Abstract
Emerging infectious diseases continue to impose unpredictable burdens on global health and economy. Infectious disease surveillance and pandemic preparedness are essential to mitigate the impact of future threats. Global surveillance networks provide unprecedented monitoring data on plant, animal and human infectious diseases. Using such sources, we report on current major One Health threats and update on their epidemiological status.
1. Text
Globally, environmental and anthropogenic changes are impacting ecosystems, and perturbing plant and animal demographics and behaviors. These changes contribute to the increasing pace of infectious disease emergence worldwide, largely driven by increasing contacts between and among species [1], [2]. Drivers of disease emergence include mobility and trade, encroachment of natural habitats and climate change, as well as intrinsic characteristics of pathogens, such as wide host range for animal pathogens and the ability of plant pathogens to hybridize [2].
The vast majority of emerging infectious diseases in humans are zoonotic in nature [3], [4]. Often, they escape their natural wildlife reservoirs and infect captive or domestic animals and humans upon cross-species transmission. While the majority of zoonotic pathogens spread limitedly among humans, occasionally some do evolve the ability to efficiently transmit [5]. These may cause devastating epidemics, if not pandemics, and may establish as novel human pathogens. Emerging infectious diseases of animals likewise have typically the ability to cross species barriers and invade new host species. In contrast, introduction of pathogens into new geographical areas and climate change play an essential role in the emergence of plant diseases, and the hybridization of plant pathogens that are not naturally sympatric is repeatedly reported to be involved in plant disease emergence events [2]. The consequences of emerging pathogens in newly infected species, be it wild or domestic, or in new geographical areas, can have dire repercussions on human welfare, for example, through the disruption of ecosystem services or from large agricultural economic losses [2], [6]. As such, emerging infectious diseases are One Health threats to the global community.
Despite progress in our understanding of the mechanisms and drivers of pathogen emergence and adaptation, infectious disease emergence and associated health and economic burdens remain essentially unpredictable. They continue to impose heavy burdens on the global community, as most recently painfully demonstrated by the emergence of MERS coronavirus in the Middle East and Ebola virus in West Africa. Because the nature, time and location of the next One Health threat cannot be forecasted, preparedness and responsiveness are essential to curb future emerging infectious disease burdens.
Surveillance is key to preparedness by identifying and monitoring new threats to plant, animal and human health, and raising early-warning flags upon changing epidemiology. Major global initiatives have profoundly revolutionized the scope of infectious disease surveillance in plants, animals and humans. These include the World Animal Health Information Database (WAHID) Interface of the OIE, the Global Animal Disease Information system EMPRES-i of the FAO, the situation assessments and reports of the WHO, and the internet-based Program for Monitoring Emerging Diseases (ProMED) of the International Society for Infectious Diseases.
Using the data collected from these different sources, we present the current status of major One Health threats. In this update, the current status of low pathogenic avian influenza virus (LPAIV) H7N9, highly pathogenic avian influenza viruses (HPAIVs) of the H5 subtype, MERS coronavirus and Ebola virus are summarized. The present report will be updated every three months, with newly acquired data on the diseases listed above, as well as with data on any new One Health threat that would have emerged during that period.
2. Low pathogenic avian influenza virus H7N9
LPAIV H7N9 was identified as a newly emerging zoonotic pathogen in early 2013. It has caused since then a total of 680 cases of zoonotic infection, with a case-fatality rate of about 20%, principally in adult and elderly individuals [7]. With an incubation time of 2–8 days, H7N9 virus infection can progress from initial symptoms of high fever and other influenza-like signs to more severe lower respiratory tract infection, respiratory distress and associated complications [8]. Exposure to infected poultry is considered the primary risk factor for human infection. A total of 556 outbreaks have been reported in domestic poultry, including chickens, ducks, geese, pigeons and pheasants, largely concurrently to zoonotic cases of infection (Table 1). A few cases were reported in wild bird species. Because of their low pathogenic nature, H7N9 viruses typically cause asymptomatic or mild infections in birds.
Table 1.
Number of animal outbreaks and zoonotic cases of low pathogenic avian influenza virus H7N9 infection from 1 January 2013 to 30 September 2015.
| Country | Humans | Poultry | Wild birds |
|---|---|---|---|
| Canada | 2 | ||
| China | 657 | 554 | 1 |
| Hong Kong SAR | 16 | 2 | |
| Malaysia | 1 | ||
| Taiwan (Province of China) | 4 | 1 | |
| Total | 680 | 556 | 2 |
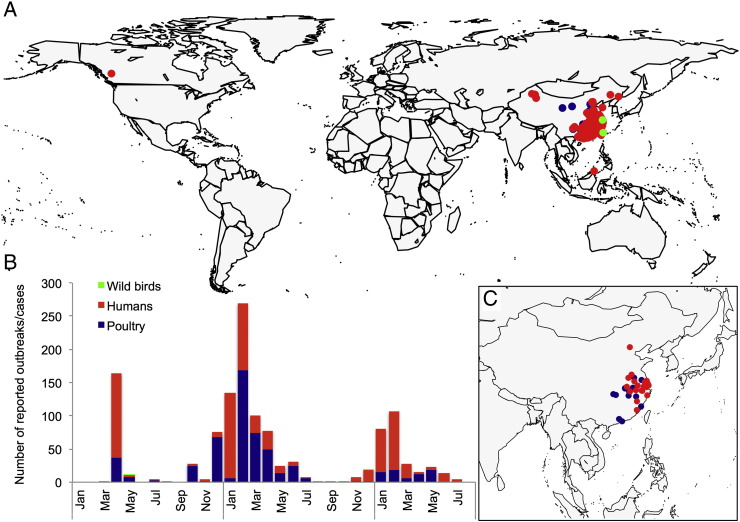
Most animal outbreaks and zoonotic cases of low pathogenic avian influenza H7N9 virus infection occurred in mainland China, while imported zoonotic cases were identified in Canada and Malaysia (Fig. 1). The first epidemic of H7N9 virus infection in poultry peaked in April 2013 soon after the first identification of the virus as a cause of a zoonotic case of infection. Epidemics subsequently re-occurred in winter 2014 and 2015, with the highest reported numbers of animal outbreaks and zoonotic cases of infection during the months of January–February of each year.
Fig. 1.
Current status of low pathogenic avian influenza virus H7N9 in animals and humans: A. Global geographical distribution of animal outbreaks and zoonotic cases of infection reported from 1 January 2013 to 30 September 2015; B. Number of reported animal outbreaks and zoonotic cases of infection from 1 January 2013 to 30 September 2015; C. Geographical distribution of animal outbreaks and zoonotic cases of infection reported from 1 April 2015 to 30 September 2015.
LPAIV H7N9, in contrast to most other avian influenza viruses, can bind to the cellular receptors used by seasonal influenza viruses [9], [10]. This ability is associated with one or two specific amino-acids in the hemagglutinin glycoprotein. Because seasonal influenza viruses and LPAIV H7N9 peak coincidentally during the winter months, they may co-infect an individual and subsequently reassort [11]. This may give rise to a transmissible variant, against which the human population has little pre-existing immunity, and may be at the origin of a new influenza pandemic. Strict monitoring and isolation measures are therefore essential to limit the risk of seasonal influenza and reassortment in individuals with zoonotic H7N9 virus infection.
3. Highly pathogenic avian influenza viruses of the H5 subtype
HPAIV H5N1 emerged in Hong Kong in 1997, causing 18 cases of zoonotic infection, including 6 fatalities [12]. After containment of the outbreak in Hong Kong, the virus re-emerged in mainland China in 2003. It infected an unmatched diversity of wild and domestic avian and mammalian species, and subsequently spread over much of Asia, Europe and Africa [13], evolving into many co-circulating antigenically-distinct clades and lineages. The severity of the disease is highly variable across animal species, ranging from asymptomatic infections, e.g., in dabbling ducks [14], to severe systemic disease with high mortality rates in other avian species as well as in most mammalian species found infected [15].
Since 2003, HPAIV H5N1 has caused a total of 834 cases of zoonotic infection in 18 countries, with a case-fatality rate of about 55% (Table 2). As for LPAIV H7N9, exposure to infected poultry is considered the primary risk factor for human infection. However, in contrast to LPAIV H7N9 infection, more than half of the cases were identified in children [7], [16]. Unusually pathogenic, HPAIV H5N1 can present a long incubation time, ranging between 2 and 17 days. Severe signs of lower respiratory tract infection and extra-respiratory symptoms, such as gastro-intestinal signs, typically rapidly supersede high fever and other influenza-like signs and symptoms.
Table 2.
Number of reported zoonotic cases of highly pathogenic avian influenza virus H5N1 infection from 1 January 2003 to 30 September 2015.
| 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Azerbaijan | 3 | 3 | ||||||||||||
| Bangladesh | 1 | 2 | 2 | 5 | ||||||||||
| Cambodia | 5 | 2 | 1 | 1 | 1 | 1 | 8 | 3 | 26 | 8 | 56 | |||
| Canada | 1 | 1 | ||||||||||||
| China | 1 | 7 | 14 | 6 | 3 | 8 | 1 | 3 | 2 | 2 | 5 | 53 | ||
| Djibouti | 1 | 1 | ||||||||||||
| Egypt | 18 | 23 | 10 | 37 | 24 | 42 | 12 | 5 | 24 | 145 | 340 | |||
| Hong Kong | 1 | 1 | ||||||||||||
| Indonesia | 16 | 59 | 40 | 21 | 23 | 10 | 11 | 10 | 3 | 1 | 194 | |||
| Iraq | 2 | 2 | ||||||||||||
| LPDRa | 2 | 2 | ||||||||||||
| Myanmar | 1 | 1 | ||||||||||||
| Nigeria | 1 | 1 | ||||||||||||
| Pakistan | 3 | 3 | ||||||||||||
| Thailand | 17 | 7 | 3 | 27 | ||||||||||
| Turkey | 8 | 8 | ||||||||||||
| Viet Nam | 3 | 31 | 68 | 8 | 5 | 6 | 7 | 4 | 2 | 2 | 136 | |||
| West Bank | 1 | 1 | ||||||||||||
| Total | 4 | 48 | 103 | 110 | 85 | 41 | 75 | 44 | 64 | 34 | 38 | 38 | 150 | 834 |
Lao People's Democratic Republic.
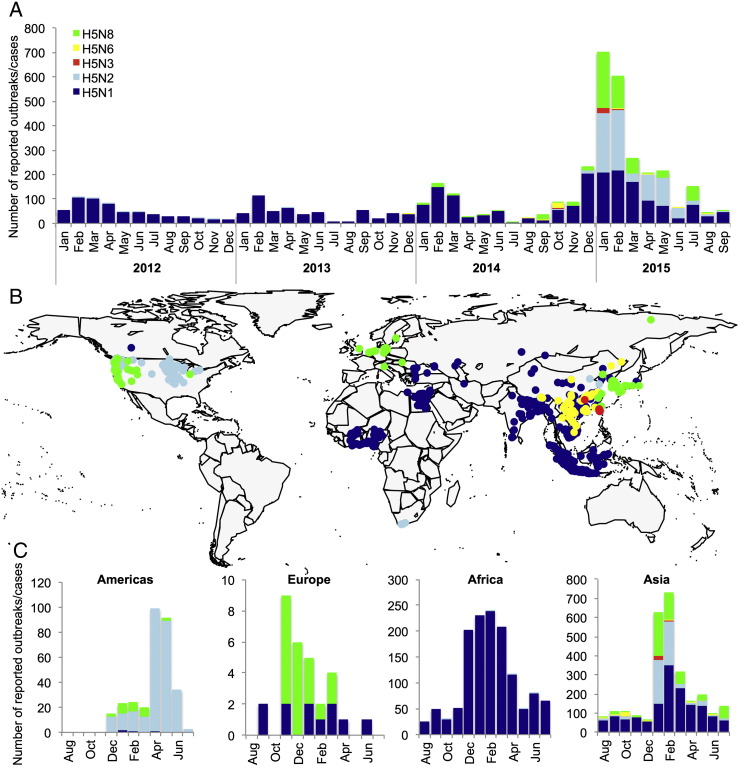
Since 2014, a wide diversity of reassortants containing the highly pathogenic H5 gene emerged and caused massive outbreaks in poultry and wild birds worldwide. Of these reassortants, only HPAIV H5N6 is reported to have caused zoonotic infections [17] (Table 3). Epidemics of the novel HPAIVs of the H5 subtype typically peaked during the winter and early spring months of December–March. A major exception is the H5N2 virus that emerged and spread in poultry in North America in 2015. Although it emerged during winter, the epidemic peaked in spring during the months of April–May (Fig. 2).
Table 3.
Number of reported animal outbreaks and zoonotic cases of infection with highly pathogenic avian influenza viruses of the H5 subtype from 1 January 2012 to 30 September 2015.
| Humans |
Poultry |
Wild birds |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| H5N1 | H5N6 | H5N1 | H5N2 | H5N3 | H5N6 | H5N8 | H5N1 | H5N2 | H5N6 | H5N8 | |
| Africa | 185 | 1162 | 7 | ||||||||
| Americas | 1 | 1 | 239 | 4 | 2 | 40 | 23 | ||||
| Asia | 72 | 3 | 1395 | 558 | 26 | 54 | 534 | 38 | 2 | 2 | 34 |
| Europe | 3 | 13 | 8 | 6 | |||||||
| Total | 258 | 3 | 2561 | 804 | 26 | 54 | 551 | 48 | 42 | 2 | 63 |
Fig. 2.
Diversity of highly pathogenic avian influenza viruses of the H5 subtype that caused outbreaks between 1 January 2012 and 30 September 2015: A. Number of reported outbreaks or cases; B. Global geographical distribution; and C. Seasonality of H5N1, H5N2, H5N3, H5N6 and H5N8 virus infection.
Outbreaks of HPAIVs of the H5 subtype were reported in poultry globally, whereas cases in wild birds were more often detected and reported in North America and Europe. Interestingly, the new reassortant viruses, which are highly pathogenic in poultry, appear to cause asymptomatic or mild infections in most wild birds found infected to date [18].
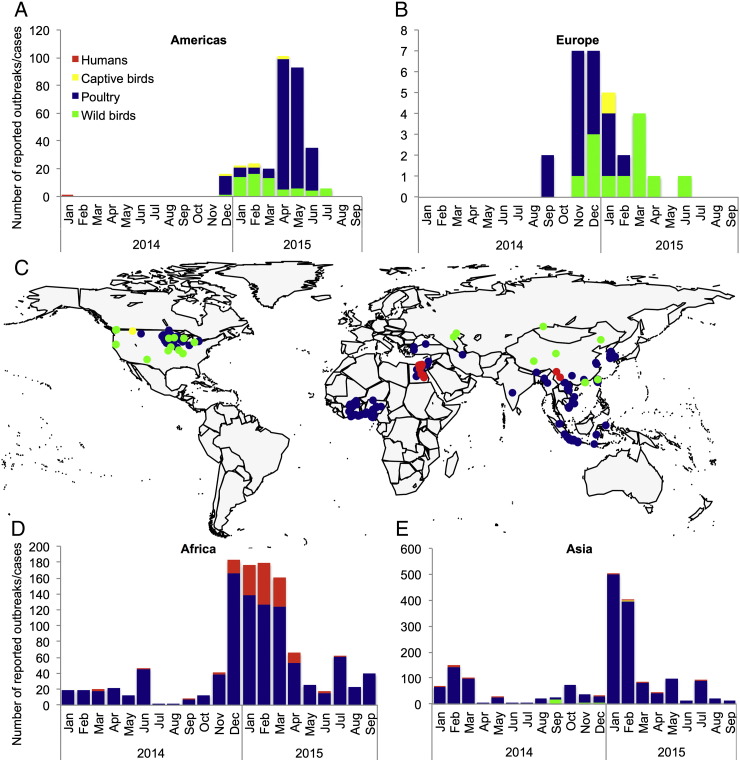
Currently, zoonotic cases of H5 virus infection chiefly occur in Egypt, which experienced last winter the largest H5N1 virus outbreak since its emergence in Africa. Zoonotic cases of H5N1 virus infection are also occasionally (and possibly under-) reported in South-East Asia (Fig. 3).
Fig. 3.
Current status of highly pathogenic avian influenza viruses of the H5 subtype in animals and humans: Number of reported animal outbreaks and zoonotic cases of infection in North America (A), Europe (B), Africa (D) and Asia (E) from 1 January 2014 to 30 September 2015; C. Global distribution of reported animal outbreaks and zoonotic cases of infection from 1 April 2015 to 30 September 2015.
The expanding diversity of HPAIVs of the H5 subtype is worrisome as it may increase opportunities for evolution towards a pandemic variant. The presence of a diverse array of reassortants in wild bird populations worldwide also indicates a major change in the epidemiology of avian influenza viruses in their bird reservoirs. Before the emergence of HPAIV H5N1, HPAIVs were thought to only evolve and spread in poultry populations, where containment and stamping-out measures contributed to their eradication. The reassortant HPAIVs of the H5 subtype represent unprecedented threats to the poultry industry. As did HPAIV H5N1, they have the potential to widely spread, if not establish, in poultry populations. Wild bird populations may represent a direct source of infection for poultry, calling for strict biosecurity measures.
4. Middle-East Respiratory Syndrome coronavirus (MERS-CoV)
MERS-CoV was first identified as a novel zoonotic pathogen in late 2012 [19]. Since then, it has caused 1138 reported cases of zoonotic infection (Table 4), with a case-fatality rate of about one-third. Infection is characterized in severe cases by symptoms of lower respiratory tract infection, respiratory distress, as well as renal failure [20]. MERS-CoV most likely originates from bat species, in which closely related viruses have been detected, causing chiefly asymptomatic infection [21], [22]. Dromedary camels, which develop mild respiratory tract infection with MERS-CoV, are increasingly recognized as the main proximate source of zoonotic infections [23], [24]. Contact with infected dromedary camels is considered the primary risk of infection in humans, while human-to-human transmission remains rare. Nonetheless, secondary cases of MERS are increasingly reported, mainly in hospitalized settings, often with mild or no signs and symptoms.
Table 4.
Number of reported cases of MERS-CoV infection from 1 September 2012 to 30 September 2015.
| Country | 2012 | 2013 | 2014 | 2015 | Total |
|---|---|---|---|---|---|
| Algeria | 2 | 2 | |||
| Austria | 1 | 1 | |||
| China | 1 | 1 | |||
| Denmark | 1 | 1 | |||
| Egypt | 1 | 1 | 2 | ||
| France | 2 | 2 | |||
| Germany | 1 | 1 | 2 | ||
| Greece | 1 | 1 | |||
| Hong Kong SAR | 1 | 1 | |||
| Iran (Islamic Republic of) | 4 | 1 | 5 | ||
| Italy | 1 | 1 | |||
| Jordan | 1 | 8 | 6 | 15 | |
| Kuwait | 2 | 1 | 3 | ||
| Lebanon | 1 | 1 | |||
| Malaysia | 1 | 1 | |||
| Netherlands | 2 | 2 | |||
| Oman | 2 | 4 | 6 | ||
| Philippines | 1 | 2 | 3 | ||
| Qatar | 1 | 7 | 2 | 4 | 14 |
| Republic of Korea | 185 | 185 | |||
| Saudi Arabia | 5 | 83 | 561 | 362 | 1011 |
| Thailand | 1 | 1 | |||
| Tunisia | 2 | 2 | |||
| Turkey | 1 | 1 | |||
| U.K. of Great Britain and Northern Ireland | 3 | 3 | |||
| United Arab Emirates | 7 | 54 | 7 | 68 | |
| United States of America | 2 | 2 | |||
| Yemen | 1 | 1 | |||
| Total | 10 | 110 | 644 | 574 | 1338 |
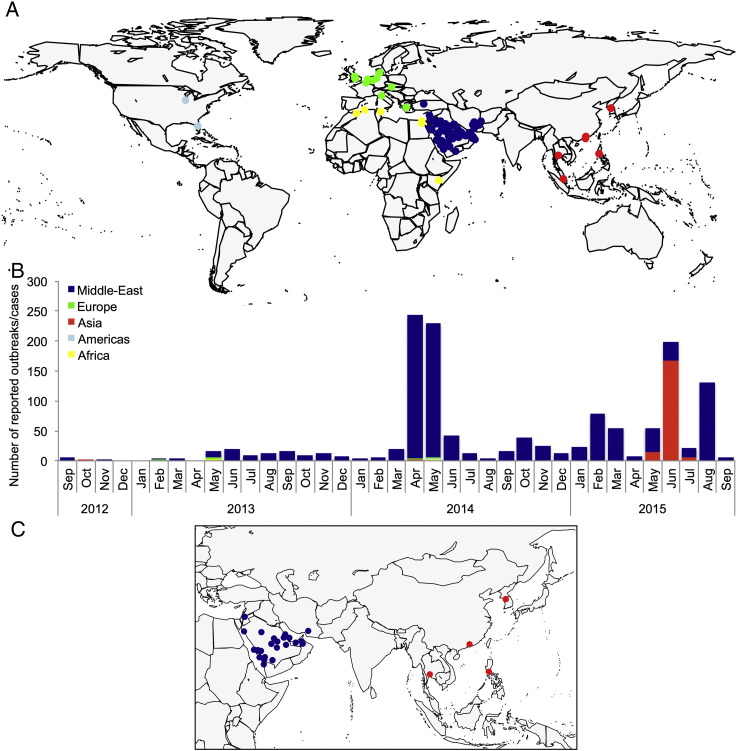
Until recently, most cases of zoonotic infection with MERS-CoV were identified in the Middle-East, while imported cases in Europe and Africa were reported in individuals who had recently traveled from that region (Fig. 4). In early May 2015, a case of zoonotic MERS-CoV infection was imported to South Korea. The hospitalization of this case initiated an outbreak of secondary infections, causing more than 180 additional cases in Asia (Table 4). The South Korea epidemic subsided in July 2015, while an outbreak of 130 cases was reported in Saudi Arabia in August 2015.
Fig. 4.
Current status of MERS-CoV infection in humans: A. Global geographical distribution of zoonotic cases of infection between 1 September 2012 and 30 September 2015; B. Number of reported zoonotic cases of infection per geographical region from 1 September 2012 to 30 September 2015; C. Geographical distribution of zoonotic cases of infection that occurred between 1 April 2015 and 30 September 2015.
The prompt characterization of the MERS-CoV as the etiological agent of the first reported case [19], of its cellular receptor [25], and of its animal reservoirs [21], [22], [23], [24] are to be commended. These are of paramount importance for the rapid diagnosis and containment of infected individuals. While the epidemiological situation has remained largely unchanged since the emergence of MERS, large outbreaks of nosocomial infections are worrisome. These call for the development of specific intervention strategies, should the virus become efficiently transmissible among humans. Prevention at the source, for example through the vaccination of dromedary camels against MERS-CoV, is essential to limit cross-species transmission of the virus from animals to humans. However, the development of specific intervention strategies to be rapidly deployed in humans is urgently needed, and should be undertaken ahead of a potential change in the ability of MERS-CoV to transmit among humans. This may necessitate profound changes in the interactions between the public and private sectors for the development of new medicines, as was recently learned upon emergence of the Ebola virus [26].
5. Ebola virus
The Ebola virus is a member of the Filoviridae, recognized as a virus family for close to half a century [27]. It was first identified in 1976 in the Democratic Republic of Congo (formerly Zaire) and Sudan, and continued to occasionally emerge in various regions of Africa. Until 2013, they caused isolated and largely self-limiting outbreaks in humans, of up to several hundred fatal cases each. The case-fatality rate of Ebola virus infections generally ranges from 50% to 90%, depending among other factors on the virus strain involved. Bats are believed to be the natural host reservoirs of Ebola and other filoviruses, and remain largely asymptomatic upon infection [28]. The viruses may spill over to other animal species, which may demonstrate high mortality rates upon infection. Infected animals that are hunted for bushmeat are considered the most likely source of zoonotic Ebola virus infections in humans. Yet, onward human-to-human transmission, via close contact with bodily fluids, contributes to the virus' further spread.
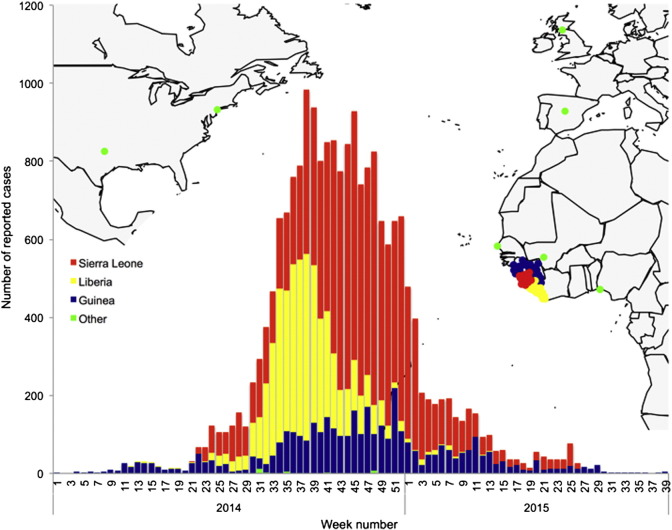
In late 2013, an unprecedented outbreak of Ebola virus infections emerged in West Africa. The virus emerged in Guinea and spread to neighboring Sierra Leone and Liberia, resulting in a massive epidemic, with close to 20,500 reported cases of infection (Table 5). Imported cases of Ebola virus infection were reported in other African countries, as well as in Europe and North America. The massive epidemic covered a period of about a year, yet cases of infection continue to be reported in Guinea in a staggering epidemic tail (Fig. 5). It remains unsure whether new epidemic flares will occur.
Table 5.
Number of reported cases of Ebola virus infection from 1 January 2014 to 30 September 2015.
| Country | 2014 | 2015 | Total |
|---|---|---|---|
| Guinea | 2767 | 1025 | 3792 |
| Liberia | 4924 | 70 | 4994 |
| Mali | 8 | 8 | |
| Nigeria | 20 | 20 | |
| Senegal | 1 | 1 | |
| Sierra Leone | 9514 | 2140 | 11,654 |
| Spain | 1 | 1 | |
| UK | 1 | 1 | |
| USA | 4 | 4 | |
| Total | 17,239 | 3236 | 20,475 |
Fig. 5.
Weekly number of reported cases of Ebola virus infection in humans from 1 January 2014 to 30 September 2015 against the geographical distribution of cases in Africa, Europe and North America.
Early Ebola virus outbreaks proved to be largely self-limiting. The devastating nature of the infection resulted in the rapid implementation of precautionary measures at the local level, thereby interrupting chains of human-to-human transmission after initial zoonotic events [29]. The reasons why this apparently failed to control the recent Ebola epidemic are unclear. However, its emergence in more uniformly and densely inhabited West African regions, poverty, larger scale movement and travel, as well as distrusting societal and behavioral responses to public health measures, are likely to have contributed to the escape of the virus into wider geographical areas, and to its associated exponential growth phase [30]. No specific medical interventions, including the use of antiviral drugs, antibodies or vaccines, were available at the time of the crisis, in spite of promising data indicating avenues for their development. One of the reasons behind the absence of specific medical interventions is related to “the industry paradox” based on a generally perceived absence of return on investment [26]. However, the scale of the epidemic, a trend towards fast-track registration for medicines against emerging pathogens, and the advent of public–private-partnerships, open prospects for the private sector to invest ahead of crises, with benefits for the entire society.
References
- 1.Gortazar C., Reperant L.A., Kuiken T. Crossing the interspecies barrier: opening the door to zoonotic pathogens. PLoS Pathog. 2014;10(6) doi: 10.1371/journal.ppat.1004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson P.K., Cunningham A.A., Patel N.G., Morales F.J., Epstein P.R., Daszak P. Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 2004;19(10):535–544. doi: 10.1016/j.tree.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 3.Taylor L.H., Latham S.M., Woolhouse M.E. Risk factors for human disease emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001;356(1411):983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woolhouse M., Gaunt E. Ecological origins of novel human pathogens. Crit. Rev. Microbiol. 2007;33(4):231–242. doi: 10.1080/10408410701647560. [DOI] [PubMed] [Google Scholar]
- 5.Lloyd-Smith J.O., George D., Pepin K.M. Epidemic dynamics at the human–animal interface. Science. 2009;326(5958):1362–1367. doi: 10.1126/science.1177345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Field H.E., Mackenzie J., Daszak P. Henipaviruses: emerging paramyxoviruses associated with fruit bats. In: Childs J., Mackenzie J., Richt J., editors. Wildlife and Emerging Zoonotic Diseases: The Biology, Circumstances and Consequences of Cross-Species Transmission SE - 7. 315. Springer Berlin Heidelberg DA - 2007/01/01; 2007. pp. 133–159. [Google Scholar]
- 7.To K.K., Chan J.F., Chen H., Li L., Yuen K.Y. The emergence of influenza A H7N9 in human beings 16 years after influenza A H5N1: a tale of two cities. Lancet Infect. Dis. 2013;13(9):809–821. doi: 10.1016/S1473-3099(13)70167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y., Liang W., Yang S. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet. 2013;381(9881):1916–1925. doi: 10.1016/S0140-6736(13)60903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiong X., Martin S.R., Haire L.F. Receptor binding by an H7N9 influenza virus from humans. Nature. 2013;499(7459):496–499. doi: 10.1038/nature12372. [DOI] [PubMed] [Google Scholar]
- 10.Tharakaraman K., Jayaraman A., Raman R. Glycan receptor binding of the influenza A virus H7N9 hemagglutinin. Cell. 2013;153(7):1486–1493. doi: 10.1016/j.cell.2013.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reperant L.A., Grenfell B., Osterhaus A.D.M.E. Quantifying the risk of pandemic influenza virus evolution by mutation and re-assortment. Vaccine. 2015;33:6955–6966. doi: 10.1016/j.vaccine.2015.10.056. [DOI] [PubMed] [Google Scholar]
- 12.de Jong J.C., Claas E.C., Osterhaus A.D., Webster R.G., Lim W.L. A pandemic warning? Nature. 1997;389(6651):554. doi: 10.1038/39218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reperant L.A., Osterhaus A.D.M.E., Kuiken T. Influenza virus infections. In: Gavier-Widen D., Duff P., Meredith A., editors. Infectious Diseases of Wild Mammals and Birds. Wiley-Blackwell; Europe: 2012. [Google Scholar]
- 14.Keawcharoen J., van Riel D., van Amerongen G. Wild ducks as long-distance vectors of highly pathogenic avian influenza virus (H5N1) Emerg. Infect. Dis. 2008;14(4):600–607. doi: 10.3201/eid1404.071016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reperant L.A., Rimmelzwaan G.F., Kuiken T. Avian influenza viruses in mammals. Rev. Sci. Tech. 2009;28(1):137–159. doi: 10.20506/rst.28.1.1876. [DOI] [PubMed] [Google Scholar]
- 16.Chen J.M., Chen J.W., Dai J.J., Sun Y.X. A survey of human cases of H5N1 avian influenza reported by the WHO before June 2006 for infection control. Am. J. Infect. Control. 2007;35(7):467–469. doi: 10.1016/j.ajic.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Yuan J., Zhang L., Kan X. Origin and molecular characteristics of a novel 2013 avian influenza A(H6N1) virus causing human infection in Taiwan. Clin. Infect. Dis. 2013;57(9):1367–1368. doi: 10.1093/cid/cit479. [DOI] [PubMed] [Google Scholar]
- 18.Verhagen J.H., Herfst S., Fouchier R.A.M. How a virus travels the world. Science. 2015;347:616–617. doi: 10.1126/science.aaa6724. [DOI] [PubMed] [Google Scholar]
- 19.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 20.Raj V.S., Osterhaus A.D., Fouchier R.A., Haagmans B.L. MERS: emergence of a novel human coronavirus. Curr. Opin. Virol. 2014;5:58–62. doi: 10.1016/j.coviro.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ithete N.L., Stoffberg S., Corman V.M. Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerg. Infect. Dis. 2013;19(10):1697–1699. doi: 10.3201/eid1910.130946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Memish Z.A., Mishra N., Olival K.J. Middle East respiratory syndrome coronavirus in bats. Emerg. Infect. Dis. 2013;19(11):1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reusken C.B., Haagmans B.L., Muller M.A. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 2013;13(10):859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raj S., Farag E., Reusken C. Isolation of MERS coronavirus from a dromedary camel, Qatar. Emerg. Infect. Dis. 2014 doi: 10.3201/eid2008.140663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raj V.S., Mou H., Smits S.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reperant L.A., van de Burgwal L.H., Claassen E., Osterhaus A.D. Ebola: public-private partnerships. Science. 2014;346(6208):433–434. doi: 10.1126/science.346.6208.433-b. [DOI] [PubMed] [Google Scholar]
- 27.Kiley M.P., Bowen E.T., Eddy G.A. Filoviridae: a taxonomic home for Marburg and Ebola viruses? Intervirology. 1982;18(1-2):24–32. doi: 10.1159/000149300. [DOI] [PubMed] [Google Scholar]
- 28.Olival K.J., Hayman D.T. Filoviruses in bats: current knowledge and future directions. Viruses. 2014;6(4):1759–1788. doi: 10.3390/v6041759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leroy E.M., Rouquet P., Formenty P. Multiple Ebola virus transmission events and rapid decline of central African wildlife. Science. 2004;303(5656):387–390. doi: 10.1126/science.1092528. [DOI] [PubMed] [Google Scholar]
- 30.Chan M. Ebola virus disease in West Africa — no early end to the outbreak. N. Engl. J. Med. 2014;371:1183–1185. doi: 10.1056/NEJMp1409859. [DOI] [PubMed] [Google Scholar]