Abstract
The N-terminal domains (NTDs) of the human immunodeficiency virus type 1 (HIV-1) capsid (CA) protein have been modeled to form hexamer rings in the mature cores of virions. In vitro, hexamer ring units organize into either tubes or spheres, in a pH-dependent fashion. To probe factors which might govern hexamer assembly preferences in vivo, we examined the effects of mutations at CA histidine residue 84 (H84), modeled at the outer edges of NTD hexamers, as well as a nearby histidine (H87) in the cyclophilin A (CypA) binding loop. Although mutations at H87 yielded infectious virions, mutations at H84 produced assembly-competent but poorly infectious virions. The H84 mutant viruses incorporated wild-type levels of CypA and viral RNAs and showed nearly normal signals in virus entry assays. However, mutant CA proteins assembled aberrant virus cores, and mutant core fractions retained abnormally high levels of CA but reduced reverse transcriptase activities. Our results suggest that HIV-1 CA residue 84 contributes to a structure which helps control either NTD hexamer assembly or the organization of hexamers into higher-order structures.
A number of functions have been attributed to the human immunodeficiency virus type 1 (HIV-1) capsid (CA) protein. As a portion of the HIV Gag precursor (PrGag) protein, the CA N-terminal domains (NTDs) and C-terminal domains (CTDs) collaborate with each other and with other Gag domains to facilitate virus assembly and budding (4, 9, 10, 13, 15, 17-22, 27, 29, 31, 35, 38-44, 46). Appropriate CA-CA contacts are necessary not only for assembly and release, but also for proper maturation and postmaturation replication steps (9, 13, 15, 20, 22, 27, 29, 38, 40, 41, 43, 44). Indeed, a variety of HIV-1 capsid mutations have manifested defects in early replication events, such as uncoating and reverse transcription (15, 20, 27, 38, 40, 41, 44). Some of these defects may be attributable to altered interactions with cellular factors, such as cyclophilin A (CypA) (1, 2, 7, 9, 41), and host susceptibility factors, such as Ref1, Trim5α, and Lv1 (6, 12, 16, 17, 24, 32, 37).
In terms of a structural role within virions, CA NTDs appear to assemble hexamer rings that are linked via CTD connections (5, 19, 20, 26, 28, 33, 34, 42); evidence suggests that the NTD rings are more tightly packed in immature than in mature virions (28), implying that more CA is assembled into particles than is necessary to build a mature virus core. In vitro experiments have demonstrated unusual pH-dependent characteristics in terms of the structures assembled by HIV-1 Gag proteins. At pH 6.0, PrGag-like proteins have been shown to assemble long tubes, whereas at pH 8.0, spheres are formed (22). The behavior of mature CA is even more complex in that CA dimers predominate at pHs below 6.6, spheres predominate at pH 6.8, and tubes are the major form at pH 7.0, while tubes and spheres may coexist at higher pHs (14). The assembly activities of CA in the pH 6.5 to 7.0 range have led to the speculation that capsid assembly or disassembly may involve a histidine switch, perhaps involving one of the three conserved (H12, H62, and H84) of the five total (H12, H62, H84, H87, and H226) capsid histidines (14).
In light of the histidine switch model, we chose to examine the effects of five substitutions at the HIV-1 CA H84. This residue was of interest because it has been modeled at the outsides of NTD hexamer rings (19, 20, 26) in a position which could modulate hexamer packing differences in immature and mature virions (28). As a control, we also tested substitution effects at the nearby CA histidine 87, a less-well-conserved residue in the CypA binding loop. As expected, a cysteine substitution at H87 had only minimal effects on viral infectivity. In contrast, four of the five H84 mutations were noninfectious, and one of these (H84A) demonstrated dominant negative effects on wild-type (wt) virus infectivity. The exceptional mutation, H84Y, produced virions that were still 30-fold less infectious than wt virions. Detailed comparison of 84A, 84Y, and wt virions showed that the mutant virions were released efficiently from cells, had normal levels of viral genomic RNA and total reverse transcriptase (RT) activity and, in contrast with other reported NTD mutants (41), carried wt levels of CypA. Although H84Y virions had wt levels in entry assays (8, 32), the H84A mutant showed slightly reduced entry signals and morphologically aberrant virus cores. Moreover, both mutants exhibited low RT-to-CA ratios in virus cores and appeared sensitive to proteolytic cleavage near NTD loop regions. Our results suggest that H84 mutations perturb aromatic interactions between HIV-1 CA NTD helices 4 and 7 that are essential to proper core morphogenesis.
MATERIALS AND METHODS
Recombinant DNA constructs.
A vesicular stomatitis virus (VSV) glycoprotein (G) expression construct, pVSV-G, was the generous gift of Randy Taplitz. The β-lactamase-vpr fusion protein expression construct BlaM-vpr (32) and the parental HIVLuc construct (pNL-LucE-R+) (11) were kindly provided by Nathaniel Landau. To make mutations at capsid residue 84 in the context of HIVLuc, the HIVLuc gag BglII (nucleotide [nt] 680)-to-HindIII (nt 1715) fragment was inserted into the homologous SP72 (Promega) sites, mutagenic double-stranded oligonucleotides were used to replace the wt sequence between the PstI (nt 1415) and SphI (nt 1443) sites, and mutations were cassetted back into HIVLuc, using the BssHII (nt 715) and SpeI (nt 1508) sites. The mutation at residue 87 was created by PCR, in a pGEM-T (Promega) vector, and BssHII-SpeI cassetted into HIVLuc. In all cases, conservative mutations also were introduced to facilitate restriction analysis. The wt and mutant sequences covering CA residues 77 to 91 are as follows, where mutated residues are underlined: wt, GCT GCA GAA TGG GAT AGA GTG CAT CCA GTG CAT GCA GGG CCT ATT; H84A, GCT GCA GAA TGG GAT AGA GTG GCG CCA GTT CAT GCA GGG CCT ATT; H84C, GCT GCA GAA TGG GAT AGA GTC TGC CCA GTT CAT GCA GGG CCT ATT; H84E, GCT GCA GAA TGG GAT AGA GTG GAA CCA GTT CAT GCA GGG CCT ATT; H84K, GCT GCA GAA TGG GAT AGA GTG AAA CCG GTT CAT GCA GGG CCT ATT; H84Y, GCT GCA GAA TGG GAT AGA GTA TAC CCA GTT CAT GCA GGG CCT ATT; H87C, GCT GCA GAA TGG GAT AGA GTG CAT CCA GTG TGC GCA GGG CCC ATT.
Cell culture and transfections.
293T and HiL cell lines were passaged at 37°C in 5% CO2 in culture medium containing Dulbecco's modified Eagle's medium supplemented with 10 mM HEPES (pH 7.4), penicillin, and streptomycin plus 10% fetal calf serum. For transfections, 10-cm plates of 293T cells were transfected by the calcium phosphate method (4, 23, 29, 30, 44, 45) with either 24 μg of HIVLuc DNA, 16 μg of HIVLuc plus 8 μg of pVSV-G DNA, or 12 μg of HIVLuc plus 6 μg of pVSV-G plus 6 μg of BlaM-vpr DNA. Briefly, confluent 10-cm dishes of 293T cells were split 1:4 the day prior to transfection. Plasmid DNAs were mixed with 1 ml of HEPES-buffered saline (pH 7.05 to 7.15; 21 mM HEPES, 137 mM NaCl, 5 mM KCl, 0.7 mM sodium phosphate, 5 mM dextrose), after which 40 μl of 2 M CaCl2 was added while vortexing. DNA solutions were incubated at room temperature for 40 min. Following this, culture medium was removed from the cells, DNA solutions were added dropwise to the cell monolayers, and then the cells were incubated at room temperature for 20 min, with gentle rocking once at 10 min. After incubations, 10 ml of culture medium containing 50 μg of gentamicin/ml was added to the cells, and plates were incubated at 37°C and 5% CO2 for 4 to 5 h. Following incubations, transfection media were removed and cells were washed with 5 ml of serum-free Dulbecco's modified Eagle's medium, incubated in 2.5 ml of 15% glycerol in HEPES-buffered saline for 3 min at 37°C, washed twice, and fed with 10 ml of culture medium plus 50 μg of gentamicin/ml.
For sample collection, virus-containing media and cell pellets washed in phosphate-buffered saline (PBS; 9.5 mM sodium potassium phosphate [pH 7.4], 137 mM NaCl, 2.7 mM KCl) were collected 3 days posttransfection and stored at −80°C prior to further processing. Virus particles in filtered (Gelman; 0.45 μm) cell-free medium were concentrated by centrifugation at 4°C through 20% sucrose cushions in PBS (2 h at 82,500 × g [25,000 rpm in an SW28 rotor, 4-ml cushions], or 45 min at 197,000 × g [40,000 rpm, SW41 rotor, 2-ml cushions]). Virus pellets were resuspended in 0.1 ml of PBS per transfected cell plate and stored in aliquots at −80°C.
Protein analysis.
For routine analysis of virus protein release, cell samples (20% of cell pellets from each plate) were suspended in IPB (20 mM Tris-hydrochloride [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.1% sodium dodecyl sulfate [SDS], 0.5% sodium deoxycholate, 1.0% Triton X-100, 0.02% sodium azide), incubated on ice for 5 min, vortexed, and cleared by centrifugation at 13,700 × g for 15 min at 4°C. Soluble material was mixed with 1 volume of 2× sample buffer (12.5 mM Tris-hydrochloride [pH 6.8], 2% SDS, 20% glycerol, 0.25% bromophenol blue) plus 0.1 volume of β-mercaptoethanol, prior to heating (3 to 5 min, 95°C) and SDS-polyacrylamide gel electrophoresis (SDS-PAGE). For virus samples, 50 μl of resuspended virus pellets was mixed with one volume of 2× sample buffer plus 0.1 volume of β-mercaptoethanol and processed as above. In some cases, virus samples were cross-linked with 1 mM bis-maleimido hexane (BMH; Pierce; diluted from a freshly made 100 mM stock in dimethyl sulfoxide) prior to processing, as described previously (23, 29, 30).
Cell and virus protein samples were fractionated by conventional 10% acrylamide Laemmli SDS-PAGE (4, 23, 29, 30, 44, 45) or 16% acrylamide Schagger and von Jagow SDS-PAGE (3, 36), electroblotted, and immunoblotted following previously described methods. Primary antibodies were as follows: Hy183 (from Bruce Chesebro) used at 1:15 from hybridoma culture medium for detection of the HIV-1 CA CTD; NEA 9306001EA (New England Nuclear) used at 1:15,000 for detection of the HIV-1 CA NTD; and SA-296 (BioMol) used at 1:15,000 for detection of CypA. Secondary reagents were alkaline phosphatase-conjugated anti-mouse antibodies (Promega S3721) used at 1:15,000 for detection of anti-HIV-CA primary antibodies and anti-rabbit immunoglobulin G (Sigma-Aldrich A3687) used at 1:2,000 for the anti-CypA polyclonal antibody. Color reactions for visualization of antibody-bound bands employed nitroblue tetrazolium plus 5-bromo-4-chloro-3-indolyl phosphate in 100 mM Tris-hydrochloride (pH 9.5), 100 mM NaCl, and 5 mM MgCl2 (4, 23, 29, 30, 44, 45). Size estimates for immunoreactive bands on immunoblots were obtained by comparison of mobilities versus those of Bio-Rad and Invitrogen size standards, assuming a linear mobility-to-log molecular weight relationship. Quantitation of band intensities was performed by densitometric scanning on an Epson model G810A scanner, followed by intensity measurement using NIH Image 1.61 software.
Sucrose gradients and core fractionations.
For sucrose density gradient fractionation, virus samples in PBS (0.1 ml) were layered on top of 5-ml 20-to-60% sucrose gradients in TSE (50 mM Tris [pH 7.4], 100 mM NaCl, 0.1 mM EDTA) and centrifuged at 4°C, 170,000 × g (average) for 18 h such that particles of ≥12S should have sedimented to equilibrium. After centrifugation, 0.4-ml fractions were collected from the gradient tops to bottoms, and Gag levels in each fraction were determined by immunoblotting as described above. Sucrose densities for fractions were determined by weighing a constant volume of each fraction from a gradient run in parallel.
For virus core fractionation, we followed a modification of the protocol of Tang et al. (41). Briefly, 0.1-ml virus samples were mixed gently with an equal volume of 0.6% NP-40 and incubated at room temperature for 10 min. Samples were layered onto 0.2-ml 20% sucrose-PBS cushions and centrifuged at 120,000 × g for 60 min at 4°C. Two 0.2-ml fractions then were collected from the top, and the pellets were resuspended in 0.2 ml of PBS to yield a third fraction. Samples were subjected to RT assays and immunoblot analysis of Gag protein content.
RT assays and RNase protections.
Exogenous RT assays were performed with poly(A) and oligo(dT) templates and primers and detergent-disrupted virions (29, 44). Gag-normalized viral samples in PBS were incubated in RT assay cocktail {50 mM Tris (pH 8.3), 20 mM dithiothreitol (DTT), 0.6 mM MnCl2, 60 mM NaCl, 0.05% NP-40, 2.5 μg of oligo(dT) (Pharmacia)/ml, 10 μg of poly(A) (Pharmacia)/ml, 10 μM dTTP (1-Ci/mM [α-32P]dTTP) } at 37°C for 2 h. As controls, dilutions of avian myeloblastosis virus RT (Roche) were run in parallel. After incubations, samples were precipitated by addition of 0.1 volume of 100% trichloroacetic acid (TCA) and incubated overnight at 4°C. TCA precipitates were pelleted by centrifugation for 10 min at 4°C, 13,600 × g, and were washed five times with 10% TCA prior to radioactivity quantitation in a scintillation counter.
RNase protections essentially followed the procedure of Wang et al. (45). Probes for RNase protection assays were made by incubation of 1 μg of EcoRI-linearized template plasmid (BlueHX 680-831) with transcription buffer (40 mM Tris [pH 7.4], 10 mM DTT, 6 mM MgCl2, 0.8 mM spermidine), 100 μCi of [α-32P]rGTP, 0.5 mM each of rATP, rCTP, and rUTP, 1 μl of RNasin (Promega), 1 mM DTT, and 20 U of T3 polymerase (Promega) at 37°C for 1 h. Probes then were ethanol precipitated, dried, resuspended, separated on 5% sequencing gels, eluted, and reprecipitated prior to use (45). For protections, Gag-normalized viral samples were precipitated with 10 μg of tRNA. Pellets were resuspended in 80% formamide, 400 mM NaCl, 40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (pH 6.4), and probe, incubated at 75 to 85°C for 5 min, and then incubated at 30°C overnight. Samples then were incubated with RNase treatment buffer (300 mM NaCl, 10 mM Tris [pH 7.5], 5 mM EDTA, 40 μg of RNase A [Roche]/ml, 2 μg of RNase T1 [Roche]/ml) and incubated at 30°C for 30 min, followed by the addition of 2.5 μl of 20-mg/ml proteinase K (Boehringer) plus 20 μl of 10% SDS and further incubation at 37°C for 15 min. Samples were phenol-chloroform extracted, ethanol precipitated, dried, fractionated on 6% acrylamide sequencing gels, and autoradiographed. Viral genomic RNA bands on autoradiographs were scanned on an Epson G810 scanner and quantitated using NIH Image 1.61 software.
Infections and entry assays.
Confluent 10-cm dishes of HiL cells (29, 44, 45) were split 1:5 the day before infections. Growth media were removed from each cell plate, and 2-ml aliquots of filtered transfection supernatants containing 8 μg of Polybrene/ml were added to the cells. Plates were incubated for 3 h at 37°C, after which 8 ml of culture medium per plate was added and plates were incubated an additional 3 days at 37°C. After infections, cells were collected in 1 ml of luciferase lysis buffer (100 mM sodium phosphate [pH 8.0], 4 mM ATP, 1 mM sodium pyrophosphate, 6 mM magnesium chloride, 0.2% Triton X-100) and either processed immediately for luciferase assays or frozen at −80°C until use. For luciferase assays, cells in luciferase lysis buffer were vortexed at room temperature and 30-μl aliquots were mixed with 0.3 ml of luciferase assay buffer (luciferase lysis buffer minus Triton X-100). Luciferase levels were measured on an EG&G Berthold Autolumat LB953 luminometer using a 0.1-ml luciferin pulse of 1 mM d-luciferin (BD Pharmingen). Raw luminometer counts were normalized versus luminometer counts obtained from the transfected cells which produced the virus samples, and mutant virus infectivities were expressed as percentages of wt HIVLuc infectivities from assays performed in parallel.
Infections for entry assays (8, 32) used 10-cm confluent plates of HiL cells which were split 1:10 onto 6-cm dishes the day before infections. Cells were infected 5 h at 37°C with 250 μl of virus in PBS plus 750 μl of serum-containing medium and 8 μg of Polybrene/ml. After incubations, viral samples were removed, cells were washed once with Hank's balanced salt solution (HBSS; without calcium or magnesium), and cells were incubated in 1.5 ml of CCF2/AM loading solution (2 μM CCF2/AM [Invitrogen], 0.8 mg of Pluronic-F127 [Invitrogen]/ml in HBSS) at 26°C, 5% CO2 overnight. Loading solution then was removed, cells were washed once in HBSS, trypsinized, pelleted, fixed (20 min, 1% formaldehyde in PBS), pelleted, washed in HBSS, pelleted, and resuspended in 1 ml of HBSS. Cells were analyzed as described previously (8, 32, 47) on a Becton-Dickinson Turbo Vantage flow cytometer to detect cleaved product as blue fluorescence and uncleaved substrate as green fluorescence. The percentages of product-positive live cells derived from flow cytometer analysis were normalized for Gag protein levels in virus samples and expressed as percentages of wt HIVLuc levels.
EM.
Concentrated virus particle samples were lifted for 2 min onto carbon-coated, UV-treated (shortwave UV, 30 to 120 min) electron microscopy (EM) grids, rinsed for 15 s in water, wicked, stained for 1 min in filtered 1.3% uranyl acetate, wicked, and dried. EM images were collected on a Philips CM120/Biotwin TEM equipped with a Gatan 794 multiscan charge-coupled device camera, searching at 2,300 to 4,000×, and collecting images at 2,300 to 34,000×. Virus particle diameters were determined with the aid of Gatan digital micrograph software, and virus particle images were ported from Gatan DM3 to TIFF or jpeg image format for presentation.
RESULTS
Release and infectivity of viral mutants.
The in vitro assembly properties of HIV-1 CA and PrGag proteins have been shown to be pH dependent. Gross et al. showed that PrGag-like proteins assembled tubes at pH 6.0 but spheres at pH 8.0 (22). Ehrlich et al. (14) observed that CA assembly properties shifted in the range of 6.0 to 7.0, near the pKa of histidines. Since HIV-1 histidine 84 is well conserved (25), modeled to be at the outer edges of NTD hexamer rings (19, 20, 26), and might influence HIV assembly or morphogenesis, we examined the effects of mutations at this residue. To do so, we initially substituted alanine, lysine, cysteine, and glutamate for histidine 84, creating the H84A, H84K, H84C, and H84E mutations. As a control, we also tested the effects of a cysteine mutation at the nearby poorly conserved histidine 87 residue in the CypA loop.
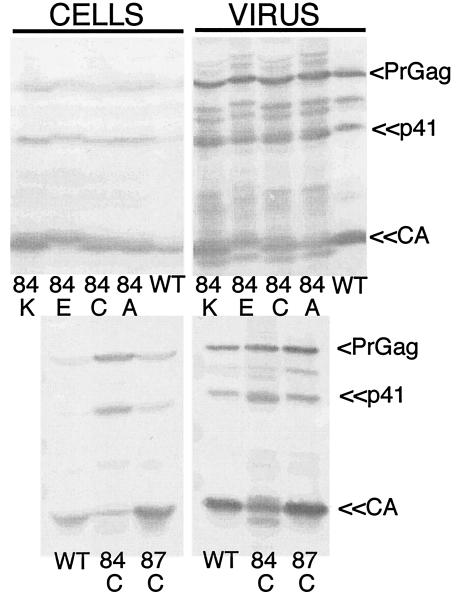
For monitoring the effects of mutations on virus assembly and release, wt and mutant HIV constructs were transfected into 293T cells and virus and cell samples were collected 3 days posttransfection. Samples were separated by SDS-PAGE and immunoblotted with an anti-HIV capsid antibody. As shown in Fig. 1, wt Gag proteins were released efficiently from cells, yielding the expected PrGag, p41, and CA bands, along with a less-pronounced processed band at 46 to 47 kDa. Relative to wt, H84K, H84E, H84C, H84A, and H87C all released Gag proteins at approximately comparable levels. These results suggest that the H84 and H87 mutations did not compromise the abilities of Gag proteins to assemble and release virus-like particles. Indeed, the only readily apparent differences from wt pertained to protein processing. In particular, for the H84C mutant, slightly higher levels of PrGag and p41 were observed in virus particle samples. Also observed were additional minor processing bands at about 46 kDa and near the CA band for the four H84 mutants; the nature of these bands is examined further below.
FIG. 1.
Virus particle release. 293T cells were transfected with wt and mutant HIV constructs. Cell lysates and pelleted virus samples were collected at 72 h posttransfection and subjected to SDS-PAGE followed by immunodetection for Gag proteins with an anti-HIV-1 CA antibody. Sample identities and bands for PrGag, CA, and the p41 processing intermediate are indicated.
Since the H84 and H87 mutant constructs efficiently released virus from transfected cells, it was of interest to test whether they were also infectious. Thus, 293T cells were cotransfected with wt or mutant HIVLuc constructs plus a plasmid encoding VSV-G. Media from transfected cells were collected and used to infect HiL cells, after which luciferase assays were performed on the infected cells to score for virus-mediated transduction of the luciferase gene. Not surprisingly, VSV-G-pseudotyped wt viruses were infectious, as were viruses produced by the H87C mutant (see below). In contrast, the infectivities of the four H84 mutants all were less than 0.2% that of wt.
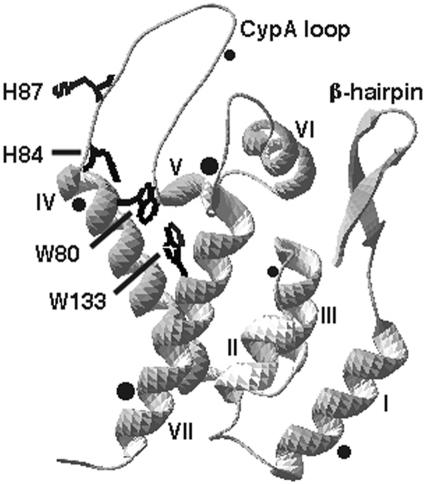
Because alanine, cysteine, lysine, and glutamate substitutions for HIV-1 CA H84 essentially abolished virus infectivities, we considered alternate substitutions that might maintain some degree of infectiousness. Inspection of the HIV-1 CA NTD structure (Fig. 2) suggested that in addition to providing a highly conserved basic residue, H84 contributes to an aromatic interaction with tryptophan residues 80 (W80) and 133 (W133) to align the base of the CypA loop and helices IV and VII in a stable tertiary structure. To examine the importance of this arrangement, we created an H84Y mutation on the hypothesis that tyrosine might preserve the residue 84-80-133 aromatic interaction.
FIG. 2.
Structural features of the HIV-1 CA NTD. Shown is the structural model (pdb 1GWP) (21, 39) for the HIV-1 CA NTD, from the N-terminal proline at the end of the β-hairpin to the C-terminal connection to the CTD (bottom left). The seven NTD helices are indicated, as are the locations of mutated residues H84 and H87 and the tryptophan residues (W80 and W133) which participate in aromatic interactions with H84. Approximate locations of anomalous processing sites (determined below in Fig. 6) are indicated with black dots, where dot sizes represent the frequencies of cleavage site usage, based on immunoblotting band intensities. Note that residue numbers reflect those of the mature capsid protein, in which the first capsid residue corresponds to codon 133 from HIV-1 gag.
Testing of the H84Y mutant assembly and release properties followed the methodology described above, and results are shown in Fig. 3. As observed in Fig. 1, the wt construct directed the efficient assembly and release of virus particles (lanes A and D). Also as observed in Fig. 1, H84A proteins were released efficiently from cells, but additional high-mobility processing bands were especially apparent (B and F). Gag assembly and release levels for H84Y (C and G) were similar to those of the wt and H84A constructs, implying no impairment of these processes. However, interestingly, the levels of high-mobility processing bands were reduced for H84Y relative to that for H84A (compare F and G).
FIG. 3.
Release of wt, H84A, and H84Y particles. 293T cells were transfected with wt and mutant HIV constructs. Cell lysates and pelleted virus samples were subjected to SDS-PAGE, followed by immunodetection for Gag proteins using an anti-HIV-1-CA antibody. Sample identities were as indicated, where M denotes the molecular weight size marker. Bands for PrGag, CA, and the p41 processing intermediate are indicated.
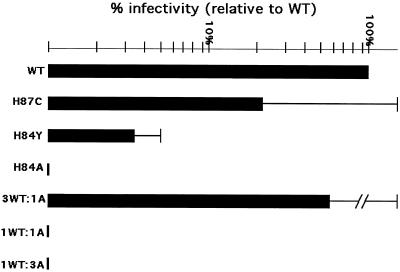
Since the H84Y mutant gave fewer anomalous CA processing bands than H84A, it seemed possible that it might prove less impaired in terms of infectivity than its counterpart. Thus, infectivity assays were performed for H84Y, along with H84A, H87C, and wt controls. As noted above and illustrated in Fig. 4, the H87C mutant had reduced but still considerable (20% of wt) infectivity, whereas H84A (as well as H84K, H84E, and H84C) showed <0.2% of wt infectivity. The H84Y mutant infectivity was reduced 30-fold compared to wt but, remarkably, it was still greater than 10 times that of the other H84 mutants. To ascertain whether mutant Gag proteins were able to impair the functions of wt Gag proteins, wt and H84A constructs were transfected into 293T cells at different ratios (3:1, 1:1, and 1:3 wt/H84A ratios) along with the VSV-G protein expression construct, and media were collected for infections. As Fig. 4 illustrates, while the 3:1 wt-H84A sample showed slightly reduced infectivity, virus particles produced from the 1:1 and 1:3 wt-H84A mixtures were as noninfectious as H84A itself. These results indicate that the H84A mutant acts as a dominant negative, diminishing the ability of wt Gag proteins to assemble infectious particles. The mechanism by which mutations at HIV-1 CA H84 inhibit infectivity is examined below.
FIG. 4.
Relative virus infectivities. HiL cells were infected with viruses derived from cotransfecting the indicated HIV-luciferase (HIVLuc) construct with the VSV-G expression construct pVSV-G. Infectivities were monitored by luciferase assay of infected cell lysates, and results were normalized to luciferase activities of the corresponding transfected cells. Infectivities were plotted relative to wt HIVLuc levels on a log scale graph and were based on two or more (H87C = 4; H84Y = 3; H84A = 7) independent sets of infections. Note that the 3WT:1A, 1WT:1A, and 1WT:3A bars correspond to cotransfections using different ratios of wt and H84A constructs and that values for the H84E, H84K, and H84C viruses were <0.2% that of wt.
CypA incorporation into viral mutants.
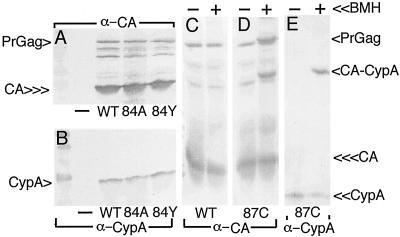
One possible cause for the noninfectivity of viral mutants might be differences in their abilities to interact with CypA. Although CypA appears to exert its major effects on HIV-1 within newly infected cells (1, 2), studies have shown that the noninfectivity of some HIV-1 NTD mutants correlates with an impaired ability to incorporate CypA into virions (7, 9, 41). Because our mutations mapped within or adjacent to the CypA loop, examination of CypA levels in virions appeared warranted. For this analysis, we chose to examine our partially infectious H84 mutant, H84Y, and one noninfectious mutant, H84A, along with negative and positive controls. Virus samples were immunoblotted for the parallel detection of CA (Fig. 5A) and CypA (Fig. 5B) proteins. As illustrated, while CypA was not released from cells in the absence of HIV-1 expression (Fig. 5B, − lane), its release from cells appeared roughly equivalent for wt, H84A, and H84Y viruses (compare Fig. 5A, CA signal, versus B, CypA signal); these results suggest that mutations at H84 did not affect CypA binding to CA.
FIG. 5.
CypA assembly into virus particles. (A and B) 293T cells were mock transfected (−) or transfected with the indicated constructs, and medium supernatants were collected 72 h posttransfection and then subjected to ultracentrifugation. Pelleted virus samples were resuspended in buffer and subjected to SDS-PAGE and parallel immunodetection with the indicated antibodies. (C to E) Samples were mock treated (−) or treated (+) with the cysteine-specific cross-linker BMH prior to SDS-PAGE and immunodetection. CypA, CA, PrGag, and CA-CypA cross-link products (D and E, cross-link lanes) are indicated. Note a putative CA-CA dimer cross-link product in the panel D cross-link lane, just above the PrGag band.
We also monitored CypA incorporation into H87C virions and found that CypA assembly into virions was not affected by this CypA loop mutation (Fig. 5E, − lane). Interestingly, treatment of H87C virus with the cysteine-specific cross-linker BMH yielded a novel anti-CypA-reactive 45-kDa band (Fig. 5E, + lane) which corresponded to an anti-CA-reactive cross-link band in H87C (Fig. 5D) but not wt (Fig. 5C) samples. These results indicate that the introduced cysteine at residue 87 readily cross-linked to a free CypA cysteine. An additional cross-link band, observed only with the anti-CA antibody (Fig. 5D), migrated slightly slower than the PrGag band. We interpret this product to represent either a CA-CA dimer or a heterodimer of CA with an undetermined cellular protein. In either case, our results with H84 and H87 did not demonstrate the correlation between loss of infectivity and loss of CypA interaction that has been observed for some other NTD mutations (41).
RNA and RT analysis.
Since CypA levels in wt and noninfectious mutant virions were equivalent, we probed for RNA and RT defects that might yield replication-defective, assembly-competent phenotypes. For RNA analysis, RNAs were isolated from equivalent amounts of virus and samples were analyzed by RNase protection, as described in Materials and Methods. Results (Table 1) demonstrated that virion-associated HIV genomic RNA levels were roughly equivalent for wt, H84Y, and H84A viruses. Similar studies also were performed on viruses to measure RT levels via exogenous template RT assays (see Materials and Methods). As shown in Table 1, RT levels were comparable in samples from WT, partially infectious H84Y, and noninfectious H84A virions. Thus, RNA encapsidation and/or RT incorporation defects do not explain the low infectivities of the H84 mutants.
TABLE 1.
RNA and RT levels in wt, H84Y, and H84A virusesa
| Construct | RNA level | RT level |
|---|---|---|
| wt | 100 | 100 |
| H84Y | 94 | 113 ± 12 |
| H84A | 109 ± 9 | 106 ± 51 |
Viral genomic RNA (RNA) and RT levels in wt, H84Y, and H84A viruses are expressed as percentages of the values obtained for wt viruses. RNA levels were determined by RNAse protection using RNA isolated from equivalent amounts of virus. Relative levels were quantitated densitometrically from one (H84Y) or two (wt, H84A) independent experiments. RT values are derived from three independent measurements for each construct and were determined using exogenous RT assays, normalized for capsid protein levels by densitometric quantitation from immunoblots.
Cleavage site analysis of mutant proteins.
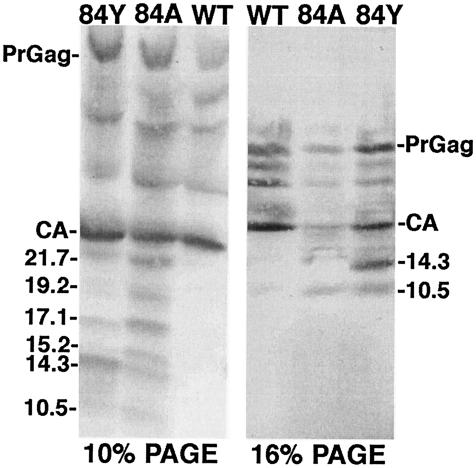
Although H84 mutant viruses gave wt levels of RNA and RT, we noted that the H84 mutants displayed a number of high-mobility CA processing bands (Fig. 1 and 3). On most gels, the number of processing bands appeared greater for the H84A mutant than for the H84Y mutant, suggesting a correlation between aberrant processing and loss of infectivity. To examine this finding in more detail, wt, H84A, and H84Y viral proteins were analyzed on 10 and 16% gels and fragment sizes were calculated from log molecular weight-versus-migration distance plots. As illustrated in Fig. 6, anti-HIV-1-CA-reactive bands were observed at calculated sizes of 21.7, 19.2, 17.1, 15.2, 14.3, and 10.5 kDa. The epitope for the antibody employed binds to the CTD, C-terminal to the CA major homology region (27, 38). Thus, if the CA-sp1 junction for these NTD mutants were cleaved appropriately, the novel processing sites within the NTD could be estimated from fragment size data. Based on this assumption, the approximate aberrant CA cleavage sites for H84A and H84Y were mapped (Fig. 2) as black dots, where the size of the dot represents the frequency of cleavage site usage. Although these cleavage site estimates are likely to be accurate only to the nearest few residues, it is interesting that one of the predicted cleavage sites maps close to residue 84 and the others tend to occur at or near loop regions in the NTD or between the NTD and CTD. These results suggest that H84A mutations perturbed tertiary or quaternary interactions such that the capsid proteins became more sensitive to proteolytic action.
FIG. 6.
Capsid protein processing products. The indicated virus samples were electrophoresed in parallel on conventional SDS-10% PAGE and SDS-16% PAGE Schagger gels. After electrophoresis, CA fragments were detected by immunoblotting with an anti-HIV-1-CA antibody which recognizes the HIV-1 CTD. Approximate fragment sizes in kilodaltons are indicated and were estimated by comparison with migration distances of known standards, assuming a linear relationship between migration and log molecular mass.
Morphologies of mutant virions.
Since H84 mutant proteins were processed anomalously in virions, we found it important to characterize the virus particles further. Particles collected by ultracentrifugation were subjected to equilibrium centrifugation on 20-to-60% sucrose gradients to analyze virion densities. As shown in Fig. 7, the peak density for wt virions (top panel) was 1.15 g/ml, within the 1.15- to 1.19-g/ml range observed for HIV-1 virions (4, 44). We also observed a smaller peak in fraction B (1.10 g/ml) and a shoulder of denser virus particles. The partially infectious H84Y mutant displayed a similar density pattern as wt, with a peak in fraction G, a smaller peak in fraction B, and a shoulder containing virions of higher density. In contrast, noninfectious H84A mutant viruses showed a slightly shifted peak fraction (fraction F) and a considerably broader range of densities with a higher proportion of lower density virions. Although our density gradient fractionation protocol is subject to some variability, the results with H84A suggested that these mutant virus particles might be morphologically anomalous.
FIG. 7.
Virus density gradient fractionation. Samples of the indicated viruses were loaded onto 20-to-60% sucrose density gradients and centrifuged at 170,000 × g (average) for 18 h in an SW 50.1 rotor such that particles of ≥12S should have sedimented to equilibrium. After centrifugation, 0.4-ml fractions were collected from the gradient tops (A) to bottoms (M) and Gag levels in each fraction were measured by densitometry of immunoblot bands. Gag levels for each fraction are plotted relative to the gradient fraction with the highest Gag signal (=100%). Sucrose densities were determined by weighing a constant volume of each fraction from a gradient run in parallel, although these values should be considered approximate, as our observed gradient-to-gradient variation was about ±0.005 g/ml in any given fraction.
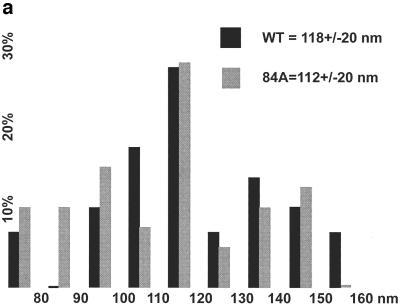
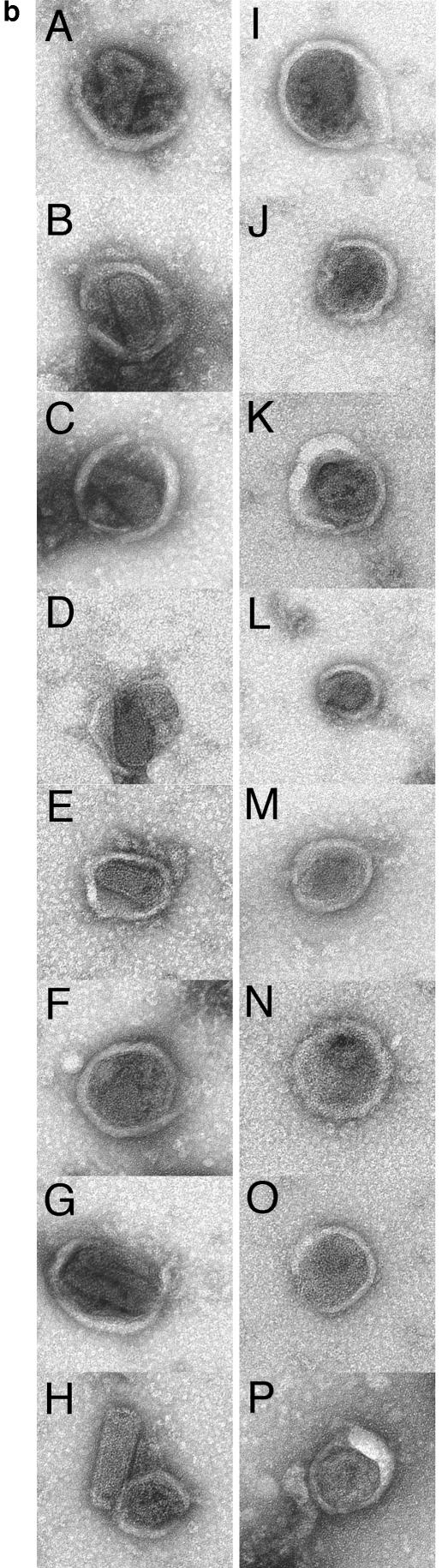
Based on sucrose density gradient results, we decided to compare virion morphologies of wt and H84A particles by EM. To do so, purified virus samples were lifted onto EM grids, washed, negatively stained, and analyzed by transmission EM. A simple examination of virus particle diameters (Fig. 8a) showed similar results for the two samples: most virus particles were in the 110- to 120-nm range, with an average wt particle diameter of 118 ± 20 nm (mean ± standard deviation) and a slightly smaller H84A average at 112 ± 20 nm. Notwithstanding the similarities in particle diameters, a significant difference was observed in particle core morphologies. Figure 8b shows a gallery of wt (panels A to H) and H84A (panels I to P) particles, stained so that lipid envelope drying and breakage revealed the internal core structure, or completely liberated cores from envelopes (Fig. 8b, panel H). It is noteworthy that all (n = 29) of the wt particles displayed visible conical or cylindrical cores (panels A to H), as is typical for HIV (10, 19, 38, 46). In contrast, H84A cores either were round, barrel shaped, or anomalously shaped (panels I to P). No particles with clearly conical or cylindrical cores were observed for the H84A mutant (n = 39).
FIG. 8.
Analysis of virus morphologies. Virus particles from wt HIVLuc- and H84A HIVLuc-transfected cells were isolated, lifted onto carbon-coated grids, stained with uranyl acetate, and imaged by transmission EM. (a) Histogram of particle diameters. Diameters of wt (n = 29) and H84A (n = 39) virus particles were plotted in 10-nm size bins relative to the frequencies (percentage of total sample) observed for each size bin. (b) Galleries of virus particle images. Images of eight wt (A to H) and H84A (I to P) virus particles are shown within 262- by 262-nm windows. Note that panel H shows a wt core, apparently released from a broken virus during preparation. Note also that all wt virions (n = 29) showed discernible cylindrical or conical cores, whereas only panel I showed a possible cylindrical core for H84A (n = 39).
Analysis of postassembly defects.
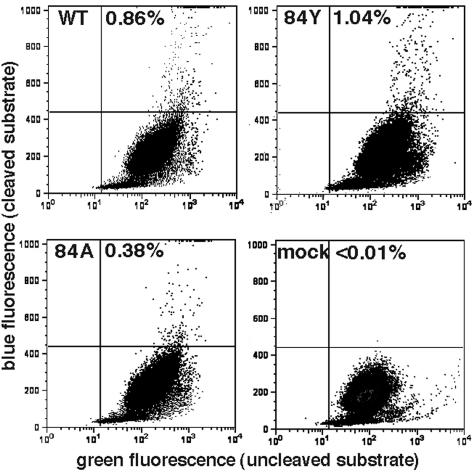
Although H84A virions had aberrant mature cores, the exact relationship between this morphological defect and the lack of infectivity was unclear. To examine postassembly defects of H84 mutants further, entry assays (8, 32) were performed to monitor the capability of virus cores to enter target cells. To generate viruses for these assays, 293T cells were cotransfected with the wt or mutant HIVLuc constructs, the VSV-G protein expression construct, and a construct encoding a vpr-β-lactamase fusion protein (32). Under our transfection and infection conditions (see Materials and Methods), we expected a multiplicity of infection in the range of 0.01%, about 10- to 20-fold higher than we have observed with HIV vectors pseudotyped with retroviral envelope proteins (29). Successful virus entry was probed as described previously (8, 32, 47) by measuring cleavage of the fluorescent β-lactamase substrate CCF2 in target cells: CCF2 cleavage is accompanied by a fluorescent shift from green to blue, and percentages of entry-positive versus -negative cells were quantitated by flow cytometry (8, 32, 47). Employing these assays with wt, H84Y, and H84A viruses gave the results shown in Fig. 9. As illustrated, while <0.01% of the mock-infected cells scored as entry positive based on blue fluorescence emission, 0.86% of the wt-infected cells gave a positive signal. Relative to wt levels, H84Y (1.04%) and H84A (0.38%) yielded higher and lower entry signals, respectively. Nevertheless, when entry results were normalized for virus particle-associated Gag protein levels in two independent experiments, entry assay results showed no impairment for H84Y (134% ± 60% wt levels) and only a slight impairment for H84A (61% ± 3% wt levels). Thus, reduced entry into target cells did not readily account for the >500-fold reduction in infectivity for H84A virions.
FIG. 9.
Virus entry assays. Virus core entry into target cells was assessed using viruses produced by cotransfection of wt, H84Y, or H84A or mock expression constructs along with expression constructs of the VSV-G (pVSV-G) and a β-lactamase-vpr (BlaM-vpr) fusion protein. Viruses were used to infect HiL cells in serum-containing medium plus 8 μg of Polybrene/ml for 5 h at 37°C. After incubations, viral samples were removed and cells were incubated an additional 18 h at 26°C in fluorescent β-lactamase substrate (CCF2/AM) loading solution. Virus entry was measured by flow cytometry detection of live cells manifesting uncleaved substrate (green fluorescence) versus cleaved product (blue fluorescence). The percentages of product-positive live cells are shown. When normalized for Gag protein levels, the average (n = 2) entry assay signals for H84A and H84Y relative to that of wt were 61% ± 3% (H84A) and 134% ± 60% (H84Y).
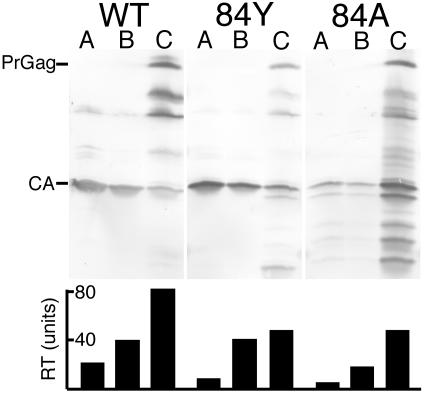
As an alternative approach to the analysis of particle defects, we decided to subject virions to a detergent treatment-fractionation protocol, since recent studies have revealed unusual fractionation profiles for certain HIV-1 CA NTD mutants (41). Consequently, wt, H84Y, and H84A viruses were detergent treated and fractionated on sucrose step gradients to obtain mature virus core-enriched pellets, as described by Tang et al. (41). Fractionation results for wt, H84Y, and H84A are shown in Fig. 10. In the case of the wt sample, the pellet fraction (lane C) preferentially contained immature and incompletely processed virions, which were not enriched in the top (lane A) or middle (lane B) fractions. As expected from prior analysis (15, 41, 46), only a small proportion of mature CA pelleted. The H84Y fractionation pattern was similar to the one observed with wt, although there was a slight increase of capsid in the pellet fraction (lane C). This trend was magnified with the H84A sample, which showed a large capsid signal in the pellet fraction. Although similar fractionation results have been observed with other HIV-1 CA NTD mutants (41), we repeated the experiment and obtained similar fractionation profiles, indicating that these were not atypical results. We also performed RT assays on each of the detergent treatment fractions. Interestingly, RT levels in all mutant fractions were lower than in their wt counterparts, suggesting a difference in wt versus mutant virus RT stabilities during the core fractionation protocol. Comparison of CA and RT levels demonstrated that although pellet-associated CA levels were increased with the H84 mutants, RT levels in the pellet (C) fractions were reduced twofold relative to wt. These observations suggest that high CA-to-RT ratios in core pellet fractions correlate with H84 mutant replication defects and are consistent with the notion that mutant virus replication is blocked after target cell entry, at a reverse transcription step.
FIG. 10.
Analysis of virus cores. Viruses collected from transfected cells were treated with 0.3% NP-40 for 10 min, layered onto an equal volume of 20% sucrose, and centrifuged at 120,000 × g for 60 min at 4°C. Top fractions (A), bottom fractions (B), and virus core-containing pellet fractions (C) were collected and subjected in parallel to RT assays (bottom panel) as well as SDS-PAGE and immunoblotting with anti-HIV-1-CA antibody. PrGag and mature CA bands are indicated on the immunoblots. RT units were determined by comparison with activities of known amounts of avian myeloblastosis virus RT.
DISCUSSION
Although the structure and some of the functions of the HIV-1 CA NTD have been clarified (9, 13, 15, 17, 20-22, 29, 35, 38-44), many structure-function relationships remain to be explained. Within mature HIV-1 cores, NTDs appear to assemble hexamer rings (5, 26, 28), and although hexamer contacts have not been determined they have been modeled to employ NTD helices I and II (19, 20, 26). In this arrangement, histidine residues H84 and 87 would occupy positions at the outer edges of hexamer rings and might be well suited to mediate putative interhexamer associations and/or histidine switch activities (14). Consequently, we probed these residues by using a mutational analysis approach.
Because HIV-1 CA H87 is not well conserved (25), it was not surprising that our cysteine substitution mutant (H87C) was infectious (Fig. 4). Our cross-linking experiments also demonstrated that H87C readily cross-linked to a free CypA cysteine, whereas the natural CA CTD cysteines did not (Fig. 5). In contrast to H87C, mutations at the highly conserved H84 substantially reduced virus infectivity (Fig. 4). Surprisingly, unlike recently described (41) mutations at CA NTD W23A and F40A at helices I and II, respectively, mutations at H84, adjacent to the CypA loop, did not reduce CypA assembly into virions (Fig. 5). However, both H84A and H84Y mutants were similar to the W23A and F40A mutants (41) in that detergent treatment experiments yielded significantly increased capsid/RT ratios in mature core fractions. These results are consistent with the interpretation that mutations at H84 reduce virus infectivity via an impairment of reverse transcription steps.
Given that H84A cores exhibited a clearly aberrant morphology (Fig. 8), a putative block at reverse transcription does not seem an unexpected phenotype. However, it is not immediately obvious why mutations at H84, which is not modeled to participate in NTD hexamer formation (19, 20, 26), should give a phenotype similar to mutations W23A and F40A in helices I and II, which have been modeled at hexamer interfaces. One possible explanation is that changes at H84 telegraph to helices I and II to alter hexamer interfaces. Alternatively, the CA NTD helix I-II and IV-VII faces may play complementary roles in assembly, perhaps in forming hexamers and in bundling hexamers into higher-order structures. Finally, mutations at CA residues 23, 40, and 84 all may have a significant impact on the folding of capsid monomers, and this common folding defect may result in related assembly anomalies.
Regardless of the role of H84 in CA oligomerization, the fact that substitution with tyrosine yielded viruses that were over 10 times more infectious than alanine, cysteine, lysine, and glutamate variants implies that the natural histidine residue is most important for its ability to participate in aromatic interactions. While this does not preclude a histidine switch (14) activity for H84, the infectivity of the H84Y mutant, albeit 30-fold less than that of wt, argues against an absolutely required H84 histidine switch. Assuming that the effect of mutations at H84 is to destabilize NTD helix IV and VII associations by weakening H84-W80-W133 aromatic interactions, the observation of aberrant CA processing products (Fig. 6) may not be surprising. However, for H84A, the combination of an increased susceptibility to processing and an increased resistance to detergent-mediated disassembly (Fig. 10) may seem contradictory. We suggest that the conformation stabilized by H84 helps control promiscuous protein oligomerization or aggregation. Our results support the concept that CA must satisfy a precarious balance between assembly and disassembly (15) and that H84 exerts a critical influence on this balance.
Acknowledgments
The HIVLuc and BlaM-vpr constructs were kindly provided by Nathaniel Landau, and the VSV-G protein expression construct was a generous gift from Randy Taplitz. We are thankful for the help and advice we've received from Ayna Alfadhli, Tenzin Choesang, and Amelia Still.
This work was supported by Public Health Service grant R01 GM060170-06 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Ackerson, B., O. Rey, J. Canon, and P. Krogstad. 1998. Cells with high cyclophilin A content support replication of human immunodeficiency virus type 1 Gag mutants with decreased ability to incorporate cyclophilin A. J. Virol. 72:303-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiken, C. 1997. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J. Virol. 71:5871-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alfadhli, A., E. Steel, L. Finlay, H. Bachinger, and E. Barklis. 2002. Hantavirus nucleocapsid protein coiled-coil domains. J. Biol. Chem. 277:27103-27108. [DOI] [PubMed] [Google Scholar]
- 4.Arvidson, B., J. Seeds, M. Webb, L. Finlay, and E. Barklis. 2003. Disruption of the retrovirus capsid interdomain linker region. Virology 308:166-177. [DOI] [PubMed] [Google Scholar]
- 5.Barklis, E., J. McDermott, S. Wilkens, S. Fuller, and D. Thompson. 1998. Organization of HIV-1 capsid proteins on a lipid monolayer. J. Biol. Chem. 273:7177-7180. [DOI] [PubMed] [Google Scholar]
- 6.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. USA 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braaten, D., E. Franke, and J. Luban. 1996. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J. Virol. 70:3551-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavrois, M., C. De Noronha, and W. Greene. 2002. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 20:1151-1154. [DOI] [PubMed] [Google Scholar]
- 9.Chiu, H.-C., F.-D. Wang, S.-Y. Yao, and C.-T. Wang. 2002. Effects of gag mutations on human immunodeficiency virus type 1 particle assembly, processing, and cyclophilin A incorporation. J. Med. Virol. 68:156-163. [DOI] [PubMed] [Google Scholar]
- 10.Coffin, J., S. Hughes, and H. Varmus (ed.). 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 11.Connor, R., B. Chen, S. Choe, and N. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 12.Cowan, S., T. Hatziioannou, T. Cunningham, M. Muesing, H. Gottlinger, and P. Bieniasz. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA 99:11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorfman, T., A. Bukovsky, A. Ohagen, S. Hoglund, and H. Gottlinger. 1994. Functional domains of the capsid protein of human immunodeficiency virus type 1. J. Virol. 68:8180-8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehrlich, L., T. Liu, S. Scarlata, B. Chu, and C. Carter. 2001. HIV-1 capsid protein forms spherical (immature-like) and tubular (mature-like) particles in vitro: structure switching by pH-induced conformational changes. Biophys. J. 81:586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forshey, B., U. von Schwedler, W. Sundquist., and C. Aiken. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 76:5667-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franke, E., H. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359-362. [DOI] [PubMed] [Google Scholar]
- 17.Gamble, T., F. Vajdos, S. Yoo, D. Worthylake, M. Houseweart, W. Sundquist, and C. Hill. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285-1294. [DOI] [PubMed] [Google Scholar]
- 18.Gamble, T., S. Yoo, F. Vajdos, U. von Schwedler, D. Worthylake., H. Wang, J. McCutcheon, W. Sundquist, and C. Hill. 1997. Structure of the carboxy-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 19.Ganser, B., S. Li, V. Klishko, J. Finch, and W. Sundquist. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80-83. [DOI] [PubMed] [Google Scholar]
- 20.Ganser-Pornillos, B., U. von Schwedler, K. Stray, C. Aiken, and W. Sundquist. 2004. Assembly properties of the human immunodeficiency virus type 1 CA protein. J. Virol. 78:2545-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gitti, R., B. Lee, J. Walker, M. Summers, S. Yoo, and W. Sundquist. 1996. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273:231-235. [DOI] [PubMed] [Google Scholar]
- 22.Gross, I., H. Hohenberg, T. Wilk., K. Wiegers, M. Grattinger, B. Muller, S. Fuller, and H. Krausslich. 2000. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 19:103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen, M., and E. Barklis. 1995. Structural interactions between retroviral Gag proteins examined by cysteine cross-linking. J. Virol. 69:1150-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatziioannou, T., S. Cowan, S. Goff, P. Bieniasz, and G. Towers. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuiken, C., B. Foley, E. Freed, B. Hahn, P. Marx, F. McCutchan, J. Mellors, S. Wolinsky, and B. Korber (ed.). 2002. HIV sequence compendium 2002. LA-UR 03-3564. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 26.Li, S., C. Hill, W. Sundquist, and J. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409-413. [DOI] [PubMed] [Google Scholar]
- 27.Mammano, F., A. Ohagen, S. Hoglund, and H. Gottlinger. 1994. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J. Virol. 68:4927-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayo, K., D. Huseby., J. McDermott, B. Arvidson, L. Finlay, and E. Barklis. 2003. Retrovirus capsid protein assembly arrangements. J. Mol. Biol. 316:667-678. [DOI] [PubMed] [Google Scholar]
- 29.McDermott, J., L. Farrell, R. Ross, and E. Barklis. 1996. Structural analysis of human immunodeficiency virus type 1 Gag protein interactions, using cysteine-specific reagents. J. Virol. 70:5106-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDermott, J., S. Karanjia, Z. Love, and E. Barklis. 2000. Crosslink analysis of N-terminal, C-terminal, and N/B determining regions of the Moloney murine leukemia virus capsid protein. Virology 269:190-200. [DOI] [PubMed] [Google Scholar]
- 31.Momany, C., L. Kovari, A. Prongay, W. Keller, R. Gitti, B. Lee, A. Gorbalenya, L. Tong, J. McClure, L. Ehrlich, M. Summers, C. Carter, and M. Rossmann. 1996. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 3:763-770. [DOI] [PubMed] [Google Scholar]
- 32.Munk, C., S. Brandt, G. Lucero, and N. Landau. 2002. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. USA 99:13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nermut, M., D. Hockley, J. Jowett, I. Jones, M. Garreau, and D. Thomas. 1994. Fullerene-like organization of HIV Gag protein shell in virus-like particles produced by recombinant baculovirus. Virology 198:288-296. [DOI] [PubMed] [Google Scholar]
- 34.Nermut, M., D. Hockley, P. Bron, D. Thomas, W. Zhang, and I. Jones. 1998. Further evidence for hexagonal organization of HIV gag protein in prebudding assemblies and immature virus-like particles. J. Struct. Biol. 123:143-149. [DOI] [PubMed] [Google Scholar]
- 35.Reicin, A., S. Paik, R. Berkowitz, J. Luban, I. Lowy, and S. Goff. 1995. Linker insertion mutations in the human immunodeficiency virus type 1 in virion morphogenesis. J. Virol. 69:642-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 37.Stremlau, M., C. Owens, M. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component Trim5α restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 38.Swanstrom, R., and J. Wills. 1997. Synthesis, assembly and processing of viral proteins, p. 263-334. In J. Coffin, S. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 39.Tang, C., Y. Ndassa, and M. Summers. 2002. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 9:537-543. [DOI] [PubMed] [Google Scholar]
- 40.Tang, S., T. Murakami, B. Agresta, S. Campbell, E. Freed, and J. Levin. 2001. Human immunodeficiency virus type 1 N-terminal capsid mutants that exhibit aberrant core morphology are blocked in initiation of reverse transcription in infected cells. J. Virol. 75:9357-9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang, S., T. Murakami, N. Cheng, A. Steven, E. Freed, and J. Levin. 2003. Human immunodeficiency virus type 1 N-terminal capsid mutants containing cores with abnormally high levels of capsid protein and virtually no reverse transcriptase. J. Virol. 77:12592-12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Schwedler, U., T. Stemmler, V. Klishko, S. Li, K. Albertine, D. Davis, and W. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Schwedler, U., K. Stray, J. Garrus, and W. Sundquist. 2003. Functional surfaces of the human immunodeficiency virus type I capsid protein. J. Virol. 77:5439-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, C., and E. Barklis. 1993. Human immunodeficiency virus gag protein mutants defective in assembly, processing, and infectivity. J. Virol. 67:4264-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, C., Y. Zhang, J. McDermott, and E. Barklis. 1993. Conditional infectivity of a human immunodeficiency virus matrix domain deletion mutant. J. Virol. 67:7067-7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Welker, R., H. Hohenberg, U. Tessmer, C. Huckhagel, and H. Krausslich. 2000. Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. J. Virol. 74:1168-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zlokarnik, G., P. Negulscu, T. Knapp, L. Mere, N. Burres, L. Feng, M. Whitney, K. Roemer, and R. Tsien. 1998. Quantitation of transcription and clonal selection of single living cells with β-lactamase as reporter. Science 279:84-88. [DOI] [PubMed] [Google Scholar]