 7.Slaughter KN, Thai T, Penaroza S, Benbrook DM, Thavathiru E, Ding K, Nelson T, McMeekin DS, Moore KN. Measurements of adiposity as clinical biomarkers for first-line bevacizumab-based chemotherapy in epithelial ovarian cancer. Gynecologic oncology. 2014;133:11–15. doi: 10.1016/j.ygyno.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Thai T, Moore K, Ding K, Mcmeekin S, Liu H, Zheng B. Quantitative measurement of adiposity using CT images to predict the benefit of bevacizumab-based chemotherapy in epithelial ovarian cancer patients. Oncology Letters. 2016;12:680–686. doi: 10.3892/ol.2016.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshizumi T, Nakamura T, Yamane M, Waliul Islam AHM, Menju M, Yamasaki K, Arai T, Kotani K, Funahashi T, Yamashita S. Abdominal Fat: Standardized Technique for Measurement at CT. Radiology. 1999;211:283–286. doi: 10.1148/radiology.211.1.r99ap15283. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Qiu Y, Thai T, Moore K, Liu H, Zheng B. Applying a computer-aided scheme to detect a new radiographic image marker for prediction of chemotherapy outcome. BMC Medical Imaging. 2016;16:52. doi: 10.1186/s12880-016-157-5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Advances in neural information processing systems. 2012:1097–1105. [Google Scholar]
- 12.Lawrence S, Giles CL, Tsoi AC, Back AD. Face recognition: A convolutional neural-network approach. Neural Networks, IEEE Transactions on. 1997;8:98–113. doi: 10.1109/72.554195. [DOI] [PubMed] [Google Scholar]
- 13.LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proceedings of the IEEE. 1998;86:2278–2324. [Google Scholar]
- 14.Simard PY, Steinkraus D, Platt JC. Best practices for convolutional neural networks applied to visual document analysis, null. IEEE. 2003:958. [Google Scholar]
- 15.Bengio Y. Learning deep architectures for AI. Foundations and trends® in Machine Learning. 2009;2:1–127. [Google Scholar]
- 16.Jones N. Computer science: The learning machines. Nature. 2014;505:146–148. doi: 10.1038/505146a. [DOI] [PubMed] [Google Scholar]
- 17.Brebisson A, Montana G. Deep Neural Networks for Anatomical Brain Segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops; 2015; pp. 20–28. [Google Scholar]
- 18.Cernazanu-Glavan C, Holban S. Segmentation of bone structure in X-ray images using convolutional neural network. Adv Electr Comput Eng. 2013;13:87–94. [Google Scholar]
- 19.Cireşan DC, Giusti A, Gambardella LM, Schmidhuber J. Mitosis detection in breast cancer histology images with deep neural networks. Medical Image Computing and Computer-Assisted Intervention–MICCAI; 2013; Springer; 2013. pp. 411–418. [DOI] [PubMed] [Google Scholar]
- 20.Davy A, Havaei M, Warde-farley D, Biard A, Tran L, Courville A, Larochelle H, Pal C, Bengio Y. Brain tumor segmentation with deep neural networks. 2014 doi: 10.1016/j.media.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Dubrovina A, Kisilev P, Ginsburg B, Hashoul S, Kimmel R. Computational Mammography using Deep Neural Networks. [Google Scholar]
- 22.Roth HR, Lu L, Farag A, Shin H-C, Liu J, Turkbey EB, Summers RM. Deeporgan: Multi-level deep convolutional networks for automated pancreas segmentation. Medical Image Computing and Computer-Assisted Intervention--MICCAI; 2015; Springer; 2015. pp. 556–564. [Google Scholar]
- 23.Shin HC, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE transactions on medical imaging. 2016;35:1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan Z, Zhan Y, Peng Z, Liao S, Shinagawa Y, Zhang S, Metaxas DN, Zhou XS. Multi-Instance Deep Learning: Discover Discriminative Local Anatomies for Bodypart Recognition. IEEE transactions on medical imaging. 2016;35:1332–1343. doi: 10.1109/TMI.2016.2524985. [DOI] [PubMed] [Google Scholar]
- 25.Chung H, Cobzas D, Birdsell L, Lieffers J, Baracos V. Automated segmentation of muscle and adipose tissue on CT images for human body composition analysis; SPIE Medical Imaging, International Society for Optics and Photonics; 2009. pp. 72610K-72610K–72618. [Google Scholar]
- 26.Hussein S, Green A, Watane A, Papadakis G, Osman M, Bagci U. Context Driven Label Fusion for segmentation of Subcutaneous and Visceral Fat in CT Volumes. 2015 arXiv preprint arXiv:1512.04958. [Google Scholar]
- 27.Kim YJ, Lee SH, Kim TY, Park JY, Choi SH, Kim KG. Body fat assessment method using CT images with separation mask algorithm. Journal of digital imaging. 2013;26:155–162. doi: 10.1007/s10278-012-9488-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mensink SD, Spliethoff JW, Belder R, Klaase JM, Bezooijen R, Slump CH. Development of automated quantification of visceral, subcutaneous adipose tissue volumes from abdominal CT scans. SPIE Medical Imaging, International Society for Optics and Photonics. 2011:79632Q-79632Q–79612. [Google Scholar]
- 29.Romero D, Ramirez JC, Mármol A. Quanification of subcutaneous and visceral adipose tissue using CT, Medical Measurement and Applications, 2006. MeMea 2006. IEEE International Workshop on, IEEE; 2006; pp. 128–133. [Google Scholar]
- 30.Zhao B, Colville J, Kalaigian J, Curran S, Jiang L, Kijewski P, Schwartz LH. Automated quantification of body fat distribution on volumetric computed tomography. Journal of computer assisted tomography. 2006;30:777–783. doi: 10.1097/01.rct.0000228164.08968.e8. [DOI] [PubMed] [Google Scholar]
- 31.Leader JK, Zheng B, Rogers RM, Sciurba FC, Perez A, Chapman BE, Patel S, Fuhrman CR, Gur D. Automated lung segmentation in X-ray computed tomography: development and evaluation of a heuristic threshold-based scheme 1. Academic Radiology. 2003;10:1224–1236. doi: 10.1016/s1076-6332(03)00380-5. [DOI] [PubMed] [Google Scholar]
- 32.Bergstra J, Breuleux O, Bastien F, Lamblin P, Pascanu R, Desjardins G, Turian J, Warde-Farley D, Bengio Y. Theano: a CPU and GPU math expression compiler. Proceedings of the Python for scientific computing conference (SciPy); Austin, TX. 2010; p. 3. [Google Scholar]
- 33.Aghaei F, Tan M, Hollingsworth AB, Qian W, Liu H, Zheng B. Computer-aided breast MR image feature analysis for prediction of tumor response to chemotherapy. Medical Physics. 2015;42:6520–6528. doi: 10.1118/1.4933198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emaminejad N, Qian W, Guan Y, Tan M, Qiu Y, Liu H, Zheng B. Fusion of quantitative image features and genomic biomarkers to improve prognosis assessment of early stage lung cancer patients. IEEE Transactions on Biomedical Engineering. 2016;63:1034–1043. doi: 10.1109/TBME.2015.2477688. [DOI] [PubMed] [Google Scholar]
- 35.Qiu Y, Tan M, McMeekin S, Thai T, Ding K, Moore K, Liu H, Zheng B. Early prediction of clinical benefit of treating ovarian cancer using quantitative CT image feature analysis. Acta Radiologica. 2016;57:1149–1155. doi: 10.1177/0284185115620947. [DOI] [PMC free article] [PubMed] [Google Scholar]