Abstract
CXCR4 is one of two physiologically relevant human immunodeficiency type 1 (HIV-1) entry coreceptors. Studies of CXCR4 mutants have not clearly identified the determinants of coreceptor function and specificity. We therefore used a panel of monoclonal antibodies to further elucidate CXCR4 expression, structure, and function. Our findings show the existence of conformational subpopulations of CXCR4 that are in equilibrium on the cell surface but are not cell type specific as previously reported. HIV-1 X4 isolates can interact with multiple CXCR4 conformations in order to gain entry into target cells.
Attachment to and entry into target cells by human immunodeficiency virus type 1 (HIV-1) are mediated by consecutive interactions of envelope glycoprotein gp120 with CD4 and a coreceptor (reviewed in reference 2). The two biologically relevant coreceptors are CCR5 and CXCR4 (reviewed in references 2 and 10). In vivo usage of the latter by HIV-1 is associated with late-stage disease progression and a rapid decline in CD4+ T lymphocytes (5, 15, 19). It is still unclear what selective pressures collude to drive the outgrowth of CXCR4-using (X4) isolates. The switch from CCR5 to CXCR4 may be the result of immune selection, fluctuations in target cell populations, and conformational constraints imposed by the structures of gp120 and the coreceptors. Understanding the details of gp120 interactions with CCR5 and CXCR4 will provide insight into the mechanism of coreceptor switching.
Our previous studies demonstrated that the stem of gp120 hypervariable loop 3 interacts with specific sulfotyrosines and negatively charged residues in the amino-terminal domain (Nt) of CCR5. The hypervariable loop 3 crown may bind residues in the second extracellular loop (ECL2) (6, 7, 20, 21). In contrast, CXCR4 domains and residues involved in gp120 attachment have proven much more difficult to identify (reviewed in reference 11). CXCR4 mutagenesis suggests that the gp120 binding site is variable and isolate specific, involving charged and tyrosine residues that are dispersed throughout the four extracellular domains (3, 4, 14, 16, 18, 22, 24). It is still unclear whether any particular amino acids or domains of CXCR4 are absolutely required for viral entry and to what extent the gp120 of any given isolate is able to utilize different residues on this coreceptor. Additionally, it has been suggested that antigenically distinct conformations of CXCR4 are present on the cell surface, perhaps only some having coreceptor function (1).
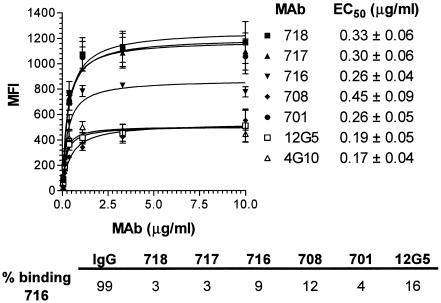
In order to further elucidate the determinants of CXCR4 coreceptor function, a panel of seven independent anti-CXCR4 monoclonal antibodies (MAbs) was characterized for binding, epitope specificity, and HIV-1 entry inhibition. Binding of MAbs 701, 708, 716, 717, 718, 12G5 (R&D Systems), and 4G10 (9) to murine L1.2 hybridoma cells (23) expressing high levels of human CXCR4 was analyzed by flow cytometry (Fig. 1). None of the MAbs cross-reacted with murine CXCR4, as evidenced by minimal binding to parental L1.2 (data not shown). Furthermore, binding of the seven MAbs to CXCR4+ L1.2 was not altered by the presence of saturating concentrations of an anti-mouse CXCR4 MAb (R&D Systems) (data not shown). All test MAbs exhibited similar half-maximal concentrations of binding (EC50s) to CXCR4+ L1.2 cells, ranging between 0.17 and 0.45 μg/ml (Fig. 1). However, maximal binding of the MAbs varied and did not correlate with the EC50s. Saturating concentrations of MAbs 701, 717, and 718 generated the highest mean fluorescence intensities (MFI, ∼1,200), whereas MAbs 708, 12G5, and 4G10 yielded the lowest signals (MFI, ∼400). Saturating binding of MAb 716 was intermediate (MFI, ∼800). Anti-CXCR4 MAbs effectively cross-competed with each other for binding to CXCR4, e.g., binding of MAb 716 to CXCR4+ L1.2 was inhibited strongly by MAbs 717, 718, and 701 and somewhat less by MAbs 708 and 12G5 (Fig. 1, bottom). The test MAbs therefore bind human CXCR4 specifically and with high affinity and may recognize subpopulations of coreceptor molecules on the basis of differences in saturating binding to CXCR4+ L1.2 cells. MAbs with the highest saturating binding presumably recognize the widest array of CXCR4 subpopulations.
FIG. 1.
Binding of anti-CXCR4 MAbs to CXCR4+ L1.2 cells. CXCR4+ L1.2 cells (5 × 105) were incubated with different concentrations of each anti-CXCR4 MAb in assay buffer (1% bovine serum albumin and 0.05% azide in Dulbecco's phosphate-buffered saline) at room temperature, and binding was measured by flow cytometry after labeling with phycoerythrin (PE)-conjugated goat anti-mouse immunoglobulin G (IgG; 1:100; Caltag). Each MFI is the mean of three independent experiments ± the standard deviation. Nonlinear regression (curve fit) was used to calculate the EC50 of each MAb. CXCR4+ L1.2 cells were incubated with biotinylated MAb 716 (1 μg/ml) in the presence of unlabeled, isotype-matched, nonspecific murine IgG or anti-CXCR4 MAbs (10 μg/ml). Binding of 716 was measured by flow cytometry after labeling with PE-conjugated streptavidin. Percent binding of 716 was calculated with the formula [(MFI 716 + anti-CXCR4 MAb)/MFI 716 alone] × 100.
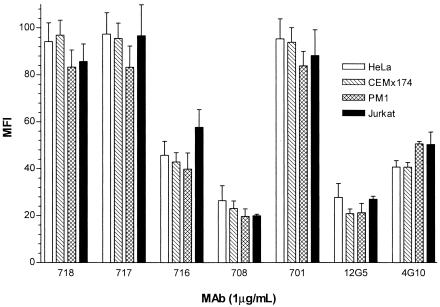
Since differential patterns of anti-CXCR4 MAb binding to CXCR4+ cells have been reported (1), we compared the binding of test MAbs to four human cell lines that naturally express CXCR4, including HeLa (CD4− endothelial cell line), CEMx174 (CD4+ T-B hybrid cell line), Jurkat (CD4+ T-cell line), and PM1 (CD4+ T-cell line). HeLa, CEMx174, and PM1 cells express similar levels of CXCR4, whereas Jurkat cells express fivefold higher levels of the coreceptor, as determined by relative MFI measured by flow cytometry (Fig. 2). We observed no differences in patterns of binding to the four cell lines at a saturating MAb concentration of 1 or 10 μg/ml (Fig. 2 and data not shown). However, the maximal MFIs generated by the different MAbs varied similarly to those measured on CXCR4+ L1.2 cells. These observations provide further evidence that anti-CXCR4 MAbs recognize subpopulations of CXCR4 molecules but different cell lines appear to comprise similar ratios of these subpopulations.
FIG. 2.
Binding of anti-CXCR4 MAbs to human cell lines. CEMx174, Jurkat, PM1, and HeLa cells were stained with a panel of anti-CXCR4 MAbs (1 μg/ml), and binding was measured by flow cytometry after labeling with PE-conjugated goat anti-mouse immunoglobulin G as described in the legend to Fig. 1. The MFIs obtained for Jurkat cells were divided by 5 so that the same y-axis scale could be used for all cell lines. Each result is the mean of three independent experiments ± the standard deviation.
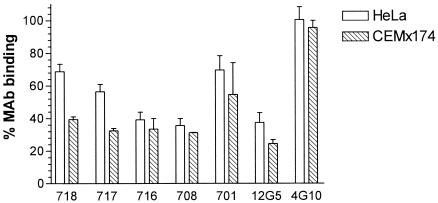
Next, we tested inhibition of MAb (1 μg/ml) binding to two of the human cell lines, HeLa and CEMx174, by the small-molecule CXCR4 antagonist AMD3100 (1 μM) (8). Binding of MAb 4G10 to both cell lines was unaffected by the presence of AMD3100. In contrast, binding of the other anti-CXCR4 MAbs varied between 25 and 75% in the presence of the inhibitor (Fig. 3). No significant differences between the inhibition patterns on HeLa and CEMx174 cells were observed (Fig. 3). There was no significant correlation between EC50 or maximal binding levels and potency of AMD3100 inhibition of MAb binding.
FIG. 3.
Inhibition of anti-CXCR4 MAb binding by AMD3100. HeLa or CEMx174 cells (5 × 105) were incubated with anti-CXCR4 MAbs (1 μg/ml) in the presence of AMD3100 (1 μM; Sigma). Binding of MAbs was measured by flow cytometry after labeling with PE-conjugated goat anti-mouse immunoglobulin G. Percent MAb binding was calculated with the formula (MFI in the presence of AMD3100/MFI in the absence of AMD3100) × 100. Each result represents the mean of three independent experiments ± the standard deviation.
Taken together, the binding and inhibition data suggest that the MAbs recognize different epitopes on CXCR4. Epitope mapping studies were therefore performed with a panel of 88 alanine point mutants of residues in all four extracellular domains of CXCR4. The CXCR4 gene was cloned into a vector comprising the T7 polymerase promoter (pcDNA3.1; Invitrogen), and alanine substitutions were introduced by site-directed mutagenesis. Wild-type and mutant CXCR4 constructs were appended with a carboxy-terminal hemagglutinin (HA) tag. BHK21 cells were lipofected with mutant or wild-type CXCR4 constructs and infected with a T7 polymerase-expressing vaccinia virus vector (vTF7-3) to boost expression as previously described (17). Cells were analyzed by flow cytometry after surface labeling with anti-CXCR4 MAbs (1 μg/ml), as well as intracellular staining with an anti-HA MAb (1:500; Covance) (17). This internal control allowed us to directly normalize binding of the anti-CXCR4 MAbs for mutant expression levels.
MAb binding to most of the CXCR4 point mutants was ≥50% of the binding to wild-type CXCR4, and we concluded that these residues do not contribute significantly to the framework of MAb epitopes (data not shown). Certain point mutations, however, reduced MAb binding by >50%, and we concluded that these residues directly contribute to MAb epitopes (Table 1). Thus, binding of 4G10 to CXCR4 depends on Nt residues E2 and I6. Surprisingly, the other anti-CXCR4 MAbs recognize similar epitopes, all of which comprise ECL2 residues E179 and D181 (Table 1). Residues that further contribute to the epitopes of individual MAbs are Y190 and W195 for MAb 717, Y184 for MAb 716, Y184 and Y190 for MAb 708, and D182 and Y190 for MAb 12G5 (Table 1). MAb 12G5 binding also requires an intact C28-C274 disulfide bond (Table 1). Mutations of C109 in ECL1 and C186 in ECL2 reduced binding of all MAbs by ≥90%, indicating that this disulfide bond profoundly affects the structure of the CXCR4 extracellular domain without directly participating in MAb binding (data not shown).
TABLE 1.
Epitope mapping of anti-CXCR4 MAbsa
| MAb | Nt
|
ECL2
|
ECL3 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| E2 | I6 | C28 | E179 | D181 | D182 | Y184 | Y190 | W195 | C274 | |
| 718 | 13 | 31 | ||||||||
| 717 | 13 | 31 | 48 | |||||||
| 716 | 12 | 25 | 19 | |||||||
| 708 | 10 | 13 | 43 | 27 | ||||||
| 701 | 11 | 30 | ||||||||
| 12G5 | 21 | 10 | 35 | 22 | 25 | 9 | ||||
| 4G10 | 2 | 31 | ||||||||
HA-tagged CXCR4 point mutants were expressed in BHK21 cells as previously described (17). Cells were harvested 24 h posttransfection and incubated with anti-CXCR4 MAbs (1 μg/ml), followed by fixation-permeabilization with Cytofix-Cytoperm (Becton Dickinson) as recommended by the manufacturer. Flow cytometry was used to detect staining with PE-conjugated goat anti-mouse immunoglobulin G, as well as intracellular staining with a fluorescein isothiocyanate (FITC)-conjugated mouse anti-HA MAb (1:500; Covance). MAb binding to CXCR4 mutants was calculated as percent binding to wild-type CXCR4 with the formula [(mutant PE MFI/wild-type PE MFI) ÷ (mutant FITC MFI/wild-type FITC MFI)] × 100%. Results are means of three independent experiments.
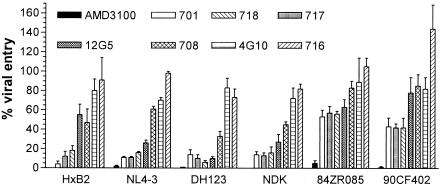
The observation that anti-CXCR4 MAbs have similar affinities for their ligand and also recognize overlapping epitopes (except for 4G10) suggested that they would similarly inhibit HIV-1 entry into target cells. We therefore characterized the ability of anti-CXCR4 MAbs to block the entry into CD4+ HeLa cells of NLluc+ env− reporter viruses pseudotyped with envelope glycoproteins of six different HIV-1 isolates: HXB2 (T-cell line adapted [TCLA], clade B), NL4-3 (TCLA, clade B), NDK (TCLA, clade D), 84ZR085 (primary X4, clade D), 90CF402 (primary X4, clade E), and DH123 (primary R5X4, clade B) as previously described (12, 20). Entry of all isolates was inhibited >90% by 1 μM AMD3100 (Fig. 4). The most potently inhibitory MAbs were 701, 718, and 717, which also generated the greatest maximal binding to CXCR4+ cells (Fig. 4). MAb 12G5 exhibited the greatest variability in potency against different HIV-1 isolates: it strongly inhibited entry mediated by envelope glycoproteins of NL4-3, DH123, and NDK but only moderately inhibited entry of HXB2, 84ZR085, and 90CF402 (Fig. 4). MAb 708 moderately inhibited the entry of all HIV-1 isolates tested, whereas MAbs 716 and 4G10 did not inhibit the entry of any of the isolates tested (Fig. 4). Furthermore, MAb 716 enhanced the entry of isolate 90CF402 by ∼50%. The potency of entry inhibition therefore generally correlated with the maximal binding levels of the MAbs. MAb 716 was the notable exception, since it exhibited intermediate maximal binding but no entry inhibition. Additionally, entry of TCLA isolates was blocked more efficiently than entry of primary X4 strains (Fig. 4). Primary R5X4 isolate DH123, however, was inhibited similarly to TCLA isolates.
FIG. 4.
Inhibition of HIV-1 pseudotype entry by anti-CXCR4 MAbs. CD4+ HeLa cells were infected with NLluc+ env− pseudovirions (∼105 relative light units/104 cells) in the presence of anti-CXCR4 MAbs (10 μg/ml) or AMD3100 (1 μM). Luciferase activity was measured in cell lysates with the Promega luciferase assay at 48 h postinfection. Percent viral entry was calculated with the formula (RLU with inhibitor/RLU without inhibitor) × 100. Each result is the mean of four independent experiments ± the standard deviation.
The determinants of CXCR4 coreceptor function remain elusive. In order to gain additional insight into CXCR4 structure-function relationships, we characterized a panel of seven independent anti-CXCR4 MAbs for binding, epitope specificity, and HIV-1 entry inhibition. MAbs bound to CXCR4 with similar EC50s ranging between 0.17 and 0.45 μg/ml. These are approximately 10-fold lower than the values reported by Baribaud et al. (1) and may be due to our use of a murine cell line that expresses high levels of CXCR4. Furthermore, the MAbs differed in maximal binding, indicating that they interact with distinct subpopulations of CXCR4 molecules. CXCR4 structure may vary as a result of posttranslational modifications, conformational fluctuations, or receptor oligomerization. Because MAbs can cross-compete for binding to CXCR4, we interpret this to mean that they are more or less restricted in their ability to interact with the different CXCR4 subpopulations. However, we were unable to detect significant differences in MAb binding to four human CXCR4+ cell lines, indicating that they express similar distributions of CXCR4 subpopulations. In contrast, Baribaud et al. reported that anti-CXCR4 MAbs exhibit differential patterns of binding to human cell lines, including the ones we tested. Differences in cell lines and assay conditions may account for these observations. Our results show the existence of CXCR4 subpopulations that do not vary in a cell-specific manner.
Epitope mapping studies show that MAb 4G10 binds to residues in the Nt of CXCR4, which was expected because this MAb was generated against an Nt peptide (9). In contrast, the other MAbs were raised against cell surface-associated CXCR4 (1) and they all recognize overlapping epitopes in ECL2. This is surprising in light of our observations that the MAbs exhibit different maximal binding and implies that this region of CXCR4 undergoes significant conformational fluctuations. The anti-ECL2 MAbs also bind to a region of CXCR4 that was previously shown to be important for its coreceptor function. For example, we demonstrated that negatively charged ECL2 residues D182, Y184, D187, and D193 participate in HIV-1 entry (14). Taken together, these findings demonstrate that ECL2—particularly residues 179 to 193—are the most prominent and immunodominant region of CXCR4 that is readily recognized by MAbs as well as HIV-1 gp120. Interestingly, the small-molecule inhibitor AMD3100 blocks binding of all anti-ECL2 MAbs but not of the anti-Nt MAb, indicating that it disrupts the conformation of the former but not the latter domain. We have previously proposed that binding of small-molecule CCR5 inhibitors TAK-779 and SCH-C disrupts the conformation of ECL2 but not the Nt, thereby disabling gp120 binding to CCR5 (13, 21). AMD3100 may act by a similar mechanism to inhibit HIV-1 entry mediated by CXCR4.
Anti-CXCR4 MAbs inhibited HIV-1 entry to various degrees. 4G10 did not inhibit entry of any test isolates, much like MAbs to the CCR5 Nt do not (or only poorly) inhibit entry of R5 isolates. These observations suggest that gp120 binding to other domains of CCR5 and CXCR4 occurs prior to the interaction with the Nt. Alternatively, the CXCR4 Nt may not participate in X4 HIV-1 entry. Other anti-CXCR4 Nt MAbs need to be tested in order to reach a definitive conclusion. Despite the fact that all of the anti-ECL2 MAbs recognize overlapping epitopes with similar affinities, there are significant differences in their abilities to inhibit HIV-1 entry. Potency of entry inhibition generally correlated with maximal binding patterns, suggesting that MAbs that recognize the widest range of CXCR4 conformations are the most efficient at blocking gp120 binding to the coreceptor. This implies that the gp120-CXCR4 interaction is not limited to a particular subset of CXCR4 molecules. Finally, the observation that all of the anti-CXCR4 MAbs are more potent against TCLA isolates than against primary X4 isolates suggests that primary gp120s bind to CXCR4 with higher affinity than TCLA gp120s do. Alternatively, gp120s of primary X4 isolates may have lower specificity for any one particular set of CXCR4 ECL2 residues, allowing them to exploit multiple binding sites on this coreceptor.
CXCR4 is the HIV-1 entry coreceptor used by R5X4 and X4 isolates that emerge late during disease progression (2). Several laboratories, including ours, have demonstrated that the interaction between gp120 and CXCR4 differs from the interaction between gp120 and CCR5 (11). In particular, gp120 binding to CCR5 requires both the Nt and ECL2. Our present work supports the notion that gp120 binding to CXCR4 is largely mediated by interactions with ECL2. In addition, our results indicate that the gp120 of any given isolate is able to recognize an array of CXCR4 conformations. Isolate-specific interactions with CXCR4 therefore may depend on recognition of different residues within CXCR4 ECL2.
Acknowledgments
We are especially thankful to Christopher C. Broder (Uniformed Services University of the Health Sciences, Bethesda, Md.) for providing MAb 4G10.
This work was supported by National Institutes of Health grants R01 AI43847 to T.D. and R43 AI40810 to W.C.O., as well as Sidaction grant AO11 to U.H.
REFERENCES
- 1.Baribaud, F., T. G. Edwards, M. Sharron, A. Brelot, N. Heveker, K. Price, F. Mortari, M. Alizon, M. Tsang, and R. W. Doms. 2001. Antigenically distinct conformations of CXCR4. J. Virol. 75:8957-8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 3.Brelot, A., N. Heveker, M. Montes, and M. Alizon. 2000. Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities. J. Biol. Chem. 275:23736-23744. [DOI] [PubMed] [Google Scholar]
- 4.Chabot, D. J., P. F. Zhang, G. V. Quinnan, and C. C. Broder. 1999. Mutagenesis of CXCR4 identifies important domains for human immunodeficiency virus type 1 X4 isolate envelope-mediated membrane fusion and virus entry and reveals cryptic coreceptor activity for R5 isolates. J. Virol. 73:6598-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cormier, E., and T. Dragic. 2002. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J. Virol. 76:8953-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cormier, E. G., D. N. Tran, L. Yukhayeva, W. C. Olson, and T. Dragic. 2001. Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J. Virol. 75:5541-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Clercq, E., and D. Schols. 2001. Inhibition of HIV infection by CXCR4 and CCR5 chemokine receptor antagonists. Antivir. Chem. Chemother. 12:19-31. [PubMed] [Google Scholar]
- 9.Dimitrov, D. S., D. Norwood, T. S. Stantchev, Y. Feng, X. Xiao, and C. C. Broder. 1999. A mechanism of resistance to HIV-1 entry: inefficient interactions of CXCR4 with CD4 and gp120 in macrophages. Virology 259:1-6. [DOI] [PubMed] [Google Scholar]
- 10.Doms, R. W. 2001. Chemokine receptors and HIV entry. AIDS 15(Suppl. 1):S34-S35. [DOI] [PubMed] [Google Scholar]
- 11.Dragic, T. 2001. An overview of the determinants of CCR5 and CXCR4 co-receptor function. J. Gen. Virol. 82:1807-1814. [DOI] [PubMed] [Google Scholar]
- 12.Dragic, T., A. Trkola, S. W. Lin, K. A. Nagashima, F. Kajumo, L. Zhao, W. C. Olson, L. Wu, C. R. Mackay, G. P. Allaway, T. P. Sakmar, J. P. Moore, and P. J. Maddon. 1998. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J. Virol. 72:279-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dragic, T., A. Trkola, D. A. Thompson, E. G. Cormier, F. A. Kajumo, E. Maxwell, S. W. Lin, W. Ying, S. O. Smith, T. P. Sakmar, and J. P. Moore. 2000. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc. Natl. Acad. Sci. USA 97:5639-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kajumo, F., D. A. D. Thompson, Y. Guo, and T. Dragic. 2000. Entry of R5X4 and X4 human immunodeficiency virus type 1 strains is mediated by negatively charged and tyrosine residues in the amino-terminal domain and the second extracellular loop of CXCR4. Virology 271:240-247. [DOI] [PubMed] [Google Scholar]
- 15.Kreisberg, J. F., D. Kwa, B. Schramm, V. Trautner, R. Connor, H. Schuitemaker, J. I. Mullins, A. B. van't Wout, and M. A. Goldsmith. 2001. Cytopathicity of human immunodeficiency virus type 1 primary isolates depends on coreceptor usage and not patient disease status. J. Virol. 75:8842-8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Labrosse, B., A. Brelot, N. Heveker, N. Sol, D. Schols, E. De Clercq, and M. Alizon. 1998. Determinants for sensitivity of human immunodeficiency virus coreceptor CXCR4 to the bicyclam AMD3100. J. Virol. 72:6381-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olson, W. C., G. E. Rabut, K. A. Nagashima, D. N. Tran, D. J. Anselma, S. P. Monard, J. P. Segal, D. A. Thompson, F. Kajumo, Y. Guo, J. P. Moore, P. J. Maddon, and T. Dragic. 1999. Differential inhibition of human immunodeficiency virus type 1 fusion, gp120 binding, and CC-chemokine activity by monoclonal antibodies to CCR5. J. Virol. 73:4145-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parolin, C., A. Borsetti, H. Choe, M. Farzan, P. Kolchinsky, M. Heesen, Q. Ma, C. Gerard, G. Palu, M. E. Dorf, T. Springer, and J. Sodroski. 1998. Use of murine CXCR-4 as a second receptor by some T-cell-tropic human immunodeficiency viruses. J. Virol. 72:1652-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Penn, M. L., J. C. Grivel, B. Schramm, M. A. Goldsmith, and L. Margolis. 1999. CXCR4 utilization is sufficient to trigger CD4+ T cell depletion in HIV-1-infected human lymphoid tissue. Proc. Natl. Acad. Sci. USA 96:663-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson, D. A., E. G. Cormier, and T. Dragic. 2002. CCR5 and CXCR4 usage by non-clade B human immunodeficiency virus type 1 primary isolates. J. Virol. 76:3059-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsamis, F., S. Gavrilov, F. Kajumo, C. Seibert, S. Kuhmann, T. Ketas, A. Trkola, A. Palani, J. Clader, J. Tagat, S. McCombie, B. Baroudy, J. Moore, T. Sakmar, and T. Dragic. 2003. Analysis of the mechanism by which the small-molecule CCR5 antagonists SCH-351125 and SCH-350581 inhibit human immunodeficiency virus type 1 entry. J. Virol. 77:5201-5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, Z., J. Berson, T. Zhang, Y. Cen, Y. Sun, M. Sharron, Z. Lu, and S. Peiper. 1998. CXCR4 sequences involved in coreceptor determination of human immunodeficiency virus type 1 tropism. J. Biol. Chem. 273:15007-15015. [DOI] [PubMed] [Google Scholar]
- 23.Wu, L., G. LaRosa, N. Kassam, C. J. Gordon, H. Heath, N. Ruffing, H. Chen, J. Humblias, M. Samson, M. Parmentier, J. P. Moore, and C. R. Mackay. 1997. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J. Exp. Med. 186:1373-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou, N., Z. Luo, J. Luo, D. Liu, J. W. Hall, R. J. Pomerantz, and Z. Huang. 2001. Structural and functional characterization of human CXCR4 as a chemokine receptor and HIV-1 co-receptor by mutagenesis and molecular modeling studies. J. Biol. Chem. 276:42826-42833. [DOI] [PubMed] [Google Scholar]