Abstract
Epstein-Barr virus (EBV) nuclear antigen 1 (EBNA1) is expressed in the majority of latency programs in EBV-infected cells and is critical for the maintenance of EBV episomes in the infected cells. EBNA1 is also known to be involved in transcriptional activation and regulates expression of the EBV latent genes, including the EBNAs and LMP1. Thus, EBNA1 is a multifunctional protein with critical functions required for the persistence of the viral genome over successive generations, producing new daughter cells from the infected cell. We identify EBNA1 here as an interacting EBNA with the known suppressor of metastasis and cell migration, Nm23-H1. Nm23-H1 inhibits cell migration when expressed in cancer cells. We show that EBNA1 associates with Nm23-H1 in EBV-infected cells in vitro, as well as in lymphoblastoid cell lines (LCLs). Nm23-H1 predominantly localizes to the cytoplasm in BJAB and 293T cells; however, upon expression of EBNA1, Nm23-H1 is translocated to the nucleus in similar compartments to EBNA1, suggesting a potential functional role that is linked to EBNA1. Convincingly, in EBV-transformed LCLs Nm23-H1 is localized predominantly to the nucleus and colocalizes to similar compartment as EBNA1. Further, we tested the effects of EBNA1 on Nm23-H1-mediated suppression of cell migration and showed that EBNA1 rescues the suppression of cell migration mediated by Nm23-H1. These in vitro studies suggest that EBNA1 plays a critical role in regulating the activities of Nm23-H1, including cell migration, through a mechanism which involves direct interaction of this major regulator in EBV-infected cells.
Epstein-Barr virus (EBV) is a ubiquitous human gammaherpesvirus associated with numerous human malignancies. EBV is predominantly associated with the infection of two target tissues in vivo: B lymphocytes, where the infection is predominantly latent, and oropharyngeal epithelial tissue, in which the infection is strictly lytic with the production of viral progeny (38, 39). Both target tissues are susceptible to EBV-associated malignant change, leading to tumors of B-cell origin, such as Burkitt's lymphoma (BL), or of epithelial cell origin, such as nasopharyngeal carcinoma (NPC) (38, 39).
An in vitro corollary of the oncogenic capacity of EBV is its ability to transform primary B cells into permanently growing lymphoblastoid cell lines (LCLs) in which every cell carries multiple episomal copies of the virus genome and expresses a limited set of EBV-encoded latent nuclear antigens (EBNAs) and latent membrane proteins (38, 39). Numerous studies over the last decade have generated an abundance of clues as to the functions of a number of these latent proteins (17, 22, 50). EBNA3C, one of the essential molecules, has been shown to be critical for B-lymphocyte transformation (22, 38, 39). A genetic knockout of the gene by introduction of a stop codon within the open reading frame renders the virus incompetent for transformation of B lymphocytes (12). Recent studies have implicated EBNA3C in regulating cell migration through the interaction with Nm23-H1, a known suppressor of cell migration (47). This interaction, surprisingly, indicated that EBV may play a role in regulating the expression of cellular molecules involved in cell migration and that EBV-positive tumors may have greater potential to be invasive. EBNA3C is typically expressed in lymphomas associated with AIDS immunocompromised patients and posttransplant patients (38, 39). However, based on current experimental evidence, EBNA3C has not been detected in EBV-associated carcinomas, including NPCs, gastric carcinomas, Hodgkin's lymphomas, and BLs to date (38, 39). However, EBNA3C may be required in the early stages of infection and initiation of transformation prior to the maintenance of the transformed state. In addition, an increase in detection capabilities as screening of tumors may result in new information regarding the presence or absence of the EBNA3 family of proteins in EBV-positive tumors.
EBNA1, known to be required for maintenance of the EBV episome, is expressed in all forms of EBV latency, a finding consistent with the central role of this protein in the maintenance and segregation of the episomal latent EBV genome (16, 38, 39). EBNA1 is a DNA-binding phosphoprotein that through binding to the plasmid origin of viral replication, oriP, is responsible for tethering the virus episome to chromosomes (26, 61). EBNA1 is also capable of transactivating the Wp/Cp promoters responsible for initiation of transcription of the six EBNAs in type III latency (38, 39) and negatively regulates the Qp during latency type I in which none of the other EBNAs is transcribed (8, 17). In addition, studies indicate that EBNA1 antisense oligodeoxyribonucleotides inhibited proliferation of EBV-immortalized cells and that EBNA1 expression in transgenic mice can induce lymphomas (58). Moreover, EBNA1 increased the tumorigenic potential and metastatic capability of a NPC cell line (43). These studies suggest that EBNA1 may have a direct role in contributing to the tumorigenic potential of EBV. However, the mechanisms and relevance to the pathogenesis of EBV-associated tumors remain unknown. Expression of EBNA1 has been demonstrated in all EBV-associated tumors, including EBV-positive NPCs and gastric adenocarcinomas (38, 39). The contribution of EBNA1 expression to the oncogenic process is thought to relate to the role of the protein in maintaining the extrachromosomal integrity of the EBV genome (15, 33). However, it is evident that additional functions are associated with EBNA1.
The nm23 gene family is highly conserved among a wide variety of eukaryotic species. Eight genes have been identified in humans: nm23-H1 (40), nm23-H2 (44), DR-nm23 (53), nm23-H4 (29), nm23-H5 (30), nm23-H6 (28, 51), nm23-H7 (52), and nm23-H8 (34). Members of the nm23 gene family are structurally and functionally conserved consisting of four to six identically folded subunits, enclosing a large (25 Å) central cavity (57). Expression of nm23 genes have been linked to suppression of tumor metastasis, differentiation, apoptosis, proliferation, and DNA mutation rate (9) and associated with nucleoside diphosphate kinase (NDPK) activity (5, 32, 55), serine phosphorylation (6, 14, 23, 31), histidine protein kinase activities (54), and transcriptional stimulatory activities (1, 4, 35, 37).
The human nm23-H1 gene product is the best-characterized member of this family of proteins. It is 152 amino acids in length with leucine repeats and alpha-helical and basic domains (Fig. 1) (39). nm23-H1 is ca. 88% identical to nm23-H2 and maps 4 kb apart at position q21.3 on chromosome 17 near the BRCA1 locus, known to be associated with early onset of familial breast and ovarian cancer (3). The nm23-H1 gene product is more closely associated with the inhibition of metastasis and signal transduction, has an acidic pI, and is identical to the A subunit of NDPK (11). Importantly, in a number of human cancers there is an inverse relationship between nm23-H1 expression and metastasis (9, 27). In neuroblastoma, mutations in the leucine zipper motif of nm23-H1 directly correlates with increased metastatic potential (13, 20). The mechanism by which Nm23-H1 regulates metastasis is not fully understood; however, the experimental data accumulated thus far strongly implicates Nm23-H1 in the regulation of metastasis in a diverse number of human cancers (24).
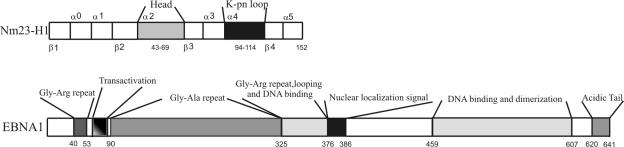
FIG. 1.
Secondary structure of the EBNA1 protein and the Nm23-H1 protein. The structure of EBNA1 includes the DNA-binding domain and the dimerization domains, as well as in internal Gly-Ala repeats. The region required for looping, the Gly-Ala repeats, and nuclear localization signal is also shown. The structure of Nm23-H1 shows the four beta sheets, the five alpha-helical regions and two regions referred to as the head and the K-pn loop.
MATERIALS AND METHODS
Constructs and cell lines.
The glutathione S-transferase (GST)-Nm23-H1 fusion protein was generated by subcloning PCR-amplified product from Nm23-H1 cDNA into pGEX-2TK vector (23). The forward primer 5′-TTAGGATCCCATATGGCCAACTGTGAGCG-3′ and reverse primer 5′-TTACCCGGGCATGGATCCTCCTCCTGTCATTCA-3′ contained BamHI sites at the ends for cloning into pGEX-2TK. The conditions used for the PCR amplifications were 1 cycle of 94°C for 5 min; 30 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min; and a final cycle of 72°C for 10 min. PCR amplification was done by using the Vent polymerase from NEB. The myc fusion protein was generated as described above except that the reverse primer 5′-GCTGCGGCCGCTTCATAGATCCAGTTCTGAGC-3′ was used. The expression vector pA3M was prepared with BamHI and NotI, and the PCR product also cut with similar enzymes was ligated and cloned for expression and fusion to the myc tag at the carboxy terminus (7). Mutant EBNA1 p367, p376, p378, p385, and p396 were kindly provided by John Yates (60).
The EBV-negative BL BJAB cell line was obtained from Elliott Kieff (56). The T-antigen-transformed human embryonic kidney cell line 293T was obtained from Jon Aster (Department of Pathology, Brigham and Women's Hospital, Harvard Medical School). The EBV-transformed LCLs LCL1 and LCL2 were generated in our laboratory. All B-cell lines were maintained in RPMI 1640 medium (Invitrogen/Gibco-BRL, Rockville, Md.) supplemented with 10% fetal bovine serum, 2 mM glutamine, and 25 U of penicillin-streptomycin (Gemini Bioproducts, Inc., Carlsbad, Calif.)/ml. All adherent cell lines were maintained in Dulbecco modified Eagle medium (DMEM) and supplemented as described above.
Immunoprecipitation experiments:.
A total of 5 × 107 cells were collected and washed with phosphate-buffered saline (PBS) before they were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 7.6], 150 mM sodium chloride, 2 mM EDTA, 1% NP-40, 1 mM phenylmethylsulfonyl fluoride, 1 μg of aprotinin/ml, 1 μg of pepstatin/ml) for 1 h on ice. The cell pellet was removed by centrifugation, and the supernatant was precleared by using protein A-Sepharose beads. The beads were collected, and the lysates were incubated with mouse monoclonal Nm23-H1 antibody (Santa Cruz, Santa Cruz, Calif.) overnight along with the protein A/G-Sepharose beads at 4°C. The immunoprecipitates were washed four times with RIPA buffer and then solubilized by using sodium dodecyl sulfate (SDS) lysis buffer. Proteins were fractionated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to 0.45-μm-pore-size nitrocellulose membrane. The membrane was blocked by using 5% milk and then incubated with EBNA1 mouse monoclonal antibodies, followed by incubation with horseradish peroxidase-conjugated secondary antibody against mouse (Amersham Biosciences, Piscataway, N.J.). The proteins were detected by using chemiluminescence detection protocols as described by the manufacturer.
In vitro protein binding assays.
GST-Nm23-H1 and GST-EBNA1 beads used for the in vitro binding experiments and in the GST pull-down assays were prepared as follows. The overnight culture of the Escherichia coli DH5α strain containing the pGEX-nm23-H1 or pGEX-EBNA1 expression constructs was used for inoculating 500 ml of Luria-Bertani medium containing 100 μg of ampicillin/ml. The fusion protein was induced for 4 h by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The induced cells were pelleted and sonicated, and the cell debris was removed by centrifugation. The supernatant was collected and rotated with glutathione-Sepharose beads at 4°C for 2 h. The beads were then washed with NETN buffer (0.5% NP-40, 20 mM Tris, 100 mM NaCl, 1 mM EDTA) supplemented with protease inhibitors (100 mM phenylmethylsulfonyl fluoride, 1 μg of pepstatin/ml, and 1 μg of aprotinin/ml) and stored at 4°C for use. For in vitro binding experiments, Nm23-H1, EBNA1, and control luciferase were incubated with 35S-labeled in vitro-translated protein in binding buffer (1× PBS, 0.1% NP-40, 0.5 mM dithiothreitol, and 10% glycerol supplemented with protease inhibitors). In vitro translation was performed with the TNT T7 QuickCoupled transcription-translation system (Promega, Inc., Madison, Wis.). The in vitro-translated proteins were first precleared by using glutathione-Sepharose beads in the binding buffer for 60 min at 4°C. The GST beads were centrifuged and washed with the same buffer, and the supernatant was rotated with glutathione-Sepharose-bound GST-Nm23-H1 or GST-EBNA1 beads overnight at 4°C. The beads were collected and washed four times with 1 ml of binding buffer. Proteins bound to both the GST and GST-Nm23-H1 or GST-EBNA1 beads were denatured by using SDS-β-mercaptoethanol lysis buffer and fractionated on SDS-PAGE. The gel was dried and analyzed by using a PhosphorImager and quantified by using the ImageQuant program (Molecular Dynamics, Inc.).
GST pull-down assays were carried out with cell lysates prepared from 100 million cells. Cells were lysed by using RIPA buffer as described above in the immunoprecipitation section. First, the cell lysates were rotated with GST beads for 60 min at 4°C. The GST beads were collected by centrifugation and then washed with the RIPA buffer, and the supernatant was incubated with GST-Nm23-H1 beads overnight at 4°C. The beads were centrifuged and then washed four times with 1 ml of RIPA buffer. As before, the proteins bound to beads were denatured by using lysis buffer and fractionated on SDS-PAGE. The proteins were transferred to 0.45-μm-pore-size nitrocellulose filters and then analyzed by Western blotting with the anti-EBNA1 mouse monoclonal antibody (or adsorbed human serum specific for ENA1).
Transfection and colocalization.
A total of 20 μg of pA3M empty vector, pA3M-nm23-H1, pSG5-EBNA1, and pSG5-EBNA1+pA3M-nm23-H1 were transfected into 10 million BJAB cells by electroporation with a Bio-Rad electroporator at 220 V and 975 μF. After 24 h, cells were collected and immunoprecipitated by using Nm23-H1 mouse monoclonal antibody and Western blotting for the detection of EBNA1. Expression of Nm23-H1 was detected by using myc ascites antibody generated from the 9E10 hybridoma.
BJAB, 293T, and LCL1 cells were fixed onto slides by using 1:1 acetone and methanol for 10 min at −20°C. The cells were blocked at room temperature for 1 h with 20% goat serum. EBNA1 rabbit polyclonal antibody at a dilution of 1:100 and Nm23-H1 mouse monoclonal antibody at a concentration of 1:150 were used as primary antibodies. The cells were incubated with primary antibody for 1 h at 37°C and then washed three times with PBS buffer. Fluorescein isothiocyanate-conjugated anti-mouse and Texas Red-conjugated anti-rabbit secondary antibodies at a concentration of 1:1,000 was then added and incubated at 37°C for 1 h. The slides were washed with PBS buffer and then mounted with Fluoromount. The colocalized proteins were visualized on an Olympus BX60 fluorescence microscope. The photographs were captured with a PixelFly camera (Cooke, Inc.).
Cell motility assays.
MDA-MB-435 cells were transfected with 10 μg of pCMV vector, pCMV-nm23-H1, pSG5EBNA1, and pCMV-nm23-H1 by using Superfect as described by the manufacturer (Qiagen, Valencia, Calif.). Then, 10 μg of p367, p376, p378, p385, and p396 were transfected by electroporation by using a Bio-Rad electroporator at 220 V and 975 μF. Cells were selected in neomycin 2 mg/ml for 72 h and then used for motility assays. Cell motility was determined by using 96-well Boyden chemotaxis chambers (NeuroProbe). The chemoattractants used were fetal bovine serum, epidermal growth factor (EGF), and fibronectin (Boehringer Mannheim, Inc., Mannheim, Germany). Dilution of the chemoattractants was carried out in DMEM containing 0.1% bovine serum albumin, 10 mM HEPES, and 100 U of penicillin-streptomycin/ml, and serial dilutions were placed in the lower part of the chamber. An 8-μm-pore-size polycarbonate polyvinylpyrrolidone-free membrane was sandwiched between the upper and lower chambers. Cells harvested 72 h after transfection were washed in PBS and resuspended in DMEM containing 0.1% bovine serum albumin, 10 mM HEPES, and 100 U of penicillin-streptomycin/ml. A total of 7.5 × 104 selected cells were added to the upper wells of the chamber, and the chamber was incubated for 2 h at 37°C in a humidifying CO2 incubator. After the removal of cells from the upper side of the membrane, it was stained with Gill's hematoxylin stain, and the migrated cells were counted at ×40 magnification with an Olympus BX40 light microscope. The data presented for each concentration represent the means of three separate experiments.
RESULTS
Previous studies have implicated the EBV nuclear antigens EBNA3C in modulating the activity of Nm23-H1 in cell migration and transcription regulation (47, 49). EBNA3C is known to be expressed in type III latency patterns and was more recently shown to be involved in regulating Nm23-H1 activity (48). We have demonstrated that EBNA3C and Nm23-H1 associate with each other in EBV-infected cells (47). Moreover, we had tested the association with the other EBNA3 family members with little association seen with Nm23-H1 (47). We decided to investigate whether or not other EBNAs expressed in all major EBV-associated tumors primarily EBNA1 can associate with Nm23-H1 in EBV-infected cells and are capable of modulating the effects of Nm23-H1 on cell migration.
EBNA1 associates with Nm23-H1 in the EBV-negative BJAB cell line.
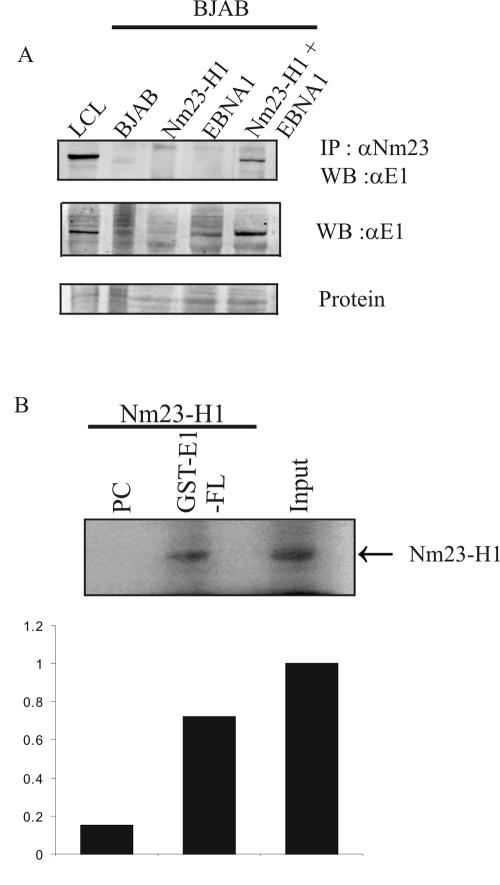
Initial studies to determine whether EBNA1 associates with Nm23-H1 were done by transient transfection of expression constructs containing the Nm23-H1 cDNA tagged with a myc epitope at its carboxy terminus and the EBNA1 cDNA, both expressed from heterologous promoters for exogenous expression in the BJAB B-cell line negative for EBV. Complexes were immunoprecipitated with anti-Nm23-H1 polyclonal antibody complexed with protein A-Sepharose beads incubated with lysates of transfected cells harvested 48 h after transfection. The complexes collected were washed and solubilized in lysis buffer, and the proteins were fractionated on SDS-PAGE gels. Western blot analysis of the fractionated proteins showed that EBNA1 signal was present, suggesting that EBNA1 was associated in complex with Nm23-H1 (Fig. 2A, top panel, last lane). In addition, Western blotting of lysates from transfected cells confirmed that EBNA1 was expressed only in the transfection with the EBNA1 expression plasmid and not in lanes without EBNA1 transfected (Fig. 2A, middle panel, compares fourth and fifth lanes with the second and third lanes). The first lane shows the positive EBNA1 signal from an EBV-positive LCL used as a control. A photograph of Ponceau S staining of membrane indicates that the levels of protein loaded were equivalent in each well (Fig. 2A, lower panel). These results suggest that EBNA1 can associate in complex with Nm23-H1 in human cells as determined by immunoprecipitation with antibodies specific to Nm23-H1.
FIG. 2.
EBNA1 coimmunoprecipitates with Nm23-H1 in transiently transfected BJAB cells and binds in vitro. Complexes with Nm23-H1 were immunoprecipitated with antibody against Nm23-H1 protein (mouse monoclonal Nm23-H1 antibody). (A) All samples were fractionated on SDS-10% PAGE gels and transferred to nitrocellulose membranes. The membranes were then probed with specific antibodies that recognize Nm23-H1 and EBNA1. The lower panel shows a protein stain intended to show similar loading of the lanes. (B) In vitro binding of full-length Nm23-H1 binding to GST fusions of EBNA1. Arbitrary units were measured by using ImageQuant.
To determine whether the association seen occurred in the transient-transfected B-cell line, we carried out in vitro binding assays with 35S-labeled Nm23-H1 and GST-EBNA1 fusion protein. Our results show that 35S-labeled Nm23-H1 banded to GST-EBNA1 in this pull-down assay with a capacity of 75% of the 10% input. Quantitation of the loading shows an affinity that is approximately 7.5 to 8% of the total based on arbitrary units from the PhosphorImager analysis.
EBNA1 associates with Nm23-H1 in EBV-transformed LCLs.
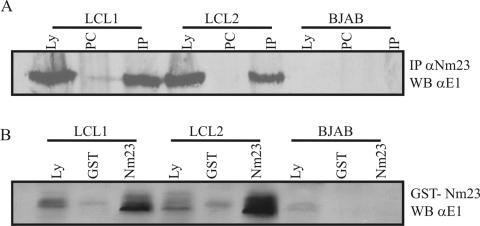
To determine whether Nm23-H1 and EBNA1 associate with each other in EBV-transformed cells, we immunoprecipitated lysates of two recently transformed cell lines, LCL1 and LCL2. Lysates from one hundred million cells were incubated with polyclonal antibody to Nm23-H1 overnight, and the complexes were collected by using protein A-Sepharose. Complexes were fractionated, transferred to nitrocellulose membranes, and then Western blotted for detection of EBNA1 protein with a polyclonal serum that specifically detects EBNA1. In both LCL1 and LCL2 we detected a specific signal for EBNA1 in the lysate lane that corresponded to the same signal in the immunoprecipitation lane (Fig. 3). The EBV-transformed human LCLs express all EBV latent proteins, including EBNA1. Therefore, this result showing an association of Nm23-H1 with EBNA1 in LCLs indicates that Nm23-H1 probably functionally associates with EBNA1 in EBV-positive cells.
FIG. 3.
Nm23-H1 coimmunoprecipitates with EBNA1 in EBV-transformed LCLs. LCL1 and LCL2 are recently transformed human B-cell lines by EBV. All of the latent genes are expressed, and they are both type III latency cell lines. Anti-Nm23-H1 antibody was used to incubate with lysates from 50 million cells for 12 to 16 h at 4°C. All complexes were collected by using protein A-Sepharose and fractionated on a SDS-12% PAGE gels. (A) Results when membranes were probed with antibodies against EBNA1 showing that EBNA1 coimmunoprecipitates in both cell lines with anti-Nm23-H1 antibodies. No signal was seen in the control EBV-negative cell line BJAB. (B) Results from a pull-down experiment with GST-Nm23-H1 fusion proteins conjugated to GST protein. Glutathione beads were bound to the fusion proteins and then incubated with lysates from the two LCLs. Complexes were again transferred to nitrocellulose membranes, and the membranes were probed with anti-EBNA1 antibodies. The results show signals for EBNA in the GST-Nm23H1 lanes (shown as Nm23-H1).
To corroborate the coimmunoprecipitation studies, we also prepared GST-Nm23-H1 beads from bacteria and incubated them with lysates from the two LCLs above. GST control beads were used as a control for specificity in our assay. In this experiment we show that GST-Nm23-H1 brought down complexes containing EBNA1. EBNA1 was seen in LCL1 and LCL2 but not in the EBV-negative B-lymphoma-cell line BJAB (Fig. 3B). These studies support the results from our immunoprecipitation with specific antibodies to Nm23-H1, strongly indicating that Nm23-H1 and EBNA1 can associate in EBV-transformed LCLs.
Nm23-H1 is localized predominantly in the cytoplasm of EBV-negative cells but translocates to the nucleus in EBV-transformed cells and in cells transiently transfected with EBNA1.
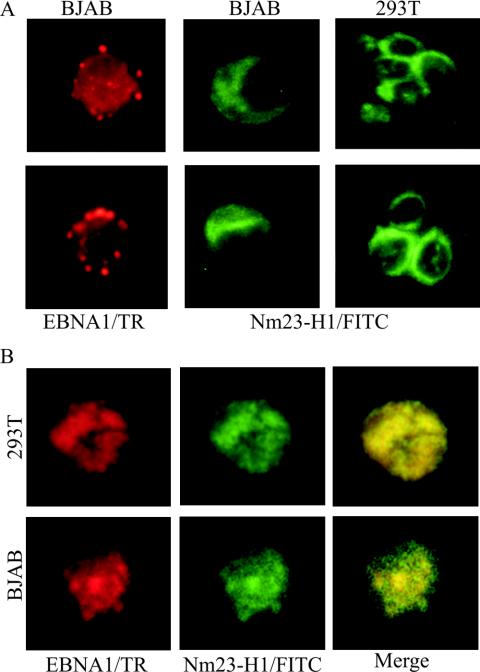
In previous experiments we determined that EBNA3C was capable of mediating translocation of Nm23-H1 to the nucleus when transiently transfected into EBV-negative BJAB cells. In addition, we showed that EBNA3C colocalizes with Nm23-H1 EBV-transformed LCLs (49). We also wanted to determine whether EBNA1 can also mediate the translocation of Nm23-H1 to the nucleus. Therefore, we transiently transfected 293T cells, as well as BJAB cells, independently with Nm23-H1 myc tagged, and BJAB cells with EBNA1, expression constructs to determine whether the localization was cytoplasmic and nuclear, respectively, based on previous studies (47). In the present study we show that EBNA1 is localized, as expected, to the nucleus in highly structured bodies, whereas Nm23-H1 was localized predominantly to the cytoplasm in both BJAB and 293T cells (Fig. 4A). In cells cotransfected with both Nm23-H1 and EBNA1 expression plasmids, Nm23-H1 signals was predominantly nuclear (Fig. 4B), strongly suggesting EBNA1-mediated nuclear translocation of Nm23-H1, since the signals for Nm23-H1 changed from being predominantly nonnuclear to nuclear in the presence of EBNA1.
FIG. 4.
Immunofluorescence analysis of Nm23-H1 signals in transfected BJAB cells and in transfected 293T cells shows the specific signals for Nm23-H1 and EBNA1. (A) EBNA1 signals were detected in the nucleus with human serum adsorbed against human B-cell lines and were previously shown to have specific signals for EBNA1 by immunofluorescence and Western blot analysis. Signals were detected by using anti-human antibody-conjugated Texas Red antibody at 1:1,000 dilutions. Mouse monoclonal antibody to Nm23-H1 was used as primary antibody and goat anti-mouse fluorescein isothiocyanate-conjugated antibody was used at 1:150 and 1:2,500 dilutions. EBNA1 signals show localization to the nucleus. (B) Signal when both the NM23-H1 expression plasmid (pA3M-Nm23-H1) and the EBNA1 expression plasmid (pSG5-EBNA1) were transfected in 293T and BJAB cells. Signals were visualized as described above. Nm23-H1 was translocated to the nucleus in the presence of EBNA1, and colocalization was observed by using a triple filter showing localization of both signals in similar nuclear compartments.
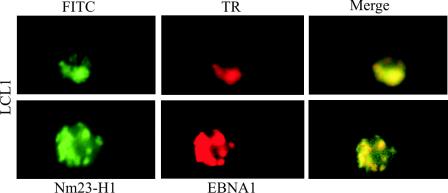
In an effort to determine whether EBNA1 and Nm23-H1 colocalize in similar nuclear compartments in the nucleus of EBV-transformed LCLs, we stained a recently transformed B-cell line LCL1 by using specific antibodies to Nm23-H1 and EBNA1. The results of these studies strongly indicate that EBNA1 mediates translocation of Nm23-H1 to the nucleus of EBV-transformed LCLs and supports a role for EBNA1 regulating the function of Nm23-H1, which may include transcriptional effects leading to changes in cell migration that are linked to nuclear translocation (Fig. 5).
FIG. 5.
EBNA1 and Nm23-H1 colocalize in the nucleus of EBV-transformed LCLs. Cells from LCL1 were fixed on slides and then probed with antibodies to EBNA1 and Nm23-H1. Signals were visualized as described above.
EBNA1 rescues the inhibition of cell migration by the suppressor of metastasis Nm23-H1.
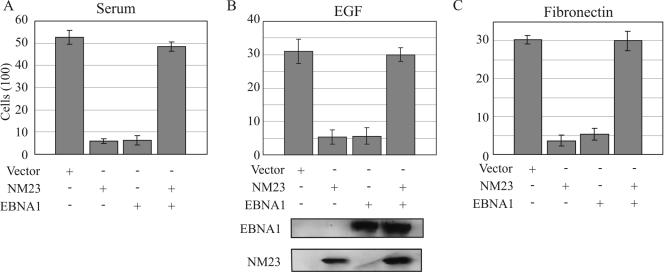
The inhibition of cell migration and metastasis by Nm23-H1 is well established in in vitro ssays and in animal studies (41, 46). We have previously shown that EBV latent antigen EBNA3C can reverse the ability of Nm23-H1 to suppress cell migration (47). In addition, increased cell motility is correlated with increased metastatic potential, leading us to investigate the migratory ability of the EBV-negative MDA-MB-435 breast carcinoma cell line in the presence of EBNA1 and Nm23-H1. The mechanism of action is yet to be understood. In our previous report we showed that EBNA3C reversed the effects of Nm23-H1 in its ability to suppress the migration of a B-cell line, as well as a breast carcinoma cell line, but that it had no effect on migration if expressed alone. However, when EBNA1 or Nm23-H1 was expressed alone, surprisingly, the number of cells migrating was dramatically reduced, but when both were coexpressed the migratory ability of the cells were restored to levels similar to that seen with control (Fig. 6). Similar results were observed when the assays were done under conditions of various chemoattractants that includes serum, EGF, and fibronectin (Fig. 6). In each case the number of cells migrating was reduced in the presence of Nm23-H1 or EBNA1, but this was effectively reversed when the two were expressed together. Interestingly, the levels seen were greater in serum than that seen with EGF and fibronectin, suggesting that the effects were more pronounced in serum with potentially multiple factors that contribute to cell migration. Nevertheless, the trends were similar in each experiment. Control Western blots showed that EBNA1 and Nm23-H1 were expressed in the cells that were used for migration assays (Fig. 6, lower panel).
FIG. 6.
EBNA1 reverses the ability of Nm23-H1 to suppress cell migration. MDA-MB-435 cells were transfected with Nm23-H1, EBNA1, or both and selected in culture. All colonies were collected and amplified, followed by analysis for expression of individual proteins by Western blotting. Cell migration assays were performed with three different chemoattractants. In each case the cells containing Nm23-H1 plus EBNA1 showed a reversal of the ability of Nm23-H1 to suppress migration. Western blots at the bottom show the signals for both EBNA1 and Nm23-H1.
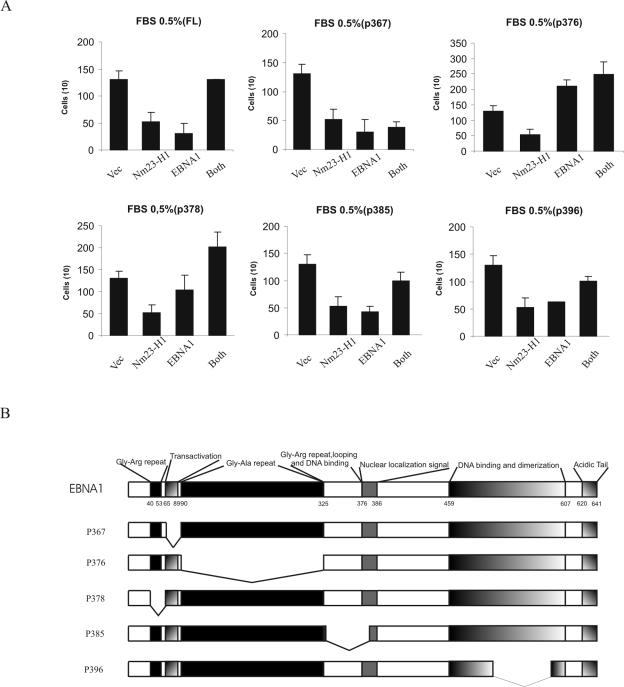
To further address and identify the responsible domain of EBNA1 that contributes to cell migration, we obtained EBNA1 mutants (60) (Fig. 7B). Our results show that EBNA1 alone had an effect in reducing motility but in the context of coexpression with Nm23-H1 resulted in increased motility. Mutants p385 and p396, which remove the looping and DNA binding and a region of the dimerization domain, respectively, did not have any significant effect on motility compared to wild-type EBNA1. However, mutant p367, in which the transactivator domain is deleted, dramatically showed no response when coexpressed with nm23-H1. The p376 mutant which lacks the Gly-Ala repeats was incapable of suppressing motility, whereas a mutation that knocks out the small Gly-Arg repeat region 5′ to the transactivation domain (p378) lacks suppression of motility but had a substantial effect on the motility when coexpressed with Nm23-H1 (Fig. 7). Taken together, these data strongly suggest that the domain that lies between amino acids 65 and 89 that is responsible for transactivation is critical for EBNA1's role in reversing the ability of Nm23-H1 to suppress motility. It is, however, evident that the mechanism by which EBNA1 itself suppresses motility is not understood and will require further study.
FIG. 7.
Cell migration assays were performed to further address and identify the responsible domain of EBNA1 that contributes to cell migration. We obtained deletion mutants of EBNA1. MDA-MB-435 cells were transfected with Nm23-H1, the deletion mutant of EBNA1, or both and selected in DMEM containing 2 mg of neomycin/ml. A cell motility assay was performed in 0.5% fetal bovine serum (FBS) chemoattractant. The p367 mutant showed suppressing motility that is critical for EBNA1's role in reversing the ability of Nm23-H1 to suppress motility.
DISCUSSION
Our previous studies indicated that Nm23-H1 interacts with EBNA3C (49). EBNA3C is one of the six essential latent genes required for EBV-mediated growth transformation of primary B lymphocytes (38, 39). In this report we showed that EBNA1, which is known to be expressed in all EBV-positive tumors, interacts with Nm23-H1 in vitro and in EBV-transformed cells. Similarly, in previous studies the association of EBNA3C with Nm23-H1 in cells mediated the translocation of Nm23-H1 to the nucleus and resulted in increased migration of breast carcinoma cells as well as a B-cell line (47). EBNA3C is not constantly detected in EBV-positive tumors and is typically seen in lymphoproliferative disorder-associated lymphomas in immunocompromised patients due to AIDS or after organ transplantation (38, 39). On the other hand, EBNA1 has been detected in the vast number of EBV-associated tumors, suggesting an important role in EBV-mediated cancers. The function of EBNA1 has been extensively studied in B cells, where in vitro infection with EBV is efficient and results in B-cell transformation (38, 39, 62). Its role with regard to epithelial EBV infection is, however, less clear. Although EBV infection of epithelial cells obviously occurs in the context of virus-associated carcinomas, the requirements for this process have not been determined. In addition, EBV is associated with a wide variety of human malignances, including NPCs, gastric carcinomas, Hodgkin's lymphoma, and BL (38, 39). EBNA1 is expressed in all forms of EBV-infected cells and plays a role in the segregation and maintenance of the episomal EBV genome (16, 38, 39).
Nm23-H1 is a 152-amino-acid, 17-kDa cellular protein (19). Suppression of cell migration and metastasis was observed in tumor cell lines transfected with Nm23-H1 (18, 19, 21, 42). Although the exact role of Nm23-H1 in metastasis is not clearly understood, a vast array of experimental data accumulated thus far implicate Nm23-H1 as playing a critical role in regulating cell migration and the metastasis of human cancer. The association with EBNA1 and EBNA3C in mediating Nm23-H1 nuclear translocation is likely to have important consequences in terms of regulating the functional effects of Nm23-H1 in cells expressing EBV latent antigens.
In this report we demonstrate that EBNA1 can interact with Nm23-H1, the suppressor of cell migration and metastasis, as a cellular target. Immunoprecipitation experiments and Western blotting confirmed that EBNA1 associates with Nm23-H1 when cotransfected in BJAB cells. In addition, we show that these molecules interact with each other in vitro, supporting the possibility that EBNA1 and Nm23-H1 directly associate with each other in EBV-infected cells. We also demonstrated that EBNA1 coimmunoprecipitates with Nm23-H1 in EBV-transformed human primary B cells (i.e., LCLs). Therefore, the evidence accumulated thus far strongly suggests that EBNA1, known to be expressed in all EBV-associated cancers, can target Nm23-H1 and modulates its function in suppressing cell migration and metastasis.
As expected, we showed that Nm23-H1 predominantly localizes to the cytoplasm in B cells and embryonic fibroblast cells. However, we showed that, when EBNA1 is coexpressed with Nm23-H1, the signal for Nm23-H1 was predominantly nuclear. We further visualized the signals of EBNA1 and Nm23-H1 by immunofluorescence with a recently transformed LCL and showed that the signals were colocalized in the nucleus of these cells. The Nm23-H1 signals in this case were predominantly nuclear, suggesting that in EBV-transformed cells EBNA1 can also influence the localization of Nm23-H1 and that this localization may have a profound influence on the function of Nm23-H1. Further, we have also shown that when fused to a GAL4 DNA-binding domain that Nm23-H1 can activate transcription of a GAL4-responsive promoter (49). Nm23-H1 sequestered to the cytoplasm may not be an effective activator of transcription. However, the translocation of Nm23-H1 to the nucleus is likely to be important in affecting the ability of Nm23-H1 to transactivate cellular promoters. We have shown that when EBNA3C was introduced the level of activation increased dramatically, suggesting that EBNA3C synergized Nm23-H1 activation (47). These studies strongly suggest that Nm23-H1 and EBV nuclear antigens can be linked to regulation of transcription activities of cellular target molecules likely to be involved in regulating a range of functions associated with Nm23-H1, including cell migration (47). The discovery that EBNA1 now can also affect localization of Nm23-H1 from a predominantly cytoplasmic to nuclear localization suggests that EBNA1 also may be involved in regulating Nm23-H1 activities and may have a substantial role in its regulation in the absence of EBNA3C in EBV-associated human cancers. Interestingly, we have also tested the interactions with the other EBV latent nuclear antigens EBNA3A and EBNA3B, and our studies showed that the predominant molecules associated with Nm23-H1 are EBNA1 and EBNA3C (47).
Nm23-H1 has numerous identified biochemical functions, including NDPK activity, DNA transactivation, nuclease activities, and serine or histidine protein kinase activities (2, 10, 25, 36, 45, 49). In addition, it associates with a number of cellular molecules and viral targets that may be crucial for regulating cell migration and metastasis.
We monitored changes in cell migration by using a number of chemoattractants (such as fetal bovine serum, EGF, and fibronectin). EBNA1- or Nm23-H1-transfected BJAB cells showed inhibition of cell migration under these conditions. Interestingly, in transfected BJAB cells, the addition of EBNA1, along with Nm23-H1, rescued the suppressive effect of Nm23-H1 to ca. 100%. Interestingly, studies with EBNA1-specific mutants show that a region of EBNA1 from amino acids 61 to 89 that is known to have transactivation activities is critical for the role of EBNA1 in affecting cell motility in the context of Nm23-H1. This suggests a role of EBNA1 in transcription activation cooperating with Nm23-H1 to affect cellar gene expression in context of cell migration. The data presented here with an in vitro binding assay and a GST pull-down assay clearly demonstrated the association of Nm23-H1 with EBNA1. Immunoprecipitation and immunofluorescence experiments also corroborated these studies in EBV-transformed human primary lymphocytes. Whether this association may be linked to the upregulation of adhesion molecules, including αv-integrins and E-cadherin is not yet known. However, current studies in our laboratory will determine whether in fact the association between the EBV molecules and Nm23-H1 can affect the transcription of cellular targets associated with modulation of cell migration. These studies strongly suggest that the essential EBV nuclear antigens play a critical role in regulating cell migration through targeting Nm23-H1, a major effector of cell migration and metastasis in EBV-associated human cancers. Further studies will be needed to determine the independent functions of the EBNA proteins associated with Nm23-H1. The specificity may be in the regulation of specific cellular targets, or it may be that these two EBV latent antigens can cooperate with Nm23-H1 to synergistically affect its function in regulating the gene expression of cellular targets that regulate cell migration.
Acknowledgments
We thank Elliott Kieff for the EBNA reagents. We especially thank Patricia S. Steeg for the nm23-H1 constructs and John Yates for the EBNA1 mutants.
This study was supported by Public Health Service grants from the National Institutes of Health (NCI CA72150-07 and NCI CA91792-01 and NIDCR grant DE14136-01 to E.S.R.). E.S.R. is a scholar of the Leukemia and Lymphoma Society of America.
REFERENCES
- 1.Arcinas, M., and L. M. Boxer. 1994. Differential protein binding to the c-myc promoter during differentiation of hematopoietic cell lines. Oncogene 9:2699-2706. [PubMed] [Google Scholar]
- 2.Aryee, D. N., I. Simonitsch, I. Mosberger, K. Kos, G. Mann, E. Schlogl, U. Potschger, H. Gadner, T. Radaszkiewicz, and H. Kovar. 1996. Variability of nm23-H1/NDPK-A expression in human lymphomas and its relation to tumour aggressiveness. Br. J. Cancer 74:1693-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Backer, J. M., C. E. Mendola, I. Kovesdi, J. L. Fairhurst, B. O'Hara, R. L. Eddy, Jr., T. B. Shows, S. Mathew, V. V. Murty, and R. S. Chaganti. 1993. Chromosomal localization and nucleoside diphosphate kinase activity of human metastasis-suppressor genes NM23-1 and NM23-2. Oncogene 8:497-502. [PubMed] [Google Scholar]
- 4.Berberich, S. J., and E. H. Postel. 1995. PuF/NM23-H2/NDPK-B transactivates a human c-myc promoter-CAT gene via a functional nuclease hypersensitive element. Oncogene 10:2343-2347. [PubMed] [Google Scholar]
- 5.Biggs, J., E. Hersperger, P. S. Steeg, L. A. Liotta, and A. Shearn. 1990. A Drosophila gene that is homologous to a mammalian gene associated with tumor metastasis codes for a nucleoside diphosphate kinase. Cell 63:933-940. [DOI] [PubMed] [Google Scholar]
- 6.Bominaar, A. A., A. D. Tepper, and M. Veron. 1994. Autophosphorylation of nucleoside diphosphate kinase on non-histidine residues. FEBS Lett. 353:5-8. [DOI] [PubMed] [Google Scholar]
- 7.Cotter, M. A., II, and E. S. Robertson. 2000. Modulation of histone acetyltransferase activity through interaction of Epstein-Barr nuclear antigen 3C with prothymosin alpha. Mol. Cell. Biol. 20:5722-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davenport, M. G., and J. S. Pagano. 1999. Expression of EBNA-1 mRNA is regulated by cell cycle during Epstein-Barr virus type I latency. J. Virol. 73:3154-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Rosa, A., R. L. Williams, and P. S. Steeg. 1995. Nm23/nucleoside diphosphate kinase: toward a structural and biochemical understanding of its biological functions. Bioessays 17:53-62. [DOI] [PubMed] [Google Scholar]
- 10.Freije, J. M., P. Blay, N. J. MacDonald, R. E. Manrow, and P. S. Steeg. 1997. Site-directed mutation of Nm23-H1: mutations lacking motility suppressive capacity upon transfection are deficient in histidine-dependent protein phosphotransferase pathways in vitro. J. Biol. Chem. 272:5525-5532. [DOI] [PubMed] [Google Scholar]
- 11.Gilles, A. M., E. Presecan, A. Vonica, and I. Lascu. 1991. Nucleoside diphosphate kinase from human erythrocytes: structural characterization of the two polypeptide chains responsible for heterogeneity of the hexameric enzyme. J. Biol. Chem. 266:8784-8789. [PubMed] [Google Scholar]
- 12.Hager, M., K. Biehler, J. Illerhaus, S. Ruf, and R. Bock. 1999. Targeted inactivation of the smallest plastid genome-encoded open reading frame reveals a novel and essential subunit of the cytochrome b(6)f complex. EMBO J. 18:5834-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hailat, N., D. R. Keim, R. F. Melhem, X. X. Zhu, C. Eckerskorn, G. M. Brodeur, C. P. Reynolds, R. C. Seeger, F. Lottspeich, J. R. Strahler, et al. 1991. High levels of p19/nm23 protein in neuroblastoma are associated with advanced stage disease and with N-myc gene amplification. J. Clin. Investig. 88:341-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemmerich, S., and I. Pecht. 1992. Oligomeric structure and autophosphorylation of nucleoside diphosphate kinase from rat mucosal mast cells. Biochemistry 31:4580-4587. [DOI] [PubMed] [Google Scholar]
- 15.Imai, S., S. Koizumi, M. Sugiura, M. Tokunaga, Y. Uemura, N. Yamamoto, S. Tanaka, E. Sato, and T. Osato. 1994. Gastric carcinoma: monoclonal epithelial malignant cells expressing Epstein-Barr virus latent infection protein. Proc. Natl. Acad. Sci. USA 91:9131-9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imai, S., J. Nishikawa, and K. Takada. 1998. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J. Virol. 72:4371-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kieff, E. 1996. Epstein-Barr virus and its replication, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 18.Leone, A., U. Flatow, C. R. King, M. A. Sandeen, I. M. Margulies, L. A. Liotta, and P. S. Steeg. 1991. Reduced tumor incidence, metastatic potential, and cytokine responsiveness of nm23-transfected melanoma cells. Cell 65:25-35. [DOI] [PubMed] [Google Scholar]
- 19.Leone, A., U. Flatow, K. VanHoutte, and P. S. Steeg. 1993. Transfection of human nm23-H1 into the human MDA-MB-435 breast carcinoma cell line: effects on tumor metastatic potential, colonization and enzymatic activity. Oncogene 8:2325-2333. [PubMed] [Google Scholar]
- 20.Leone, A., R. C. Seeger, C. M. Hong, Y. Y. Hu, M. J. Arboleda, G. M. Brodeur, D. Stram, D. J. Slamon, and P. S. Steeg. 1993. Evidence for nm23 RNA overexpression, DNA amplification, and mutation in aggressive childhood neuroblastomas. Oncogene 8:855-865. [PubMed] [Google Scholar]
- 21.Lin, L. I., P. H. Lee, C. M. Wu, and J. K. Lin. 1998. Significance of nm23 mRNA expression in human hepatocellular carcinoma. Anticancer Res. 18:541-546. [PubMed] [Google Scholar]
- 22.Longnecker, R., C. L. Miller, B. Tomkinson, X. Q. Miao, and E. Kieff. 1993. Deletion of DNA encoding the first five transmembrane domains of Epstein-Barr virus latent membrane proteins 2A and 2B. J. Virol. 67:5068-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacDonald, N. J., A. de la Rosa, M. A. Benedict, J. M. Freije, H. Krutsch, and P. S. Steeg. 1993. A serine phosphorylation of Nm23, and not its nucleoside diphosphate kinase activity, correlates with suppression of tumor metastatic potential. J. Biol. Chem. 268:25780-25789. [PubMed] [Google Scholar]
- 24.MacDonald, N. J., A. de la Rosa, and P. S. Steeg. 1995. The potential roles of nm23 in cancer metastasis and cellular differentiation. Eur. J. Cancer 31A:1096-1100. [DOI] [PubMed] [Google Scholar]
- 25.MacDonald, N. J., J. M. Freije, M. L. Stracke, R. E. Manrow, and P. S. Steeg. 1996. Site-directed mutagenesis of nm23-H1: mutation of proline 96 or serine 120 abrogates its motility inhibitory activity upon transfection into human breast carcinoma cells. J. Biol. Chem. 271:25107-25116. [DOI] [PubMed] [Google Scholar]
- 26.Marechal, V., A. Dehee, R. Chikhi-Brachet, T. Piolot, M. Coppey-Moisan, and J. C. Nicolas. 1999. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 73:4385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin, K. K., and G. J. Pilkington. 1998. Nm23: an invasion suppressor gene in CNS tumours? Anticancer Res. 18:919-926. [PubMed] [Google Scholar]
- 28.Mehus, J. G., P. Deloukas, and D. O. Lambeth. 1999. NME6: a new member of the nm23/nucleoside diphosphate kinase gene family located on human chromosome 3p21.3. Hum. Genet. 104:454-459. [DOI] [PubMed] [Google Scholar]
- 29.Milon, L., M. F. Rousseau-Merck, A. Munier, M. Erent, I. Lascu, J. Capeau, and M. L. Lacombe. 1997. nm23-H4, a new member of the family of human nm23/nucleoside diphosphate kinase genes localized on chromosome 16p13. Hum. Genet. 99:550-557. [DOI] [PubMed] [Google Scholar]
- 30.Munier, A., C. Feral, L. Milon, V. P. Pinon, G. Gyapay, J. Capeau, G. Guellaen, and M. L. Lacombe. 1998. A new human nm23 homologue (nm23-H5) specifically expressed in testis germinal cells. FEBS Lett. 434:289-294. [DOI] [PubMed] [Google Scholar]
- 31.Munoz-Dorado, J., S. Inouye, and M. Inouye. 1993. Eukaryotic-like protein serine/threonine kinases in Myxococcus xanthus, a developmental bacterium exhibiting social behavior. J. Cell Biochem. 51:29-33. [DOI] [PubMed] [Google Scholar]
- 32.Munoz-Dorado, J., S. Inouye, and M. Inouye. 1990. Nucleoside diphosphate kinase from Myxococcus xanthus. II. Biochemical characterization. J. Biol. Chem. 265:2707-2712. [PubMed] [Google Scholar]
- 33.Murray, P. G., G. Niedobitek, E. Kremmer, F. Grasser, G. M. Reynolds, A. Cruchley, D. M. Williams, N. Muller-Lantzsch, and L. S. Young. 1996. In situ detection of the Epstein-Barr virus-encoded nuclear antigen 1 in oral hairy leukoplakia and virus-associated carcinomas. J. Pathol. 178:44-47. [DOI] [PubMed] [Google Scholar]
- 34.Padma, P., A. Hozumi, K. Ogawa, and K. Inaba. 2001. Molecular cloning and characterization of a thioredoxin/nucleoside diphosphate kinase related dynein intermediate chain from the ascidian, Ciona intestinalis. Gene 275:177-183. [DOI] [PubMed] [Google Scholar]
- 35.Postel, E. H., S. J. Berberich, S. J. Flint, and C. A. Ferrone. 1993. Human c-myc transcription factor PuF identified as nm23-H2 nucleoside diphosphate kinase, a candidate suppressor of tumor metastasis. Science 261:478-480. [DOI] [PubMed] [Google Scholar]
- 36.Postel, E. H., S. J. Berberich, J. W. Rooney, and D. M. Kaetzel. 2000. Human NM23/nucleoside diphosphate kinase regulates gene expression through DNA binding to nuclease-hypersensitive transcriptional elements. J. Bioenerg. Biomembr. 32:277-284. [DOI] [PubMed] [Google Scholar]
- 37.Postel, E. H., and C. A. Ferrone. 1994. Nucleoside diphosphate kinase enzyme activity of NM23-H2/PuF is not required for its DNA binding and in vitro transcriptional functions. J. Biol. Chem. 269:8627-8630. [PubMed] [Google Scholar]
- 38.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 39.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2446. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 40.Rosengard, A. M., H. C. Krutzsch, A. Shearn, J. R. Biggs, E. Barker, I. M. Margulies, C. R. King, L. A. Liotta, and P. S. Steeg. 1989. Reduced Nm23/Awd protein in tumour metastasis and aberrant Drosophila development. Nature 342:177-180. [DOI] [PubMed] [Google Scholar]
- 41.Russell, R. L., K. R. Geisinger, R. R. Mehta, W. L. White, B. Shelton, and T. E. Kute. 1997. nm23—relationship to the metastatic potential of breast carcinoma cell lines, primary human xenografts, and lymph node negative breast carcinoma patients. Cancer 79:1158-1165. [DOI] [PubMed] [Google Scholar]
- 42.Russell, R. L., A. N. Pedersen, J. Kantor, K. Geisinger, R. Long, N. Zbieranski, A. Townsend, B. Shelton, N. Brunner, and T. E. Kute. 1998. Relationship of nm23 to proteolytic factors, proliferation and motility in breast cancer tissues and cell lines. Br. J. Cancer 78:710-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheu, L. F., A. Chen, C. L. Meng, K. C. Ho, W. H. Lee, F. J. Leu, and C. F. Chao. 1996. Enhanced malignant progression of nasopharyngeal carcinoma cells mediated by the expression of Epstein-Barr nuclear antigen 1 in vivo. J. Pathol. 180:243-248. [DOI] [PubMed] [Google Scholar]
- 44.Stahl, J. A., A. Leone, A. M. Rosengard, L. Porter, C. R. King, and P. S. Steeg. 1991. Identification of a second human nm23 gene, nm23-H2. Cancer Res. 51:445-449. [PubMed] [Google Scholar]
- 45.Steeg, P. S. 2003. Metastasis suppressors alter the signal transduction of cancer cells. Nat. Rev. 3:55-63. [DOI] [PubMed] [Google Scholar]
- 46.Steeg, P. S., A. de la Rosa, U. Flatow, N. J. MacDonald, M. Benedict, and A. Leone. 1993. Nm23 and breast cancer metastasis. Breast Cancer Res. Treat. 25:175-187. [DOI] [PubMed] [Google Scholar]
- 47.Subramanian, C., M. A. Cotter II, and E. S. Robertson. 2001. Epstein-Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23-H1: a molecular link to cancer metastasis. Nat. Med. 7:350-355. [DOI] [PubMed] [Google Scholar]
- 48.Subramanian, C., J. S. Knight, and E. S. Robertson. 2002. The Epstein-Barr nuclear antigen EBNA3C regulates transcription, cell transformation, and cell migration. Front. Biosci. 7:d704-d716. [DOI] [PubMed] [Google Scholar]
- 49.Subramanian, C., and E. S. Robertson. 2002. The metastatic suppressor Nm23-H1 interacts with EBNA3C at sequences located between the glutamine- and proline-rich domains and can cooperate in activation of transcription. J. Virol. 76:8702-8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomkinson, B., E. Robertson, and E. Kieff. 1993. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J. Virol. 67:2014-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsuiki, H., M. Nitta, A. Furuya, N. Hanai, T. Fujiwara, M. Inagaki, M. Kochi, Y. Ushio, H. Saya, and H. Nakamura. 1999. A novel human nucleoside diphosphate (NDP) kinase, Nm23-H6, localizes in mitochondria and affects cytokinesis. J. Cell Biochem. 76:254-269. [DOI] [PubMed] [Google Scholar]
- 52.van Golen, K. L., S. Risin, A. Staroselsky, D. Berger, M. A. Tainsky, S. Pathak, and J. E. Price. 1996. Predominance of the metastatic phenotype in hybrids formed by fusion of mouse and human melanoma clones. Clin. Exp. Metastasis 14:95-106. [DOI] [PubMed] [Google Scholar]
- 53.Venturelli, D., R. Martinez, P. Melotti, I. Casella, C. Peschle, C. Cucco, G. Spampinato, Z. Darzynkiewicz, and B. Calabretta. 1995. Overexpression of DR-nm23, a protein encoded by a member of the nm23 gene family, inhibits granulocyte differentiation and induces apoptosis in 32Dc13 myeloid cells. Proc. Natl. Acad. Sci. USA 92:7435-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wagner, P. D., and N. D. Vu. 1995. Phosphorylation of ATP-citrate lyase by nucleoside diphosphate kinase. J. Biol. Chem. 270:21758-21764. [DOI] [PubMed] [Google Scholar]
- 55.Wallet, V., R. Mutzel, H. Troll, O. Barzu, B. Wurster, M. Veron, and M. L. Lacombe. 1990. Dictyostelium nucleoside diphosphate kinase highly homologous to Nm23 and Awd proteins involved in mammalian tumor metastasis and Drosophila development. J. Natl. Cancer Inst. 82:1199-1202. [DOI] [PubMed] [Google Scholar]
- 56.Wang, F., C. Gregory, C. Sample, M. Rowe, D. Liebowitz, R. Murray, A. Rickinson, and E. Kieff. 1990. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J. Virol. 64:2309-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Webb, P. A., O. Perisic, C. E. Mendola, J. M. Backer, and R. L. Williams. 1995. The crystal structure of a human nucleoside diphosphate kinase, NM23-H2. J. Mol. Biol. 251:574-587. [DOI] [PubMed] [Google Scholar]
- 58.Wilson, J. B., J. L. Bell, and A. J. Levine. 1996. Expression of Epstein-Barr virus nuclear antigen-1 induces B-cell neoplasia in transgenic mice. EMBO J. 15:3117-3126. [PMC free article] [PubMed] [Google Scholar]
- 59.Yao, Q. Y., R. J. Tierney, D. Croom-Carter, D. Dukers, G. M. Cooper, C. J. Ellis, M. Rowe, and A. B. Rickinson. 1996. Frequency of multiple Epstein-Barr virus infections in T-cell-immunocompromised individuals. J. Virol. 70:4884-4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yates, J. L. 1988. Dissection of DNA replication and enhancer activation functions of Epstein-Barr virus nuclear antigen 1. Cancer Cells 6:197-205. [Google Scholar]
- 61.Yates, J. L., S. M. Camiolo, S. Ali, and A. Ying. 1996. Comparison of the EBNA1 proteins of Epstein-Barr virus and herpesvirus papio in sequence and function. Virology 222:1-13. [DOI] [PubMed] [Google Scholar]
- 62.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]