Abstract
Background: Previously we identified a DNA damage response–deficient (DDRD) molecular subtype within breast cancer. A 44-gene assay identifying this subtype was validated as predicting benefit from DNA-damaging chemotherapy. This subtype was defined by interferon signaling. In this study, we address the mechanism of this immune response and its possible clinical significance.
Methods: We used immunohistochemistry (IHC) to characterize immune infiltration in 184 breast cancer samples, of which 65 were within the DDRD subtype. Isogenic cell lines, which represent DDRD-positive and -negative, were used to study the effects of chemokine release on peripheral blood mononuclear cell (PBMC) migration and the mechanism of immune signaling activation. Finally, we studied the association between the DDRD subtype and expression of the immune-checkpoint protein PD-L1 as detected by IHC. All statistical tests were two-sided.
Results: We found that DDRD breast tumors were associated with CD4+ and CD8+ lymphocytic infiltration (Fisher’s exact test P < .001) and that DDRD cells expressed the chemokines CXCL10 and CCL5 3.5- to 11.9-fold more than DNA damage response–proficient cells (P < .01). Conditioned medium from DDRD cells statistically significantly attracted PBMCs when compared with medium from DNA damage response–proficient cells (P < .05), and this was dependent on CXCL10 and CCL5. DDRD cells demonstrated increased cytosolic DNA and constitutive activation of the viral response cGAS/STING/TBK1/IRF3 pathway. Importantly, this pathway was activated in a cell cycle–specific manner. Finally, we demonstrated that S-phase DNA damage activated expression of PD-L1 in a STING-dependent manner.
Conclusions: We propose a novel mechanism of immune infiltration in DDRD tumors, independent of neoantigen production. Activation of this pathway and associated PD-L1 expression may explain the paradoxical lack of T-cell-mediated cytotoxicity observed in DDRD tumors. We provide a rationale for exploration of DDRD in the stratification of patients for immune checkpoint–based therapies.
The presence of an immune response is recognized to be a prognostic factor in breast cancer (1,2). The underlying mechanisms driving this response are unclear. It has been proposed that DNA released from apoptotic cells or tumor neoantigen production may be responsible for this immune response; however, these mechanisms do not explain the absence of response in other tumors (3). Previously (4) we used unsupervised hierarchical clustering of gene expression data to identify a DNA damage response–deficient (DDRD) molecular subtype in breast cancer and demonstrated that this represented loss of the S-phase-specific DNA damage response mechanism, the Fanconi Anemia (FA)/BRCA pathway. Based on this, we developed a 44-gene expression assay that could prospectively identify this group of tumors and demonstrated that it could predict benefit from DNA-damaging chemotherapy, presumably because of inherent defects in DNA repair capacity (4). Importantly, upregulation of interferon-related genes was observed in the DDRD molecular subtype, and DDRD assay–positive tumors were associated with lymphocytic infiltration. However, the key pathways driving this biology were unknown. In this current study, we explore the activation of immune genes identified in the DDRD molecular subtype.
Methods
Further details of methods can be found in Supplementary Materials (available online).
Cell Lines
MDA-MB-436-EV and MDA-MB-436 -BRCA1 were a kind gift from Ms. Paula Haddock (Queens University Belfast, UK) and were generated by transfecting the BRCA1-mutant MDA-MB-436 cells with either empty Rc/CMV or Rc/CMV-BRCA1, followed by selection in 300 μg/mL G418 (Roche, Basel, Switzerland). HCC1937-EV and HCC1937-BRCA1 have been described previously (5). These isogenic cell lines were used to model the immune effects of BRCA1 deficiency. Hela cells (ATCC, Manassas, VA) were used to investigate the effects of exogenous DNA damage.
Immunohistochemistry
Immunohistochemistry (IHC) was performed in the Northern Ireland Molecular Pathology Laboratory using the Ventana Discovery-XT Automated Stainer. A tissue microarray of a previously described cohort (4) of 184 N0-N1 estrogen receptor (ER)–positive and ER-negative formalin-fixed, paraffin-embedded (FFPE) breast tumor samples (ethics number NIB12-0043) was scored in triplicate. CD4 (4B12, M7310, Dako, Ely, UK) and CD8 antibodies (C8/144B, M7103, Dako) were used at 1:50, PD-L1 antibody (SP142, Roche) at 1:40 with an amplification step using OptiView Amplification Kit (Roche). A semiquantitative scoring system was employed for CD4+ and CD8+ characterization. A score of 3 indicates strong CD4+ or CD8+ expression, 2 moderate expression, 1 low or weak expression, 0 absence. Scores were determined by two independent observers. For PD-L1, previously published cutoffs of 1% or greater and 5% or greater were used (6).
Migration Assay
Peripheral blood mononuclear cells (PBMCs) were obtained from buffy coats of 12 healthy donors, with written informed consent obtained and ethical approval granted from the Northern Ireland Blood Transfusion Service. Using Corning Transwell polycarbonate membrane 5 μm inserts (Sigma Aldrich, St Louis, MO), PBMCs were resuspended in Optimem 0.5% BSA, and 5x105 PBMCs placed in the top chamber. In the bottom chamber, conditioned media (from indicated cell lines ± transfected knockdowns) was placed. Media was changed to optimem on day 1, and collected on day 3. For CXCR3 inhibition, 100 nM SCH546738 (7), synthesized in-house, was added to the PBMCs. After four hours, the migration of PBMCs to the bottom chamber was measured using a Cell Titre Glo assay (Promega, Madison, WI) with a standard curve to calculate cell number.
Immunofluorescence
MDA-MB-436-EV, HCC1937-EV, and their BRCA1-complemented isogenic pairs were seeded on coverslips. HeLa cells were treated with IC50 doses of cisplatin, hydroxyurea and paclitaxel, and DMSO control for 48 hours. Cells were washed with phosphate-buffered saline prior to the addition of fresh medium containing PicoGreen DNA stain (4 µg/mL). Following incubation at 37 °C for two hours, fixation was performed with 4% paraformaldehyde. Coverslips were mounted using Prolong Gold with DAPI (Thermo Fisher, Renfrew, PA). Cytosolic dsDNA staining was assessed by selection of random fields of view using a Nikon Eclipse Ti Fluorescent Microscope.
Statistical Analysis
Each experiment was carried out at least in triplicate. To determine statistical significance, the unpaired, two-tailed Student’s t test was calculated using the t test calculator available on GraphPad Prism 5.0 software. Fisher’s exact test was calculated using the online calculator available at http://in-silico.net/tools/statistics/fisher_exact_test. Statistical significance is defined as a P value of less than .05. All statistical tests were two-sided.
Results
Analysis of CD4+ and CD8+ T-Lymphocytes in DDRD Assay–Positive and –Negative Tumors
As we had observed upregulation of interferon-related genes including T-cell-specific ligands in the epithelial component of tumors classified within the DDRD molecular subtype (Supplementary Table 1, available online) (4), we asked if these were associated with a T-cell immune response. The presence of intratumoral and stromal CD4+ and CD8+ T-lymphocytes were assessed by IHC using a semiquantitative score in a previously described cohort of 184 N0-N1 ER-positive and ER-negative breast tumor samples, scored as DDRD molecular subtype–positive or –negative using the DDRD assay according to the cut-off value defined in Mulligan et al. (4). Patient characteristics and histopathological factors are given in Supplementary Table 2 (available online).
A statistically significant association of intratumoral and stromal CD4+ and CD8+ T-lymphocytes with DDRD assay–positive tumors was identified (P < .001) (Table 1). In the DDRD-positive cohort (n = 65), 53.9% of cores scored as having moderate/high intratumoral CD8+ lymphocytic infiltrate, compared with 16.2% of DDRD assay–negative tumors (n = 117). Similarly, a strong association of CD4+ intratumoral lymphocytic infiltration was identified with DDRD assay–positive tumors compared with DDRD assay–negative (24.6% vs 3.4% scoring moderate/high). In assessment of the stromal component of the tumor, 89.3% of DDRD assay–positive tumors were associated with moderate/high CD8+ stromal expression, compared with 56.4% of DDRD assay–negative, and 80.0% of DDRD assay–positive tumors were associated with stromal CD4+ expression vs 32.8% DDRD-negative (Table 1, Figure 1A). In DDRD assay–positive samples, a statistically significant association between CD4+ and CD8+ lymphocytes in the intratumoral and stromal compartments was identified (P < .001).
Table 1.
CD8+ and CD4+ intratumoral and stromal lymphocytic infiltrate assessed by IHC in DDRD-positive and DDRD-negative breast tumors
| Score | DDRD positive, No. (%) | DDRD negative, No. (%) | P* |
|---|---|---|---|
| Intratumoral | |||
| CD8+ | 65 (100) | 117 (100) | <.001 |
| 3 | 6 (9.2) | 0 (0.0) | |
| 2 | 29 (44.6) | 19 (16.2) | |
| 1 | 25 (38.5) | 68 (58.1) | |
| 0 | 5 (7.7) | 30 (25.6) | |
| CD4+ | 65 (100) | 119 (100) | <.001 |
| 3 | 3 (4.6) | 1 (0.8) | |
| 2 | 13 (20.0) | 3 (2.5) | |
| 1 | 43 (66.2) | 75 (63.0) | |
| 0 | 6 (9.2) | 40 (33.6) | |
| Stromal | |||
| CD8+ | 65 (100) | 117 (100) | <.001 |
| 3 | 20 (30.8) | 8 (6.8) | |
| 2 | 38 (58.5) | 58 (49.6) | |
| 1 | 7 (10.8) | 44 (37.6) | |
| 0 | 0 (0.0) | 7 (6.0) | |
| CD4+ | 65 (100) | 119 (100) | <.001 |
| 3 | 21 (32.3) | 7 (5.9) | |
| 2 | 31 (47.7) | 32 (26.9) | |
| 1 | 13 (20.0) | 66 (55.5) | |
| 0 | 0 (0.0) | 14 (11.8) |
P values were calculated using a two-sided Fisher’s exact test. DDRD = DNA damage response–deficient; IHC = immunohistochemistry.
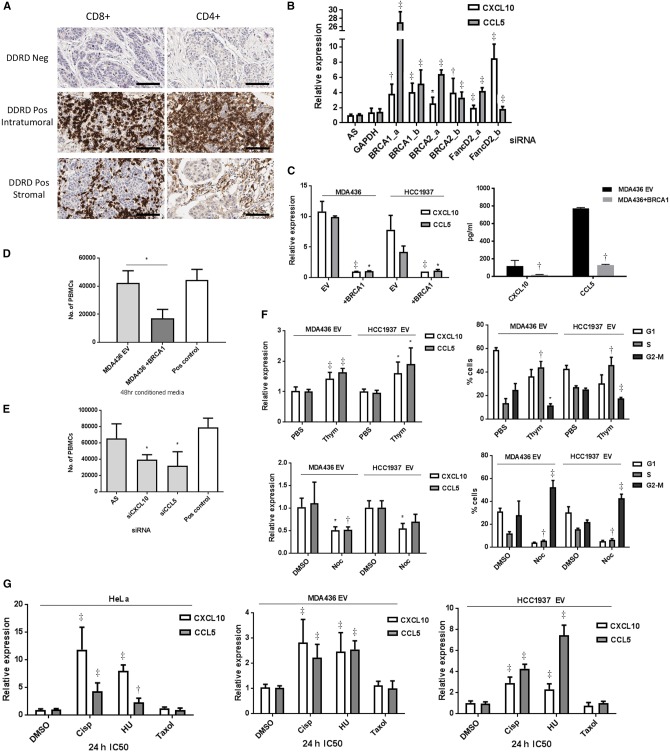
Figure 1.
Immune gene expression in FA/BRCA DNA repair pathway loss. A) Immunohistochemistry (IHC) images (x40) showing absence of CD8+ and CD4+ lymphocytes in DDRD assay–negative tumors, and both intratumoral and stromal CD8+ and CD4+ lymphocytes in DDRD assay–positive breast tumors. Scale bar represents 100 µm. B) Quantitative polymerase chain reaction (qPCR) measurement of CCL5 and CXCL10 chemokine mRNA expression following knockdown of BRCA1, BRCA2, and FANCD2 in HeLa cells using two independent siRNAs (a and b) compared with control siRNA (AS). GAPDH siRNA has also been used as a negative control. C) qPCR measurement (left panel) of CCL5 and CXCL10 chemokine mRNA expression in MDA-MB-436-EV and HCC1937-EV BRCA1-mutant cell lines compared with BRCA1-corrected MDA-MB-436 and HCC1937-BRCA1 cell lines. CXCL10 and CCL5 quantification by enzyme-linked immunosorbent assay (ELISA) of media collected from MDA-MB-436 BRCA1 and empty vector counterparts (EV) are shown in the right panel. D) Migration of activated peripheral blood mononuclear cells (PBMCs) toward conditioned media from MDA-MB-436-EV compared with MDA-MB-436-BRCA1 using a Boyden Invasion Chamber Assay. E) Migration of activated PBMCs toward conditioned media from MDA-MB-436-EV cells treated with CXCL10 and CCL5 siRNA compared with nontargeting siRNA control (AS) using a Boyden invasion chamber assay. See also Supplementary Figure 1, C and D (available online). F) qPCR measurement of CXCL10 and CCL5 mRNA in MDA-MB-436-EV and HCC1937-EV cells arrested in S-phase using double-thymidine block (2.5 mM) (top left panel) and M-phase using nocodazole (100 ng/mL) (bottom left panel). Cell cycle analysis following double-thymidine treatment as compared with phosphate-buffered saline (PBS) control is shown in the top right panel, and nocodazole-treated cells as compared with DMSO control in the bottom right panel. G) qPCR measurement of CXCL10 and CCL5 mRNA expression in HeLa (left panel), MDA-MB-436-EV (center panel), and HCC1937-EV (right panel) cells 24 hours following treatment with cisplatin, HU, or paclitaxel at IC50 concentration for each cell line, and the data was normalized to PUM1 expression. Data are represented as mean ± SD, and P values were calculated using the unpaired, two-tailed Student’s t test. *P < .05, †P < .01, ‡P < .001. DDRD = DNA damage response–deficient; PBMC = peripheral blood mononuclear cell; PBS = phosphate-buffered saline.
Assessment of Chemokine Production in the Context of DNA Damage Response Deficiency
CXCL10 is the most discriminative gene by weight in the DDRD assay (4) and has previously been reported as a prognostic factor in breast cancer (4,8). CCL5 was identified as the top differentially expressed chemokine in DDRD assay–positive ER-negative tumors compared with DDRD-negative ER-negative tumors (Supplementary Table 3, available online). As the CXCL10/CXCR3 axis has been reported as key for the chemotaxis of CD4+ and CD8+ T-lymphocytes (9), and CXCL10 and CCL5 overexpression is associated with the presence of CD8+ lymphocytes in melanoma, gastric, and colorectal cancers (10–12), we investigated the link between the DDRD molecular subtype and chemokine expression. We used siRNA to knock down the expression of the FA-BRCA pathway genes BRCA1, BRCA2, and FANCD2 in HeLa cells and observed statistically significant upregulation of CXCL10 and CCL5 (P < .05) (Figure 1B;Supplementary Figure 1A, available online). Next we assessed CXCL10 and CCL5 expression in BRCA1-mutant DNA damage response–deficient cell lines HCC1937 and MDA-MB-436, stably transfected with either an empty expression vector (EV) or the vector containing wild-type BRCA1 cDNA (BRCA1) (Supplementary Figure 1B, available online). Using these isogenic models, we observed statistically significant repression of CXCL10 and CCL5 in the BRCA1-complemented lines compared with their DDRD counterparts at the mRNA level and protein level from conditioned media (P < .05) (Figure 1C). Importantly, because the phenotype was observed in cell lines, this indicated that the observed production of chemokines in tumors within the DDRD molecular subtype could arise from the epithelial component.
To test if the upregulation of CXCL10 and CCL5 in cancer cells could result in lymphocytic infiltration, migration of peripheral blood mononuclear cells into conditioned media from MDA-MB-436-EV and BRCA1-corrected cells was assessed. This demonstrated a statistically significant increase in migration into media conditioned by MDA-MB-436-EV cells compared with their BRCA1-corrected counterparts (P = .02) (Figure 1D). Additionally, siRNA-mediated knockdown of CXCL10 and CCL5 reduced PBMC migration, indicating their importance for lymphocytic infiltration (P < .05) (Figure 1E;Supplementary Figure 1C, available online). Using the CXCR3 inhibitor SCH546738 (targeting the CXCL10 receptor), we also observed reduced PBMC migration (P < .001) (Supplementary Figure 1D, available online).
Together these data suggest that DNA damage response deficiency in breast cancer cells can result in the production of chemokines that stimulate lymphocytic migration.
DNA Damage–Associated Chemokine Expression in Relation to Phase of the Cell Cycle
As the DDRD molecular subtype in breast cancer was previously reported to be associated with loss of the FA/BRCA pathway (4), an S-phase-specific DNA repair mechanism, and endogenous DNA damage would therefore be expected to be maximal in the S-phase, we asked if chemokine release would be related to the phase of the cell cycle. MDA-MB-436-EV and HCC1937-EV cells were synchronized at S-phase or M-phase of the cell cycle. Quantitative polymerase chain reaction (qPCR) analysis of S-phase cells showed an increase in CXCL10 and CCL5 expression, whereas M-phase cells predominantly showed a reduction in chemokine expression (Figure 1F). As the type of sporadic DNA damage that would be expected following the loss of the FA/BRCA pathway is predominantly S-phase in nature, we further investigated the role of cell cycle–specific DNA damage. We assessed the effect of cisplatin or hydroxyurea on CXCL10 and CCL5 expression in a DNA repair–competent cell (Hela) using predetermined IC50 doses (Supplementary Figure 1E, available online). The microtubule stabilizer paclitaxel was used as an M-phase control. mRNA expression of CXCL10 and CCL5 was statistically significantly upregulated following treatment with cisplatin and HU, but not with paclitaxel (P < .01) (Figure 1G). Together these data support an S-phase-specific signal for activation of the immune response to DNA damage. A similar induction of cytokines in response to S-phase-specific DNA damage was noted in DDRD-positive HCC1937-EV and MDA-MB-436-EV cells vs their corrected isogenic pair (Figure 1G;Supplementary Figure 1G, available online).
Chemokine Expression and Relationship to DNA Damage Response Kinases
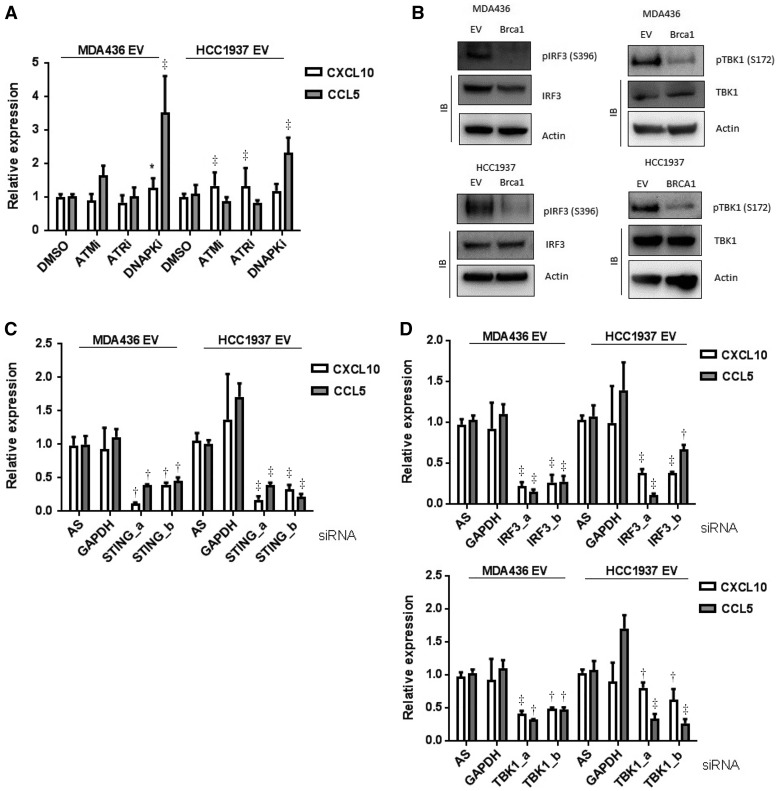
Next we wanted to investigate whether ATM, ATR, or DNAPK, the three main kinases activated in response to DNA damage, were necessary for the chemokine upregulation in DDRD cells. Activation of ATM has previously been reported to result in the upregulation of immune genes, suggesting that this kinase may be required for chemokine production in DNA damage response deficiency (13). We therefore treated DDRD cells (MDA-MB-436-EV and HCC1937-EV) with small molecule inhibitors of ATM, ATR, and DNA-PK and observed no reduction in CXCL10 and CCL5 expression (Figure 2A;Supplementary Figure 2A, available online). Together these data indicate that these classic DNA damage response kinases are not required for the chemokine response observed in DDRD cells.
Figure 2.
The role of STING, TBK1, and IRF3 in DNA damage–dependent chemokine expression. A) quantitative polymerase chain (qPCR) measurement of CXCL10 and CCL5 mRNA extracted from MDA-MB-436-EV and HCC1937-EV cells treated with small molecule inhibitors to ATM (1 µm Ku60019), ATR (5 µm ETP46464), and DNA-PK (5 µm Nu7441) for 24 hours. B) Immunoblots of immunoprecipitate from whole cell lysate from MDA-MB-436-EV, MDA-MB-436- BRCA1, HCC1937-EV, and HCC1937-BRCA1 cells for phosphorylation of IRF3 (ser396) and TBK1 (ser172) are shown. IRF3, TBK1, and Actin from whole cell lysates are also shown. C) qPCR measurement of CXCL10 and CCL5 mRNA 72 hours following knockdown of STING using two independent siRNAs (a and b) in MDA-MB-436-EV and HCC1937-EV cells normalized to a nontargeting siRNA control (AS). D) qPCR measurement of CXCL10 and CCL5 mRNA 72 hours following knockdown of TBK1 or IRF3 using two independent siRNAs (a and b) in MDA-MB-436-EV (top panel) and HCC1937-EV cells (bottom panel) normalized to a nontargeting siRNA control (AS), and GAPDH siRNA was used as a negative control. All data are representative of mean ± SD, and P values were calculated using the unpaired, two-tailed Student’s t test. *P < .05, †P < .01, ‡P < .001.
Activation of the STING/TBK1/IRF3 Pathway in DDRD Cancer Cells
To identify potential transcription factors that could activate the immune response observed in DDRD cells, we performed in silico analysis of differentially expressed genes from DDRD assay–positive ER-negative tumors compared with DDRD assay–negative using Toppfun analysis software (https://toppgene.cchmc.org/enrichment.jspp). Interferon regulatory factors (IRFs) were identified as the predominant transcription factors binding the promoter regions of genes differentially expressed between the DDRD assay–positive and –negative tumors (Supplementary Figure 2B, available online). Both CXCL10 and CCL5 had an IRF3 binding motif within the promoter region. IRF3 is activated in response to DNA-damaging agents (14), is a major effector of the STING/TBK1 signaling pathway, and has been reported to regulate expression of CXCL10 (15). STING-deficient mice do not demonstrate CD8+ expansion in response to stimulation with exogenous DNA and lack an inflammatory infiltrate following treatment with the powerful carcinogen DMBA (16,17). We therefore hypothesized that the STING/TBK1/IRF3 pathway is active in DDRD cancer cells. Supporting this, we observed constitutive phosphorylation (activation) of IRF3 from whole cell lysates of BRCA1-mutant MDA-MB-436-EV and HCC1937-EV cells, compared with their BRCA1-corrected isogenic lines (Figure 2B). Similarly, TBK1 was constitutively phosphorylated in the DDRD MDA-MB-436-EV and HCC1937-EV cell lines compared with the isogenic corrected cell lines (Figure 2B). siRNA-mediated knockdown of STING, TBK1, and IRF3 resulted in reduced expression of chemokines in BRCA1-mutant cells, indicating that this pathway is required for chemokine production in DDRD cells (Figure 2, C and D).
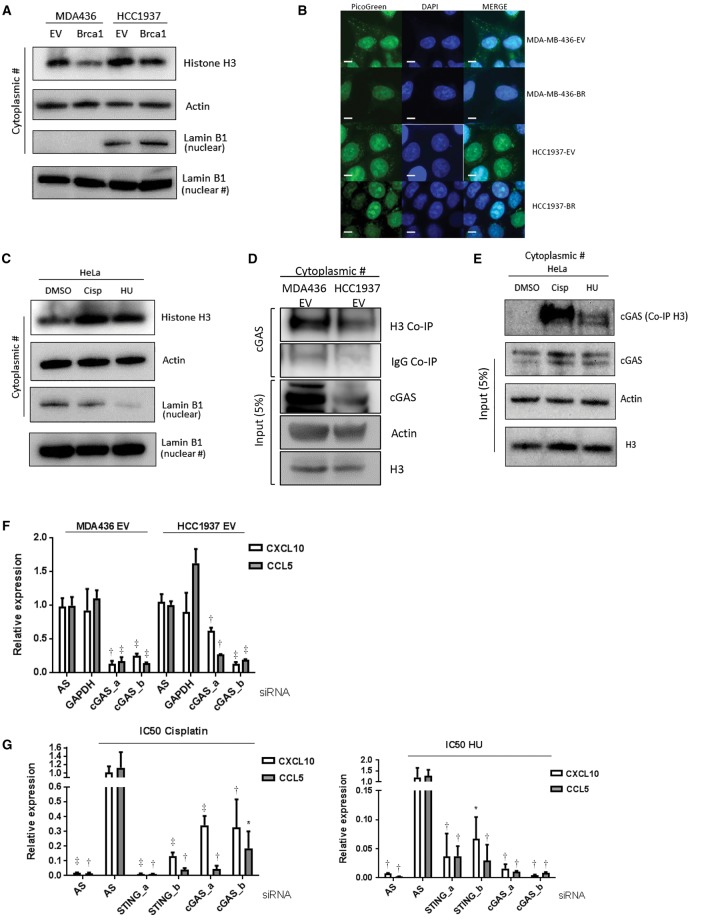
Identification of Cytosolic DNA in the Presence of DNA Damage
The cytosolic DNA sensor cGAS has been described as the most potent activator of the STING pathway (18). We therefore asked if cytosolic DNA detected by cGAS was associated with the observed immune response in DDRD cells. Firstly, we analyzed cytosolic fractions of MDA-MB-436-EV and -BRCA1, and HCC1937-EV and -BRCA1 cells, for the presence of Histone H3 (a marker of DNA) and found Histone H3 cytosolic protein expression was increased in the DDRD BRCA1-mutant cells (Figure 3A; confirmed with densitometry in Supplementary Figure 3B, available online). To confirm the presence of cytosolic DNA in DDRD cells, the HCC1937-EV and MDA-MB-436-EV cells and their BRCA1-corrected isogenic pairs were stained with PicoGreen. This confirmed increased DNA within the BRCA1-deficient cells compared with the corrected cell lines (Figure 3A). We confirmed that the increased expression of Histone H3 was specific to S-phase DNA damage by treating HeLa cells with cisplatin and HU (Figure 3B). Furthermore, PicoGreen staining of HeLa cells treated with IC50 doses of cisplatin and HU revealed increased cytosolic DNA under confocal microscopy (Supplementary Figure 3A, available online). This increase in cytosolic DNA was not observed in response to treatment with paclitaxel.
Figure 3.
Analysis of cytosolic DNA in DNA damage response deficiency. A) Immunoblot for Histone H3 in the cytosolic fraction of DNA damage repair–deficient cell lines (MDA-MB-436-EV and HCC1937-EV) compared with isogenic BRCA1–corrected cells (MDA-MB-436-BRCA1 and HCC1937-BRCA1). Actin is included as a loading control. Nuclear Lamin B1 shows degree of fractionation achieved. Densitometry has been performed on Histone H3 expression in Supplementary Figure 3B (available online). B) PicoGreen staining of MDA-MB-436 and HCC1937 BRCA1-complemented and empty vector counterparts. Scale bar represents 10 µm. C) Immunoblot for Histone H3 in the cytosolic of HeLa cells treated with IC50 dose cisplatin (1.46 µM) and hydroxyurea (326 µM) compared with a DMSO control. Actin and Lamin B1 are included as loading controls. Densitometry has been performed on Histone H3 expression in Supplementary Figure 3B (available online). D) Immunoblot for cGAS using Histone H3 immunoprecipitate from the cytoplasmic fraction of MDA-MB-436-EV and HCC1937-EV cells. Total cGAS, Histone H3 input, and Actin loading controls are shown. E) Immunoblot for cGAS using Histone H3 immunoprecipitate from the cytoplasmic fraction of HeLa cells treated with cisplatin and HU compared with DMSO control. Total cGAS, Histone H3 input, and Actin loading controls are shown. F) Quantitative polymerase chain reaction (qPCR) measurement of CXCL10 and CCL5 mRNA expression in MDA-MB-436-EV and HCC1937-EV cells using two independent siRNAs targeting cGAS (a and b), compared with nontargeting siRNA control (AS). G) qPCR measurement of CXCL10 and CCL5 mRNA 48 hours after IC50 dose cisplatin (1.46 µM) (left panel) and hydroxyurea (326 µM) (right panel treatment in HeLa cells). cGAS expression was knocked down 24 hours prior to drug treatment using two independent siRNAs targeting cGAS (a and b). A nontargeting siRNA control (AS) is included. All data are representative of mean ± SD, and P values were calculated using the unpaired, two-tailed Student’s t test. *P < .05, †P < .01, ‡P < .001.
Next we asked if cGAS was bound to DNA following endogenous or exogenous DNA damage. We demonstrated that cGAS and Histone H3 were bound in the cytosolic fraction of the DDRD-positive cells MDA-MB-436-EV and HCC1937-EV (Figure 3C). To confirm the relationship to S-phase-specific DNA damage, HeLa cells were treated with IC50 doses of cisplatin and HU, cGAS was again bound to Histone H3, and this interaction was not observed in the DMSO-treated control (Figure 3D). siRNA-mediated knockdown of cGAS resulted in statistically significant reduction in CCL5 and CXCL10 expression in BRCA1-mutant cells (P < .01) (Figure 3E). Importantly, knockdown of STING or cGAS reduced upregulation of chemokine expression in response to cisplatin or HU, reinforcing the importance of this pathway in signaling S-phase-specific DNA damage (Figure 3F). These data support the requirement of cGAS for the expression of chemokines in response to DNA damage.
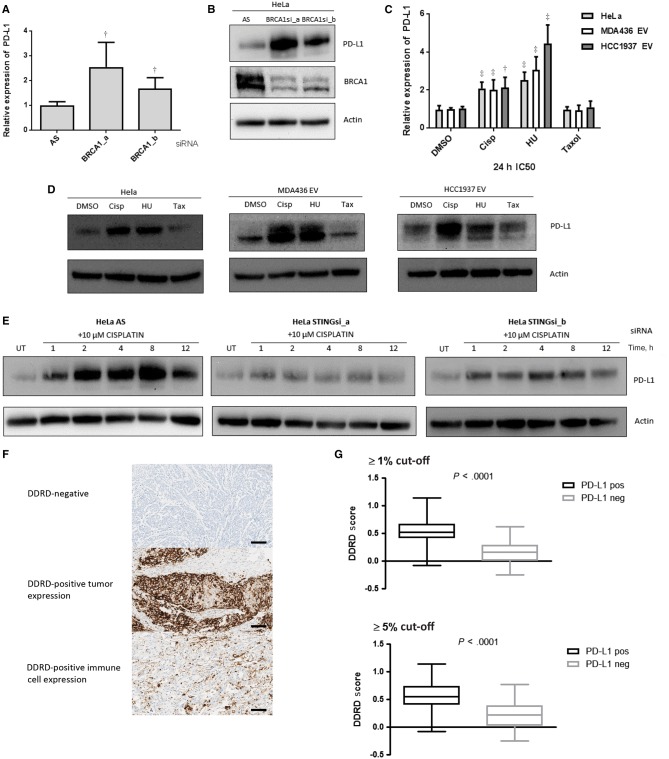
Analysis of PD-L1 Expression in the Context of S-Phase DNA Damage
As PD-L1 is one of the genes assessed in the DDRD assay and its upregulated expression is associated with the DDRD assay–positive molecular subtype, we hypothesized that increased expression of this immune checkpoint gene may provide an explanation for the lack of immune-mediated cytotoxicity observed in cancers within the DDRD molecular subtype despite lymphocytic infiltration (5). Consistent with this hypothesis, siRNA knockdown of BRCA1 in HeLa cells upregulated PD-L1 expression as measured by qPCR (Figure 4A) and Immunoblotting (Figure 4B).
Figure 4.
PD-L1 expression in DNA damage response deficiency. A) Quantitative polymerase chain reaction (qPCR) measurement of PD-L1 expression in HeLa cells using two independent siRNAs targeting BRCA1 (a and b), compared with nontargeting siRNA control (AS). B) Immunoblot of PD-L1 and BRCA1 protein expression in HeLa cells following two independent siRNAs targeting BRCA1 (a and b). Actin is shown as a loading control. C) qPCR measurement of PD-L1 mRNA expression in HeLa, MDA-MB-436-EV, and HCC1937-EV cells 24 hours following treatment with cisplatin, HU, and paclitaxel at the IC50 dose appropriate for each cell line. All data are representative of mean ± SD, and P values were calculated using the unpaired, two-tailed Student’s t test. *P < .05, †P < .01, ‡P < .001. D) Immunoblot for PD-L1 protein expression in HeLa, MDA-MB-436-EV, and HCC1937-EV cells 24 hours following treatment with IC50 doses of cisplatin, HU, and paclitaxel. Actin is included as a loading control. E) Immunoblot for PD-L1 in HeLa cells after siRNA-mediated knockdown of STING using two independent siRNAs (a and b) from one to 12 hours following cisplatin treatment (10 µM). A nontargeting siRNA control (AS) is included. Actin is included as a loading control. F) Immunohistochemistry (IHC) images (x20) representing immunostaining for PD-L1 in no staining (upper panel), epithelial staining (middle panel), and immune staining (lower panel). Scale bar represents 100 µm. G) Boxplot graph showing relationship between DDRD assay score and PD-L1 expression as assessed by IHC at ≥ 1% (upper panel) and ≥5% (lower panel) cut-offs. DDRD = DNA damage response–deficient.
To confirm the relationship between PD-L1 expression and S-phase-specific DNA damage, HeLa, HCC1937-EV, and MDA-MB-436-EV cell lines were exposed to cisplatin and hydroxyurea. Enhanced expression of PD-L1 at mRNA and protein level was observed in response to S-phase DNA damage but not paclitaxel (Figure 4, C and D). These data are consistent with PD-L1 induction being predominantly related to S-phase-specific DNA damage. Additionally, siRNA knockdown of STING abrogated upregulation of PD-L1 in response to cisplatin, indicating the requirement of this cytosolic DNA-sensing pathway for immune checkpointing following DNA damage (Figure 4E).
Analysis of Expression of PD-L1 in Breast Tumors Within the DDRD Molecular Subtype
To determine if PD-L1 expression was observed in breast cancers within the DDRD molecular subtype, we performed IHC analysis of the cohort of breast tumors previously scored for CD4+ and CD8+ lymphocytic infiltration. Previously published cut-offs of 1% or greater and 5% or greater were used to define PD-L1 positivity (Figure 4F) (6). A statistically significant association between PD-L1 expression and a positive score for the DDRD assay was observed at both of the predefined cut-offs (P < .001). Interestingly, both tumor epithelial cell PD-L1 positivity and infiltrating immune cell PD-L1 expression were statistically significantly associated with DDRD assay positivity (Table 2, Figure 4G), indicating that DDRD epithelial cells are not only associated with PD-L1 expression but may also induce PD-L1 expression in infiltrating lymphocytes.
Table 2.
PD-L1 expression assessed by IHC in DDRD-positive and DDRD-negative breast tumors
| PD-L1 expression | DDRD positive, No. (%) | DDRD negative, No. (%) | P* |
|---|---|---|---|
| ≥1% score | |||
| Tumor | 64 (100) | 111 (100) | <.001 |
| PD-L1 ≥ 1% | 29 (45.3) | 5 (4.5) | |
| PD-L1 < 1% | 35 (54.7) | 106 (95.5) | |
| Immune cells | 64 (100) | 111 (100) | <.001 |
| PD-L1 ≥ 1% | 48 (75.0) | 9 (8.1) | |
| PD-L1 < 1% | 16 (25.0) | 102 (91.9) | |
| Tumor/immune cells | 64 (100) | 111 (100) | <.001 |
| PD-L1 ≥ 1% | 48 (75.0) | 9 (8.1) | |
| PD-L1 < 1% | 16 (25.0) | 102 (91.9) | |
| ≥5% score | |||
| Tumor | 64 (100) | 111 (100) | <.001 |
| PD-L1 ≥ 5% | 13 (20.3) | 4 (3.6) | |
| PD-L1 < 5% | 51 (79.7) | 107 (96.4) | |
| Immune cells | 64 (100) | 111 (100) | <.001 |
| PD-L1 ≥ 5% | 25 (39.1) | 3 (2.7) | |
| PD-L1 < 5% | 39 (79.7) | 108 (97.3) | |
| Tumor/immune cells | 64 (100) | 111 (100) | <.001 |
| PD-L1 ≥ 5% | 28 (43.8) | 6 (5.4) | |
| PD-L1 < 5% | 36 (56.3) | 105 (94.6) |
P values were calculated using a two-sided Fisher’s exact test. DDRD = DNA damage response–deficient; IHC = immunohistochemistry.
Discussion
The DDRD molecular subtype represents tumors that have loss of function of the FA/BRCA pathway, an important response mechanism to stalled DNA replication in the S-phase of the cell cycle. Our new data suggest that in the absence of a functional FA/BRCA pathway or as a result of exogenous DNA damage, there is a mechanism through which an accumulation of cytosolic DNA activates the STING/TBK1/IRF3 innate immune response.
Genomically unstable breast cancers such as BRCA1-mutant are characterized by lymphocytic infiltration (19). Previous studies have suggested that genomic instability may activate immune signaling through the production of neoantigens (3). Our model proposes cytosolic DNA in the epithelial component of the cancer as an important immune stimulus that does not require recognition of abnormal proteins. Although it is unclear why S-phase DNA damage should result in cytosolic DNA, we hypothesize that this may be a byproduct of re-establishing DNA replication. There is some evidence that the cell may actively export DNA fragments from the nucleus, possibly to prevent misincorporation into genomic DNA (20). Normally cytosolic DNA is processed by cytoplasmic DNase II; however, it may be that this mechanism is overwhelmed by a failure to respond to endogenous DNA damage or following exogenous DNA damage, thereby triggering the cGAS-mediated innate immune response. Indeed, similar activation of the STING pathway in response to accumulation of cytosolic DNA has been observed in systemic lupus erythematosus (21).
Our DDRD gene assay contains two immune checkpointing genes that represent therapeutic targets, PD-L1 and IDO1 (4). Inhibition of the PD-1/PD-L1 axis has resulted in dramatic responses in a subset of patients with advanced solid tumors including melanoma and non–small cell lung cancer (22). Importantly, our observation that DDRD-positive tumors associate with PD-L1 expression provides a rationale for exploration of immune checkpoint treatments in this molecular subtype. In support of this approach is the recent report for the utility of PD-1 inhibitors in mismatch repair–deficient colorectal cancer (23). Mismatch repair proteins have been reported to have a role in the response to S-phase replication fork stalling (24), which our study suggests should activate the cGAS/STING pathway, upregulating PD-L1 expression. Whereas the ligands CXCL10 and CCL5 are known to be direct targets of IRF3, PD-L1 is not known to have an IRF3-binding region in the promoter. Other groups, however, have reported a dependence on STING and TBK1 for PD-L1 upregulation consistent with our findings (25,26). Therefore, there may be an indirect mechanism of regulation, possibly through targets of IRF3 such as interferon beta (27). Interestingly, where we observed tumor cell expression of PD-L1, PD-L1 was also expressed on infiltrating immune cells, suggesting the cytokine release from tumor cells may also activate PD-L1 expression in the tumor microenvironment.
The S-phase-specific nature of the immune signal also raises a potentially important issue around combination therapies with immune checkpoint agents. Interestingly, direct activation of the STING pathway using synthetic cyclic dinucleotide molecules has been reported to enhance responses to PD-1 antibodies, which is in keeping with our data (28). Another logical combination may be an S-phase-specific DNA-damaging agent such as cisplatin along with a PD-1/PD-L1 antibody. Antimicrotubule agents, however, may antagonize PD-1/PD-L1 antibodies by causing cell cycle arrest in the mitotic phase of the cell cycle, thereby preventing the STING-mediated immune response.
An important limitation of this study is the absence of immune infiltration in tissue culture models, which means we were unable to study the effects of immune checkpointing on tumor cytotoxicity. We therefore intend to extend this work into syngeneic animal model systems to test the sensitivity of tumors with S-phase-specific DNA damage to PD-L1 inhibition.
In summary, we have identified a mechanism of immune response in breast tumors deficient in DNA repair. Activation of the innate immune STING-mediated pathway is responsible for chemokine production in response to DNA damage in vitro, resulting in an inflammatory microenvironment in DDRD breast tumors. Expression of PD-L1 is associated with tumors deficient in DNA damage repair, and we provide a rationale for investigating the role of immune treatments in the context of endogenous or exogenous S-phase DNA damage.
Funding
EEP is funded by a grant from Cancer Research UK.
This work was supported by Invest NI (reference ST263) through the European Sustainable Competitiveness Programme 2007–2013, European Regional Development Fund.
The samples used in this research were received from the Northern Ireland Biobank, which is funded by Health and Social Care (HSC) Research and the Development Division of the Public Health Agency in Northern Ireland and Cancer Research UK through the Belfast CR-UK Centre and the Northern Ireland Experimental Cancer Medicine Centre; additional support was received from the Friends of the Cancer Centre. The Northern Ireland Molecular Pathology Laboratory, which is responsible for creating resources for the Northern Ireland Biobank (NIB), has received funding from Cancer Research UK, the Friends of the Cancer Centre, and the Sean Crummey Foundation.
Notes
The funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Author contributions: designed and performed experiments: EEP, LET, RW, MTH; designed experiments: SMW, NMcC, KIS, PBM, RDK; wrote paper: EEP, SMW, NMcC, KIS, PBM, DPH, RDK; confocal imaging: KIS, KDMcC, EEP, RW; bioinformatics: LH; staining, scoring, analysis of IHC: MS-T, SMcQ, NEB, EEP.
The buffy coat samples used in this study were provided by the Northern Ireland Blood Transfusion Service, with thanks to Dr. Kathryn Maguire.
Supplementary Material
References
- 1. Loi S, Sirtaine N, Piette F, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J Clin Oncol. 2013;31(7):860–867. [DOI] [PubMed] [Google Scholar]
- 2. Denkert C, Loibl S, Noske A, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2010;28(1):105–113. [DOI] [PubMed] [Google Scholar]
- 3. Woo SR, Corrales L, Gajewski TF. Innate immune recognition of cancer. Annu Rev Immunol. 2015;33:445–474. [DOI] [PubMed] [Google Scholar]
- 4. Mulligan JM, Hill LA, Deharo S, et al. Identification and validation of an anthracycline/cyclophosphamide-based chemotherapy response assay in breast cancer. J Natl Cancer Inst. 2014;106(1):djt335. doi: 10.1093/jnci/djt335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quinn JE, Kennedy RD, Mullan PB, et al. BRCA1 functions as a differential modulator of chemotherapy-induced apoptosis. Cancer Res. 2003;63(19):6221–6228. [PubMed] [Google Scholar]
- 6. Soria J, Gettinger S, Gordon MS, et al. Biomarkers associated with clinical activity of PD-L1 blockade in non-small cell lung carcinoma (NSCLC) patients (pts) in a phase I study of MPDL3280A. Ann Oncol. 2014;25(suppl 4):iv426, Abstract 1322 P. [Google Scholar]
- 7. Jenh CH, Cox MA, Cui L, et al. A selective and potent CXCR3 antagonist SCH 546738 attenuates the development of autoimmune diseases and delays graft rejection. BMC Immunol. 2012;13:2-2172-13-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mulligan AM, Raitman I, Feeley L, et al. Tumoral lymphocytic infiltration and expression of the chemokine CXCL10 in breast cancers from the Ontario familial breast cancer registry. Clin Cancer Res. 2013;19(2):336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Groom JR, Luster AD. CXCR3 in T cell function. Exp Cell Res. 2011;317(5):620–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kunz M, Toksoy A, Goebeler M, Engelhardt E, Brocker E, Gillitzer R. Strong expression of the lymphoattractant C-X-C chemokine mig is associated with heavy infiltration of T cells in human malignant melanoma. J Pathol. 1999;189(4):552–558. [DOI] [PubMed] [Google Scholar]
- 11. Ohtani H, Jin Z, Takegawa S, Nakayama T, Yoshie O. Abundant expression of CXCL9 (MIG) by stromal cells that include dendritic cells and accumulation of CXCR3+ T cells in lymphocyte-rich gastric carcinoma. J Pathol. 2009;217(1):21–31. [DOI] [PubMed] [Google Scholar]
- 12. Muthuswamy R, Berk E, Junecko BF, et al. NF-kappaB hyperactivation in tumor tissues allows tumor-selective reprogramming of the chemokine microenvironment to enhance the recruitment of cytolytic T effector cells. Cancer Res. 2012;72(15):3735–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brzostek-Racine S, Gordon C, Van Scoy S, Reich NC. The DNA damage response induces IFN. J Immunol. 2011;187(10):5336–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim T, Kim TY, Song YH, Min IM, Yim J, Kim TK. Activation of interferon regulatory factor 3 in response to DNA-damaging agents. J Biol Chem. 1999;274(43):30686–30689. [DOI] [PubMed] [Google Scholar]
- 15. Motani K, Ito S, Nagata S. DNA-mediated cyclic GMP-AMP synthase-dependent and -independent regulation of innate immune responses. J Immunol. 2015;194(10):4914–4923. [DOI] [PubMed] [Google Scholar]
- 16. Woo SR, Fuertes MB, Corrales L, et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41(5):830–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ahn J, Xia T, Konno H, Konno K, Ruiz P, Barber GN. Inflammation-driven carcinogenesis is mediated through STING. Nat Commun. 2014;5:5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ablasser A, Goldeck M, Cavlar T, et al. cGAS produces a 2'-5'-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498(7454):380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Turner N, Tutt A, Ashworth A. Hallmarks of 'BRCAness' in sporadic cancers. Nat Rev Cancer. 2004;4(10):814–819. [DOI] [PubMed] [Google Scholar]
- 20. Lan YY, Londono D, Bouley R, Rooney MS, Hacohen N. Dnase2a deficiency uncovers lysosomal clearance of damaged nuclear DNA via autophagy. Cell Rep. 2014;9(1):180–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ahn J, Gutman D, Saijo S, Barber GN. STING manifests self DNA-dependent inflammatory disease. Proc Natl Acad Sci U S A. 2012;109(47):19386–19391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Svendsen JM, Smogorzewska A, Sowa ME, et al. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138(1):63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Corrales L, Glickman LHH, McWhirter SMM, et al. Direct Activation of STING in the Tumor Microenvironment Leads to Potent and Systemic Tumor Regression and Immunity. Cell Rep. 2015;11(7):1018–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang S, Imamura Y, Jenkins RW, et al. Autophagy Inhibition Dysregulates TBK1 Signaling and Promotes Pancreatic Inflammation. Cancer Immunol Res. 2016;4(6):520–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sharpe AH, Wherry EJ, Ahmed R, Freeman GJ. The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nat Immunol. 2007;8(3):239–245. [DOI] [PubMed] [Google Scholar]
- 28. Fu J, Kanne DB, Leong M, et al. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci Transl Med. 2015;7(283):283ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.