Abstract
A porcine enteric calicivirus (PEC), strain Cowden in the genus Sapovirus of the Caliciviridae family, can be propagated in a porcine kidney continuous cell line (LLC-PK) in the presence of bile acids in the cell culture medium. A full-length cDNA copy of the Cowden PEC genome was cloned into a plasmid vector directly downstream from the T7 RNA polymerase promoter, and capped RNA transcripts derived from this clone were infectious when transfected into LLC-PK cells. The recovery of PEC after transfection of RNA transcripts was dependent on the presence of bile acids, consistent with our recent identification of a bile acid-mediated signaling pathway required for PEC replication (Chang et al., Proc. Natl. Acad. Sci. USA 101:8733-8788, 2004). Recovery of virus was verified by detection of PEC antigen in transfected cells by immunofluorescence and enzyme-linked immunosorbent assays, direct observation of recovered viral particles by electron microscopy, and partial sequence analysis of their genomes (first 1,070 nucleotides) to differentiate them from tissue culture-adapted parental virus. The recovered virus retained its ability to infect piglets when administered by the oral route and showed an attenuated phenotype similar to that of the tissue culture-adapted parental virus. This reverse genetics system for PEC provides a new tool to study the molecular basis of replication and pathogenesis for caliciviruses associated with diarrheal disease.
Caliciviruses (family Caliciviridae) are small, nonenveloped viruses of 27 to 35 nm in diameter (3). They possess a single strand, plus-sense genomic RNA of 7 to 8 kb, and four genera have been established in the family: Vesivirus, Lagovirus, Norovirus, and Sapovirus (9). Caliciviruses are veterinary and human pathogens associated with a wide range of economically important diseases in their respective hosts. Feline calicivirus (FCV), a member of the genus Vesivirus, causes respiratory disease in cats (4, 8) and is controlled by vaccination. A member of the genus Lagovirus, rabbit hemorrhagic disease virus, is associated with a fatal liver disease in rabbits (17, 18). Caliciviruses in the genera Norovirus and Sapovirus are predominantly enteric pathogens in humans and animals. Recent studies estimate that the noroviruses are responsible for more than 90% of nonbacterial gastroenteritis outbreaks (5, 10). Sapoviruses have also been associated with gastroenteritis outbreaks and with disease in pediatric patients (10), but the extent of their role in diarrheal disease is not yet known. Because most enteric caliciviruses cannot be grown in cell culture, it has been difficult to study pathogenesis and immunity to these ubiquitous pathogens.
Porcine enteric calicivirus (PEC), a member of the genus Sapovirus, can be grown in cell culture and serves as a model for studies of replication and enteropathogenesis of the enteric caliciviruses. The polyadenylated single-stranded RNA genome is 7,320 nucleotides (nt) in length [excluding the poly(A) tail] and organized into two major open reading frames (ORFs), ORF1 (nt 10 to 6774) and ORF2 (nt 6603 to 7265) (11). ORF1 encodes a large polyprotein that contains coding sequences for the nonstructural proteins and the major capsid protein, VP1. ORF2 encodes the minor structural protein, VP2.
The Cowden PEC strain was adapted to grow in a continuous cell line (LLC-PK) (6, 19), and growth was dependent on the presence of an intestinal content (IC) fluid filtrate from uninfected gnotobiotic pigs in the virus growth medium. We recently identified bile acids as active factors in IC that support the replication of PEC in LLC-PK cells and proposed that the bile acid-associated induction of a cellular cyclic AMP (cAMP)/protein kinase A (PKA) pathway was involved in the down-regulation of STAT1 (2). It is possible that enteric viruses such as PEC may have evolved in the bile acid-rich environment of the gut to replicate efficiently by the utilization of cell-signaling pathways and a down-regulation of innate immunity. The availability of a bile acid-dependent cell culture system and an animal model for PEC provides an opportunity to elucidate the mechanisms responsible for the pathogenesis of calicivirus diarrheal disease in a natural host, as well as viral replication in cells. In this study, we report the development of a reverse genetics system for PEC and show that bile acids are essential for recovery of progeny viruses in cultured porcine kidney cells following transfection of RNA transcripts derived from a full-length cDNA clone of the Cowden PEC genome. A reverse genetics system for PEC should provide an important tool for the study of this virus both in vitro and in vivo.
MATERIALS AND METHODS
Cells, viruses, and antisera.
LLC-PK cells (American Type Culture Collection, Manassas, Va.) were maintained in a mixture (50:50) of Eagle's minimal essential medium (E-MEM) and Opti-MEM (Invitrogen, Carlsbad, Calif.) containing 5% fetal bovine serum and antibiotics (amphotericin B, 2.5 μg/ml; chlortetracycline, 25 μg/ml; penicillin, 250 U/ml; and streptomycin, 250 μg/ml). The cell culture-adapted Cowden PEC (Po/SV/Cowden/1980/US) was serially passaged at least 27 times in LLC-PK cells in the presence of IC (1%) from uninfected gnotobiotic pigs in the cell culture medium (6, 19). The IC was collected from uninfected gnotobiotic pigs, prepared as a 0.45-μm-pore-size filtrate, and stored in aliquots at −20°C as described previously (6, 19). The bile acids taurochenodeoxycholic acid (TCDCA) and glycochenodeoxycholic acid (GCDCA) were purchased from Sigma (St. Louis, Mo.) and resuspended in distilled water. Hyperimmune antiserum specific for the PEC capsid protein was prepared by intramuscular immunization of guinea pigs with recombinant virus-like particles (rVLPs) obtained by expression of the PEC capsid-encoding gene in the baculovirus system (13). Hyperimmune antiserum specific for native PEC virions was prepared by the administration of virulent Cowden PEC to pigs by the oral route, followed by intramuscular injection of purified cell culture-adapted Cowden PEC virions (7). Hyperimmune sera specific for other Cowden PEC-encoded proteins (VPg, RNA polymerase, and ORF2) were raised in guinea pigs by immunization as described previously with recombinant proteins generated in a bacterial expression system (detailed below) (23). The specificity of each region-specific antiserum was shown by radioimmunoprecipitation assay (RIPA) with PEC-infected cell lysates (data not shown).
PEC detection assays.
The detection of PEC was accomplished with one or more of the following assays: (i) reverse transcription-PCR (RT-PCR), (ii) immunofluorescence assay (IFA), and (iii) enzyme-linked immunosorbent assay (ELISA).
(i) RT-PCR.
The PEC RNA was extracted by using an RNeasy kit (QIAGEN, Valencia, Calif.) according to the instructions provided by the manufacturer. The extracted RNA was reverse transcribed with primer PEC5kA (Table 1) by using a first-strand cDNA synthesis kit (Invitrogen) at 42°C for 1 h. The cDNA was then amplified by PCR with Taq DNA polymerase (Invitrogen) and the primer pair of PEC4kM and PEC5kA (Table 1), producing an approximately 700-bp fragment. The PCR was performed for 30 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and elongation at 72°C for 90 s, followed by an extension at 72°C for 7 min.
TABLE 1.
Primers used in construction of full-length clone, detection of PEC, and sequence analysis in this study
| Primer | Sequencea | Polarity | Location in PEC (nt)b |
|---|---|---|---|
| PEC5′/NheI/T7 | tcactggctagcCTGGTAATACG | + | 5′ end |
| ACTCACTATAGTGATCGTGATGGC | |||
| TAATTGCCGTCCG | |||
| PEC3′ (T30)/ NotI | gagtgaccgcggccgcT(35)GCCCCA CAGCCGCCACATGTGTAAGG | − | 3′ end |
| PEC/ORF1/3′/ NotI | gagtgaccgcggccgcTCATCGTG | − | 6751-6774 |
| AGCTGTGAATGGACCTTCCTGATG | |||
| TCCCACTTCGTCCAATGAGGGG | |||
| PEC0kM | ACATCACTGTTGCATGAGGTG | + | 301-321 |
| PEC1kS | ACGCCGCAGGCAGGCAATGAT | + | 931-951 |
| PEC1kA | AGCCACCGTCGAGGTGATAA | − | 1052-1072 |
| PEC1kM | ACCTCTGGCTGGGTTGATAC | + | 1281-1300 |
| PEC2kS | CTGATTAAAGAGAAGTACAAC | + | 1930-1950 |
| PEC2kA | CTGTGTTCGGGTAGGAGTGGAA | − | 2041-2062 |
| PEC2kM | TCAACAAGGTCATGATGCAGG | + | 2279-2299 |
| PEC3kA | ACTTGCCGATGATGTCCACAA | − | 3050-3070 |
| PEC3kS | AGTTCCTGGATCTCAAGGAA | + | 2930-2949 |
| PEC3kM | GAGGTGGAGTTCAAGGTCGCA | + | 3271-3291 |
| PEC4kA | AACACCACGGTGCGCCATAT | − | 4052-4071 |
| PEC4kS | TCGAATCAATGGCCCATGACA | + | 3932-3952 |
| PEC4kM | TGAACAAATGAACCAAGCCT | + | 4281-4300 |
| PEC5kA | GTTGTACGTTGCCACTGCG | − | 5052-5070 |
| PEC5kS | CTGGAAGTGATGCTTGCATAT | + | 4930-4950 |
| PEC5kM | CGAGCAACCCAGAGGGCACT | + | 5271-5290 |
| PEC6kA | ATGTTGGGGTAAAGGGGCCCATT | − | 6052-6074 |
| PEC6kS | TTTTCAGCTGAGGGCACCAC | + | 5929-5948 |
| PEC6kM | GCAACATCAATGGCAGTATGC | + | 6274-6294 |
| PEC7kA | TGTTTGTTGCCCACCAAGGTA | − | 7053-7073 |
| VPg-pF | GCGAAAGGGAAAAACAAACGC | + | 2812-2831 |
| VPg-pR | CTCACGCCTGTGGCCTGACTTGC | − | 3065-3087 |
| Pol-pF | ATGCCTGTGGTGGTCAAGAACC | + | 4684-4705 |
| Pol-pR | CACGAACACTTCTGGCTCTTCATC | − | 5116-5139 |
| ORF2-F | ATGAGTTGGATTGCAGGAGCAATG | + | 6771-6794 |
| ORF2-R | TCACACTTTGCTGTGAGTGGTGG | − | 7243-7265 |
The nonviral sequences of the oligonucleotides are shown in lowercase, and restriction sites used for subcloning are underlined. T7 promoter sequences are in uppercase italic.
nt, nucleotides.
(ii) IFA.
PEC capsid antigen expression in LLC-PK cells was detected by IFA. At various times after infection or transfection, cells were fixed with 100% cold methanol followed by incubation with hyperimmune sera specific for rVLPs or native virions and fluorescein isothiocyanate-conjugated anti-guinea pig or pig immunoglobulin G (Kirkegaard & Perry Laboratories, Gaithersburg, Md.).
(iii) ELISA.
ELISAs were performed as described previously (1). Briefly, 96-multiwell plates (Nunc, Rochester, N.Y.) were coated with hyperimmune guinea pig antiserum specific for the Cowden PEC rVLPs as capture antibody. Reagents were added to duplicate wells in the following sequence: 1:10 dilutions of test samples; pig hyperimmune antiserum against Cowden PEC virions; goat anti-pig immunoglobulin G-conjugated horseradish peroxidase; and substrate tetramethylbenzidine (Kirkegaard & Perry Laboratories). The absorbance was measured at 630 nm. Each cutoff value was expressed as an absorbance at 630 nm that was at least 3 standard deviations above the absorbance in LLC-PK cell control wells.
Generation of a full-length cDNA clone of Cowden PEC.
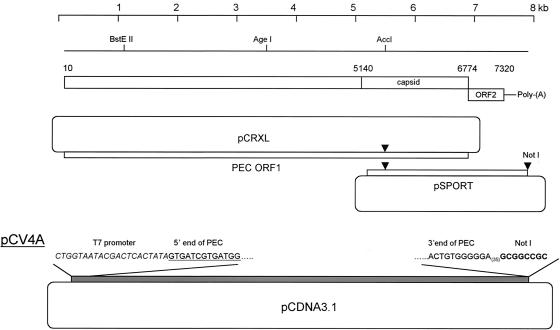
The cell culture-adapted Cowden PEC (passage number 27) grown in LLC-PK cells in the presence of IC was concentrated by ultracentrifugation (100,000 × g for 2 h), and viral RNA was extracted by using the RNeasy Kit (QIAGEN). The cDNA of the full-length genome or ORF1 of PEC was synthesized by using a first-strand cDNA synthesis kit (Invitrogen) with primer PEC3′(T30)/NotI or PEC/ORF1/3′/NotI (Table 1), respectively. PCR was performed with the Elongase Kit (Invitrogen) to amplify either the full-length genome or ORF1 of PEC with the primer pairs of PEC5′/NheI/T7 and PEC3′(T30)/NotI or PEC5′/NheI/T7 and PEC/ORF1/3′/NotI, which generated an approximately 7.4-or 6.8-kbp product, respectively. The primer PEC5′/NheI/T7 contained an NheI enzyme site, T7 promoter, and the first 27 nt of PEC. The amplified products were cloned into the pCRXL vector (Invitrogen). At least three clones containing either the full-length genome or ORF1 were identified, and sequence analysis of each clone revealed the presence of sporadic point mutations (up to 20 nt) in comparison to the tissue culture-adapted PEC sequence in the GenBank database (accession number AF182760). A cDNA library of PEC was generated by using the SuperScript plasmid system (Invitrogen) according to the instructions provided by the supplier. Screening of the cDNA library (in pSPORT vector) identified a recombinant clone containing a consensus sequence of the subgenomic region of Cowden PEC. With the use of unique restriction sites (AccI and NotI), the subgenomic region of Cowden PEC in pSPORT was engineered into the corresponding sites of the pCRXL/ORF1 cDNA clone. The selected clone was used to engineer the full-length PEC genome into the plasmid pCDNA3.1 (Invitrogen) downstream of the cytomegalovirus (CMV) promoter by using NheI/NotI restriction sites (Fig. 1). The resulting intermediate plasmid, pCV4, contained two T7 promoters (one engineered immediately upstream of the Cowden genome and the other present in the pCDNA3.1 vector). The CMV and T7 promoters in the vector sequence of pCV4 were removed by BglII and NheI digestion. Following T4 DNA polymerase treatment and ligation with T4 DNA ligase, plasmid pCV4A was selected that contained one T7 promoter immediately upstream of the Cowden PEC genome (Fig. 2). Both ends of the Cowden PEC RNA genome (passage 27) were examined again by 5′- and 3′-end rapid amplification of cDNA ends (Invitrogen) and compared to the previously determined sequence (GenBank accession number AF182760). The 5′-end sequence was confirmed, but the 3′-end terminal nucleotide immediately upstream of the poly(A) tail was identified as a G residue (with C in the reference sequence). This G residue was present in the subgenomic clone selected from the cDNA library used in the construction of the full-length clone.
FIG. 1.
Genomic organization of Cowden PEC and the construction of full-length cDNA clone, pCV4A. ORF1 of PEC was amplified by RT-PCR and cloned into pCRXL. A second cDNA clone containing the subgenomic region of PEC in pSPORT was identified in a cDNA library of the PEC genome. With the use of unique enzyme sites (AccI and NotI), the subgenomic region of Cowden PEC in pSPORT was engineered into the pCRXL/ORF1 cDNA clone. The full-length PEC genome was then cloned into pCDNA3.1 to generate intermediate plasmid pCV4 (not shown). The plasmid pCV4A was generated by removing pCMV and T7 promoter (provided by pCDNA3.1) by digestion with BglII and NheI and religation of the plasmid.
FIG. 2.
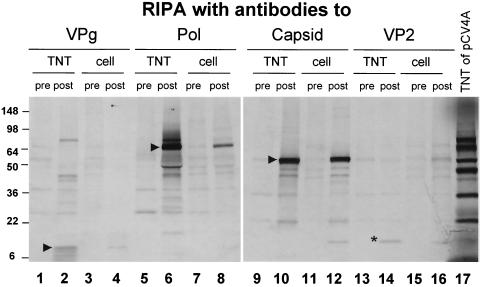
Comparison of proteins synthesized from clone pCV4A in a TNT reaction with those produced in PEC-infected cells. At 24 h after PEC infection in the presence of IC, cells were [35S]methionine-labeled for 4 h, and cell lysates were prepared in RIPA buffer. RIPA results of the TNT reaction and cell lysates of PEC infection with the hyperimmune guinea pig sera (pre- and postimmunization) against PEC VPg (lanes 1 to 4), polymerase (lanes 5 to 8), capsid (VLPs) (lanes 9 to 12), or VP2 (lanes 13 to 16) are shown. The tentative final cleavage products consistent with the expected masses of PEC VPg, ProPol, and capsid are marked with arrows. The VP2 protein is indicated with an asterisk. Lane 17, radiolabeled proteins from a coupled TNT reaction of pCV4A.
Expression of the PEC genome fragments encoding the predicted PEC VPg, polymerase region, and VP2 proteins in Escherichia coli BL21(DE3) cells was performed as previously described (23). Briefly, DNA fragments corresponding to each gene region was amplified by RT-PCR with primer pairs VPg-pF/VPg-pR (for VPg), Pol-pF/Pol-pR (polymerase), and ORF2-F/ORF2-R (VP2) (Table 1) and cloned into pET-28a(+) (Novagen) so that the C-terminal part of the encoded sequence was fused to a His6-tag. Ni-nitrilotriacetic acid agarose (QIAGEN) was used for the purification of each His6-tagged protein.
The full-length Cowden PEC cDNA clone (pCV4A) was analyzed in an in vitro transcription and translation reaction (TNT) (T7 Coupled Reticulocyte Lysate System; Promega, Madison, Wis.). Briefly, 2 μg of plasmid DNA was used as a template, and the reaction mixture was incubated at 30°C for 4 h. For radiolabeling of synthesized protein, [35S]methionine (>1,000 Ci/mmol) from ICN (Irvine, Calif.) or Amersham (Piscataway, N.J.) was used at a concentration of 1.5 mCi/ml. Radioimmunoprecipitation of in vitro translation mixtures was performed as described previously (23). Aliquots (15 μl) of the TNT reaction were diluted with 80 μl of RIPA buffer, and the mixtures were then incubated for 1 h at room temperature with 5 μl of the hyperimmune sera. The immune complexes were precipitated with protein A beads (Sigma). The binding and washing conditions were the same as those described previously (23). For radiolabeling of viral proteins, the LLC-PK cells (grown in six-well plates) were infected (or mock infected) with PEC at a multiplicity of infection (MOI) of 0.5. After 24 h at 37°C, the infected cells were washed and incubated in methionine- or cysteine-free growth medium for an additional 30 min. [35S]methionine (>1,000 Ci/mmol; Amersham) or [35S]cysteine (>800 Ci/mmol; ICN) was then added at a concentration of 100 μCi/ml, and the cells were incubated for an additional 4 h. The monolayer was washed with phosphate-buffered saline before lysis in 500 μl of RIPA buffer. Immunoprecipitation of radiolabeled proteins was performed with 100-μl aliquots of the cell lysates.
In vitro transcription and RNA purification.
Plasmid DNA was linearized with NotI, and capped RNA transcripts were synthesized from the linearized template with an mMessage in vitro transcription kit (Ambion, Austin, Tex.). A typical 20-μl reaction contained ATP, CTP, and UTP (7.5 mM each), 1.5 mM GTP, 6 mM m7G(5′)ppp(5′)G cap analog (Ambion), T7 reaction buffer, 1 to 2 μg of linearized pCV4A, and T7 enzyme mix. The reaction mixtures were incubated at 37°C for 3 h, followed by treatment with DNase I at 37°C for 1 h. Uncapped RNA was synthesized in a RiboMax large-scale RNA production system (Promega, Madison, Wis.) in the absence of cap analog. All RNA transcripts were purified with an RNeasy kit (QIAGEN) and analyzed by electrophoresis in an agarose (1%) gel under denaturing conditions with formaldehyde.
Transfection of DNA or RNA.
Plasmid DNA or RNA transcripts were transfected into LLC-PK cells with Lipofectamine 2000 (Invitrogen). Briefly, DNA (1 to 3 μg) or RNA (0.1 to 3 μg) (or mock buffer control) was incubated with 12 μl of Lipofectamine 2000 for 30 min at room temperature in Opti-MEM I (Invitrogen). The transfection mixture was then placed on cells containing E-MEM alone or E-MEM supplemented with IC (1%), TCDCA (200 μM), or GCDCA (200 μM). Medium containing TCDCA and GCDCA included the addition of α-tocopherol (100 μM) to control cytotoxicity (2). In experiments that utilized the vaccinia virus expression system (26), the cells were incubated with MVA/T7 (modified vaccinia virus Ankara strain expressing T7 polymerase) at an MOI of 10, 5, or 2 for 1 h prior to transfection of plasmid DNA. All transfected cells were incubated at 37°C for up to 10 days. Evidence for PEC replication was monitored by the appearance of cytopathic effect (CPE) and the detection of capsid antigen by IFA and ELISA.
Recovery and analysis of virus progeny.
At 5 to 6 days following transfection of RNA or DNA, the cells and medium were subjected to three rounds of freeze-thawing, and the material was concentrated to 1/10 of its original volume by ultracentrifugation (100,000 × g for 2 h). The pelleted material was mixed with 1% phosphotungstic acid for negative staining and observation by electron microscopy (EM). Recovered viruses were isolated and characterized following three rounds of plaque purification. The plaque-forming assay was performed with medium containing E-MEM, AgarPlaque agarose (1%) (Pharmingen, San Diego, Calif.), neutral red (0.1%), and IC (1%) or GCDCA (200 μM). The RNA of parental Cowden PEC or the plaque-purified recovered viruses was extracted, and the first 1,070 nt of the viral genome were amplified by RT-PCR by using primers PEC5′/NheI/T7 and PEC1kA (Table 1). Direct sequence analysis of the amplified RT-PCR products was performed by using the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit. This region contained three nucleotide substitutions that were distinct between the parental Cowden PEC strain and the sequence encoded in the full-length cDNA clone (Table 2). For the comparison of growth kinetics between the parental Cowden PEC and the recovered viruses propagated in the presence of IC (1%) or GCDCA (200 μM), ELISAs were performed on cell lysates collected at 24, 48, 72, 96, and 120 h after virus inoculation.
TABLE 2.
Differences between the tissue culture-adapted Cowden PEC and the full-length cDNA construct pCV4A
| Position (nucleotide no.) | Nucleotide
|
Amino acida
|
Region of genomeb | ||
|---|---|---|---|---|---|
| Cowden PEC | pCV4A | Cowden PEC | pCV4A | ||
| 59 | T | C | F | S | N-term |
| 62 | A | G | D | G | N-term |
| 450 | T | C | L | − | p28 |
| 1075 | A | G | I | V | NTPase |
| 1109 | A | G | K | R | NTPase |
| 1644 | T | C | S | − | NTPase |
| 3888 | C | T | I | − | ProPol |
| 5367 | A | G | G | − | Capsid |
| 5532 | T | C | V | − | Capsid |
| 7320 | C | G | 3′ NTR | ||
−, indicates no predicted amino acid change.
Predicted proteins of PEC. N-term, N-terminal protein; ProPol, protease-polymerase; NTR, nontranslated region.
Inoculation of the recovered viruses (pCV4A) into gnotobiotic pigs.
Animal protocols were approved by The Ohio State University Institutional Laboratory Animal Care and Use Committee. Recombinant viruses recovered from plasmid pCV4A were administered orally to two gnotobiotic piglets at a dose of 5 × 106 50% tissue culture infective dose in a preliminary study to examine their infectivity in animals. Two gnotobiotic pigs were challenged orally with wild-type (gnotobiotic pig-passaged) Cowden PEC (7) as a control. After inoculation, pigs were examined daily for clinical signs and development of diarrhea, and fecal consistency was scored (0, normal; 1, pasty; 2, semiliquid; and 3, liquid). Pigs with scores of 2 or more were considered to have diarrhea. Feces (rectal swabs) were collected daily and stored at −20°C until tested. Each rectal swab sample was tested for virus shedding by ELISA as described above.
RESULTS
Generation of full-length clone of Cowden PEC.
A full-length cDNA copy of the Cowden PEC genome was engineered into the vector pCDNA3.1 (Fig. 1) to generate the plasmid pCV4A. Sequence analysis of pCV4A identified 10 nucleotide changes (resulting in four amino acid substitutions) in the cloned PEC genome compared to that of the tissue-culture adapted Cowden PEC sequence deposited in the GenBank database (AF182760) (Table 2). Two amino acid substitutions were located in the predicted N-terminal protein, and two were in the predicted NTPase protein.
In vitro TNT of pCV4A.
The proteins encoded in plasmid pCV4A were expressed in a TNT reaction and compared by RIPA to those produced in virus-infected cells. Antisera specific for the VPg, polymerase region, capsid (VP1), or VP2 proteins were used in the analysis. The protein profiles of the TNT products and PEC-infected cell lysates were similar in the RIPA (Fig. 2), indicating that the proteins encoded in the pCV4A ORF1 (which includes the capsid coding sequence) underwent authentic proteolytic processing when expressed in vitro. The polymerase-specific antibodies precipitated an approximately 76-kDa protein both in the TNT reaction (Fig. 2, lane 6) and in infected cells (Fig. 2, lane 8), indicating that the major form of the PEC polymerase present in virus-infected cells, like that of FCV (24), is the proteinase-polymerase precursor, ProPol. Cleavage products consistent with the expected masses of the fully processed PEC VPg (13 kDa), ProPol (73 kDa) precursor, and capsid (58 kDa) proteins are indicated with arrows (Fig. 2). Of interest, the VP2 protein (18 kDa) encoded in ORF2 was expressed from the full-length RNA transcript (Fig. 2, marked by the asterisk in lane 14) and had an observed mass similar to that of the protein expressed in infected cells (Fig. 2, lane 16).
In vitro synthesis of full-length RNA and transfection into LLC-PK cells.
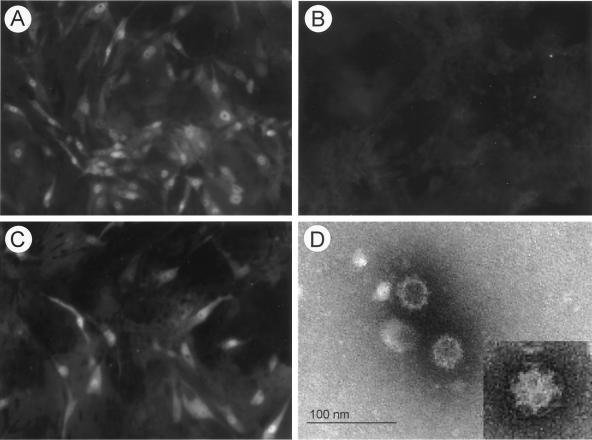
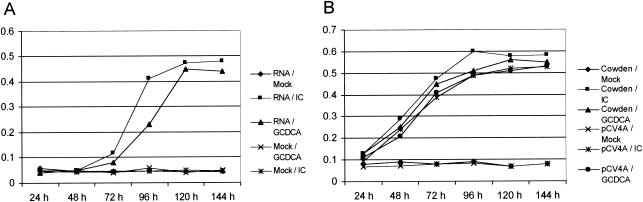
In vitro transcription of RNA molecules derived from Not1-linearized pCV4A with T7 polymerase in the presence of cap analog produced RNA molecules of approximately 7.4 kb (data not shown). First, we transfected these RNA transcripts into LLC-PK cells in the presence or absence of IC, since we had shown previously that native viral RNA was infectious only when transfected in the presence of IC (1). The RNA transcripts derived from pCV4A were infectious in the presence of IC as measured by the appearance of CPE and capsid antigen expression in cells by IFA staining (Fig. 3A). Direct EM observation of concentrated cell lysates collected at 96 h following transfection and incubation with IC showed the presence of PEC virions with classical calicivirus morphology as well as assembled empty capsid structures lacking packaged viral RNA (Fig. 3D). Growth kinetics after RNA transfection with IC showed that viral antigen was detected beginning 72 h after transfection (Fig. 4A), and infectious virus titers characteristically reached ∼106 50% tissue culture infective dose per ml (data not shown). The recovered viruses were further purified by three sequential plaque isolations. Transfection of pCV4A transcripts without IC produced no evidence of virus growth or viral capsid protein expression (Fig. 3B), and noncapped RNA transcripts from pCV4A did not yield evidence for virus replication, even in the presence of IC (data not shown). In addition, capped RNA transcripts derived from pCV4 (the intermediate plasmid with two T7 promoters) were not infectious.
FIG. 3.
IFA with hyperimmune guinea pig serum against VLP of PEC at 72 h after transfection of RNA transcripts from pCV4A and incubation with IC (1%) (A), mock MEM (B), or GCDCA (200 μM) (C). (D) EM observation of cell lysates at 96 h after transfection of RNA transcripts from pCV4A and incubation in the presence of IC is shown in panel C. Cell lysates were concentrated 100 times by ultracentrifugation, and virus particles were directly observed by EM after negative staining.
FIG. 4.
(A) Growth kinetics of PEC after transfection of RNA transcripts from pCV4A in the presence of IC (1%) or GCDCA (200 μM) by ELISA. RNA transcripts (1 μg) were transfected in LLC-PK cells with Lipofectamine 2000, and the transfected cells were incubated with IC or GCDCA for the desired period. (B) Growth kinetics of Cowden PEC and pCV4A viruses in the presence of IC (1%) or GCDCA (200 μM) by ELISA. The viruses were inoculated at an MOI of 0.5.
We examined whether bile acids alone could support recovery of virus from the infectious RNA transcripts derived from pCV4A. Viruses were recovered from the transfection of RNA transcripts of pCV4A in the presence of bile acids GCDCA (200 μM) (Fig. 3C) or TCDCA (200 μM) (data not shown). The growth kinetics of viruses recovered after RNA transfection in the presence of GCDCA (200 μM) was similar to that of IC (Fig. 4A), and the plaque morphology was indistinguishable (Fig. 5). Viruses recovered in the presence of GCDCA did not grow in the absence of IC or GCDCA. The growth kinetics of the parental tissue-culture adapted Cowden PEC and viruses recovered from pCV4A in the presence of IC or GCDCA were similar (Fig. 4B). Transfection of pCV4A plasmid DNA after MVA-T7 infection did not yield evidence for virus growth even in the presence of IC, TCDCA, or GCDCA (data not shown).
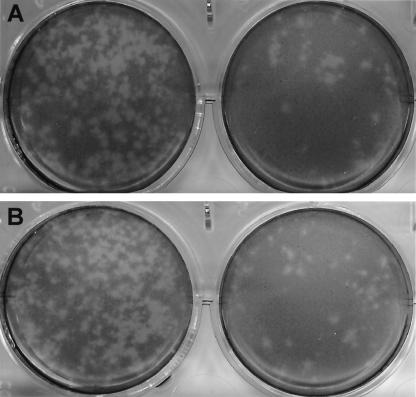
FIG. 5.
Plaque-forming assay of pCV4A viruses measured at dilutions of 10−4 (left cultures) and 10−5 (right cultures) in the presence of IC (1%) (A) or GCDCA (200 μM) (B). After LLC-PK cells were infected for 1 h with the viruses recovered from pCV4A, the cells were placed under medium containing MEM, agarose (1%), neutral red (0.002%), and IC (1%) or GCDCA (200 μM). Pictures were taken 96 h after virus inoculation.
The recovered pCV4A viruses were administered by the oral route to two gnotobiotic pigs. The viruses induced a limited amount of virus shedding after 7 days postinoculation for 2 to 4 days without apparent clinical signs (animal number 3) or with mild (+/−) diarrhea (animal number 4) (Table 3). Wild-type Cowden PEC was associated with virus shedding 3 days postinoculation in two animals and with mild or moderate diarrhea (Table 3).
TABLE 3.
Pathogenesis of wild-type (gnotobiotic pig-passaged) PEC and recovered PEC from pCV4A in gnotobiotic pigs
| Animal | Inoculum | Fecal consistency by no. of days postinoculationa
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||
| 1 | wtPEC | ||||||||||||
| Diarrhea | − | − | +/− | +/− | EUT | ||||||||
| Virus sheddingb | − | − | + | + | |||||||||
| 2 | wtPEC | ||||||||||||
| Diarrhea | − | − | + | + | − | − | − | EUT | |||||
| Virus shedding | − | − | + | + | + | + | + | ||||||
| 3 | pCV4A | ||||||||||||
| Diarrhea | − | − | − | − | − | − | − | − | − | − | − | − | |
| Virus shedding | − | − | − | − | − | − | − | − | + | + | − | − | |
| 4 | pCV4A | ||||||||||||
| Diarrhea | − | − | − | − | − | +/− | +/− | +/− | EUT | ||||
| Virus shedding | − | − | − | − | − | − | + | + | |||||
+, fecal consistency of ≥2; +/−, questionable or mild diarrhea (fecal consistency of 1); −, no diarrhea (normal feces). EUT, euthanized.
Virus shedding determined by ELISA.
DISCUSSION
The first reverse genetics system for the Caliciviridae was developed in 1995 for FCV, a member of the genus Vesivirus (22). We report here the development of a reverse genetics system for a second genus, Sapovirus, in the family Caliciviridae. PEC causes diarrhea in pigs and replicates in cell culture, which has led to its development as a model system for other enteric caliciviruses. A reverse genetics system for PEC will provide an important new tool for its study.
We previously reported that the RNA purified from Cowden PEC virions was infectious when transfected into LLC-PK cells in the presence of IC (1). In this study, capped RNA transcripts derived from plasmid pCV4A encoding a full-length cDNA copy of the PEC genome were also infectious in the presence of IC. Furthermore, bile acids, active components of IC (2), could support the recovery of PEC from the capped transcripts. However, unlike results with FCV infectious cDNA clones (22, 25), the transfection of pCV4A plasmid in MVA-T7-infected cells did not yield virus recovery. The MVA-T7 virus caused significant CPE in LLC-PK cells, and the expression efficiency of a green fluorescent protein reporter plasmid was low (10 to 20%) in MVA-T7-infected LLC-PK cells (data not shown). Following transfection of the pCV4A plasmid DNA into MVA-T7-infected LLC-PK cells, we detected PEC capsid protein (from the expression of ORF1) by IFA and RIPA (with or without IC or bile acids) within 24 h. After 48 h, the majority of cells died (presumably due to MVA-T7 CPE and toxicity of the transfection reagent), and no PEC was recovered. When RNA transcripts derived from pCV4A were transfected, PEC capsid protein was detected only after 48 h in the presence of IC or bile acids, and live progeny viruses were recovered and passaged. Therefore, transfection of RNA transcripts was utilized to generate the data presented in this paper. Recovered pCV4A viruses infected gnotobiotic pigs when administered by the oral route and showed attenuated virulence as evidenced by delayed virus shedding and mild or no diarrhea, which is consistent with previous pig infectivity data showing attenuation of the tissue culture-adapted PEC (12). This preliminary study established a basis for fine mapping of viral virulence determinants (11) by using the infectious cDNA clone and the gnotobiotic pig animal model.
We recently identified bile acids as an active factor in IC that could independently support PEC growth in cell culture (2). Here we demonstrate that the reverse genetics system for PEC is dependent on the presence of bile acids or IC. Bile acids are synthesized in the liver, stored in the gall bladder, and secreted into the duodenum. They are adsorbed from the intestine and returned to the liver through the portal vein before being reexcreted into the gall bladder (14). This enterohepatic circulation is essential in maintaining an effective concentration of bile acids and cholesterol homeostasis (14). Recent studies indicate that bile acids not only play an essential role as carriers of dietary lipids for absorption but also may function as hormone-like regulatory molecules with specific cellular receptors (14, 15, 21). Previously, we reported evidence for a connection between bile acid-mediated signaling via cAMP/PKA and interferon-mediated STAT1 pathways: the induction of cAMP by bile acids down-regulated interferon-mediated STAT1 activation, a key element for innate immunity against virus infection (2). Although at present we do not have direct evidence that this PKA/cAMP/STAT1 pathway functions in the gut where PEC replicates, it has been reported that certain bile acids could induce an increase in cAMP levels in animal and human intestine in vivo (16, 20). The availability of a cell culture system, reverse genetics, and an animal model will allow further investigation of both host and viral determinants involved in the pathogenesis of PEC diarrheal disease.
Acknowledgments
We thank Albert Z. Kapikian for continuing support of our work and Tanaji Mitra for technical assistance and helpful discussions.
This work was supported in part by a grant from the National Institutes of Health, NIAID grant RO1AI 49716 (L.J.S.).
REFERENCES
- 1.Chang, K. O., Y. Kim, K. Y. Green, and L. J. Saif. 2002. Cell-culture propagation of porcine enteric calicivirus mediated by intestinal contents is dependent on the cyclic AMP signaling pathway. Virology 304:302-310. [DOI] [PubMed] [Google Scholar]
- 2.Chang, K. O., S. V. Sosnovtsev, G. Belliot, Y. Kim, L. J. Saif, and K. Y. Green. 2004. Bile acids are essential for porcine enteric calicivirus replication in association with down-regulation of signal transducer and activator of transcription 1. Proc. Natl. Acad. Sci. USA 101:8733-8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clarke, I., and P. R. Lambden. 1997. The molecular biology of caliciviruses. J. Gen. Virol. 78:291-301. [DOI] [PubMed] [Google Scholar]
- 4.Cubitt, D. 1994. Caliciviruses, p. 549-568. In A. Z. Kapikian (ed.), Viral infections of the gastrointestinal tract. Marcel Dekker Inc., New York, N.Y.
- 5.Fankhauser, R. L., J. S. Noel, S. S. Monroe, T. Ando, and R. I. Glass. 1998. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 178:1571-1578. [DOI] [PubMed] [Google Scholar]
- 6.Flynn, W. T., and L. J. Saif. 1988. Serial propagation of porcine enteric calicivirus-like virus in primary porcine kidney cell cultures. J. Clin. Microbiol. 26:206-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flynn, W. T., L. J. Saif, and P. D. Moorhead. 1988. Pathogenesis of porcine enteric calicivirus-like virus in four-day-old gnotobiotic pigs. Am. J. Vet Res. 49:819-825. [PubMed] [Google Scholar]
- 8.Geissler, K., K. Schneider, G. Platzer, B. Truyen, O. R. Kaaden, and U. Truyen. 1997. Genetic and antigenic heterogeneity among feline calicivirus isolates from distinct disease manifestations. Virus Res. 48:193-206. [DOI] [PubMed] [Google Scholar]
- 9.Green, K. Y., T. Ando, M. S. Balayan, T. Berke, I. N. Clarke, M. K. Estes, D. O. Matson, S. Nakata, J. D. Neill, M. J. Studdert, and H. J. Thiel. 2000. Taxonomy of the caliciviruses. J Infect. Dis. 181(Suppl. 2):S322-S330. [DOI] [PubMed] [Google Scholar]
- 10.Green, K. Y., R. M. Chanock, and A. Z. Kapikian. 2001. Human caliciviruses, p. 841-874. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 11.Guo, M., K. O. Chang, M. E. Hardy, Q. Zhang, A. V. Parwani, and L. J. Saif. 1999. Molecular characterization of a porcine enteric calicivirus genetically related to Sapporo-like human caliciviruses. J. Virol. 73:9625-9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo, M., J. Hayes, K. O. Cho, A. V. Parwani, L. M. Lucas, and L. J. Saif. 2001. Comparative pathogenesis of tissue culture-adapted and wild-type Cowden porcine enteric calicivirus (PEC) in gnotobiotic pigs and induction of diarrhea by intravenous inoculation of wild-type PEC. J. Virol. 75:9239-9251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo, M., Y. Qian, K. O. Chang, and L. J. Saif. 2001. Expression and self-assembly in baculovirus of porcine enteric calicivirus capsids into virus-like particles and their use in an enzyme-linked immunosorbent assay for antibody detection in swine. J. Clin. Microbiol. 39:1487-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, L. R. 1998. Secretion, p. 445-472. In L. R. Johnson (ed.), Essential medical physiology, 2nd ed. Lippincott-Raven, New York, N.Y.
- 15.Kawamata, Y., R. Fujii, M. Hosoya, M. Harada, H. Yoshida, M. Miwa, S. Fukusumi, Y. Habata, T. Itoh, Y. Shintani, S. Hinuma, Y. Fujisawa, and M. Fujino. 2003. A G protein-coupled receptor responsive to bile acids. J. Biol. Chem. 278:9435-9440. [DOI] [PubMed] [Google Scholar]
- 16.Krag, E., and S. F. Phillips. 1974. Active and passive bile acid absorption in man. Perfusion studies of the ileum and jejunum. J. Clin. Investig. 53:1686-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohlinger, V. F., B. Haas, G. Meyers, F. Weiland, and H. J. Thiel. 1990. Identification and characterization of the virus causing rabbit hemorrhagic disease. J. Virol. 64:3331-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohlinger, V. F., and H. J. Thiel. 1991. Identification of the viral haemorrhagic disease virus of rabbits as a calicivirus. Rev. Sci. Technol. 10:311-323. [DOI] [PubMed] [Google Scholar]
- 19.Parwani, A. V., W. T. Flynn, K. L. Gadfield, and L. J. Saif. 1991. Serial propagation of porcine enteric calicivirus in a continuous cell line. Effect of medium supplementation with intestinal contents or enzymes. Arch. Virol. 120:115-122. [DOI] [PubMed] [Google Scholar]
- 20.Potter, G. D., J. H. Sellin, and S. M. Burlingame. 1991. Bile acid stimulation of cyclic AMP and ion transport in developing rabbit colon. J. Pediatr. Gastroenterol. Nutr. 13:335-341. [DOI] [PubMed] [Google Scholar]
- 21.Redinger, R. N. 2003. The role of the enterohepatic circulation of bile salts and nuclear hormone receptors in the regulation of cholesterol homeostasis: bile salts as ligands for nuclear hormone receptors. Can. J. Gastroenterol. 17:265-271. [DOI] [PubMed] [Google Scholar]
- 22.Sosnovtsev, S., and K. Y. Green. 1995. RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VpG for infectivity. Virology 210:383-390. [DOI] [PubMed] [Google Scholar]
- 23.Sosnovtsev, S. V., S. A. Sosnovtseva, and K. Y. Green. 1998. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J. Virol. 72:3051-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sosnovtseva, S. A., S. V. Sosnovtsev, and K. Y. Green. 1999. Mapping of the feline calicivirus proteinase responsible for autocatalytic processing of the nonstructural polyprotein and identification of a stable proteinase-polymerase precursor protein. J. Virol. 73:6626-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thumfart, J. O., and G. Meyers. 2002. Feline calicivirus: recovery of wild-type and recombinant viruses after transfection of cRNA or cDNA constructs. J. Virol. 76:6398-6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wyatt, L. S., B. Moss, and S. Rozenblatt. 1995. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology 210:202-205. [DOI] [PubMed] [Google Scholar]