Abstract
Virions of mouse leukemia virus spread on glass substrates were visualized by atomic force microscopy. The size distribution mode was 145 nm, significantly larger than that for human immunodeficiency virus particles. The distribution of particle sizes is broad, indicating that no two particles are likely identical in content or surface features. Virions possess knoblike protrusions, which may represent vestiges of budding from cell membranes. Particles which split open allowed imaging of intact cores with diameters of 65 nm. They also permitted estimation of viral shell thickness (35 to 40 nm) and showed the presence of a distinct trough between the shell and the core surface.
Mouse leukemia virus (MuLV) is among the most thoroughly studied of the oncogenic retroviruses, first shown to produce leukemias in mice by Gross in 1951 (3, 4). Its properties have been reviewed (9, 11, 13, 15), and models for its structure have been described (14, 16). From electron microscopy (EM) studies, the C-type virus was shown to be roughly spherical, with a diameter of 80 to 120 nm, but to have a pleiomorphic surface. Our earlier atomic force microscopy (AFM) investigation of MuLV emerging from infected cells was consistent with those descriptions (6). In this study, virions were isolated from the culture media by centrifugation and examined by AFM after they were spread on poly-l-lysine-coated coverslips. Thus each particle is independent of others and the cell which produced it.
The production of Moloney MuLV in NIH 3T3 cells (strain 43-D) has been previously described (2, 6, 7). Virions were purified from the media by centrifugation as for human immunodeficiency virus (HIV), fixed with 0.5% glutaraldehyde, and dehydrated in stages (8). Virions were also recorded in physiological buffer to evaluate the degree of shrinkage due to dehydration. AFM was carried out using a Nanoscope IIIa atomic force microscope (Digital Instruments, Santa Barbara, Calif.) operated under 70% propanol in tapping mode (5) with oscillation frequencies of about 9.2 kHz and a scanning frequency of 1 Hz. All other procedures were essentially the same as we previously reported for MuLV emerging from host cells (6, 7) and for HIV (8). All diameters of particles were based strictly on their measured height above background.
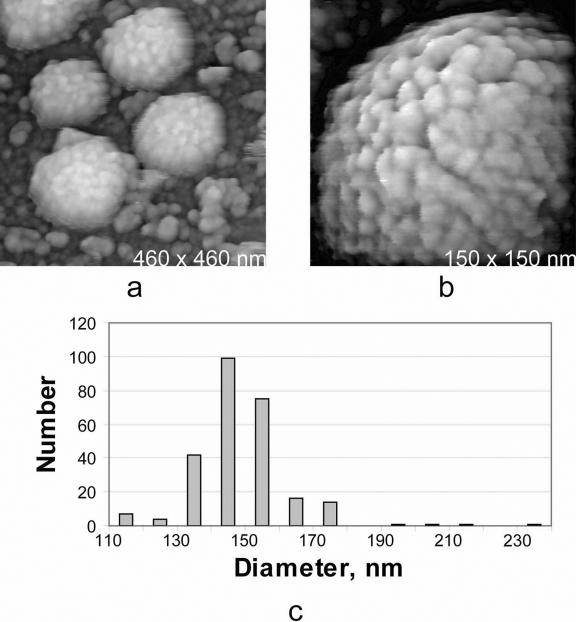
A low-resolution image of wild-type particles is seen in Fig. 1a, and a higher-magnification image is seen in Fig. 1b. The particles shown in Fig. 1 closely resemble those observed on host cell surfaces, as previously presented (6, 7). The virions are roughly spherical, and their surfaces are heavily studded with protein “tufts.”
FIG. 1.
(a) AFM image of MuLV virions isolated from culture media and spread on poly-l-lysine-coated glass substrates. (b) A single virion at higher magnification. The appearance of the particles is essentially the same as that of particles associated with host cell surfaces (6). The tufted protein arrangement seen on the virion surfaces is characteristic of both MuLV and HIV. (c) Histogram of particle sizes (corrected for shrinkage due to dehydration) for wild-type MuLV isolated from the culture media of virus-infected NIH 3T3 cells (strain 43-D) and spread on glass substrates for analysis by AFM. The mode of the distribution is 145 nm.
Figure 1c is a histogram showing the distribution of particle sizes measured from 500 virions from culture media which had been spread on poly-l-lysine-coated glass coverslips. The mode of the distribution is 145 nm, but the distribution is very broad, extending from particle sizes as low as 100 nm up to sizes exceeding 200 nm. The breadth of the distribution is representative of the variation in size of the particles and is not due to measurement error, which is of the order of a few nanometers. This implies that MuLV virions are by no means identical to one another, but rather assume sizes and appearances dependent on their contents and the vagaries of the budding process. A similar conclusion emerged from electron microscopy investigations of both HIV and MuLV, where similar, broad distributions were found (1, 16).
A similar statistical analysis of sizes was also carried out for MuLV observed on cell surfaces. The mode of that distribution was again 145 nm, the same as for free particles, and the shape of the distribution was almost the same. We also examined MuLV-based vector particles produced by two NIH 3T3-based packaging cell lines, ψ2/BAG and PA317/BAG (10, 12). The ψ2 and PA317 packaging cells produce all of the structural proteins of MuLV from mRNAs lacking an MuLV packaging signal. The vector particles exhibit the same external appearance and distribution on the host cell surfaces as wild-type virus and appear indistinguishable from wild-type virions. The size distribution for the vector particles was the same as that for normal virions.
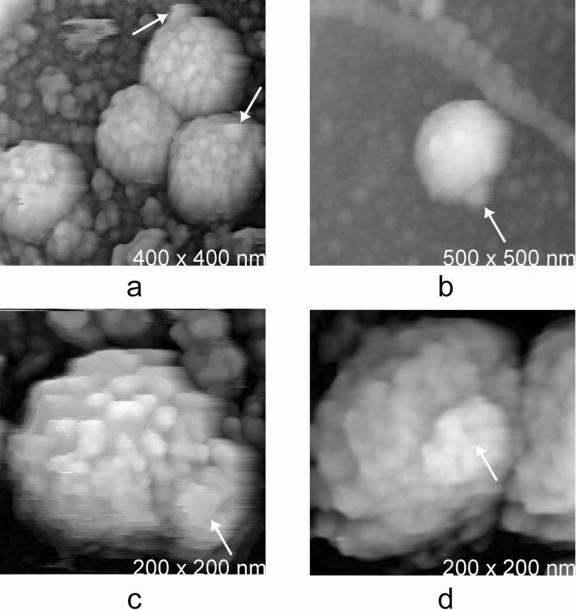
Common to virtually all of the isolated virions was a distinctive protrusion, or knob. Images illustrating these structures are presented in Fig. 2. Similar appendages have also been noted on MuLV particles by cryoelectron microscopy (16) and were referred to as “blebs.” Particles on the surfaces of cells only rarely exhibit such protrusions, leading us earlier to describe the virions as spherical (6); the virions are in fact not exactly so, but rather are anisotropic as a consequence of the protrusions. We believe that the blebs are likely a consequence of the budding process and may be “bud scars” resulting from the pinching off of the membrane. If so, that would explain their absence in earlier studies where they were associated with cell surfaces. The point of attachment lies between the virion and cell surface; therefore, it would not have been accessible to the AFM tip, being in the AFM shadow of the particle.
FIG. 2.
Images of MuLV isolated from media and spread on glass substrates showing the appearance of the protruding blebs (arrows), never more than one per virion, which mark their surfaces. These are likely to be scars resulting from the budding process.
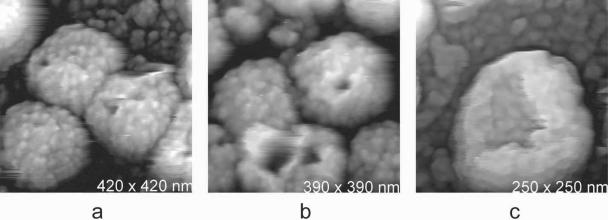
We previously noted, for cell-attached virions, the occasional presence of particles which appeared to have lost a sector of surface protein and consequently exposed a deep pit or channel into the interior (6). Examples of such particles on a glass substrate are shown in Fig. 3. Such particles are significantly more frequent among those isolated from the media, suggesting that they may result from damage in preparation. If virions from the media are fixed with glutaraldehyde before centrifugation and attachment to the glass substrate, the occurrence of the pits is greatly reduced.
FIG. 3.
(a and b) MuLV particles on glass substrates which exhibit deep pits on their surfaces. These are likely due to loss of surface proteins or sectors of proteins incurred in the preparation process. Similar defects have, however, also been seen on particles still associated with host cell surfaces (6). (c) The shell remaining after loss of the nucleocapsid from an MuLV virion was produced by centrifugation during sample preparation. The shell retains its structural integrity in spite of the loss of a large sector of its surface.
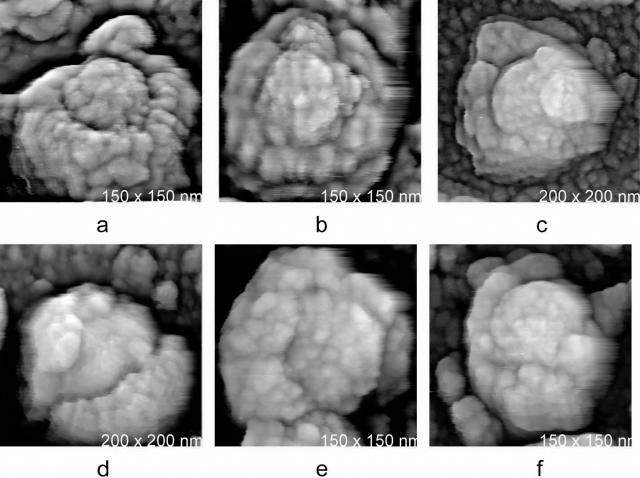
Although most of the isolated MuLV particles were intact, we also recorded virions which were damaged, probably by centrifugation, or which were otherwise incomplete. These particles allowed us to visualize virion interiors. In Fig. 4 are AFM images of partially disrupted MuLV virions, where a portion of the viral envelope was shorn away to reveal the nucleic acid-containing cores. Evident in these images is that the MuLV cores are indeed spherical in shape and that they have diameters of about 65 nm. While the virus particles themselves display a rather wide variation in size, the cores seem to be relatively uniform.
FIG. 4.
Gallery of AFM images of MuLV particles which have been isolated from culture media and spread on glass substrates and which have been damaged in the process. Large portions of the shell-like envelope have been displaced, and the nucleic acid-containing cores of the particles can be seen enclosed inside or emerging from the envelopes. The cores are roughly spherical, are marked by an irregular arrangement of proteins on their surfaces, and have a diameter of about 65 nm. There is a distinctive demarcation, or gap, between the cores and the inside surfaces of the shells. Accessory proteins appear to inhabit this space between the cores and shells.
It may be noteworthy that we were able to visualize the cores of MuLV particles in this study. Using virtually the same procedures, we failed to record intact cores from HIV virions (8).
The shell surrounding the core presents itself as a single unit with a thickness of about 35 to 40 nm, approximately the same as we found for the envelope of HIV using similar methods (8). To account for the observed thickness, the shell must be composed of the exterior and transmembrane portions of the envelope protein, the virus lipid membrane, and the interior matrix (MA) protein. These components seem to be contiguous with one another and to form a discreet and distinctive shell around the core. Evidence for a tight association of MA protein with the interior of the lipid membrane has also come from cryoelectron microscopy for both MuLV (16) and HIV (1). There is a clear demarcation between the shells and the cores. This is quite evident in Fig. 4 and is also consistent with what is seen by cryoelectron microscopy (1). The matrix protein is closely integrated into the viral shell and is separated by a gap or space from the surface of the core.
In the region between the core and the MA protein on the interior of the shell, there are also other proteins which appear as discrete globular units in the AFM images. These proteins are smaller, in general, than those on the exterior of virions, principally the SU protein, and are probably in the range of 20 to 50 kDa. These proteins do not appear to be closely associated with either the shell or core.
The images presented here represent the first direct observations by AFM of isolated MuLV particles independent of their host cells. In addition, these are the first recordings of the interiors of MuLV particles revealing the nucleic acid-containing cores of the particles, the virion shells, and the interface between them.
Acknowledgments
We thank Aaron Greenwood for his assistance in producing the figures.
This work was supported by NIH grant CA32455 to H.F., NIH grant GM58868 to A.M., contract number NAS800017 from NASA, and assistance from the UCI Cancer Center.
REFERENCES
- 1.Briggs, J. A. G., T. Wilk, R. Welker, H.-G. Krausslich, and S. D. Fuller. 2003. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 22:1707-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan, H., H. Chute, E. Chao, and M. Feuerman. 1983. Construction and characterization of Moloney murine leukemia virus mutants unable to synthesize glycosylated gag polyprotein. Proc. Natl. Acad. Sci. USA 80:5965-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gardner, M. B. 1980. Historical background, p. 2-46. In J. R. Stephenson (ed.), Molecular biology of RNA tumor viruses. Academic Press, New York, N.Y.
- 4.Gross, L. 1970. Oncogenic viruses. Pergamon, Elmsford, N.Y.
- 5.Hansma, H. G., K. J. Kim, D. E. Laney, R. A. Garcia, M. Argaman, M. J. Allen, and S. M. Parsons. 1997. Properties of biomolecules measured from atomic force microscope images: a review. J. Struct. Biol. 119:99-108. [DOI] [PubMed] [Google Scholar]
- 6.Kuznetsov, Y. G., S. Datta, N. H. Kothari, A. Greenwood, H. Fan, and A. McPherson. 2002. Atomic force microscopy investigation of fibroblasts infected with wild type and mutant murine leukemic virus (MuLV). Biophys. J. 83:3665-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuznetsov, Y. G., A. Low, H. Y. Fan, and A. McPherson. 2004. Atomic force microscopy investigation of wild type Moloney murine leukemia virus particles and virus particles lacking the envelope protein. Virology 323:189-196. [DOI] [PubMed] [Google Scholar]
- 8.Kuznetsov, Y. G., J. G. Victoria, W. E. J. Robinson, and A. McPherson. 2003. Atomic force microscopy investigation of human immunodeficiency virus (HIV) and HIV-infected lymphocytes. J. Virol. 77:11896-11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy, J. A. 1992. The Retroviridae. Plenum Press, New York, N.Y.
- 10.Miller, A. D., and C. Baltimore. 1986. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol. Cell. Biol. 6:2895-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pincus, T. 1980. The endogenous murine type C viruses, p. 78-130. In J. R. Stephenson (ed.), Molecular biology of RNA tumor viruses. Academic Press, New York, N.Y.
- 12.Price, J., D. Turner, and C. Cepko. 1987. Lineage analysis in the vertebrate nervous system by retrovirus-mediated gene transfer. Proc. Natl. Acad. Sci. USA 84:156-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephenson, J. R. (ed.). 1980. Molecular biology of RNA tumor viruses. Academic Press, New York, N.Y.
- 14.Vogt, V. M. 1997. Retroviral virions and genomes, p. 27-69. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed] [Google Scholar]
- 15.Weiss, R. A., N. Teich, H. E. Varmus, and J. M. Coffin (ed.). 1982. Molecular biology of tumor viruses: RNA tumor viruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Yeager, M., E. M. Wilson-Kubalek, S. G. Weiner, P. O. Brown, and A. Rein. 1998. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc. Natl. Acad. Sci. USA 95:7299-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]