Abstract
The product of the vlf-1 (very late factor 1) gene is required for expression of very late genes during the final phase of infection. To determine whether VLF-1 functions as a transcriptional activator, VLF-1 was overexpressed and purified by affinity and cation exchange chromatography. The addition of purified protein to transcription assays containing baculovirus RNA polymerase stimulated transcription of the very late polyhedrin promoter but not the late 39k promoter. Furthermore, construction and analysis of chimeric templates identified sequences within the polyhedrin promoter that were necessary for enhancement.
Baculoviruses are unusual in having two distinct phases of late gene expression. The standard late genes encode virion structural proteins and are expressed following the onset of DNA replication. The very late genes, which encode proteins required for the formation of occluded virions, are maximally expressed after the formation of budded virus. A viral gene required for the burst of very late gene transcription, called very late expression factor 1 (vlf-1), was identified by marker rescue of the temperature-sensitive mutant virus tsB837 (4). This mutant was viable at the nonpermissive temperature but was defective in the production of occluded virus. The failure of tsB837 to produce occluded virions was due to a significant reduction in the level of polyhedrin (polh) mRNA, which codes for the major occlusion body protein. Transcription and expression of p10, another highly expressed very late gene, was also reduced at nonpermissive temperature, although to a lesser extent than polyhedrin.
The specific stimulation of very late promoters by VLF-1 has also been observed in transient expression assays (10, 14). The addition of plasmids encoding VLF-1 stimulated expression from the two very late promoters, polh and p10, but had little or no effect on two late promoters, vp39 and p6.9. Late and very late promoters differ primarily by the presence or absence of a burst sequence, a sequence downstream of the transcriptional start site that is 90% A+T (3, 5, 7, 9). The burst sequence of a very late promoter is needed in order to respond to stimulation by VLF-1 in transient-expression assays, and gel mobility shift and footprinting assays also indicated that VLF-1 specifically bound to the burst sequence in vitro (12).
The mechanism by which VLF-1 stimulates very late genes, however, is unknown. When VLF-1 was originally described, the authors who described it noted sequence similarity with the integrase family of tyrosine recombinases (4). While the homology is intriguing, it lends few clues as to the function of VLF-1. The other members of this family are primarily involved in DNA integration and DNA replication, and none of them are known to act as transcription factors. Furthermore, all integrases and tyrosine recombinases are characterized by an essential active-site tyrosine which forms a covalent protein-DNA intermediate (1, 6). VLF-1 has this conserved tyrosine but it is not required for VLF-1 to stimulate transcription (14). Thus, it seems unlikely that the mechanism of VLF-1 function in transcription is related to integration or recombination.
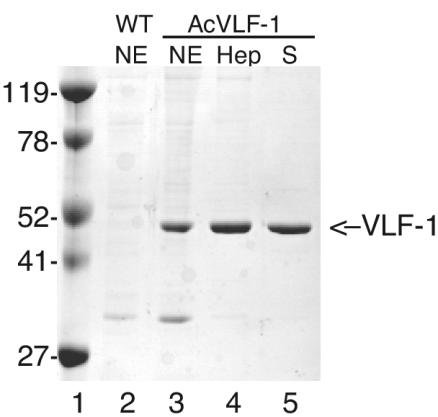
To characterize the mechanism of VLF-1 function, we purified VLF-1 by using a baculovirus overexpression system in order to examine its activity in transcription reactions containing purified baculovirus RNA polymerase (RNApol). A recombinant virus, called AcVLF-1, containing the vlf-1 gene under the control of the polyhedrin promoter was constructed according to standard procedures (8). A 1,685-bp SpeI-SspI fragment of an AcNPV HindIII-B genomic clone was inserted into the XbaI and SmaI sites of pVL1393. Spodoptera frugiperda (Sf9) cells were cotransfected with 5 μg of the resulting transfer vector, pVLF-1, and 2 μg of BakPak viral DNA previously digested with Bsu36I. Individual plaques were further purified, and the correct construction of AcVLF1 was verified by restriction digestion and sequence analysis. Nuclear extracts prepared from AcVLF1-infected cells at 60 h postinfection contained a highly expressed protein compared to extracts infected with wild-type virus (Fig. 1). This protein migrated at the position expected for VLF-1. Crude extracts were precipitated with saturated ammonium sulfate and dialyzed for loading onto a heparin affinity column. VLF-1 was purified by affinity chromatography on a heparin agarose column by using column buffers as previously described for the baculovirus protein PP31 (2). Fractions were monitored by Coomassie blue staining of sodium dodecyl sulfate (SDS)-polyacrylamide gels. Peak fractions were pooled, diluted, and loaded onto a Mono S cation exchange column. Fractions containing purified VLF-1 were pooled and stored in 50 mM HEPES (pH 7.2)-400 mM KCl-50% glycerol-1 mM dithiothreitol-0.1 mM EDTA. The yield of purified VLF-1 ranged from 50 to 200 μg per liter of infected cells. The purified protein was subjected to peptide mass fingerprinting. Masses from 15 tryptic peptides were fit to the masses of predicted VLF-1 peptides, representing 39.3% coverage, which is sufficient to confirm that the purified protein was indeed VLF-1.
FIG. 1.
Purification of VLF-1. Baculovirus expression. Nuclear extracts (NE) were prepared at 60 h postinfection from S. frugiperda cells infected with wild-type (WT) AcNPV (lane 2) or the recombinant Ac-VLF1 (lane 3). Nuclear extracts were purified on a heparin (Hep) column, and peak fractions (lane 4) were further purified on a Mono S column (S) (lane 5). Each lane contains 2 μg of protein. Lane 1, crude extract; lane 2, protein eluted from Talon matrix with 100 mM imidazole; lane 3, peak fraction from a Mono S column. The position of VLF-1 is indicated by an arrow on the right, and the positions of relevant molecular markers are indicated on the left.
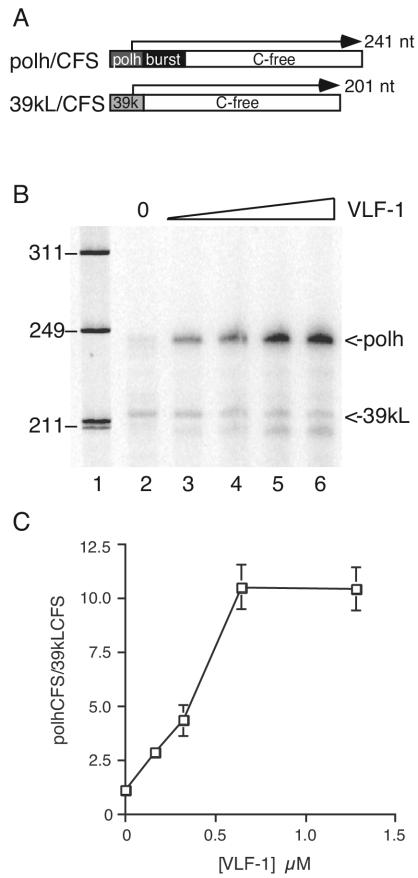
To determine whether VLF-1 acts directly at the level of transcription, in vitro transcription assays were conducted by using purified baculovirus RNA polymerase and VLF-1. Transcription reactions contained equimolar amounts of two templates (Fig. 2A). The plasmid pPolh/CFS is a C-free version of the polyhedrin promoter driving transcription of a 191-nucleotide C-free cassette, while p39kL/CFS is a C-free version of the late 39k promoter driving the same C-free cassette (11). The transcripts produced from these two templates differ in size; therefore, both templates can be added to the same reaction mixture to determine which is preferred by polymerase under different conditions. In the absence of VLF-1, the late and very late templates produced approximately equal amounts of RNA transcript (Fig. 2B), indicating that RNApol cannot distinguish between the two types of promoter, as previously shown (11). The addition of VLF-1 resulted in a dramatic increase in the amount of transcript originating from the pPolh/CFS template but did not significantly affect the p39kL/CFS product. Since both templates were added to the same reaction, the preference can best be demonstrated by plotting the ratio of polh transcript to 39kL transcript (Fig. 2C). This ratio increased proportionately with the concentration of VLF-1 added. The highest concentration tested produced a more-than-10-fold increase in transcription from the very late polh promoter compared to the level for the late 39k promoter. Similar results were obtained with His-tagged VLF-1 produced in bacteria (data not shown).
FIG. 2.
VLF-1 enhances transcription from the polyhedrin promoter. (A) Schematic diagram of the transcriptional templates pPolh/CFS (polh/CFS) and 39kL/CFS. The open box represents the C-free cassette. The darkly shaded box represents the polyhedrin (polh) promoter, the lightly shaded box represents the 39k late promoter, and the black box represents the burst sequence. (B) Transcription assay. Lane 1, φX174/HinfI marker; lanes 2 to 6, transcripts produced from reaction mixtures containing 0.04 μM purified RNA polymerase; 0.4 pmol of each template in the absence of VLF-1 (lane 2) or increasing amounts of purified VLF-1 (0.16 to 1.28 μM in twofold increments [lanes 3 to 6]) were incubated under standard transcription reaction conditions at 30°C for 12 min. The RNA transcripts were resolved on 6% polyacrylamide-8 M urea gel and exposed on PhosphorImager plates. The positions of the transcripts are indicated on the right. The positions of relevant molecular size markers are indicated in nucleotides on the left. (C) Transcripts were quantitated by using ImageQuant software and plotted as the ratio of the polh transcript to the 39k transcript as a function of input VLF-1 concentration.
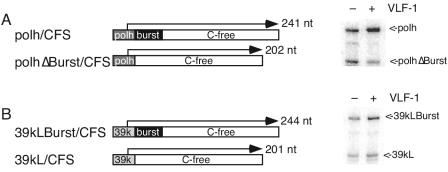
To test whether stimulation of pPolh/CFS was due to the burst sequence, the plasmid pPolhΔburst/CFS was constructed by inserting a double-stranded oligonucleotide containing the polyhedrin promoter lacking the burst sequence (ATCATGGAGATAATTAAAATGATAACCATCTCGCAAATAAATAAGTATTTT) into the SmaI site of pCFS, the parental plasmid for pPolh/CFS (11). In transcription assays involving equimolar amounts of pPolh/CFS and pPolΔburst/CFS in the absence of VLF-1, the levels of transcripts from both templates were equivalent. The addition of VLF-1 at a concentration yielding half-maximal stimulation increased the amount of pPolh/CFS transcript relative to that of pPolhΔburst/CFS added to the same tube by a factor of 4.4 ± 0.3 in triplicate reactions (Fig. 3A). This result indicates that the burst sequence is a major determinant of VLF-1 action.
FIG. 3.
Transcriptional enhancement by VLF-1 requires a burst sequence. (A) Transcription from the polyhedrin promoter in the presence and absence of a burst sequence. Transcripts were produced from in vitro transcription reaction mixtures containing pPolh/CFS and pPolhΔburst/CFS. Transcripts were quantified by using ImageQuant software and calculated as the ratio of pPolh/CFS to pPolhΔburst/CFS transcript. (B) Transcription from the chimeric p39kLburst/CFS template is stimulated by VLF-1. Reactions contained equimolar amounts of p39kL/CFS and pp39kLburst/CFS. Transcripts were quantified by using ImageQuant software.
This model also suggests that it should be possible to make a late promoter responsive to VLF-1 by inserting the polyhedrin burst sequence downstream of the transcription start site. To test this hypothesis, a C-free version of the burst sequence (GTGTTTTTGTAAAAGTTTTGTAATAAAAAAAATTATTAAATAG) was inserted into the EcoRV site of p39kL, an intermediate in the construction of pp39kL/CFS (11). The two templates p39kL/CFS and pp39kLburst/CFS were assayed in competition reactions (Fig. 3B). In the absence of VLF-1, transcript levels were comparable. The addition of VLF-1 stimulated transcription from pp39kLburst/CFS but had no effect on transcription from p39kL/CFS, as expected. In triplicate reactions, the amount of 39kL/CFS transcript was 2.3 ± 0.06-fold higher than that of 39kL/CFS. Although this was less than the amount seen with polyhedrin, this result confirms that the burst sequence is important for VLF-1 activity. This lower activity of the synthetic very late promoter could be due to a loss of some secondary structure in the leader region or other unknown sequence features of late promoters compared to very late ones (3).
In vivo, the timing of expression and high level of synthesis of polyhedrin appear to be regulated by the concentration of VLF-1. VLF-1 is a late protein and so is not present during the early stages of infection (4). VLF-1 appears by 15 h postinfection and increases through 24 h postinfection (14). Polyhedrin protein is synthesized at low levels during the late stage of infection but increases significantly thereafter, reaching maximum levels at 36 to 48 h postinfection. Furthermore, the timing of polyhedrin expression can be modified by varying the onset of VLF-1 expression (14). Construction of recombinant viruses with vlf-1 cloned under the control of a strong late promoter resulted in higher levels of VLF-1 and accelerated expression of polyhedrin. In contrast, production of a recombinant virus with vlf-1 under the control of a minimal promoter produced lower levels of VLF-1 and reduced expression of polyhedrin. Together, these experiments suggest that polyhedrin expression is regulated by the intracellular concentration of VLF-1 (13). The results presented here are consistent with these observations, as they show that templates containing a burst sequence were stimulated by VLF-1 in a concentration-dependent manner. Questions still remain regarding the mechanism of VLF-1 function, but the results presented here suggest that it can be studied by using this in vitro system, which is amenable to biochemical analyses.
Acknowledgments
This research was supported by a grant from the National Science Foundation (MCB 0110925).
REFERENCES
- 1.Esposito, D., and J. J. Scocca. 1997. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res. 25:3605-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guarino, L. A., T. A. Mistretta, and W. Dong. 2002. DNA binding activity of the baculovirus late expression factor PP31. Virus Res. 90:187-195. [DOI] [PubMed] [Google Scholar]
- 3.Mans, R. M., and D. Knebel-Morsdorf. 1998. In vitro transcription of pe38/polyhedrin hybrid promoters reveals sequences essential for recognition by the baculovirus-induced RNA polymerase and for the strength of very late viral promoters. J. Virol. 72:2991-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McLachlin, J. R., and L. K. Miller. 1994. Identification and characterization of vlf-1, a baculovirus gene involved in very late gene expression. J. Virol. 68:7746-7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris, T. D., and L. K. Miller. 1994. Mutational analysis of a baculovirus major late promoter. Gene 140:147-153. [DOI] [PubMed] [Google Scholar]
- 6.Nunes-Duby, S. E., H. J. Kwon, R. S. Tirumalai, T. Ellenberger, and A. Landy. 1998. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 26:391-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ooi, B. G., C. Rankin, and L. K. Miller. 1989. Downstream sequences augment transcription from the essential initiation site of a baculovirus polyhedrin gene. J. Mol. Biol. 210:721-736. [DOI] [PubMed] [Google Scholar]
- 8.O'Reilly, D. R., L. K. Miller, and V. A. Luckow. 1992. Baculovirus expression vectors: a laboratory manual. W.H. Freeman & Co., New York, N.Y.
- 9.Rankin, C., B. G. Ooi, and L. K. Miller. 1988. Eight base pairs encompassing the transcriptional start point are the major determinant for baculovirus polyhedrin gene expression. Gene 70:39-49. [DOI] [PubMed] [Google Scholar]
- 10.Todd, J. W., A. L. Passarelli, A. Lu, and L. K. Miller. 1996. Factors regulating baculovirus late and very late gene expression in transient-expression assays. J. Virol. 70:2307-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu, B., S. Yoo, and L. A. Guarino. 1995. Differential transcription of baculovirus late and very late promoters: fractionation of nuclear extracts by phosphocellulose chromatography. J. Virol. 69:2912-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang, S., and L. K. Miller. 1999. Activation of baculovirus very late promoters by interaction with very late factor 1. J. Virol. 73:3404-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang, S., and L. K. Miller. 1998. Control of baculovirus polyhedrin gene expression by very late factor 1. Virology 248:131-138. [DOI] [PubMed] [Google Scholar]
- 14.Yang, S., and L. K. Miller. 1998. Expression and mutational analysis of the baculovirus very late factor 1 (vlf-1) gene. Virology 245:99-109. [DOI] [PubMed] [Google Scholar]