Abstract
Intense efforts are underway to identify inhibitors of the enzyme gamma‐glutamyl transpeptidase 1 (GGT1) which cleaves extracellular gamma‐glutamyl compounds and contributes to the pathology of asthma, reperfusion injury and cancer. The glutamate analog, 6‐diazo‐5‐oxo‐norleucine (DON), inhibits GGT1. DON also inhibits many essential glutamine metabolizing enzymes rendering it too toxic for use in the clinic as a GGT1 inhibitor. We investigated the molecular mechanism of human GGT1 (hGGT1) inhibition by DON to determine possible strategies for increasing its specificity for hGGT1. DON is an irreversible inhibitor of hGGT1. The second order rate constant of inactivation was 0.052 mM −1 min−1 and the K i was 2.7 ± 0.7 mM. The crystal structure of DON‐inactivated hGGT1 contained a molecule of DON without the diazo‐nitrogen atoms in the active site. The overall structure of the hGGT1‐DON complex resembled the structure of the apo‐enzyme; however, shifts were detected in the loop forming the oxyanion hole and elements of the main chain that form the entrance to the active site. The structure of hGGT1‐DON complex revealed two covalent bonds between the enzyme and inhibitor which were part of a six membered ring. The ring included the OG atom of Thr381, the reactive nucleophile of hGGT1 and the α‐amine of Thr381. The structure of DON‐bound hGGT1 has led to the discovery of a new mechanism of inactivation by DON that differs from its inactivation of other glutamine metabolizing enzymes, and insight into the activation of the catalytic nucleophile that initiates the hGGT1 reaction.
Keywords: enzyme inactivation, enzyme kinetics, crystal structure, protein conformation, human gamma‐glutamyl transpeptidase, gamma‐glutamyl transferase
Short abstract
PDB Code(s): 5V4Q
Introduction
Gamma‐glutamyl transpeptidase (GGT1, a.k.a. gamma‐glutamyl transferase) is a cell surface enzyme that cleaves extracellular gamma‐glutamyl compounds including oxidized and reduced glutathione and glutathione S‐conjugates.1 Inhibitors of the enzyme are being sought due to their therapeutic potential in the treatment of asthma, reperfusion injury, cardiovascular disease and cancer.1, 2, 3, 4
To aid in the development of novel inhibitors of hGGT1 we have investigated the molecular mechanisms by which known inhibitors block enzymatic activity.5 A series of glutamate analogs including 6‐diazo‐5‐oxo‐L‐norleucine (DON) inhibit GGT1 activity, but these inhibitors lack specificity.6, 7 DON also inhibits glutamine metabolizing enzymes including glutaminases and L‐asparagine synthetase and has significant toxic side effects.8, 9, 10, 11, 12, 13, 14
DON is a gamma‐glutamyl diazocompound (Fig. 1). Previous studies have shown DON to be an irreversible inhibitor of human GGT1 (hGGT1), binding stoichiometrically to the small subunit of the enzyme.15 No structures have been reported for DON bound to any GGT, prokaryotic or eukaryotic. We investigated the binding mode of DON to hGGT1 and found that it differed from the interactions described for any other enzyme which has provided insight into its mechanism of inhibition of hGGT1. We also conducted kinetic analysis of the mechanism of inhibition of purified hGGT1 by DON and are the first to report the K i and rate constant of inactivation of hGGT1 by DON.
Figure 1.
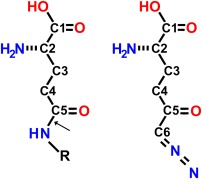
Chemical structures of a generic gamma‐glutamyl compound (left) and DON (right). Arrow indicates the gamma‐glutamyl bond in the generic gamma‐glutamyl compound.
Results
Inactivation of hGGT1 by DON
Preincubation of hGGT1 with DON resulted in time‐ and dose‐dependent inhibition with a K i of 2.7 ± 0.7 mM and the maximum k inact was 0.13 min−1 (Fig. 2). The data for the inactivation were first‐order [Fig. 2(B)]. DON was an irreversible inhibitor of hGGT1 with the second order rate constant of inactivation, k inact/K i = 0.052 mM −1 min−1. Thermofluor studies showed that inactivation with DON stabilized the structure of hGGT1 in solution. We determined the melting temperature of hGGT1 inactivated with DON at six different pHs in six different buffers. The data showed that among the conditions tested the enzyme is most stable at pH 6.7 in sodium citrate buffer (Table 1). Inactivating hGGT1 with DON further stabilized the structure of the enzyme under all conditions tested. It increased the melting temperature by 13°C to 16°C and reduced the effect of pH. Previously, we found that GGsTop, a glutamate analog that inactivates hGGT1 by forming one covalent and 11 hydrogen bonds with the enzyme, increased the melting temperature by 20°C.5 The data in Table 1 indicate that DON forms multiple interactions with the enzyme thereby stabilizing the structure in solution.
Figure 2.
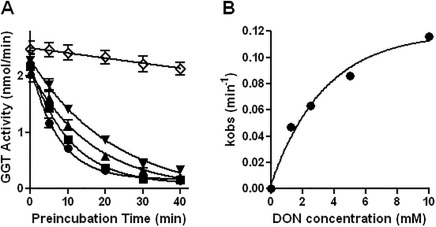
Inhibition of hGGT1 by DON. (A) hGGT1 was preincubated at 37°C with 10 mM (closed circles), 5 mM (closed squares), 2.5 mM (closed triangles), 1.25 mM (inverted closed triangles) or 0 mM DON (open diamonds) for the time indicated. Immediately following the preincubation, the enzyme was assayed for activity. Data points are average of mean values for four experiments ± standard error. For some data points the error bars are smaller than the symbol. (B) The data for the inactivation of hGGT1 by DON were first‐order.
Table 1.
Melting Temperatures from Thermofluor Graphs for Deglycosylated hGGT1
| Buffers\Protein | hGGT1 (°C)a | hGGT1‐bound DON (°C) |
|---|---|---|
| BisTris pH 5 | 55 | 70 |
| NaAc pH 5.5 | 53 | 69 |
| HEPES pH 6.5 | 56 | 71 |
| Na Citrate pH 6.7 | 58 b | 71 |
| HEPES pH 7.0 | 57 | 72 b |
| Tris‐HCl pH 7.0 | 56 | 71 |
| 2M K/NaPO4 pH 7.0 | 55 | 71 |
| Tris‐HCl pH 7.9 | 54 | 71 |
Data for hGGT1 published previously.5
Boldface type indicates the conditions under which hGGT1 is most stable.
Structure of hGGT1‐DON Complex
hGGT1 is synthesized as a single polypeptide then autocleaves into an enzymatically active heterodimer with a large and a small subunit.16 We solved the structure of the DON‐hGGT1 complex at 2.2 Å resolution by the difference Fourier method using the apo‐form of hGGT1 with the water molecules and cofactor atoms removed as the starting model (Table 2, Fig. 3). The overall structure of the complex resembles our previously published structure of the hGGT1 apo‐enzyme (4Z9O).5 The enzyme is composed of two central β‐sheets surrounded by α‐helices. LSQ superposition of the CA atoms of the enzyme in the apo‐form and in complex with DON showed no significant differences between the two structures (rmsd deviation was 0.2 Å for the 519 CA atoms of the whole molecule, 0.2 Å for the large subunit, and 0.2 Å for the small subunit). The largest movement was detected in the loop forming the oxyanion hole (residues 472–475). In the DON‐bound hGGT1, the loop was shifted toward the bound inhibitor molecule relative to the position of the loop in the apo‐enzyme, displacing the CA atoms of Gly473 and Gly474 by 0.9 Å and 1.2 Å, respectively. Other noticeable movements of the main chain of the enzyme in the complex included shifts in the lid loop (residues 427–437), the loop connecting strands β16 and β17 (residues 506–512) and C terminal part of helix α14 (residues 319–332). These three elements form the upper cover of the entrance to the active site. None of these shifts is more than 1 Å and therefore would not block access to the active site. In the structure of the hGGT1‐DON complex [Fig. 4(A)], the catalytic residue Thr381 was detected in only one conformation that resembles one of two conformations of this residue (CHI = −169) in the apo‐hGGT1 structure (4Z9O). But, Thr381 is further rotated around the C‐CA bond changing the corresponding torsion angle from −40.9° to −31.1°. There was also a small change in the CHI angle of the side chain of Thr381 from −166.5° to −175.5°. This orientation is almost identical to the orientation of Thr381 in our structures of hGGT1 with substrate or inhibitors bound in the active site via an acyl bond with the side chain oxygen of Thr381. These structures include hGGT1‐serine‐borate (4ZC6) and hGGT1‐GGsTop (4ZBK).5
Table 2.
Diffraction Data and Refinement Statistics for DON‐Bound hGGT1
| Data Collection | |
| Protein Data Bank code | 5V4Q |
| Space Group | C2221 |
| Unit cell (Å) | 105.5, 127.3, 103.1 |
| Resolution (Å) | 100‐2.2 (2.24–2.20) |
| No. of reflections | 34792 (1661) |
| Data cutoff | −3σ |
| Completeness (%) | 99.7 (97.6) |
| Redundancy | 5.5 (4.0) |
| /<σ> | 12.6 (2.3) |
| R merge(%)a | 11.5 (59.6) |
| Bwilsonb | 39.7 |
| Refinement | |
| Resolution high (Å) | 2.2 (2.26–2.2) |
| No. of reflections | |
| Work set | 33034 (1912) |
| Free set | 1738 (95) |
| Overall | 34772 (2007) |
| No. of atoms | 4476 |
| R work (%) | 16.33 (35.0) |
| R free (%) | 21.89 (36.8) |
| R overall (%) | 16.60 |
| Figure of merit | 82.44 |
| Correlation coefficient | 0.97 |
| Mean B (Å2) | |
| All | 44.00 |
| Subunit A | 44.26 |
| Subunit B | 41.46 |
| Inhibitor | 49.10 |
| Water | 51.62 |
| Cofactors (Cl, Na) | 61.63 |
| Carbohydrate | 77.2 |
| Estim coord error based on likelihood (Å) | 0.14 |
| Estim B value error (Å2) | 5.71 |
| RMS from ideal values | |
| Bonds | 0.013 |
| Angles | 1.48 |
| Ramachandran Plotc | |
| Favored (%) | 97.7 |
| Allowed (%) | 2.3 |
R merge = (Σ H Σ j |I hj – <I H >|)/(Σ H Σ j <I H >), where I Hj is the jth observation of reflection H.
Bwilson, B‐factor determined from Wilson plot.
Ramachandran analysis was performed with Rampage in CCP4 suite.
Values in parenthesis refer to the highest resolution shell.
Figure 3.
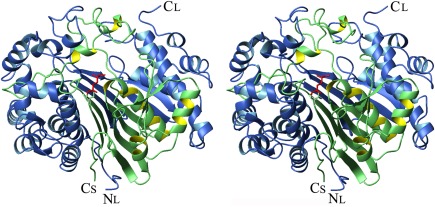
Crystal structure of DON‐bound hGGT1. A stereo ribbon presentation of the hGGT1 heterodimer with DON bound in the active site (5V4Q). The large subunit (chain A) is colored blue, and the small subunit (chain B) is colored green. Thr381, the N‐terminus of the small subunit, is shown as a green stick figure. The DON atoms are colored red (the diazo group of DON is released when DON binds to the enzyme and is not present in the final structure).
Figure 4.
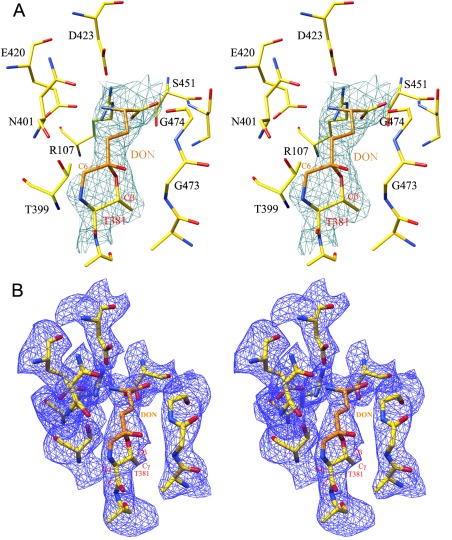
DON‐bound hGGT. (A) Stereo presentation of the model of DON with diazo nitrogen atoms removed fitted into initial F o – F c density map (contoured at 3σ level). (B) Final 2F o – F c map for DON molecule (contoured at 1.5σ level). Enzyme carbon atoms are colored yellow, DON carbon atoms are colored orange, oxygens are red, and nitrogens are blue.
The initial 2F o – F c and F o – F c difference Fourier maps calculated after 10 cycles of rigid body refinement of the starting model revealed a clear density for a molecule bound in the active site of the hGGT1 (Figs. 3 and 4). A molecule of DON was modeled into the density. The electron density maps showed a clear density between atoms C5 of DON and OG of Thr381 and density between the C6 of DON and N terminal nitrogen of small subunit (α‐nitrogen of Thr381). The distances between the pairs of atoms are 1.4 Å and 1.3 Å, respectively, which indicated a covalent bond. The existence of a covalent bond between C6 of DON and the N terminal nitrogen was so unexpected that the initial cycles of refinement were carried out with a molecule of DON lacking the diazocarbon (C6) and the diazonitrogen atoms modeled into the density. This approach allowed us to confirm that the DON molecule had been cleaved between the diazocarbon and diazonitrogens and detect its position without introducing bias into calculated maps. Further refinements were carried out with a molecule of DON lacking only the diazonitrogen atoms. Final R work and R free values were 16.3% and 21.9%, respectively and the final 2F o – F c map is shown in Figure 4(B). The two covalent bonds between DON and hGGT1 resulted in the formation of a six member ring consisting of the N, CA, CB, OG atoms of Thr381 and the C5 and C6 atoms of DON (Fig. 5). Such a complex is stable and, in agreement with the biochemical data, would irreversibly inhibit hGGT1.
Figure 5.
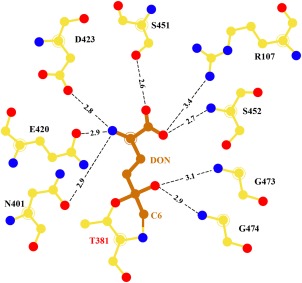
DON‐bound hGGT. Schematic representation of interactions of DON in the active site of hGGT1. Enzyme carbon atoms are colored yellow, DON carbon atoms are colored orange, oxygens are red, and nitrogens are blue. The distances shown are in Angstroms.
In the refined structure the α‐carboxy and the α‐amino groups of the DON molecule occupy the same position as the corresponding groups of glutamate in the glutamate‐bound structure (4ZCG), and participate in the same extensive network of hydrogen bonds and charge interactions with enzyme (Fig. 5). The α‐carboxyl group of DON formed hydrogen bonds with Ser‐451 OG (2.6 Å), the main chain nitrogen of Ser‐452 (2.7 Å) and a weak hydrogen bond with Arg‐107 NH1 (3.4 Å). The α‐amino group of DON formed bonds with Asn‐401 OD1 (2.9 Å), Glu‐420 OE1 (2.9 Å), and Asp‐423 OD2 (2.8 Å). The oxyanion (O3 atom of DON) hydrogen bonds with the main chain nitrogens of Gly473 and Gly474, at distances of 3.1 Å and 2.9 Å, respectively.
Discussion
Kinetic data showed DON to be an irreversible inhibitor of hGGT1. It inactivates the enzyme, but it is a weak inhibitor (K i 2.7 ± 0.7 mM) with a slow rate constant of inactivation (0.052 mM −1 min−1). Prior to crystallizing the enzyme we incubated it for 48 hours with DON under conditions that completely inactivated the enzyme. Our structure of hGGT1‐DON, shows a cleaved product of DON covalently bound in the active site. DON forms two covalent bonds with the enzyme, one between the OG atom of Thr381 and the C5 of DON and the second between the N‐terminal nitrogen of Thr381 and C6 of DON. This is the first structure of any enzyme inhibited by DON in which covalent bonds form between two atoms of enzyme and two different carbons of DON.
All reported structures of DON‐enzyme complexes show different interactions from those that we observed in the hGGT1‐DON complex. DON also inhibits glutamine metabolizing enzymes including glutaminases and L‐asparagine synthetase. A search of the protein data bank yielded crystal structures of 11 enzymes with DON or its cleaved product 5‐oxo‐L‐norleucine (ONL) bound in the active site (1DJP, 1OFE, 1ECC, 1ECG, 2J6H, 2OSU, 2Q3Z, 3BRM, 3DLA, 4O7D, 4ZDK). Ten of these structures have been published.9, 11, 17, 18, 19, 20, 21, 22, 23, 24 The structures of seven glutamine metabolizing enzymes co‐crystalized with DON showed a covalent bond between diazocarbon (C6) of DON and either the side chain oxygen of a serine nucleophile or the sulfur atom of a cysteine nucleophile in the active site of the enzyme. Those enzymes include: human glutaminase (4O7D), human transglutaminase 2 (2Q3Z), E. coli glutamine phosphoribosylpyrophosphate amidotransferase (1ECC, 1ECG), E. coli glucosamine‐6‐phosphate synthase (2J6H), Bacillus subtilis glutaminase YbgJ (3BRM), Mycobacterium glutamine‐dependent NAD+ synthetase (3DLA), and M. tuberculosis CTP synthase PyrG (4ZDK).2, 9, 11 0–24 The structure of glutamate synthase from the cyanobacterium synechocystis sp (1OFE) did not show any covalent bonds with DON.18 The crystal structure of Pseudomonas 7A glutaminase‐asparaginase‐DON complex revealed the carbonyl carbon of DON (C5) formed covalent bonds with the side chain oxygen of both a Thr and a Tyr.17 In addition, the structure of the hGGT1‐DON complex that we observed differs from the structure proposed by Tate and Meister, who predicted that the C6 carbon of DON forms a covalent bond with the side chain oxygen of a nucleophile in the active site of GGT1.25
Based on our structure, we propose a novel mechanism of enzyme inactivation by DON (Fig. 6), in which the initial interactions between DON and the enzyme are the same as those that occur when a gamma‐glutamyl substrate binds to the enzyme. The α‐nitrogen and α‐carboxy oxygens of the gamma‐glutamyl substrate or DON form hydrogen bonds with multiple atoms of the enzyme thereby binding the substrate or DON in the active site and aligning the C5 of the substrate or DON with Thr381. We propose that the amine of Thr381 (the N‐terminus of the small subunit of hGGT1) activates the nucleophile by accepting a proton from the OG atom of Thr381 via the OG atom of Thr399. The OG atom of Thr381 then initiates a nucleophilic attack on the C5 atom of the gamma‐glutamyl substrate or DON which results in the formation of a tetrahedral intermediate (Fig. 6). The formation of the tetrahedral intermediate and its stabilization is aided by the interaction of the carboxy‐oxygen of the substrate or DON with the main chain nitrogen atoms of Gly473 and Gly474, which form the oxyanion hole within the active site. Upon formation of the tetrahedral intermediate, we propose that the proton from the amine of Thr381 is transferred to the gamma‐glutamyl nitrogen of the substrate or the C6 of DON. When a gamma‐glutamyl substrate such as glutathione is bound in the active site, the tetrahedral intermediate collapses. The C‐N gamma‐glutamyl bond is cleaved and the first product (cysteinyl‐glycine) is released. Subsequent attack of a water molecule hydrolyzes the acyl bond (between the OG of Thr381 and the C5 of the gamma‐glutamyl substrate) with release of glutamate, the second product of the reaction, and free enzyme. DON does not contain a C‐N gamma‐glutamyl bond, rather there is a diazocarbon in place of the nitrogen. Upon formation of the DON‐tetrahedral intermediate, we propose an electron pair on N7 of DON migrates to generate an N‐N triple bond as the C6 of DON is protonated by the α‐amine of Thr381. The resulting neutral α‐amine of Thr381 then attacks the C6 of DON. N2 is released and a covalent bond is formed between the C6 of DON and the α‐nitrogen of Thr381. The formation of the second covalent bond between DON and the enzyme results in a six membered ring consisting of the N, CA, CB, OG of Thr381 and the C5 and C6 atoms of DON as observed in our structure (Figs. 4, 5, 6). Upon formation of this stable complex hGGT1 is irreversibly inhibited.
Figure 6.
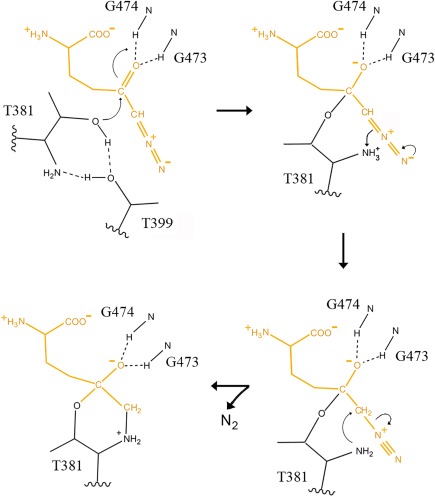
Proposed mechanism of hGGT1 inactivation by DON. The amine of Thr381 activates the nucleophile by accepting a proton from the OG oxygen of Thr381 via OG atom of Thr399. The OG atom of Thr381 attacks the carbonyl carbon (C5) of the DON molecule (top left structure). These results in the formation of a tetrahedral adduct which is stabilized by the interaction between the carboxy‐oxygen of DON and the main chain nitrogen atoms of Gly473 and Gly474 (top right structure). An electron pair on N7 of DON migrates to generate an N‐N triple bond and a proton is transferred from the amine of Thr381 to the C6 of DON (bottom right structure). Attack of the α‐nitrogen of Thr381 on the C6 of DON cleaves the single C‐N bond of the DON molecule releasing N2 and results in the formation of a covalent bond between the α‐nitrogen of Thr381 and C6 of DON. The final product (detected in the x‐ray structure, Figs. 3–5) contains a six‐membered ring composed of N, CA, CB, OG of the Thr381 and the C5 and C6 atoms of DON (bottom left structure).
Based on the structure of the hGGT1‐DON complex we suggest the activation of the OG atom of Thr381, the catalytic nucleophile that initiates the reaction, is via transfer of a proton from the OG oxygen of Thr381 to the α‐amine of Thr381 via the OG atom of Thr399. The proton on the α‐amine of Thr381 is the source of the proton that protonates the initial product of the hGGT1 reaction. When DON is bound to hGGT1 the proton protonates the leaving group of DON. This is consistent with our observation that in the structure of apo‐hGGT1 the OG of Thr399 is within hydrogen bond distance of both the side chain hydroxyl of Thr381 and the α‐amine of Thr381.5 Menard and colleagues conducted in depth kinetic studies of the reaction catalyzed by rat GGT1.26 They were not aware that the N‐terminal Thr of the small subunit of rat GGT1 (Thr380) was the catalytic nucleophile. Their kinetic data led them to propose that the tetrahedral intermediate undergoes a general‐acid catalyzed breakdown in which a proton is transferred to the nitrogen of the gamma‐glutamyl C‐N bond. Their data were consistent with an ammonium ion acting as a general acid. They hypothesized that a nearby His imidazolium or primary ammonium group was acting as the general acid catalyst. Our data are consistent with the proposal that the proton from the OG oxygen of Thr381 transferred via the OG atom of Thr399 to the α‐amine of Thr381 is the source of the proton.
Azaserine, a diazo compound, is structurally similar to DON differing by only one atom, an oxygen in place of the C4 of DON. Azaserine also inhibits GGT1.27 Structures of E. coli GGT1 in complex with azaserine have been reported by Wada and colleagues (2Z8I, 2Z8J).28 The structures showed an acyl bond between the carbonyl carbon (C4) of azaserine and the OG atom of Thr391 (the nucleophilic residue of E. coli GGT1) similar to the covalent bond we observed between the carbonyl carbon of DON (C5) and the OG of Thr381 of hGGT1. In one of their structures, 2Z8I, Wada and colleagues detected azaserine in the active site lacking the diazo‐nitrogens. They proposed that azaserine forms a tetrahedral intermediate with E. coli GGT that would collapse with the release of a degraded molecule of azaserine and free enzyme. This differs from our structure of the hGGT1‐DON complex which shows a stable structure consistent with inactivation of hGGT1 by DON. There are several possible explanations for the differences in the crystal structures of the E. coli‐azaserine complex and the hGGT1‐DON complex. First: there are differences in electronic states of carbonyl and diazo carbons in the two inhibitor molecules as a result of substitution of the C4 atom of DON with O in azaserine. Second: there are differences in the environment surrounding the catalytic Thr residue in E. coli GGT1 versus hGGT1. LSQ superposition of apo forms of the human and E. coli GGT1 structures show that in hGGT1 the catalytic Thr is closer to the oxyanion loop (residues 473–475 in hGGT1) than it is in E. coli GGT1 due to replacement of Ala472 of the human enzyme by more bulky and rigid Pro482 in E. coli. In addition, the loop on the opposite side of the catalytic Thr differs between hGGT1 and E.coli: AHSMG (residues 80–84) in hGGT1 and PQAGN (residues 88–92) in E. coli GGT. This loop is further from the catalytic Thr in E. coli than it is in hGGT and the main chain torsion angles differ between the two enzymes. These shifts result in different networks of interactions between the catalytic Thr and other amino acids in the two enzymes.
The data from this study on the interactions between DON and hGGT1 will aid in the design of new generations of inhibitors that are more specific for hGGT1 and are less toxic.
Materials and Methods
hGGT1 Expression and Purification
For kinetic and crystallization studies, the natural variant V272A of hGGT1 (P19440) was expressed in Pichia pastoris strain X‐33 and purified as described previously.29 The purified protein was deglycosylated prior to the thermal stability studies and crystallization.29
GGT1 Activity
A transpeptidation assay was used to quantify hGGT1 activity. The assay has been described previously.30 The concentration of the substrate L‐gamma‐glutamyl para‐nitroanalide (L‐GpNA, Sigma) was 3 mM with 40 mM GlyGly (Sigma) as the acceptor. The assay buffer contained: 100 mM Na2HPO4, 3.2 mM KCl, 1.8 mM KH2PO4, and 27.5 mM NaCl pH 7.4. To initiate the reaction, 2 milliunits of hGGT1 were added. One unit of GGT activity was defined as the amount of enzyme that released 1 μmol of p‐nitroaniline/min at 37°C at pH 7.4. The K m of L‐GpNA in the transpeptidation reaction is 1.2 ± 0.1 mM.31
Inhibition of hGGT1 by DON
Two milliunits of hGGT1 and DON were preincubated at 37°C in a total volume of 20 µL assay buffer. The concentrations of DON in the preincubation mixture were 0, 1.25, 2.5, 5.0, and 10 mM. Immediately following the preincubation, the reaction mix (80 µL, 37°C) containing L‐GpNA and GlyGly was added and the amount of product released was monitored continuously at OD405 for 5 min.
Data Analysis
The pseudo‐first order rate constant of inactivation (k inact) of hGGT1 by DON was obtained as the slope of a plot of the natural log of hGGT1 activity versus time of inactivation.32 The k obs for each dose of DON was calculated from the initial data of DON concentration versus GGT activity. To determine the K i and second order rate constant, the reciprocal of the absolute value of k obs was plotted versus the reciprocal of the DON concentration. The reciprocal of the slope was the second order rate constant for inactivation starting with free enzyme and free DON.32 The slope divided by the Y‐intercept provided the K i for DON.32 Graphs were prepared with GraphPad Prism Software (San Diego, CA).
Thermal Stability of hGGT1
The protein sample alone or complexed with DON consisted of 0.1 mg/mL deglycosylated hGGT1 in 10 mM HEPES buffer, pH 7.5, 150 mM NaCl, and 5x SYPRO Orange. Each well of a 96‐well plate contained 12 μL of the protein sample and 4 μL of 0.1 M screening buffer. Six buffers with 6 different pH levels were used. The fluorescence of the protein‐bound SYPRO Orange was measured over a temperature range of 25–95°C as previously described.5
Crystallization Conditions and Cryopreservation
The protein stock solution contained 4.3 mg/mL hGGT1 in 50 mM HEPES, pH 8.0, 0.5 mM EDTA, and 0.02% sodium azide. Two µl of 0.5 M DON (in water) was mixed with 50 µL of protein solution. The mixture was incubated for 48 h at 4°C. Under these conditions the enzyme was found to be completely inactivated. Crystals of the hGGT1‐DON complex were prepared at room temperature. The crystallization drops were composed of 2 μL protein solution, 1.7 μL H2O, and 2 μL reservoir solution. They were equilibrated against 500 μL of reservoir solution containing 20–25% PEG3350, 0.1 M Na Cacodylate buffer pH 6.0 and 0.1 M ammonium chloride. Microseeding with previously grown crystals of apo‐hGGT1 facilitated crystal growth. Crystals appeared in 1 or 2 days after seeding. After an additional week, the crystals reached their final size of 0.05 mm × 0.1 mm × 0.5 mm. Crystals were quickly dragged through a cryoprotectant solution consisting of the reservoir solution with 15% PEG 1500 and 20 mM DON added. The crystals were rapidly frozen by immersion in liquid nitrogen.
Data Collection
hGGT1‐DON crystals were screened at the Laboratory of Biomolecular Structure and Function at the University of Oklahoma Health Sciences Center (Oklahoma City, OK). The x‐ray diffraction data were collected at 100 K at beam line X25 at the National Synchrotron Light Source (Brookhaven, NY). The beam line was equipped with a Pilatus 6M detector, and data were collected at 1.1 Å wavelength. The diffraction data were processed with HKL‐2000 suite.33
Structural Determination, Refinement, and Analysis
The unit cell parameters for the hGGT1‐DON crystals were isomorphous with those of the apo‐form of hGGT1 (4Z9O), therefore rigid body refinement with REFMAC 5 was used to position the apo‐form structure (without alternative conformations, water molecules or Cl and Na ions) in the unit cell.34, 35 Weighted difference 2F o – F c and F o – F c maps were used for detection of a bound inhibitor molecule. Computer graphics program COOT was used to correct the model.34, 36 In the last stages of refinement cofactor atoms (chlorine and sodium) and water molecules were added to the structure using COOT. The refinement statistics and Protein Data Bank accession code are listed in Table 2. The figures were made with Molmol, LIGPLOT, and Chimera.37, 38, 39
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors gratefully acknowledge Ms. Nancy Wakeham's assistance during the purification of hGGT1 and in making Figure 2.
IMPACT STATEMENT: The cell surface enzyme gamma‐glutamyl transpeptidase (GGT1) has been implicated in the progression of asthma reperfusion injury and cancer. There are no inhibitors that can be used clinically to inhibit GGT1. This study focused on the mechanism by which 6‐diazo‐5‐oxo‐norleucine inhibits GGT1. The data revealed a novel mechanism of inhibition and new insight in to the activation of the catalytic nucleophile that initiates the reaction. These data will aid in the design of novel GGT1 inhibitors.
REFERENCES
- 1. Hanigan MH (2014) Gamma‐glutamyl transpeptidase: redox regulation and drug resistance. Advan Cancer Res 122:103–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tuzova M, Jean JC, Hughey RP, Brown LA, Cruikshank WW, Hiratake J, Joyce‐Brady M (2014) Inhibiting lung lining fluid glutathione metabolism with ggstop as a novel treatment for asthma. Front Pharmacol 5:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yamamoto S, Watanabe B, Hiratake J, Tanaka R, Ohkita M, Matsumura Y (2011) Preventive effect of ggstop, a novel and selective gamma‐glutamyl transpeptidase inhibitor, on ischemia/reperfusion‐induced renal injury in rats. J Pharmacol Exp Ther 339:945–951. [DOI] [PubMed] [Google Scholar]
- 4. Hanigan MH, Gallagher BC, Townsend DM, Gabarra V (1999) Gamma‐glutamyl transpeptidase accelerates tumor growth and increases the resistance of tumors to cisplatin in vivo. Carcinogenesis 20:553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Terzyan SS, Burgett AW, Heroux A, Smith CA, Mooers BH, Hanigan MH (2015) Human gamma‐glutamyl transpeptidase 1: structures of the free enzyme, inhibitor‐bound tetrahedral transition states, and glutamate‐bound enzyme reveal novel movement within the active site during catalysis. J Biol Chem 290:17576–17586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ahluwalia GS, Grem JL, Hao Z, Cooney DA (1990) Metabolism and action of amino acid analog anti‐cancer agents. Pharmacol Ther 46:243–271. [DOI] [PubMed] [Google Scholar]
- 7. Griffith OW, Meister A (1979) Translocation of intracellular glutathione to membrane‐bound gamma‐glutamyl transpeptidase as a discrete step in the gamma‐glutamyl cycle: glutathionuria after inhibition of transpeptidase. Proc Natl Acad Sci USA 76:268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hartman SC, McGrath TF (1973) Glutaminase a of escherichia coli. Reactions with the substrate analogue, 6‐diazo‐5‐oxonorleucine. J Biol Chem 248:8506–8510. [PubMed] [Google Scholar]
- 9. Brown G, Singer A, Proudfoot M, Skarina T, Kim Y, Chang C, Dementieva I, Kuznetsova E, Gonzalez CF, Joachimiak A, Savchenko A, Yakunin AF (2008) Functional and structural characterization of four glutaminases from escherichia coli and bacillus subtilis. Biochemistry 47:5724–5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rosenbluth RJ, Cooney DA, Jayaram HN, Milman HA, Homan ER (1976) Don, conv and donv‐ii. Inhibition of l‐′asparagine synthetase in vivo. Biochem Pharmacol 25:1851–1858. [DOI] [PubMed] [Google Scholar]
- 11. Thangavelu K, Chong QY, Low BC, Sivaraman J (2014) Structural basis for the active site inhibition mechanism of human kidney‐type glutaminase (kga). Sci Rep 4:3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alt J, Potter MC, Rojas C, Slusher BS (2015) Bioanalysis of 6‐diazo‐5‐oxo‐l‐norleucine in plasma and brain by ultra‐performance liquid chromatography mass spectrometry. Anal Biochem 474:28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cervantes‐Madrid D, Romero Y, Duenas‐Gonzalez A (2015) Reviving lonidamine and 6‐diazo‐5‐oxo‐l‐norleucine to be used in combination for metabolic cancer therapy. BioMed Res Intl 2015:690492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rais R, Jancarik A, Tenora L, Nedelcovych M, Alt J, Englert J, Rojas C, Le A, Elgogary A, Tan J, Monincova L, Pate K, Adams R, Ferraris D, Powell J, Majer P, Slusher BS (2016) Discovery of 6‐diazo‐5‐oxo‐l‐norleucine (don) prodrugs with enhanced csf delivery in monkeys: a potential treatment for glioblastoma. J Med Chem 59:8621–8633. [DOI] [PubMed] [Google Scholar]
- 15. Tate SS, Ross ME (1977) Human kidney gamma‐glutamyl transpeptidase. Catalytic properties, subunit structure, and localization of the gamma‐glutamyl binding site on the light subunit. J Biol Chem 252:6042–6045. [PubMed] [Google Scholar]
- 16. West MB, Wickham S, Quinalty LM, Pavlovicz RE, Li C, Hanigan MH (2011) Autocatalytic cleavage of human {gamma}‐glutamyl transpeptidase is highly dependent on n‐glycosylation at asparagine 95. J Biol Chem 286:28876–28888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ortlund E, Lacount MW, Lewinski K, Lebioda L (2000) Reactions of pseudomonas 7a glutaminase‐asparaginase with diazo analogues of glutamine and asparagine result in unexpected covalent inhibitions and suggests an unusual catalytic triad thr‐tyr‐glu. Biochemistry 39:1199–1204. [DOI] [PubMed] [Google Scholar]
- 18. van den Heuvel RH, Svergun DI, Petoukhov MV, Coda A, Curti B, Ravasio S, Vanoni MA, Mattevi A (2003) The active conformation of glutamate synthase and its binding to ferredoxin. J Mol Biol 330:113–128. [DOI] [PubMed] [Google Scholar]
- 19. Krahn JM, Kim JH, Burns MR, Parry RJ, Zalkin H, Smith JL (1997) Coupled formation of an amidotransferase interdomain ammonia channel and a phosphoribosyltransferase active site. Biochemistry 36:11061–11068. [DOI] [PubMed] [Google Scholar]
- 20. Kim JH, Krahn JM, Tomchick DR, Smith JL, Zalkin H (1996) Structure and function of the glutamine phosphoribosylpyrophosphate amidotransferase glutamine site and communication with the phosphoribosylpyrophosphate site. J Biol Chem 271:15549–15557. [DOI] [PubMed] [Google Scholar]
- 21. Mouilleron S, Badet‐Denisot MA, Golinelli‐Pimpaneau B (2006) Glutamine binding opens the ammonia channel and activates glucosamine‐6p synthase. J Biol Chem 281:4404–4412. [DOI] [PubMed] [Google Scholar]
- 22. Pinkas DM, Strop P, Brunger AT, Khosla C (2007) Transglutaminase 2 undergoes a large conformational change upon activation. PLoS Biol 5:e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. LaRonde‐LeBlanc N, Resto M, Gerratana B (2009) Regulation of active site coupling in glutamine‐dependent nad(+) synthetase. Nat Struct Mol Biol 16:421–429. [DOI] [PubMed] [Google Scholar]
- 24. Mori G, Chiarelli LR, Esposito M, Makarov V, Bellinzoni M, Hartkoorn RC, Degiacomi G, Boldrin F, Ekins S, de Jesus Lopes Ribeiro AL, Marino LB, Centárová I, Svetlíková Z, Blaško J, Kazakova E, Lepioshkin A, Barilone N, Zanoni G, Porta A, Fondi M, Fani R, Baulard AR, Mikušová K, Alzari PM, Manganelli R, de Carvalho LP, Riccardi G, Cole ST, Pasca MR (2015) Thiophenecarboxamide derivatives activated by etha kill mycobacterium tuberculosis by inhibiting the ctp synthetase pyrg. Chem Biol 22:917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tate SS, Meister A (1978) Serine‐borate complex as a transition‐state inhibitor of gamma‐glutamyl transpeptidase. Proc Natl Acad Sci USA 75:4806–4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Menard A, Castonguay R, Lherbet C, Rivard C, Roupioz Y, Keillor JW (2001) Nonlinear free energy relationship in the general‐acid‐catalyzed acylation of rat kidney gamma‐glutamyl transpeptidase by a series of gamma‐glutamyl anilide substrate analogues. Biochemistry 40:12678–12685. [DOI] [PubMed] [Google Scholar]
- 27. Tate SS, Meister A (1977) Affinity labeling of gamma‐glutamyl transpeptidase and location of the gamma‐glutamyl binding site on the light subunit. Proc Natl Acad Sci USA 74:931–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wada K, Hiratake J, Irie M, Okada T, Yamada C, Kumagai H, Suzuki H, Fukuyama K (2008) Crystal structures of escherichia coli gamma‐glutamyltranspeptidase in complex with azaserine and acivicin: novel mechanistic implication for inhibition by glutamine antagonists. J Mol Biol 380:361–372. [DOI] [PubMed] [Google Scholar]
- 29. West MB, Chen Y, Wickham S, Heroux A, Cahill K, Hanigan MH, Mooers BH (2013) Novel insights into eukaryotic gamma‐glutamyl transpeptidase 1 from the crystal structure of the glutamate‐bound human enzyme. J Biol Chem 288:31902–31913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wickham S, Regan N, West MB, Kumar VP, Thai J, Li PK, Cook PF, Hanigan MH (2012) Divergent effects of compounds on the hydrolysis and transpeptidation reactions of gamma‐glutamyl transpeptidase. J Enzyme Inhib Med Chem 27:476–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wickham S, Regan N, West MB, Thai J, Cook PF, Terzyan SS, Li PK, Hanigan MH (2013) Inhibition of human gamma‐glutamyl transpeptidase: development of more potent, physiologically relevant, uncompetitive inhibitors. Biochem J 450:547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cook PF, Cleland WW (2007) Enzyme kinetics and mechanism. London: Garland Press. [Google Scholar]
- 33. Otwinowski Z, Minor W (1997) Processing of x‐ray diffraction data collected in oscillation mode. Macromol Cryst A 276:307–326. [DOI] [PubMed] [Google Scholar]
- 34. Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA (2011) Refmac5 for the refinement of macromolecular crystal structures. Acta Cryst D67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum‐likelihood method. Acta Cryst D53:240–255. [DOI] [PubMed] [Google Scholar]
- 36. Emsley P, Cowtan K (2004) Coot: Model‐building tools for molecular graphics. Acta Cryst D60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 37. Koradi R, Billeter M, Wuthrich K (1996) Molmol: a program for display and analysis of macromolecular structures. J Mol Graph 14:51–55. 29‐32. [DOI] [PubMed] [Google Scholar]
- 38. Wallace AC, Laskowski RA, Thornton JM (1995) Ligplot: a program to generate schematic diagrams of protein‐ligand interactions. Protein Engin 8:127–134. [DOI] [PubMed] [Google Scholar]
- 39. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. [DOI] [PubMed] [Google Scholar]


