Abstract
Protozoans within the genus Plasmodium are well-known as the causative agents of malaria in humans. Numerous Plasmodium species parasites also infect a wide range of non-human primate hosts in tropical and sub-tropical regions worldwide. Studying this diversity can provide critical insight into our understanding of human malarias, as several human malaria species are a result of host switches from non-human primates. Current spillover of a monkey malaria, Plasmodium knowlesi, in Southeast Asia highlights the permeability of species barriers in Plasmodium. Also recently, surveys of apes in Africa uncovered a previously undescribed diversity of Plasmodium in chimpanzees and gorillas. Therefore, we carried out a meta-analysis to quantify the global distribution, host range, and diversity of known non-human primate malaria species. We used published records of Plasmodium parasites found in non-human primates to estimate the total diversity of non-human primate malarias globally. We estimate that at least three undescribed primate malaria species exist in sampled primates, and many more likely exist in unstudied species. The diversity of malaria parasites is especially uncertain in regions of low sampling such as Madagascar, and taxonomic groups such as African Old World Monkeys and gibbons. Presence–absence data of malaria across primates enables us to highlight the close association of forested regions and non-human primate malarias. This distribution potentially reflects a long coevolution of primates, forest-adapted mosquitoes, and malaria parasites. The diversity and distribution of primate malaria are an essential prerequisite to understanding the mechanisms and circumstances that allow Plasmodium to jump species barriers, both in the evolution of malaria parasites and current cases of spillover into humans.
Keywords: Primates, Plasmodium knowlesi, Zoonosis, Malaria richness, Spillover, Species richness estimates
Highlights
-
•
Primates are infected with at least 30 species of malaria parasites.
-
•
6 more proposed Plasmodium species have been described from a small number of hosts.
-
•
We predict there are up to 5 additional malaria species in undersampled primates.
-
•
Increasing taxonomic breadth of host samples will improve estimates of parasite richness.
-
•
Research should focus on identity of vectors to understand distribution of parasites.
Introduction
Malaria is arguably the most important infectious disease in humans. Annually, it causes an estimated 584,000 deaths and leads to over 198 million cases globally [1]. Plasmodium parasites that cause malaria have also had a significant impact on the human evolution, as evidenced by hundreds of mutations that have arisen to high frequency due to selection by malaria [2], [3].
While there are four Plasmodium species that primarily infect humans, there are over 250 Plasmodium species that infect other animals, including birds, lizards, snakes and mammals [4], [5], [6]. Studying these species can provide a critical insight into human malaria, especially malaria parasites of non-human primates (NHPs). Two of the most important human malaria parasites, Plasmodium falciparum and Plasmodium vivax, originated from a cross species transmission event of a NHP malaria to humans [7], [8], [9], [10]. Additionally, zoonotic spillover of wild primate malaria is an emerging global public health concern [11], [12], [13], [14]. This is especially important in Malaysian Borneo, where the majority of human malaria cases are caused by a monkey malaria, Plasmodium knowlesi [15], [16], [17]. Several authors have implicated changing land use in the region as the cause for the recent emergence of P. knowlesi, but the mechanisms underlying the recent rise in infections in humans remain unknown. Understanding the diversity and distribution of NHP malarias is an important first step to predict the potential zoonotic risk of NHP malarias.
The term ‘malaria’ has historically referred to disease caused by species from the Apicomplexan genera Leucocytozoon, Haemosporidia, Plasmodium, and Hepatocystis. Here, we will use just ‘malaria’ to describe disease caused by members of the genus Plasmodium and ‘malaria-like’ parasites to describe Leucocytozoon, Haemosporidia, and Hepatocystis[18]. All malaria and malaria-like parasites have a digenic (two-stage) life cycle that requires an intermediate vertebrate host and a definitive insect hosts. Insects in the order Diptera are typically utilized for sexual reproduction and transmission among vertebrate hosts. Although four species of Plasmodium utilize humans as intermediate hosts, at least twenty-six additional NHP parasites exist outside Homo sapiens hosts (Table A.1).
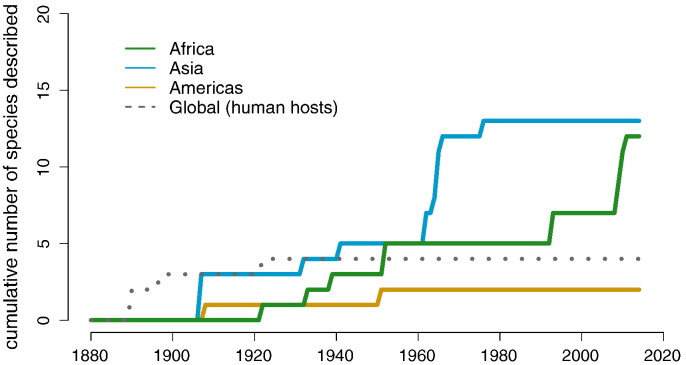
The first account of a NHP malaria parasite was recorded by Laveran (1905) in an orangutan, Pongo pygmaeus. The species Plasmodium pitheci was described a few years later, along with Plasmodium inui and Plasmodium cynomolgi, that infect sympatric monkey species, Macaca fascicularis and Macaca nemestrina, in Borneo [19]. Discovery of primate malarias continued as parasitologists examined an increasing diversity of apes, monkeys, and lemurs from across the globe (Fig. 1, Table A.1). Sample sizes were often small and restricted to animals captured for zoos or killed for examination. The infection of a human with a monkey malaria in 1965 [20] initiated a resurgence of NHP malaria research focused on Southeast Asia, resulting in nine newly described malaria species in Asian apes and monkeys [21]. More recently, identification of a cluster of a monkey malaria, P. knowlesi, within humans in Southeast Asia [15], and discoveries of novel parasite species in apes [7], [8], [9], [10], [22], [23], [24] and monkeys [25], [26], [27] in Central Africa has reignited research interest in primate malarias. This is in part due to improved sampling techniques, leading to the identification of at least four novel clades in the Laverania subgenus of Plasmodium that are awaiting morphological description [28].
Fig. 1.
Discovery of primate Plasmodium species. Colored lines are used to indicate geographic regions where the parasites are endemic. Aside from human malarias (‘Global’), all primate malarias are restricted to the region in which they were described. We included parasites in Table A.1 that have been morphologically described by at least two groups or belong to the Laverania clade (see Methods for distinction of species). The last reviews of primate malarias were conducted in 1933 [58], 1941 [33], 1966 [4], and 1971 [21]. Fooden [75] also reviewed Macaca literature in 1994. The dataset gathered here nearly doubles the number of individuals examined in previous reviews and presents the most updated taxonomic distinctions. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Malariologists have spent decades characterizing these parasite species, and over a century of surveys are available to inform our understanding of the distribution and host range of primate malarias. This presents the opportunity to revisit the global distribution and diversity of NHP malarias for the first time in over 30 years, this time using modern analytical techniques developed to understand patterns of biodiversity accumulation.
In this paper, we critically review all published surveys of primate malarias between 1905–2015 to present an updated global characterization of Plasmodium in primates. Whenever possible, we integrate historical literature with newer molecular work. Using species accumulation curves, we are able to predict expected species richness in primate taxa that have been sampled; and by quantifying sampling effort in each species, we highlight undersampled taxonomic and geographic areas that may harbor a hidden diversity of malaria. We also utilize primate host ranges and presence–absence parasite data to create a global map of non-human primate malarias, and draw attention to regions that may be at risk for zoonotic malaria.
Methods
Primate malaria database
To create a database of primate malarias, we used the search terms Plasmodium and/or malaria and all genera of primates in PubMed and Web of Science until January 1, 2015. We also used key reference books [4], [21] and historical literature reviews to find obscure and non-English publications [31], [32], [33] to supplement online searches. We included recent ape Plasmodium species within the Laverania subgenus with the naming established by Rayner et al.[28]. These species have only been identified molecularly, and await morphological characterization to solidify their species status [34]. However we include these species because they have been isolated by several groups that have confirmed the genetic distinction as species and ensure their identity doesn't overlap with already named species. Other authors have reported novel malaria species in lemurs [35], [36] and African Old World Monkeys [27], [37] based on molecularly characterized samples. These species are listed in Table A.1 but we did not consider these as species in host species accumulation calculations, however they were included in group calculations few isolates or few groups have isolated the parasites, and it is unknown whether their identity overlaps with morphologically described species. For example, almost every lemur malaria parasite found has been named a unique Plasmodium species [35], [36], [38], but there is no corresponding molecular and morphological data from most of these parasites.
For each published account, the location, host species name (in the paper), subspecies (if applicable), number of hosts sampled, sampling method, identification method, Plasmodium species found, and any notes of interest were recorded (full data available in Table A.2). Because genus and species names have changed over the century the data was collected, we used locations and descriptions to update host names to the Mammals of the World 2005 nomenclature [39], which we cross-validated with species synonym files [40]. When possible, we contacted authors for missing information from publications to complete database entries.
A prerequisite for the data to be included in the geographic analysis was that a specific country of origin must be known. Occasionally authors utilized samples from zoos with unknown origin, or worked with samples that had been exported without a country of origin specified, so these were excluded from our presence/absence maps. For data to be included in the species accumulation analysis, the surveys they came from had to utilize methods that did not obviously bias species discovery. For example, if specific primers for a sub genus or particular Plasmodium species were used, then these were excluded from species accumulation analyses. These exclusions are identified for each entry in Table A.2.
The methods used to detect and identify malaria parasites in primates vary in their sensitivity and specificity. Historically, Plasmodium was identified by erythrocytic morphological characteristics using microscopy. Malaria parasites can also be identified in blood samples using molecular markers (i.e., a nested PCR protocol with species-specific primers). Microscopy has a limit of detection of approximately 40 parasites/μL, meaning well-trained malariologists must check at least 100 fields of thick blood films to detect parasites in an individual. Information on screening protocol was rarely given, so we are unable to compare sensitivities among microscopy studies. PCR using DNA isolated from blood is the most sensitive method for detection of active infections [41]. The last method is parasite detection from fecal samples. Although there are benefits to using non-invasively-collected samples, they degrade quickly in the field and range in their sensitivity (30–95%; [42]). Nevertheless, all of these data are informative for determining relative frequencies of infection, and predicting species richness in primates.
Predicting malaria species diversity
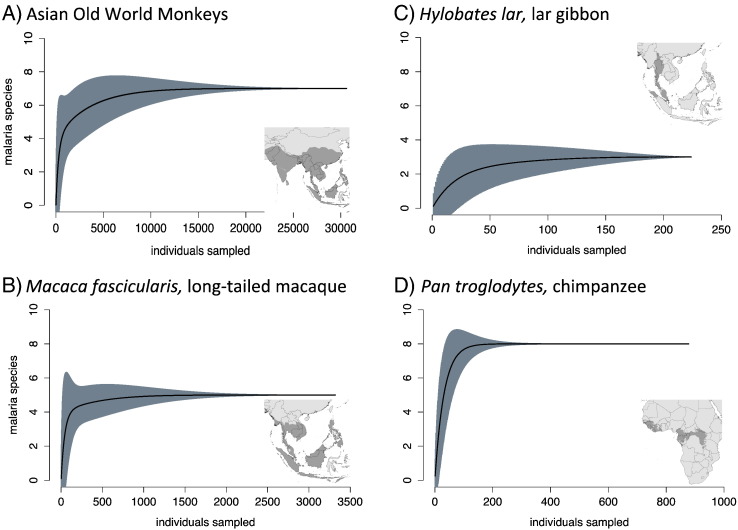
Datasets were separated into individual host species for analysis in R computing software version 3.0.2 [44]. To quantify sampling effort, we assumed that individual hosts belonging to the same species represented equivalent sampling units. Some surveys reported unknown Plasmodium species; these records were excluded from the analysis. From the vegan package (vs. 2.0), the functions “specaccum” and “specpool” were used to generate species accumulation curves and estimate total richness, respectively [45] (Fig. 2). We also used the Jackknife1 algorithm to provide the robust estimates for parasite species richness [46]. This algorithm relies on the presence of rare species to estimate total richness, and is a better estimation of presence–absence for records that have a negative binomial distribution.
Fig. 2.
Species accumulation curves for parasites of well-sampled primate hosts and groups. Although Asian Old World Monkeys may yet have undiscovered species in unsampled primate species, the sampled hosts are well covered (A), this includes several individual species theat have been well-sampled, such as Macaca fascicularis (B). Several ape parasite accumulation curves saturate, i.e. Hylobates lar (C), but potentially overlapping species identities among Pan troglodytes underscores that more sampling needs to be done to marry molecular and morphological surveys (D).
Sampling effort within each species was determined by estimating the number of undiscovered parasite species within a given host. Undiscovered malaria species in hosts were determined by subtracting observed Plasmodium species from expected species calculated with the Jackknife1 algorithm. Species accumulation curves could not be constructed for species without malaria parasites, so we calculated the minimum sample sizes needed to detect one species of malaria with 95% confidence. All hosts from a given region that were naturally infected with malaria were used to create regional species accumulation curves, which in turn were used to estimate the sample size needed to declare a primate species free from malaria with high confidence.
To estimate global NHP malaria species richness, NHP hosts were pooled based on the host range of their parasites. Apart from human malaria parasites, Plasmodium species are restricted to Africa, the Americas (South and Central America), and Asia. NHP host records were pooled according to region of origin and broad taxonomic group (i.e., ape, monkey, prosimian), and species accumulation curves and estimates were created with the resulting datasets.
Geographic analysis
For geographic distributions of NHP malaria parasites, we were limited to presence–absence at the country level. There were sufficient state-level data from Brazil and India to reduce the geographic scale for those countries. Survey data was merged with species ranges from the International Union for Conservation of Nature Red List spatial data [47], [48] to present a global map of sampled primate ranges and suspected malaria distributions based on sampling effort.
Forest cover data from 2000 was obtained from Hansen et al.[49]. This dataset can distinguish closed canopy forests from savannah ecosystems. Lastly, vector data was available for Asian primate malarias. Several mosquito species within the Anopheles Leucosphyrus group have been found with Plasmodium sporozoites of primate species, although they may not be the only vectors in the region. Vector distribution of the Anopheles Leucosphyrus group was first described by Colless in the 1950s [50]. Although nomenclature and species divisions have recently been overhauled [51], [52], [53], the distribution of this group of Anopheles is consistent with Colless' original work.
Results
Malaria sampling effort across primate taxa
The final dataset included 48,225 individual records derived from 117 primate species. Of these data, 43,437 records fit the criteria for inclusion in the species accumulation analysis. These data represent almost a third of the known primate species (117/376). Only seven publications examined species in Strepsirrhini, the suborder of primates including lemurs, galagos, and lorises, and we could not locate any surveys that examined tarsiers (suborder Haplorrhini, infraorder Tarsiiformes). Research has focused mostly on simian species (suborder Haplorrhini, infraorder Simiiformes), with a higher coverage of 37% (105/281). Of the primate species sampled, approximately half (n = 63) have been found naturally infected with one or more Plasmodium species in at least part of their range.
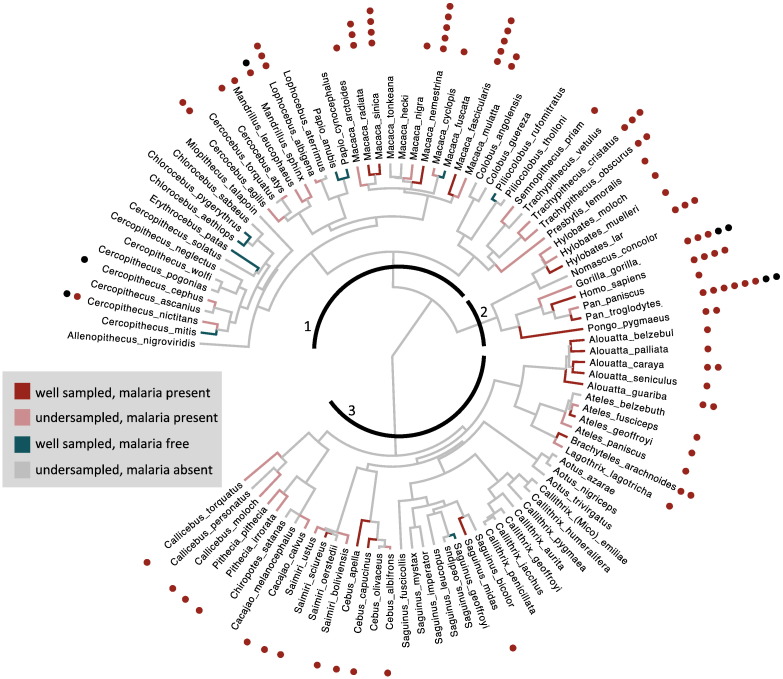
Sampling effort is heterogeneous across taxa. Each primate species was placed into one of four categories based on the difference between observed and expected Plasmodium species (Table A.3): infected with malaria and under-sampled, infected with malaria and well-sampled, uninfected with malaria but undersampled, and free from malaria (well-sampled and uninfected). Well-studied species that had malaria had less than 0.5 undiscovered Plasmodium parasites (N = 18, i.e. Fig. 2b,c) and are indicated with dark red edges on the primate phylogeny (without prosimians Fig. 3; full phylogeny Fig. A.1). In Latin America, we adapted these criteria to include monkeys positive for Plasmodium brasilianum with a sample size of 344 individuals, the minimum number of individuals needed to detect 2 species 95% of the time in Plasmodium simium/P. brasilianum hosts (Alouatta caraya, Alouatta belzebul, and Brachyteles arachnoides)
Fig. 3.
Simian phylogeny with malaria parasite sampling effort. Phylogeny truncated from 10K Trees [92] and includes Old World Monkeys (1), apes (2), and New World Monkeys (3). (Fig. A1 includes prosimians too). Dark red edges are well-sampled primates with malaria, light red edges are undersampled species that have been found with malaria parasites, and gray edges are species that have been sampled but have not been found with malaria parasites. Species that are not infected with Plasmodium in the wild are highlighted in blue (see also Fig. A.2). The number of morphologically described Plasmodium species in each primate are given by the red circles next to the species name, whereas molecularly described are represented by black circles. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Under-studied species are primate species that have greater than 0.5 undiscovered malaria parasite species (N = 45 i.e. Fig. 2d) or have not been sampled sufficiently to be malaria free (N = 45). To be considered malaria free in the wild (N = 9), a host species had to have a minimum number of individuals surveyed without a Plasmodium infection. The sample effort varied by region: 28 individuals in Africa, 108 individuals in Asia, and 421 individuals in America (Fig. A.2). Adequate sampling was determined as the minimum number of individuals necessary to detect one parasite species with 95% confidence in naturally infected primate species from a given region (Asia, Americas, Africa; Fig. A.2). The identity of Plasmodium species in a given host is given in supplementary tables (Tables A.4-A.6), but is simplified as circles next to the primate host on the phylogeny (Fig. 3).
There is a large variation in sampling effort among the Old World Monkeys, family Cercopithecidae. At least 30,603 Macaca individuals (subfamily Cercopithecinae) have been sampled for malaria, whereas only 479 leaf monkeys and langurs (subfamily Colobinae) have been sampled across South and Southeast Asia. Among Asian monkey species, there are at least seven malarias, ranging from host specific species (Plasmodium simiovale, infecting one macaque species, Macaca sinica) to generalist species (P. inui has at least 10 monkey hosts). Oftentimes, sympatric monkeys in Asia will share parasites, although molecular work suggests that lineages may actually be host-specific [56].
In Africa, there is a significantly greater sampling effort among Cercopithinae (n = 2371) as opposed to Colobinae members (n = 159). The reported parasite diversity in Africa is much lower than their counterparts in Asia monkeys. Only three species of Plasmodium have been morphologically described from African OWM hosts (Plasmodium gonderi[58], Plasmodium georgesi, and Plasmodium petersi[25], [26]), but species have recently been detected from all representative genera in the subfamily Cercopithinae. A malaria parasite, known as Plasmodium sp. DAJ-2004, has been isolated from, mandrills (Mandrillus sphinx) and some guenon species (Cercopithecus nictitans and Cercopithecus cephus) [27], [37]. More research on African monkey malaria that reflects the huge diversity of monkey hosts should reveal up to two novel species.
Recent discoveries of ape malarias in Africa are not unsurprising, considering historical work was mostly restricted to morphological techniques that relied on opportunistic samples from zoos and captive individuals. Nevertheless, these studies have altered our understanding of malaria parasite evolution and have greatly expanded the known species in Laverania subgenus (reviewed by [28], [59]). The diversity of African simian malarias includes species in the Laverania subgenus (shared with P. falciparum) and other parasites that are like human P. vivax, Plasmodium ovale, and Plasmodium malariae (Fig. 2D). The confusion over species distinctions in apes dates back a century [60], [61], [62], [63] and requires a thorough study using paired molecular and morphological data (see Text A.1 for more detail).
Asian apes have also been the subjects of several parasitological surveys, but the diversity appears to be much lower. Orangutans in Borneo (P. pygmaeus) have two malaria species, but their Sumatran sister species (Pongo abelli) have not yet been surveyed. There is not enough data on gibbons, but we suspect they will harbor an interesting diversity of malaria parasites. Four species (Plasmodium eylesi, Plasmodium hylobati, Plasmodium jefferyi, Plasmodium youngi) have been described, mostly through case reports. Less than 250 individuals have been checked in the wild and all but four were Hylobates lar (Fig. 2C). An individual Hylobates moloch and three Hylobates muelleri have been recorded with P. hylobati infections but this sampling effort represents only a small sample of gibbon species (3/18) and genera (1/6) across Asia.
Prosimians are also infected by malaria parasites, but estimates of the true diversity have large errors because of the uncertainty in the distribution and host range of their parasites. At least five species of Plasmodium have been identified morphologically in prosimians [38] and five species have been proposed based on molecular evaluation [35], [36]. It is unknown whether these species overlap, as there is no body of work that has successfully used both microscopy and molecular methods. Surveys have covered several areas of Madagascar, but do not yet offer extensive evaluations of the Lemuridae community, nor large sample sizes (N = 154 tested individuals, 15 positive for malaria; [35], [36], [38], [64]).
Estimating global species richness of non-human primate malarias
Host range varies greatly within primate malarias, which affects estimations of parasite species richness. Malaria parasites of monkeys have a much higher average host range (mean = 5.38 host species, mean = 2.54 host genera) than ape malaria parasites (mean = 1.47 host species, mean = 1.47 host genera), even when excluding the extreme generalist P. brasilianum (monkeys: mean = 3.42 host species, mean = 1.83 host genera). While we acknowledge these trends are influenced by sampling bias and a larger diversity of monkey hosts, the generalism of monkey malarias is consistent across continents. To make estimates of global species richness among sampled primates, we used extrapolated species estimates from the following groups: Asian Old World Monkeys, African Old World Monkeys, lemurs, New World Monkeys, and all species of apes (Table 1). We estimate there are three undiscovered malaria species among sampled primates, however the greatest uncertainty in these estimates are for lemurs, followed by African Old World Monkeys. New World Monkeys and Asian Old World Monkeys are relatively well-surveyed, although the host range of existing Plasmodium species is likely larger than is currently known.
Table 1.
Global primate malaria species estimates. Among primates sampled, we estimated total species richness in prosimians and monkeys, as these primate malarias are generalist and often shared among sympatric hosts. However, ape malarias tend to be more specific and species accumulation data were presented for four ape species that have sufficient sample sizes. All estimates are calculated with the Jackknife1 (see Analysis). We used molecularly described species in these group estimates for lemurs [35], [36] and African Old World Monkeys [37].
| Primates | Individuals sampled | Observed malaria species | Estimated malaria species | Standard error | Undescribed species |
|---|---|---|---|---|---|
| Prosimians | 154 | 7 | 8.9868 | 1.404 | 1.99 (0.58–3.39) |
| New World Monkeys | 8850 | 2 | 2.000 | – | 0 |
| African Old World Monkeys | 1782 | 5 | 5.9994 | 0.9994 | 1.00 (0–2.00) |
| Asian Old World Monkeys | 30,448 | 7 | 7.000 | – | 0 |
| Apes | |||||
| Hylobates lar | 225 | 3 | 3.000 | – | 0 |
| Gorilla gorilla | 165 | 5 | 4.000 | – | 0 |
| Pan paniscus | 42 | 1 | – | – | – |
| Pan troglodytes | 885 | 8 | 8.00 | – | 0 |
| Pongo pygmaeus | 209 | 2 | 2.00 | – | 0 |
Not all wild primates have natural Plasmodium infections. A large number of species sampled were never recovered with active malaria infections (N = 54 species). Sample sizes within each species ranged from 1–799 (mean = 66.63, SD = 141.75), but only a handful of species (N = 9) had sample sizes large enough to conclude that they are indeed free from malaria in the wild. Across Latin America, 364 owl monkeys from several species (Aotus), and 256 Callithrix species were examined for malaria without finding a single active infection, however these sample sizes are not sufficient to conclude they are not infected in the wild. Across East and Central Africa, 457 baboons (Papio species) have been checked for malaria, without success. Although there is still a chance that these species do have malaria in the wild, it is unlikely, given the sample size (Fig. A.2). No other African savanna monkey species has been found with Plasmodium infections, despite sampling populations of vervets (N = 180, Chlorocebus pygerythrus), patas (N = 66, Erythrocebus patas), and green monkeys (N = 252, Chlorocebus aethiops; N = 16, Chlorocebus sabaeus) in semi-arid regions around Africa.
Biogeography of primate malarias
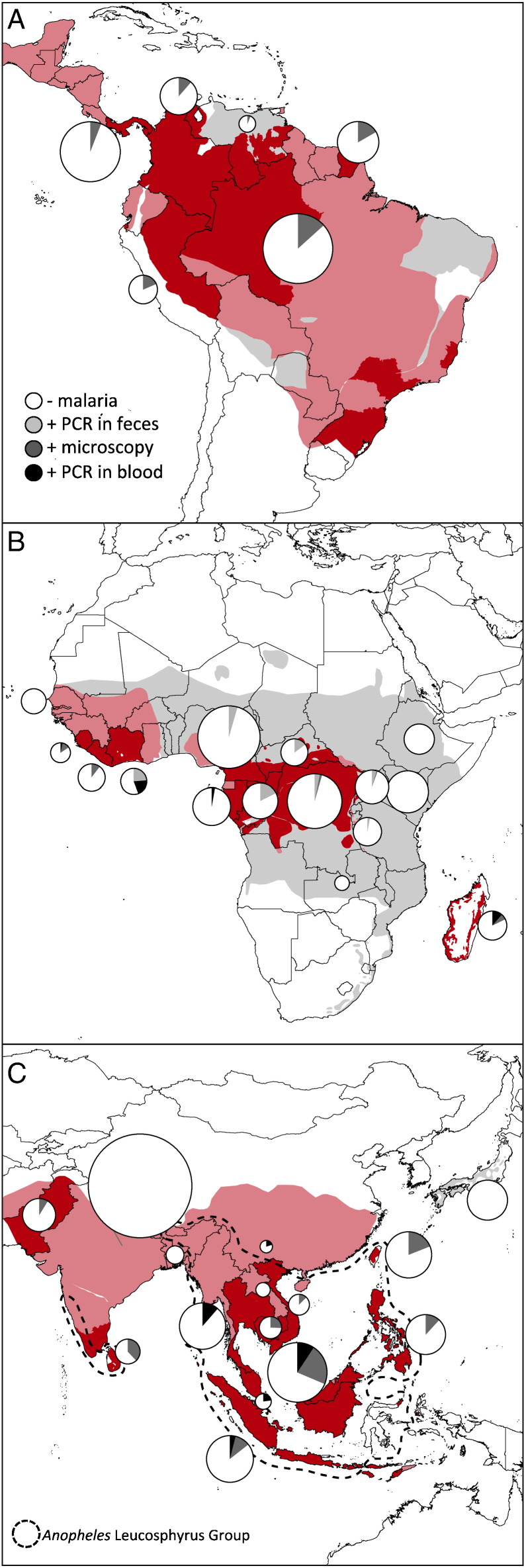
Geographically, the 138 published primate malaria surveys cover thirty-six countries in four continents (Asia, South America, North America, Africa). 1909 samples didn't have a geographic origin or the individuals represented were resampled from the existing literature, leaving 46,316 georeferenced samples. In addition to unequal sampling effort across taxa, sampling effort varied across region (Fig. 5A–C), but coverage was sufficiently high: samples originated from almost every country where primates are endemic.
Fig. 5.
Prevalence of primate malarias by region. Dark red shading indicates the intersection of countries where simian malaria has been recorded and the range of natural primate hosts, whereas light red areas include primates ranges that can be infected with malaria in other parts of their range but have not been found infected (or sampled) in the location (species ranges, [47]). Gray shading indicates ranges of primates that have been sampled for malaria but have never been found infected in any part of their range. All known surveys of simian malaria were pooled into countries for the Americas (A), Africa (B), and Asia (C). Non-white portions of pie charts represent proportion infected and reflect sampling method — microscopy (dark gray) or PCR-based methods in blood (black) or fecal (light gray) samples. Size of pie charts is scaled by the square root of the total sample size from the county. The range of Anopheles Leucosphyrus group of vectors is shown in Asia (C).
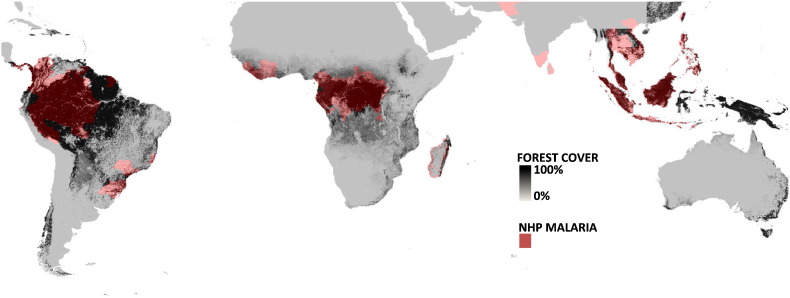
As with human malaria, distribution of malaria is most likely dependent on several local factors, such as microclimate, vector competence, and host densities. We do not attempt to make claims about relative prevalence in countries. Fig. 4 illustrates the NHP malaria presence as it relates to forest cover. Areas where NHP malaria has been found almost exclusively overlaps with regions of high forest cover. Focusing on regions (Fig. 5A–C), it is noticeable that sampling effort varies across the maximum possible extant of primate malarias. The total range of primates naturally infected with malarias (light red) is given as a contrast to the current knowledge of primate host range areas where only malaria parasites have been found (dark red). Lastly, regions where primate species are free from malaria is illustrated (gray).
Fig. 4.
Global distribution and prevalence of NHP malarias. Global distribution of the ranges of susceptible primate species to Plasmodium and the countries and states in which they have been found infected. The map is overlaid with the percentage forest cover, data on coverage is from Hansen et al. [49].
Although these maximum extent maps represent a coarse estimate of the distribution, we can begin to look at factors that influence non-human primate malaria distributions. The countries and states where primate malaria has been found is tightly linked with areas of high forest cover (Fig. 4A). Sampling efforts in eastern Brazil, East Africa, and central India are sufficient to conclude that primate malarias are absent from regions in these countries where forest cover is < 55%. Savannah regions have been sampled, in some cases extensively, and no Plasmodium species have been found in non-human primates.
Discussion
Thanks to the wealth of parasitological accounts of primates malaria parasites, we are able to estimate that there are approximately three undiscovered malaria parasite species within primates that have been screened based on sampling effort and parasite host ranges. Malaria is present in non-human primates throughout tropical regions around the globe, with the greatest diversity of NHP malaria in the Old World. Malaria in New World Monkeys is likely a much more recent event following human colonization of these regions. All NHP malarias described to date are closely linked to areas of high forest cover. The updated maps and hosts ranges developed from the meta-analysis provides context for recent research on NHP malarias and directs focus of future research to areas of geographic and taxonomic importance when evaluating the risk for zoonotic malaria.
More precise estimates of the time and location of the first cross-over of malaria parasites from non-human primates to humans could help give context for current and future zoonotic malaria spillovers. Twenty-six primate malaria species, each infecting between 1–29 primate hosts have been formally described. An additional eleven species are genetically characterized though awaiting morphological description. Ape malarias are typically specialist parasites, whereas malaria parasites infecting monkeys are transmitted across genera and even families. There may be specific lineages circulating within each primate host, as molecular data of P. inui in sympatric long-tailed (M. fascicularis) and pig-tailed (Macaca nemestrina) macaques suggest [56]. Sympatric ape species rarely share parasite communities despite utilizing similar habitats (i.e. Pan troglodytes and Gorilla gorilla or P. pygmaeus and H. lar). P. knowlesi is the only confirmed malaria that transmits to both monkeys and humans. This malaria circulates in the monkey species M. fascicularis, M. nemestrina, Presbytis femoralis, and Trachypithecus obscurus. More research on the transmission dynamics and mechanisms underlying these cross-species transmission events will be essential in understanding the evolution of primate malarias.
There are clear taxonomic biases in sampling effort. The well-sampled species belong to: hominids (great apes: Homo, Pan, Gorilla, Pongo), macaques (Macaca), baboons (Papio) and New World Monkeys (i.e. Ateles, Cebus, Saimiri, Aotus). This bias is likely a reflection of a taxonomic group's relatedness to humans (apes), concern for causing zoonosis (macaques), and easy access to samples (baboons). Interestingly, these patterns differ from sampling bias observed across other parasite taxa [69]. Focusing on Old World Monkeys, especially the Colobinae subfamily (i.e. Colobus, Procolobus, Presbytis, Pygathrix, Nasalis species), will immensely improve the phylogenetic and geographic coverage of primate malaria surveys. This area of the malaria phylogenetic tree is also very important-as the fossil split between Asian and African OWM is very often used for estimating divergence times of their malaria parasites. Understanding the true diversity of Old World Monkey malarias will vastly improve the understanding of the radiation of malarias outside of Africa. Increasing sampling effort of prosimians globally, but especially in Madagascar, would lend an essential missing piece in the diversity and evolution of primate malarias. Very little can be said about the true biodiversity of malarias in these primates, as the sampling is so sparse.
Many of the New World Monkeys are considered well-sampled (Fig. 3). This region has the lowest diversity of non-human primate malaria species, with only two species present: P. brasilianum and P. simium (Table A.5). P. brasilianum is closely related to human P. malariae and P. simium is closely related to Old World strains of human P. vivax[48]. Both may reflect a recent anthroponosis, resulting from cross-species transmission of the human parasite to New World Monkeys, but more genetic data is needed before the direction of transfer is substantiated [14], [54], [55]. P. brasilianum is the most generalist mammalian parasite and infects at least 29 species of Atelidae and Cebidae monkeys. It is found throughout the Amazon, Central America and Andean lowlands (Fig. 4). In contrast, only A. caraya, A. guariba, and B. arachnoides are infected with P. simium around Sao Paulo and Espirito Santo in Southern Brazil. Although the complete host range of these parasites is unknown, we can be confident that Latin America has been thoroughly sampled and it is unlikely that there are more than two non-human primate malaria species. More genetic work on these parasites would help determine the transmission dynamics and whether these species are pan-American or if there are unique lineages within certain hosts or regions. Genetic data will also help uncover the direction of host transfer of these species and how much transmission occurs with humans in the present day [14], [67], [68].
Uncovering additional NHP malaria species will impact our understanding of the evolution of malarias. Estimates for when malaria crossed-over from non-mammalian hosts into primates ranges from thirteen million years to seventy-eight million years ago, with several estimates in between [71], [72]. Inclusion of orangutan, gorilla, and lemur malarias has shifted the timeline of the evolution of primate malarias [9], [35], [73] — demonstrating the important effect of taxonomic bias in the reconstruction of phylogenies. Improving the genetic data and resources for NHP malarias will only aid our understanding of the evolution and extant diversity of all malaria parasites.
Our findings are not only relevant to the study of host–parasite evolution, but are also important for global health. Malaria parasites are an immense global health burden. Recent emergence of macaque malaria in Southeast Asia highlights the importance of understanding how often these cross-species transmission events occur and what determines the underlying distributions of potential zoonosis. Humans can be experimentally infected with several species of NHP malaria [11], but the extent to which malaria is shared among primates is still up for debate. In Latin America, primates potentially serve as reservoirs for human infection with P. malariae/P. brasilianum[67]. Although apes may not be a significant source of infection for humans in Africa [70], they can infect travelers [8]. Additional epidemiological and genetic data will improve our understanding of the role of these parasites in human cycles.
A true map of non-human primate malaria would require more thorough geographic sampling and knowledge of vector distributions, which is limited to South and Southeast Asia [57]. We included vector distribution, albeit at a very coarse scale and from only one region, as a foundation for future work. We are not the first to show that primates found outside the broadleaf evergreen forests where these mosquitoes breed are rarely infected with malaria (Fig. 5C, distribution from Colless [50]). Within forested regions of Central Africa, malaria is absent from some regions (i.e. south of the Congo River [10]), which could be due to vector distributions. Information on vector identity and distribution in Asia has been important in explaining the absence of Plasmodium in central India, and could be a key factor in the patchy distribution of Plasmodium in apes. Increasing entomological surveys will improve estimates of distributions and allow more accurate assessments of zoonotic risk.
Within the tropics, the distribution of primate malarias is closely linked with high forest cover Fig. 4. In Asia, macaque malaria parasites are restricted to tropical broadleaf evergreen rain forest, the habitat of the Anopheles Leucosphyrus group of vectors [50], [57], [75], [76], rather than the distribution of primate hosts. Although there are is less known about American and African vectors of primate malaria [77], [78], the distribution of surveys would lead us to believe that Anopheles vectors responsible for non-human primate malarias transmission are likely restricted to forested habitats. All other mammalian wildlife malaria (Plasmodium species) has been recorded in forest or woodland species (i.e. rodents, colugo, duiker) [79], [80]. Of course, human malaria transmission is not restricted to forested areas but the evolution of anthropophilic mosquitoes that have adapted to human-altered habitats could conceivably lead to malaria's emigration out of forests.
A move of primates into savannah ecosystems might have coincided with the evolution of Hepatocystis, or at least radiation, as Plasmodium species have never been found in savannah [81], [82], [83]. Baboons (Papio species) are not infected with Plasmodium parasites, despite their close relatedness to other susceptible Old World Primates and their ability to be infected in the laboratory. Troops of baboons instead have high infection rates of Hepatocystis kochi, a malaria-like Apicomplexan parasite [81], [84], [85]. Both Plasmodium and Hepatocystis replicate in the liver, while Plasmodium also replicates in red blood cells whereas Hepatocystis only infects blood cells as gametocytes (sexual stage) [4]. Vectors of Hepatocystis belong to the genus Culicoides, more commonly referred to as the biting midges. Their larvae grow and emerge from large lakes and other bodies of sunlit, still water. The home range and sleeping behavior of baboons may make them more susceptible to Culicoides vectors that carry Hepatocystis, than forest-dwelling great apes and monkeys of Central and West Africa that are exposed to Anopheles vectors. At least fifty species of Hepatocystis parasites infect other species of monkeys and bats that live in forests, but more arboreal species are less infected [27], [84], [85], [86], [87] which again suggests a role of vector preference. More investigations of Dipteran vectors and the parasites they harbor will lead to a better understanding of the community composition of haemosporidia and the distribution of parasite species that affect primates. The low prevalence of malaria in these vectors make this a non-trivial logistic challenge, although one that might be overcome by modern molecular methods.
While primate malarias have been relatively well-studied, knowledge of vectors is sparse and, in our opinion, is the most important limiting factor in understanding the distribution and host range of primate malaria species. The earlier evolutionary host jump of malaria from birds or squamates to mammals not only drastically changed the parasites erythrocytic environment (nucleated to anucleated red blood cells) but is also associated with a change in vector species. Mammalian Plasmodium are exclusively spread by Anopheles mosquito vectors, a distinction between other Plasmodium of birds and squamates that are spread by genera within Culicidae: Aedes, Culex, Culiseta, Armigeres, Wyeomyia, Psorphora, Mansonia, and some Anopheles). Because Plasmodium did not arise within primates, understanding the distribution of its vectors will provide vital future insights into the distribution and diversity of NHP malarias. Understanding the behavior, diversity and distribution of vectors will improve the knowledge base of evolution of multi-host pathogens and human malaria.
Determining species distinctions has been an active subject of research for malariologists historically. The concept of species within Plasmodium is now further complicated by a disconnect between species defined morphologically and recent molecular discoveries. Novel Plasmodium species in apes, monkeys, and lemurs have only been characterized molecularly and lack a morphological description to compare to historical accounts. Equally, many rare or hard to access Plasmodium species have not been molecularly characterized (i.e. P. jefferyi, P. georgesi, Plasmodium giradi). Therefore, we caution readers that some molecular species may actually represent previous morphologically described species, i.e. P. vivax in apes may reflect previous descriptions of Plasmodium schwetzi[8]. We have explicitly labeled species that have only been identified with one method and have only included parasites that have been molecularly described by more than one group in the full species list (Table A.1). Especially in older reports, it is impossible for us to know whether authors have correctly identified Plasmodium species. Excluding these uncertain surveys increases our estimates of species richness but does not drastically change our understanding of the distribution. In some of the more isolated areas (i.e. islands in Southeast Asia), predictions of the distribution of malaria parasites would greatly benefit from broader molecular surveys to uncover cryptic infections and those present at low parasitemias.
The analysis is not without limitations: primate samples are difficult to obtain and non-invasive samples struggle from reduced sensitivity of detection. This is complicated by chronic malaria infections, common in adult primates, that have very low parasitemia and make detection difficult [88]. Splenectomizing can increase the chance of finding chronically low and mixed species infections [89], while the advent of molecular methods is improving detection and allowing for ethical evaluation of this cryptic diversity. For example, it is now known that apes can be infected with multiple lineages of Plasmodium that often include several strains co-occurring within a population [9]. Advancements based on using PCR to detect malaria parasites have yielded much higher prevalence of co-infection than previously observed, suggesting a high force of infection and repeated exposures and/or long lasting infections that can only be detected via PCR [9], [90]. Future surveys can build upon existing methods for the detection of malaria parasites from fecal samples; but because sensitivity is very low, conclusions about population prevalence are difficult, if not impossible. It will be important in the coming years to develop a standard method of evaluation to properly discern malaria species using molecular methods.
Research on primate malarias is rapidly progressing with the advent of non-invasive molecular methods. It is an essential field of research, both in terms of the evolutionary ecology of malaria but also the public health concerns of zoonotic malarias. While there is little evidence that apes are a large-scale reservoirs for zoonotic Laverania infections in humans, an ape-related isolate of P. vivax has been recorded in a traveler returning from Central African Republic [8]. Spillover of monkey malaria in Southeast Asia demonstrates that primate malarias have the ability to cause widespread public health concerns, as zoonotic hotspots report hundreds of human P. knowlesi (reviewed most recently by Antinori et al.[91]). The mechanisms for spillover in the latter are inevitably complex and will only benefit from a more thorough understanding of primate malarias globally.
The following are the supplementary data related to this article.
Details of primate malaria survey. This word document includes all references for the parasite surveys and includes details of the methodology used in this meta-analysis. It includes interpretation of some of the studies that did not have data on number of individuals surveyed and number of individuals infected and how these were recorded in the database. It also includes notes of some unusual and potentially misleading case reports.
Full primate phylogeny with sampling efforts. Phylogeny of all primates that extends the main text phylogeny by including prosimians. As in the main text, well-sampled primates are denoted with red branches, undersampled species are golden, and species free from malaria in the wild are colored green. The number of Plasmodium species that have been recorded in each primate host is given by red circles (morphologically described) and gray circles (molecularly characterized, pending taxonomic description).
Minimum sampling effort to detect Plasmodium infection by region. Records from all primate species from a region (a. Africa, b. Asia, c. Americas) that were infected in the wild were combined to create species accumulation curves for each region. The minimum sampling effort necessary to detect one malaria species with 95% confidence was considered the cut off for declaring a primate species free of malaria in the wild.
Extant simian malarias and descriptions. This table summarizes all the known species of simian malarias, including recently identified ape malarias in Central Africa that have not yet been described morphologically. Nomenclature of parasite species follows names used by the main texts in primate malaria and a recent review [4], [21], [28]. Primate host species are updated to the current binomial using Wilson and Reed [39]. Periodicity refers to the length of one erythrocytic cycle in the blood (quotidian — 24 h, tertian — 48 h, quartan — 72 h).
Historical surveys of Plasmodium in simian species. A csv file with all the summaries for all known published cases of Plasmodium in primate species. Data includes records include individuals that were surveyed for Plasmodium but did not have any species. The full reference list can be found in the A.1 Text.
Parasite species estimates for each primate species with parasite data. A csv file with all the observed number of Plasmodium species and expected number of species estimated using Jackknife1. We were unable to estimate species richness for two reasons: 1) only one parasite species was found in the host and 2) no malaria species have been found in any individuals. Refer to the main text for how we determined that a host is likely not infected with malaria parasites.
Table A.4. African primate hosts, countries positive, citations for first records. A table that includes citations for each first record of a Plasmodium species in a new primate host and/or country in Africa.
Table A.5. Latin American primate hosts, countries positive, citations for first records. A table that includes citations for each first record of a Plasmodium species in a new primate host and/or country in the Americas.
Table A.6. Asian primate hosts, countries positive, citations for first records. A table that includes citations for each first record of a Plasmodium species in a new primate host and/or country in Asia.
Acknowledgments
The authors are grateful to J.K. Peterson, C. Standley, A. van Leeuwen, A. C. Staver, and three anonymous reviewers for helpful comments and discussions on versions of the manuscript. We would also like to extend out gratitude to researchers that were willing to provide us with additional data to complete missing sections of the database. CLF was funded by the National Defense Science and Engineering Graduate Fellowship and a National Geographic Young Explorer Grant (#9125-12).
References
- 1.World Health O . World Health Organization; Geneva: 2014. World Malaria Report 2014 — Full Report; pp. 1–242. [Google Scholar]
- 2.Weatherall D.J., Clegg J.B. Inherited haemoglobin disorders: an increasing global health problem. Bull. World Health Organ. 2001;79:704–712. [PMC free article] [PubMed] [Google Scholar]
- 3.Patrinos G.P. Improvements in the HbVar database of human hemoglobin variants and thalassemia mutations for population and sequence variation studies. Nucleic Acids Res. 2004;32:537D–541D. doi: 10.1093/nar/gkh006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garnham P.C.C. Blackwell Scientific; Oxford, UK: 1966. Malaria Parasites and Other Haemosporidia. [Google Scholar]
- 5.Telford S.R. CRC Press; 2009. Hemoparasites of the Reptilia. [Google Scholar]
- 6.Valkiūnas G. CRC Press; Boca Raton, FL: 2005. Avian Malaria Parasites and Other Haemosporidia. [Google Scholar]
- 7.Prugnolle F., Durand P., Neel C., Ollomo B., Ayala F., Arnathau C. African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. PNAS. 2010;107:1458–1463. doi: 10.1073/pnas.0914440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prugnolle F., Rougeron V., Becquart P., Berry A., Makanga B., Rahola N. Diversity, host switching and evolution of Plasmodium vivax infecting African great apes. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8123–8128. doi: 10.1073/pnas.1306004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W., Li Y., Learn G.H., Rudicell R.S., Robertson J.D., Keele B.F. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature. 2010;467:420–425. doi: 10.1038/nature09442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W., Li Y., Shaw K.S., Learn G.H., Plenderleith L.J., Malenke J.A. African origin of the malaria parasite Plasmodium vivax. Nat. Commun. 2014;5:1–10. doi: 10.1038/ncomms4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baird J. Malaria zoonoses. Travel Med. Infect. Dis. 2009;7:269–277. doi: 10.1016/j.tmaid.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Cox-Singh J., Davis T., Lee K., Shamsul S., Matusop A., Ratnam S. Plasmodium knowlesi malaria in humans is widely distributed and potentially life-threatening. Clin. Infect. Dis. 2008;46:165. doi: 10.1086/524888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duval L., Arley F. Ape Plasmodium parasites as a source of human outbreaks. Clin. Microbiol. Infect. 2012;18:528–532. doi: 10.1111/j.1469-0691.2012.03825.x. [DOI] [PubMed] [Google Scholar]
- 14.Guimaraes L., Bajay M., Wunderlich G., Bueno M., Rohe F., Catao-Dias J. The genetic diversity of Plasmodium malariae and Plasmodium brasilianum from human, simian and mosquito hosts in Brazil. Acta Trop. 2012;124:27–32. doi: 10.1016/j.actatropica.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 15.Singh B., Sung L.K., Matusop A., Radhakrishnan A., Shamsul S.S.G., Cox-Singh J. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–1024. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 16.William T., Rahman H.A., Jelip J., Ibrahim M.Y., Menon J., Grigg M.J. Increasing incidence of Plasmodium knowlesi malaria following control of P. falciparum and P. vivax malaria in Sabah, Malaysia. PLoS Negl. Trop. Dis. 2013;7:e2026. doi: 10.1371/journal.pntd.0002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vythilingam I., Lim Y.A.L., Venugopalan B., Ngui R., Leong C., Wong M. Plasmodium knowlesi malaria an emerging public health problem in Hulu Selangor, Selangor, Malaysia (2009–2013): epidemiologic and entomologic analysis. Parasites Vectors. 2014;7:436. doi: 10.1186/1756-3305-7-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valkiunas G., Anwar A., Atkinson C., Greiner E., Paperna I., Peirce M. What distinguishes malaria parasites from other pigmented haemosporidians? Trends Parasitol. 2005;21:357–358. doi: 10.1016/j.pt.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Halberstaedter L., Von Prowazek S. Untersuchunger uiberdie malariaparasiten der affen. Arb. Geshundhtsamt. 1907;26:37–43. [Google Scholar]
- 20.Chin W., Contacos P., Coatney G., Kimball H. A naturally acquired quotidian-type malaria in man transferable to monkeys. Science. 1965;149:865. doi: 10.1126/science.149.3686.865. [DOI] [PubMed] [Google Scholar]
- 21.Coatney G.R., Collins W.E., Warren M., Contacos P.G. US Government Printing Office; Washington DC: 1971. The Primate Malarias. [Google Scholar]
- 22.Ollomo B., Durand P., Prugnolle F., Douzery E., Arnathau C., Nkoghe D. A new malaria agent in African hominids. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rich S.M., Leendertz F.H., Xu G., LeBreton M., Djoko C.F., Aminake M.N. The origin of malignant malaria. Proc. Natl. Acad. Sci. 2009;106:14902–14907. doi: 10.1073/pnas.0907740106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duval L., Nerrienet E., Rousset D., Mba S.A.S., Houze S., Fourment M. Chimpanzee malaria parasites related to Plasmodium ovale in Africa. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poirriez J., Del-Cas E., Landau I. Further description of blood stages of Plasmodium petersi from Cercocebus albigena monkey. Folia Parasitol. 1994;41:168–172. [PubMed] [Google Scholar]
- 26.Poirriez J., Baccam D., Dei-Cas E., Brogan T., Landau I. Description de Plasmodium petersi n. sp. et Plasmodium georgesi n. sp., parasites d'un Cercocebus albigena originale de Republique Centrafricaine. Ann. Parasitol. Hum. Comp. 1993;68:203–210. [Google Scholar]
- 27.Prugnolle F., Ollomo B., Durand P., Yalcindag E., Arnathau C., Elguero E. African monkeys are infected by Plasmodium falciparum nonhuman primate-specific strains. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11948–11953. doi: 10.1073/pnas.1109368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rayner J.C., Liu W., Peeters M., Sharp P.M. A plethora of Plasmodium species in wild apes: a source of human infection? Trends Parasitol. 2011;27:222–229. doi: 10.1016/j.pt.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinton J.A., Mulligan H.W. A critical review of the literature relating to the identification of the malarial parasites recorded from monkeys of the families Circopithecidae and Colobidae. Rec. Malar. Surv. India. 1933;3:357–380. [Google Scholar]
- 32.Sinton J.A., Mulligan H.W. Mixed infections in the malaria of the lower monkeys. II. The probable occurrence of mixed infections in some of the older records of monkey malaria. Rec. Malar. Surv. India. 1933;3:769–808. [Google Scholar]
- 33.Aberle S.D. National Research Council: Division of Medical Sciences; 1945. Primate Malaria; pp. 1–193. [Google Scholar]
- 34.Valkiūnas G., Ashford R.W., Bensch S., Killick-Kendrick R., Perkins S. A cautionary note concerning Plasmodium in apes. Trends Parasitol. 2011;27:231–232. doi: 10.1016/j.pt.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Pacheco M.A., Battistuzzi F.U., Junge R.E., Cornejo O.E., Williams C.V., Landau I. Timing the origin of human malarias: the lemur puzzle. BMC Evol. Biol. 2011;11:299. doi: 10.1186/1471-2148-11-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duval L., Fourment M., Nerrienet E., Rousset D., Sadeuh S., Goodman S. African apes as reservoirs of Plasmodium falciparum and the origin and diversification of the Laverania subgenus. Proc. Natl. Acad. Sci. 2010;107:10561–10566. doi: 10.1073/pnas.1005435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Escalante A.A., Freeland D.E., Collins W.E., Lal A.A. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc. Natl. Acad. Sci. 1998;95:8124–8129. doi: 10.1073/pnas.95.14.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landau I., Lepers J.P., Rabetafika L., Baccam D., Peters W., Coulanges P. Plasmodies de Lemuriens Malgaches. Ann. Parasitol. Hum. Comp. 1989;64:171–184. doi: 10.1051/parasite/1989643171. [DOI] [PubMed] [Google Scholar]
- 39.Mammal Species of the World . John Hopkins University Press; 2005. A Taxonomic and Geographic Reference. [Google Scholar]
- 40.Jones K.E., Bielby J., Cardillo M., Fritz S.A., O'Dell J., Orme C.D.L. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology. 2009;90:2648. [Google Scholar]
- 41.Pacheco M.A., Cranfield M., Cameron K., Escalante A.A. Malarial parasite diversity in chimpanzees: the value of comparative approaches to ascertain the evolution of Plasmodium falciparum antigens. Malar. J. 2013;12:1. doi: 10.1186/1475-2875-12-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siregar J., Faust C., Murdiyarso L., Rosmanah L., Saepuloh U., Dobson A. Non-invasive surveillance for Plasmodium in reservoir macaque species. Malar. J. 2015;14:404. doi: 10.1186/s12936-015-0857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Team R.C. R Foundation for Statistical Computing; Vienna, Austria: 2013. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 45.Oksanen J., Blanchet F.G., Kindt R., Legendre P., Minchin P.R., O'Hara R.B. 2015. vegan: Community Ecology Package; pp. 1–12. [Google Scholar]
- 46.Walther B.A., Morand S. Comparative performance of species richness estimation methods. Parasitology. 1998;116(Pt 4):395–405. doi: 10.1017/s0031182097002230. [DOI] [PubMed] [Google Scholar]
- 47.IUCN . 2012. IUCN Red List of Threatened Species. Version 2012. http://www.iucnredlist.org. Downloaded on Oct 23 2012. [Google Scholar]
- 48.Li J., Collins W., Wirtz R., Rathore D., Lal A.A., McCutchan T. Geographic subdivision of the range of the malaria parasite Plasmodium vivax. Emerg. Infect. Dis. 2001;7:35–42. doi: 10.3201/eid0701.010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen M.C., Potapov P.V., Moore R., Hancher M., Turubanova S.A., Tyukavina A. High-resolution global maps of 21st-century forest cover change. Science. 2013;342:850–853. doi: 10.1126/science.1244693. [DOI] [PubMed] [Google Scholar]
- 50.Colless D.H. The Anopheles Leucosphyrus group. Trans. R. Ent. Soc. Lond. 1956;108:37–116. [Google Scholar]
- 51.Sallum M.A.M., Foster P.G., Li C., Sithiprasasna R., Wilkerson R.C. Phylogeny of the Leucosphyrus group of Anopheles(Cellia) (Diptera: Culicidae) based on mitochondrial gene sequences. Ann. Entomol. Soc. Am. 2007;100:27–35. [Google Scholar]
- 52.Sallum M., Peyton E., Harrison B., Wilkerson R. Revision of the Leucosphyrus group of Anopheles (Cellia)(Diptera, Culicidae) Rev. Bras. Entomol. 2005;49:1–152. [Google Scholar]
- 53.Sallum M.A.M., Peyton E.L., Wilkerson R.C. Six new species of the Anopheles Leucosphyrus group, reinterpretation of An. elegans and vector implications. Med. Vet. Entomol. 2005;19:158–199. doi: 10.1111/j.0269-283X.2005.00551.x. [DOI] [PubMed] [Google Scholar]
- 54.Tazi L., Ayala F.J. Unresolved direction of host transfer of Plasmodium vivax v. P. simium and P. malariae v. P. brasilianum. Infect. Genet. Evol. 2011;11:209–221. doi: 10.1016/j.meegid.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 55.Fandeur T., Volney B., Peneau C., De Thoisy B. Monkeys of the rainforest in French Guiana are natural reservoirs for P. brasilianum/P. malariae malaria. Parasitology. 2000;120:11–21. doi: 10.1017/s0031182099005168. [DOI] [PubMed] [Google Scholar]
- 56.Putaporntip C., Jongwutiwes S., Thongaree S., Seethamchai S., Grynberg P., Hughes A.L. Ecology of malaria parasites infecting Southeast Asian macaques: evidence from cytochrome b sequences. Mol. Ecol. 2010;19:3466–3476. doi: 10.1111/j.1365-294X.2010.04756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warren M.W., Wharton R.H. The vectors of simian malaria: identity, biology, and geographical distribution. J. Parasitol. 1963:892–904. [PubMed] [Google Scholar]
- 58.Sinton J.A., Mulligan H.W. A critical review of the literature relating to the identification of the malarial parasites recorded from monkeys of the families Cercopithecidae and Colobidae. Rec. Malar. Surv. India. 1933;3:381–443. [Google Scholar]
- 59.Prugnolle F., Durand P., Ollomo B., Duval L., Ariey F., Arnathau C. A fresh look at the origin of Plasmodium falciparum, the most malignant malaria agent. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reichenow E. Ueber das Vorkommen der Malariaparasiten des Menschen bei den afrikanischen Menschenaffen. Centralbl f Bakt. 1920;85:207–216. [Google Scholar]
- 61.Adler S. Malaria in chimpanzees in Sierra Leone. Ann. Trop. Med. Parasitol. 1923;17:13–18. [Google Scholar]
- 62.Schwetz J. Sur les parasites malariens (Plasmodium) des Singes superieurs (Anthropoides Africains. Soc. Belge Biol. 1922:710–711. [Google Scholar]
- 63.Schwetz J. 1922. Sur une infection malarienne triple d'un chimpanze; pp. 105–109. [Google Scholar]
- 64.Garnham P.C., Uilenberg G. Malaria parasites of lemurs. Ann. Parasitol. Hum. Comp. 1975;50:409–418. doi: 10.1051/parasite/1975504409. [DOI] [PubMed] [Google Scholar]
- 67.Lalremruata A., Magris M., Vivas-Martínez S., Koehler M., Esen M., Kempaiah P. Natural infection of Plasmodium brasilianum in humans: man and monkey share quartan malaria parasites in the Venezuelan Amazon. EBioMedicine. 2015;2:1186–1192. doi: 10.1016/j.ebiom.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Volney B., Pouliquen J.F., De Thoisy B., Fandeur T. A sero-epidemiological study of malaria in human and monkey populations in French Guiana. Acta Trop. 2002;82:11–23. doi: 10.1016/s0001-706x(02)00036-0. [DOI] [PubMed] [Google Scholar]
- 69.Cooper N., Nunn C.L. Identifying future zoonotic disease threats: where are the gaps in our understanding of primate infectious diseases? Evol. Med. Publ. Health. 2013;2013:27–36. doi: 10.1093/emph/eot001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sundararaman S.A., Liu W., Keele B.F., Learn G.H., Bittinger K., Mouacha F. Plasmodium falciparum-like parasites infecting wild apes in southern Cameroon do not represent a recurrent source of human malaria. Proc. Natl. Acad. Sci. 2013;110:7020–7025. doi: 10.1073/pnas.1305201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ricklefs R.E., Outlaw D.C. A molecular clock for malaria parasites. Science. 2010;329:226–229. doi: 10.1126/science.1188954. [DOI] [PubMed] [Google Scholar]
- 72.Silva J.C., Egan A., Arze C., Spouge J.L., Harris D.G. A new method for estimating species age supports the co-existence of malaria parasites and their mammalian hosts. Mol. Biol. Evol. 2015;32:1354–1364. doi: 10.1093/molbev/msv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pacheco M.A., Reid M.J.C., Schillaci M.A., Lowenberger C.A., Galdikas B.M.F., Jones-Engel L. The origin of malarial parasites in orangutans. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0034990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fooden J. Malaria in macaques. Int. J. Primatol. 1994;15:573–596. [Google Scholar]
- 76.Garnham P.C.C. Distribution of malaria parasites in primates, insectivores, and bats. Symp. Zool. Soc. Lond. 1973;33:377–404. [Google Scholar]
- 77.Paupy C., Makanga B., Ollomo B., Rahola N., Durand P., Magnus J. Anopheles moucheti and Anopheles vinckei are candidate vectors of ape Plasmodium parasites, including Plasmodium praefalciparum in Gabon. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0057294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Deane L., Neto J., Deane M.P., Silveira I. Anopheles (Kerteszia) cruzi, a natural vector of the monkey malaria parasites, Plasmodium simium and Plasmodium brasilianum. Trans. R. Soc. Trop. Med. Hyg. 2012;64:647–648. doi: 10.1016/0035-9203(70)90088-x. [DOI] [PubMed] [Google Scholar]
- 79.Garnham P.C.C. Malaria parasites in mammals excluding man. Adv. Parasitol. 1967;5:139–204. doi: 10.1016/s0065-308x(08)60377-2. [DOI] [PubMed] [Google Scholar]
- 80.Keymer I.F. Investigations on the duiker (Sylvicapra grimmia) and its blood protozoa in Central Africa. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1969;255:33–108. [Google Scholar]
- 81.Tung J., Primus A., Bouley A.J., Severson T.F., Alberts S.C., Wray G.A. Evolution of a malaria resistance gene in wild primates. Nature. 2009;460:388–391. doi: 10.1038/nature08149. [DOI] [PubMed] [Google Scholar]
- 82.Pourrut X., Robert V. Absence d'infection plasmodiale chez les singes sauvages du Nord-Sénégal? Bull. Soc. Pathol. Exot. 2009;102:17–18. doi: 10.3185/pathexo3185. [DOI] [PubMed] [Google Scholar]
- 83.Garnham P.C.C., Heisch R.B., Minter D.M. The vector of Hepatocystis (= Plasmodium) kochi; the successful conclusion of observations in many parts of tropical Africa. Trans. R. Soc. Trop. Med. Hyg. 1961;55:497–502. doi: 10.1016/0035-9203(61)90071-2. [DOI] [PubMed] [Google Scholar]
- 84.Keymer I. Blood protozoa of insectivores, bats and primates in Central Africa. J. Zool. 1971;163:421–441. [Google Scholar]
- 85.Kuntz R.E., Myers B.J. Parasites of the Kenya baboon: Arthropods, blood protozoa and helminths (Kenya, 1966) Primates. 1967;8:75–82. [Google Scholar]
- 86.Olival K.J., Stiner E.O., Perkins S.L. Detection of Hepatocystis sp. in Southeast Asian flying foxes (Pteropodidae) using microscopic and molecular methods. J. Parasitol. 2007;93:1538–1540. doi: 10.1645/GE-1208.1. [DOI] [PubMed] [Google Scholar]
- 87.Seethamchai S., Putaporntip C., Malaivijitnond S., Cui L., Jongwutiwes S. Malaria and Hepatocystis species in wild macaques, southern Thailand. Am.J.Trop. Med. Hyg. 2008;78:646–653. [PubMed] [Google Scholar]
- 88.Krief S., Escalante A.A., Pacheco M.A., Mugisha L., Andre C., Halbwax M. On the diversity of malaria parasites in African apes and the origin of Plasmodium falciparum from bonobos. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dissanaike A.S. Simian malaria parasites of Ceylon. Bull. World Health Organ. 1965;32:593–597. [PMC free article] [PubMed] [Google Scholar]
- 90.Lee K.-S., Divis P.C.S., Zakaria S.K., Matusop A., Julin R.A., Conway D.J. Plasmodium knowlesi: reservoir hosts and tracking the emergence in humans and macaques. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Antinori S., Galimberti L., Milazzo L., Corbellino M. Plasmodium knowlesi: the emerging zoonotic malaria parasite. Acta Trop. 2013;125:191–201. doi: 10.1016/j.actatropica.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 92.Arnold C., Matthews L.J., Nunn C.L. The 10kTrees website: a new online resource for primate phylogeny. Evol. Anthropol. Issues News Rev. 2010;19:114–118. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of primate malaria survey. This word document includes all references for the parasite surveys and includes details of the methodology used in this meta-analysis. It includes interpretation of some of the studies that did not have data on number of individuals surveyed and number of individuals infected and how these were recorded in the database. It also includes notes of some unusual and potentially misleading case reports.
Full primate phylogeny with sampling efforts. Phylogeny of all primates that extends the main text phylogeny by including prosimians. As in the main text, well-sampled primates are denoted with red branches, undersampled species are golden, and species free from malaria in the wild are colored green. The number of Plasmodium species that have been recorded in each primate host is given by red circles (morphologically described) and gray circles (molecularly characterized, pending taxonomic description).
Minimum sampling effort to detect Plasmodium infection by region. Records from all primate species from a region (a. Africa, b. Asia, c. Americas) that were infected in the wild were combined to create species accumulation curves for each region. The minimum sampling effort necessary to detect one malaria species with 95% confidence was considered the cut off for declaring a primate species free of malaria in the wild.
Extant simian malarias and descriptions. This table summarizes all the known species of simian malarias, including recently identified ape malarias in Central Africa that have not yet been described morphologically. Nomenclature of parasite species follows names used by the main texts in primate malaria and a recent review [4], [21], [28]. Primate host species are updated to the current binomial using Wilson and Reed [39]. Periodicity refers to the length of one erythrocytic cycle in the blood (quotidian — 24 h, tertian — 48 h, quartan — 72 h).
Historical surveys of Plasmodium in simian species. A csv file with all the summaries for all known published cases of Plasmodium in primate species. Data includes records include individuals that were surveyed for Plasmodium but did not have any species. The full reference list can be found in the A.1 Text.
Parasite species estimates for each primate species with parasite data. A csv file with all the observed number of Plasmodium species and expected number of species estimated using Jackknife1. We were unable to estimate species richness for two reasons: 1) only one parasite species was found in the host and 2) no malaria species have been found in any individuals. Refer to the main text for how we determined that a host is likely not infected with malaria parasites.
Table A.4. African primate hosts, countries positive, citations for first records. A table that includes citations for each first record of a Plasmodium species in a new primate host and/or country in Africa.
Table A.5. Latin American primate hosts, countries positive, citations for first records. A table that includes citations for each first record of a Plasmodium species in a new primate host and/or country in the Americas.
Table A.6. Asian primate hosts, countries positive, citations for first records. A table that includes citations for each first record of a Plasmodium species in a new primate host and/or country in Asia.